Abstract
Networks are often used to incorporate heterogeneity in contact patterns in mathematical models of pathogen spread. However, few tools exist to evaluate whether potential transmission pathways in a population are adequately represented by an observed contact network. Here, we describe a novel permutation-based approach, the network k-test, to determine whether the pattern of cases within the observed contact network are likely to have resulted from transmission processes in the network, indicating that the network represents potential transmission pathways between nodes. Using simulated data of pathogen spread, we compare the power of this approach to other commonly used analytical methods. We test the robustness of this technique across common sampling constraints, including undetected cases, unobserved individuals and missing interaction data. We also demonstrate the application of this technique in two case studies of livestock and wildlife networks. We show that the power of the k-test to correctly identify the epidemiologic relevance of contact networks is substantially greater than other methods, even when 50% of contact or case data are missing. We further demonstrate that the impact of missing data on network analysis depends on the structure of the network and the type of missing data.
Keywords: clustering, social network analysis, livestock movement, wildlife epidemiology, missing data, pathogen transmission
1. Introduction
Social network approaches are a common method in infectious disease ecology to document contact patterns among interacting individuals and for modelling the spread of pathogens [1–6]. In a network approach, the epidemiological units of infection (e.g. individuals, herds, farms) are defined as nodes and inter-linked according to who is in contact with whom, where contact is assumed to represent transmission opportunities between the two nodes [2,3]. Theoretical work has repeatedly demonstrated that incorporating contact pattern heterogeneity into epidemiological models can substantially alter model predictions [6–8], while empirical studies show that network connectivity influences the risk of an individual acquiring an infection [9–14]. Therefore, for network-based epidemiological investigations or predictive infectious disease modelling, it is important that the network through which an infection is assumed to spread correctly reflects the contact patterns that are, in fact, opportunities for pathogen transmission.
However, a pathogen may be transmitted through multiple types of contact and the relative contribution of different contact types in facilitating transmission opportunities may not be well understood. For example, gastrointestinal pathogens can be transmitted among individuals either during direct social interactions or through shared space leading to environmental exposure; the relative contribution of each type of contact to transmission can be difficult to tease apart, and the duration of contact adequate for exposure and transmission is usually unknown [9,15,16]. Thus, a foundational question that often goes unaddressed is how to determine whether the observed pattern of cases is consistent with pathogen spread through an observed network that is presumed to represent potential transmission pathways between nodes [17]. Such validation of a network's ability to explain observed infection patterns is critical if we are to use these networks to develop predictive models of pathogen spread [3].
Current approaches for determining whether an observed contact network has epidemiological significance for a pathogen of interest focus on statistically relating the occurrence of a pathogen (i.e. which nodes are infected) to the network connectivity of those individuals [2,9,13,15,18]. The most common approach is to compare the connectivity of infected and uninfected individuals in the network. An individual's connectivity can be quantified through a number of established centrality metrics [19,20]. Degree is among the most common metric used, and is defined as the total number of contacts in which an individual engages [21]. If the network contributes to infection patterns, then we predict that individuals engaging in a large number of interactions (i.e. having high degree) would be more likely to be exposed to a pathogen. While this pattern is frequently reported [10,13,22], high centrality can also correlate with a number of other factors, such as age, social dominance, hormone levels, etc., that may also influence an individual's susceptibility to infection [23–25]. Thus, higher infection rates observed in well-connected nodes do not directly answer whether an infection was transmitted along network edges. In addition, approaches that distil network data into individual-based measures cannot account for global clustering patterns of cases within the network, as would be expected for a pathogen propagating across network connections.
The social learning literature describes more sophisticated approaches for evaluating the significance of an observed contact network for creating transmission opportunities, where the goal is to identify if information or learned behaviours were acquired from social contacts [17,26–29]. In network-based diffusion analysis, the order or time in which individuals acquire a learned behaviour is statistically related to their position in the social network in order to assess whether these behaviours are acquired through learning from social contacts [17]. In epidemiology, however, temporal data about the order or time of infection acquisition are often inaccurate or completely lacking, particularly, in the case of chronic infections, infections with latent or asymptomatic phases, and pathogens with poor diagnostics. In wildlife systems, serology data obtained from cross-sectional sampling are often used to define cases, and the date of infection is usually unknown [30]. Uncertainties about the date of infection also exist for livestock data, especially for chronic infections or for infections that are only detected through periodic surveillance [31]. In this paper, we focus on the types of data most frequently used in SNA studies (e.g. static networks with unknown dates of pathogen acquisition).
Not only are temporal data often unavailable, but also other forms of data inaccuracy often arise due to missing data [32]. Collection of data for constructing networks and identifying infected nodes may be incomplete. Interactions among individuals or even the individuals themselves can be undocumented. Sampling constraints can also lead to transient cases being missed, or individuals showing few clinical signs to go undetected. Even when case definitions are based on diagnostics rather than observation, diagnostic tests often have poor sensitivity. Thus, missing data can create a situation where the contact network and infection patterns are only partially observed.
For these reasons, new tools are needed to evaluate whether the observed pattern of cases is consistent with pathogen spread through the observed network. New approaches should consider the global infection pattern in the network, be robust to missing data and not rely on temporal ordering or time of infection. In this paper, we describe a novel permutation-based technique for assessing whether the pattern of observed cases is likely to have resulted from transmission processes in an observed network, which we refer to as the ‘epidemiologic relevance’ of the contact network. We compare the power of this technique in determining the epidemiologic relevance of the observed network to other commonly used analytical methods. We then test the robustness of this technique across common sampling constraints, including undetected cases, unobserved individuals and missing interaction data. Lastly, we demonstrate the utility of this method in two real-word datasets.
2. Network k-test procedure
To determine whether an observed contact network has epidemiologic relevance for a specific pathogen, we developed a permutation-based procedure loosely based on spatial clustering methods [33]. Here, we define the k-statistic as the mean number of cases observed to occur within one step of an infected case in the network, which is synonymous with an individual's direct contacts in the network (i.e. degree). To determine significance, the observed k-statistic is compared to a permuted distribution of k-statistics, in which the locations of cases are randomly re-allocated within the network (i.e. node-label swapping). A p-value is calculated as the number of permutations that produce k-statistics more extreme than the observed k-statistic. For example, a p-value of 0.05 signifies that only 5% of the random permutations resulted in a k-statistic that exceeded the observed k-statistic. If the mean number of cases within k steps is significantly greater than expected if cases were randomly distributed in the network, this suggests that the occurrence of cases is a result of propagation of the pathogen through network links. We refer to this procedure as the network k-test.
2.1. Evaluation of the network k-test on simulated datasets
We evaluated the accuracy of the network k-test in correctly identifying the epidemiologic relevance of a contact network for a variety of network types and infection patterns. We generated hypothetical datasets by simulating the spread of a pathogen through various theoretical network structures, assuming a simple susceptible–infected model of infection. We applied the network k-test procedure to these hypothetical datasets and calculated power as the proportion of simulations, in which the k-test correctly detected a significant relationship (p < 0.05) between the observed network and the distribution of cases across the network. That is, power was taken to be equal to (1 – type II error rate). We compared the power of the network k-test to the Kruskal–Wallis test, which compares the degree (number of contacts) of infected and uninfected nodes, and logistic regression, which tests whether node degree is a significant predictor of infection status. These methods were selected because they represent commonly used approaches for assessing the importance of network connectivity on infection patterns [2,10,34,35]. Monte Carlo p-values based on random re-assortments of individuals across nodes were used in both tests.
Datasets were generated for four different network structures: Bernoulli, modular, scale-free and small world. All networks were undirected, consisted of 100 nodes, and were constructed to have approximately the same density (0.04–0.06) using network generation algorithms in the R package igraph [36]. Bernoulli networks were constructed with an edge probability of 0.05 [37]. Modular networks were constructed using the ‘inter-connected island’ algorithm with five communities. The probability of edges between members of the same community was set to 0.22. Two edges connected each pair of communities yielding an average modularity approximately 0.7, indicating strong community structure [38]. Scale-free networks were constructed using the Barabasi algorithm with linear preferential attachment and the number of edges added per additional node set to three [39]. Small-world networks were constructed using the Watts–Strogatz network model with each node having two neighbours and edges randomly re-wired with probability 0.05 [40].
For a given network, pathogen spread was initiated by randomly infecting a single node. Transmission from an infected to an uninfected node was simulated stochastically and occurred with probability β per time-step, where higher values of β indicate a more transmissible pathogen. Pathogen spread was simulated until a pre-defined prevalence cut-off value was reached, allowing statistical methods to be applied to different network structures while keeping the number of cases constant. Prevalence cut-offs of 0.05, 0.25, 0.50 and 0.75 were considered. For each network type, we also considered two epidemic scenarios to evaluate the detection methods over different patterns of infection: a moderately infectious pathogen (β = 0.04) and a highly infectious pathogen (β = 0.133).
We simulated pathogen spread 100 times for all combinations of network type, pathogen infectiousness and prevalence cut-off values. Statistical tests were applied to each simulation and power was calculated for each scenario across the 100 simulations.
2.2. Robustness of network k-test to missing data
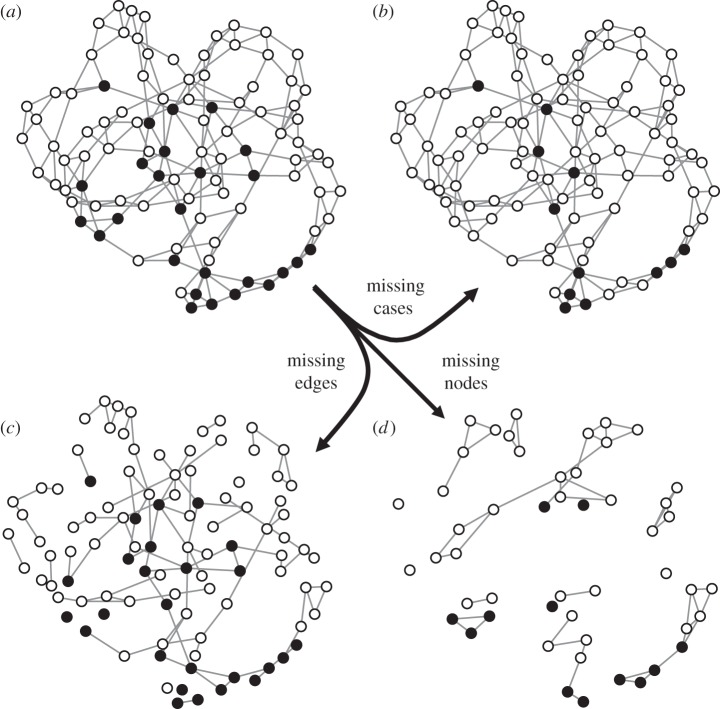
We evaluated the robustness of the network k-test across three common types of missing data in epidemiological studies: missing edges, missing nodes and missing cases. Following the same methodology as in our primary analysis, we first simulated the spread of a pathogen through hypothetical networks. We then explored the robustness of each analytical method (k-test, Kruskal–Wallis test, and logistic regression) across increasing levels of missing data by randomly eliminating 25% or 50% of edges, cases or nodes before running statistical tests (figure 1; electronic supplementary material, figures S1–S3). β and the prevalence cut-off were held constant across all simulations at 0.04 and 0.25, respectively. For each network type, 100 simulations were run for each level and type of missing data. Power was calculated for each scenario.
Figure 1.
(a) A simulated infectious disease outbreak (25% prevalence) in a small-world network. The same network with differing types of missing data, including (b) 50% of cases undetected, (c) 50% of edges unobserved and (d) 50% of nodes unobserved. Nodes are plotted in the same position in each panel. Black nodes indicate infected individuals. See electronic supplementary material, figures S1–S3 for a visual representation of other network types.
3. Simulated results
3.1. Network k-test implementation
The k-test was implemented in R and made publically available with a graphical user interface in html format at https://stemma.shinyapps.io/k-test/. The user is prompted to provide an edge list and attribute table that includes the infection status of each node in the network. Outputs of this method include a spreadsheet specifying the mean and median number of infected nodes within k steps for each permutation; the k-statistic and corresponding p-value for the observed data; and a density plot depicting the k-statistic's permuted distribution (figure 2). For the small-world network in figure 1a, for example, the corresponding density plot depicts the permuted distribution of k-statistics (i.e. the expected number of infected nodes within k = 1 steps of each infected node if cases are distributed randomly in the network). The vertical line indicates the observed k-statistic (figure 2).
Figure 2.
Graphical results of the k-test for scenarios, in which (a) the pattern of cases within a small-world network was determined by pathogen propagation along network edges (prevalence = 0.25) and (b) cases were randomly assigned to nodes. Both plots depict the null distribution of the k-statistic when the location of infected nodes was randomized within the network. The vertical line indicates the observed number of cases within one step of an infected node. For (a), the pattern of cases within the network is extremely unlikely to have emerged by chance (p < 0.001). For (b), the k-test fails to reject the null hypothesis that the cases were randomly distributed (p = 0.73).
3.2. Comparison of the k-test to other analytical approaches
The k-test had substantial power (low type II error rates) to detect the epidemiologic relevance of an observed network, correctly rejecting the null hypothesis that the infection was distributed randomly in the network. Across a range of prevalence levels and network types, the power of the k-test was consistently close to one (figure 3). By contrast, the power of the Kruskal–Wallis and logistic regression to detect an effect of the network on infection patterns was often less than 0.5, especially for small-world networks, indicating that these tests would fail to detect a pattern around 50% of the time. The performance of these degree-based tests varied markedly based on pathogen prevalence and network type. For Bernoulli, modular and scale-free networks, degree-based tests performed adequately (power > 0.75) if the prevalence was 75%. However, power rapidly dropped off as prevalence declined. For low-prevalence pathogens (5%), the statistical power of degree-based tests was approximately 0.1. Degree-based tests performed very poorly for all prevalence levels for small-world networks (figure 3). There were no apparent differences in the performance of any statistical tests for pathogens with low and high transmissibility (electronic supplementary material, figure S4). When the size of the network was increased to 1000 nodes, all statistical approaches were performed with power close to 1. However, when the network size was reduced to 20 nodes, the k-test's power declined to 0.4 and 0.34 for Bernoulli and scale-free networks, respectively, though the power of the k-test was consistently 2–10 times higher than for the corresponding degree-based tests.
Figure 3.

Power of three statistical tests for detecting associations between the observed network and the distribution of cases across a range of pathogen prevalence levels (0.05–0.75) and network types (Bernoulli, modular, scale-free and small-world). Power was calculated as the proportion of simulations where a significant effect (p < 0.05) was detected (1 – type II error rate).
To assess the k-test's ability to discriminate scenarios where the network had no relationship with pathogen spread, we repeated our analysis on networks where the observed cases were randomly distributed. We found that the k-test had high discriminatory abilities. Type I error rates were generally below 0.05, indicating that the frequency with which the k-test incorrectly rejects the null hypothesis was rare (electronic supplementary material, figure S5). Degree-based tests also exhibited low type I error rates, though logistic regression tended to have higher error rates particularly for small world and scale-free networks. However, when 50% of cases resulted from transmission through a scale-free network, and 50% were randomly distributed (i.e. transmitted through unknown mechanisms), the k-test still indicated that the network was epidemiologically relevant with power close to one, though one limitation is that the test does not give an estimation of the relative strength of the network's influence on transmission dynamics.
3.3. Robustness of k-test across common sampling constraints
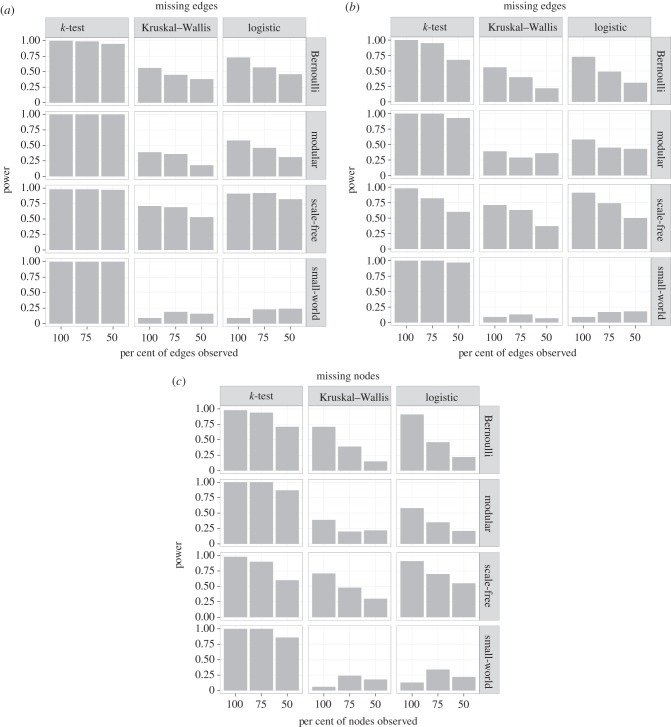
The performance of the k-test was highly robust to missing edges (figure 4a). Statistical power was close to one even when only 50% of edges were observed, regardless of network type. By contrast, the power of degree-based tests declined by nearly half when only 50% of edges were observed in Bernoulli and modular networks. The power of degree-based tests was less sensitive to missing edges in scale-free and small-world networks; however, the power was already quite low even when the network was fully observed.
Figure 4.
Power of three statistical tests for detecting whether the contact network influenced the pattern of infection across common sampling constraints. (a) Missing edges: not all interactions were observed. (b) Missing cases: infection was not detected in all infected individuals. (c) Missing nodes: not all individuals were observed. Power was calculated as the proportion of simulations where a significant effect was detected (p < 0.05).
The power of all three tests was reduced by missing cases and missing nodes (figure 4). For the k-test, declines in power were generally only observed when missing data reached 50%, whereas the other tests experienced performance reductions when only 25% of data were missing. Generally, even with only 50% of the data available, the power of the k-test still matched or exceeded the power of the other tests when applied to complete data. The k-test was more robust to missing cases and nodes for small-world networks, whereas scale-free networks were more susceptible to missing data. We also explored a more likely scenario where a 100-node scale-free network was missing multiple types of data concurrently (25% of each type). In this case, the k-test's power dropped to 0.66, whereas the power of the Kruskal–Wallis test and logistic regression decreased to 0.32 and 0.38, respectively. Thus, all tests experienced declines in power when excluding multiple types of data, but the performance of the k-test still exceeded that of the degree-based tests.
4. Applications to real-word datasets
To demonstrate the utility of our technique, we use the k-test to evaluate the epidemiologic relevance of two real-world contact networks. The first is based on bovine tuberculosis (bTB) in a cattle movement network in Uruguay, where edges were fully observed and cases were partially observed due to limited diagnostic sensitivity. The second example examines the occurrence of canine distemper virus (CDV) in a contact network based on spatial overlap of prides of African lions (Panthera leo).
4.1. Bovine tuberculosis in Uruguay
The first example uses data from a fully observed network of between-farm cattle movements in Uruguay, a country with a comprehensive animal traceability programme [41]. Here, nodes in the network represent farms (N = 62 767 farms), and edges between nodes represent the movement of a batch of cattle (figure 5a). Uruguay experiences a very low farm-level incidence of bTB, with typically fewer than 30 new infected farms detected annually. Dairy farms in Uruguay are tested annually for bTB using a tuberculin skin test, which has limited sensitivity to detect infected animals [42]. Movement of animals between farms is a commonly cited source of between-farm spread of livestock pathogens [18,42,43], though local factors such as wildlife reservoirs and fence-line transmission have also contributed to bTB transmission in other countries [44]. If animal movements were a major contributor to bTB transmission in Uruguayan cattle, then we would expect that bTB-positive farms would be significantly more inter-linked in the network than expected by chance.
Figure 5.

(a) Network of cattle movement between farms in Uruguay. Red nodes indicate farms that were positive for bovine tuberculosis (bTB). For visualization purposes, only infected farms and their immediate neighbours are depicted (1178/62 767 farms). Node positions do not correspond to geographical location. (b) Graphical results of the k-test (p < 0.001). The blue shaded region represents the null distribution of the k-statistic when the location of infected nodes was randomized within the network. The vertical line indicates the observed number of cases within one step of an infected node. (Online version in colour.)
Owing to low transmissibility of the pathogen and lags in the detection of infected farms [31], movements from several years prior to the detection of an infected farm may be responsible for between-farm transmission events. Thus, we used the k-test to assess the epidemiological relevance of the movement network from July 2008 to June 2013 to the distribution of bTB-positive farms observed in years 2011–2013 (n = 58 infected farms). We also directly compared the relative roles of animal movement and geographical proximity in creating epidemiological links between farms by additionally contrasting spatial clustering with network clustering of cases. Parallel to the network permutations, we also calculated the number of bTB-positive farms that were within a 10 km radius of infected farms and compared this with the expected number of bTB-farms if the infected farms were randomly re-distributed across the population. This allowed us to simultaneously address clustering of cases across two hypothesized transmission pathways: local spatial spread and animal movements. This two-dimensional test is an extension of the basic k-test.
Infected farms were connected to a mean of approximately one other infected farms, which according to the k-test, is substantially greater than expected if bTB was distributed randomly in the network (p < 0.001, figure 5b). Infected farms were also significantly more likely to have an infected neighbour within 10 km than expected if cases were randomly distributed in space, suggesting that both spatial and network processes interact to determine transmission patterns.
4.2. Canine distemper virus in African lions
In 1993–1994, an outbreak of CDV killed approximately one-third of the African lion population in Serengeti National Park [45]. Our second example explores the role of spatial overlap among territorial prides of lions in CDV transmission during this outbreak [46]. Infection was likely introduced into the Serengeti from domestic dogs bordering the park, and then spread through wild carnivore populations. Because this outbreak affected multiple host species, the network of interactions among lion prides was not thought to correlate with the pattern of infection [46]. Here, we apply the k-test to this outbreak to assess if our technique provides results that are consistent with previous conclusions on the lack of epidemiologic relevance of the inter-pride contact network.
In this analysis, nodes correspond to lion prides. Cases (i.e. infected prides) were nferred through a pride member's death or disappearance, and 90% of cases were detected only by serology [47]. Edges represent whether the territories of two prides overlapped in space. Territory boundaries were determined using a fixed-kernel utilization distribution of pride sightings over a 2-year period [48,49]. A 75% probability contour was used to exclude outlying observations and produce a core range used by prides [50]. Data used to construct pride territories included all observations of each pride from 1991 to 1992, which was deemed to best represent the space use of each pride before the onset of the CDV outbreak in December 1993 [45,46]. Here, we focus on the prides infected in the first six weeks of the outbreak (seven infected prides), which represents the early, rapid growth phase of the outbreak that was subsequently followed by a three-week lag in new cases (figure 6a). We chose to focus on this period because all prides eventually became infected, thus eliminating variation in the k-statistic. As with the Uruguay bTB example, we also calculated the number of cases within 10 km of each infected pride in the observed and permuted data to assess geographical clustering of cases as an alternative hypothesis for transmission. Distances between prides were calculated from the territory centroid. Additional detail on these data can be found in Craft et al. [46,51].
Figure 6.

(a) Network of lion prides where nodes represent prides and are labelled with pride name, and edges represent prides with overlapping territories. Nodes are positioned according to the geographical centre of their territories. Black nodes indicate prides that became infected in the first 6 weeks of the 1993–1994 CDV outbreak. (b) Graphical results of the k-test (p = 0.474). The blue-shaded region represents the null distribution of the k-statistic when the location of infected nodes was randomized within the network. The vertical line indicates the observed number of cases within one step of an infected node. (Online version in colour.)
The conclusions of our analysis are in agreement with the previous work and confirm that the contact network was unrelated to the pattern of CDV spread in this host–pathogen system; the k-test failed to reject the null hypothesis that cases were distributed randomly in the network (p = 0.496). Furthermore, there was no evidence of spatial clustering. Infected prides were not more likely to be within 10 km of other infected prides than expected if CDV were distributed randomly in space (p = 0.474).
5. Discussion
In this paper, we present a novel approach, the network k-test, for determining whether an observed contact network is epidemiologically relevant given that transmission may occur by processes not captured by the observed data. The intuition behind the network k-test is that if the network connections represent transmission pathways, then infected nodes will be more likely to be connected to other infected nodes in the network than expected by chance. Using simulated data for a 100-node network, we showed that the k-test correctly identifies the epidemiologic relevance of the contact network nearly 100% of the time when the network is fully observed (figure 3), and consistently outperforms other commonly used statistical tests. Our results strongly suggest that an analytic approach focusing on the connectivity of infected nodes relative to other infected nodes will yield more statistical power than degree-based statistical approaches, which rely on comparisons of the degree of infected and uninfected nodes.
Unlike degree-based tests, such as Kruskal–Wallis tests and logistic regression, the power of the network k-test was not affected by pathogen prevalence or network type. Degree-based tests operate under the hypothesis that nodes with high degree are more likely to become infected. Thus, the poor performance of degree-based tests at low prevalence levels (5%) is related to the fact that the most highly connected nodes do not necessarily become infected due to the limited extent of the epidemic. In addition, network types with low variation in the degree distribution, such as small-world networks, may have insufficient variation to discern differences in connectivity among infected and uninfected nodes. Given that many real-world networks have small-world properties [40,51–54], it is important to take into account network structure and pathogen prevalence when selecting appropriate statistical methods. Our results suggest that the k-test performs well across a diversity of scenarios.
The network k-test was highly robust to missing data (figure 4). Even with 50% of edges, cases or nodes missing, the k-test often achieved higher power than degree-based tests with complete data. Missing edges had less impact on power than other types of missing data, which is reassuring given that interaction data are often the most under-sampled type of data in practice [32,55]. For the k-test, missing cases resulted in a greater reduction in power than missing nodes. While both may fragment infection chains, missing cases provide incorrect information (false-negative nodes), which may introduce more noise into the analysis than simply missing the node entirely.
The application of the k-test to two real-world datasets demonstrates the ability of the k-test to correctly discriminate between scenarios where the network did or did not influence the spread of infectious disease. In the first example, the k-test indicated that movement of cattle between farms in Uruguay played a significant role in determining the observed pattern of bTB cases in the country. In this example, the k-test performed effectively even with extremely low prevalence (less than 0.005%) and an unknown proportion of missing cases (not all bTB-positive farms were directly connected to other infected farms). In the second example, the k-test failed to reject the null hypothesis that CDV cases were randomly distributed in a contact network based on spatial overlap between lion prides. This is consistent with previous epidemiological models, which concluded that the pattern of spread of CDV in lions could not be explained by pride contact networks [46]. CDV is multi-host pathogen, and other carnivore species present in the ecosystem likely contributed to its spread [47]. Thus for CDV, we can consider a lion-only network as suffering from a large number of missing nodes (i.e. other carnivore species) or potentially an inappropriate definition of inter-pride contact.
The k-test provides a method to quantify whether the contact network has epidemiological relevance, which is the goal of many social network analysis studies. The k-test could also be applied as a first step in the process of developing predictive mathematical models of pathogen spread through networks based on empirical data, verifying that the assumed relationship between network connections and transmission is in fact consistent with the data. Furthermore, different types of contact may contribute to transmission, such as spatial proximity, fomites or physical contact, and it is important to verify that the contact definitions used in the empirical network are indeed relevant for pathogen spread [1]. An insignificant p-value in the k-test indicates that there is not sufficient support to conclude that the observed network plays a role in determining transmission opportunities.
The network k-test and network-based diffusion analysis (NBDA) have similar objectives of detecting whether the pattern of cases is consistent with transmission or diffusion through a network [17,26,27]. While NBDA methods are robust tools for examining the extent to which the network influences diffusion/transmission processes, data on the time or order in which nodes become infected are often not available for cross-sectional sampling approaches, or when there exist delays in detection or dependence on serological tests that only indicate prior exposure. Thus, dates (and even order) of detection may not correspond to the date of infection. NBDA may be used if longitudinal sampling has been conducted, whereas the k-test may be more appropriate for other study designs. However, adapting the maximum-likelihood approaches used by NBDA for cross-sectional data could be a fruitful area of further methodological development of the k-test, especially if researchers are interested in incorporating individual-level variation in susceptibility.
One extension of the k-test could include incorporating two dimensions of contact to directly contrast alternative hypotheses about the definition of contact relevant for transmission. These two dimensions could contrast between network connectivity and geographical distances between cases, as explored by the case studies, or they could include two different contact networks with the same nodes as long as an adjacency matrix can represent each dimension. Indeed, a more general extension involving quantifications of the relative importance of each network would be highly useful.
The current version of the k-test is limited by its reliance on an unweighted network (i.e. network edges are binary and take on the values of either 1 or 0). Incorporating data on the relative strength of contact among nodes (such as the number of animals moved between farms or the frequency of contact between individuals) could be achieved with a path-based approach. The summed weight of the edges along the shortest path connecting each pair of infected nodes could be used as an alternative to the k-statistic. A path-based approach could easily be adapted for dynamic networks, where patterns of contact change through time [56,57]. These extensions are currently under development.
While the epidemiologic importance of an observed network can also be validated by comparing observed case data with predictions made by network-based epidemiological models, such models rely on a number of assumptions about transmissibility, incubation periods, etc., all of which can make outputs difficult to interpret. By contrast, the k-test is a data-driven approach that relies on few assumptions. The k-test outperforms other statistical approaches that compare the degree of infected and uninfected nodes, with high power across a diversity of network types, pathogen prevalence levels, and missing data constraints. Thus, our approach will likely be broadly applicable for analysing how observed contact networks contribute to transmission processes in populations.
Supplementary Material
Supplementary Material
Acknowledgements
We thank A. Cheeran, S. Wells, A. Perez, J. Alvarez, A. Mosser and N. Fountain-Jones for their contributions to the development and implementation of this procedure on the real-world case studies. Data for the Uruguay case study were provided by the Directory of Animal Identification System (SIRA in Spanish), Ministry of Livestock, Agriculture and Fisheries, Montevideo, Uruguay. Data for the African lion case study were provided by the Serengeti Lion Project, University of Minnesota, St Paul, MN, USA.
Data accessibility
The datasets supporting the lion CDV case study have been uploaded as part of the electronic supplementary material.
Authors' contributions
K.V.W. developed the network k-test, designed the simulation study, performed the epidemiological modelling, analysed data and wrote the manuscript. E.A.E. participated in the development of the k-test, study design and helped draft the manuscript. M.E.C. participated in the development of the k-test and study design, provided data, contributed to the design and interpretation for the lion case study and helped draft the manuscript. Cr.P. collected and provided data and contributed to the design and interpretation of the lion case study. Ca.P. contributed data and epidemiological expertise for the Uruguay case study. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This research was supported by USDA-NIFA AFRI Foundational Program grant no. 2013-01130, the National Science Foundation (DEB-1413925), the University of Minnesota's Institute on the Environment, the Office of the Vice President for Research and the Cooperative State Research Service, US Department of Agriculture, under project nos. MINV-62-044 and 62-051.
Disclaimer
Any opinions, findings, conclusions or recommendations expressed in this publication are those of the author and do not necessarily reflect the view of the US Department of Agriculture.
References
- 1.Craft ME. 2015. Infectious disease transmission and contact networks in wildlife and livestock. Phil. Trans. R. Soc. B 370, 20140107 ( 10.1098/rstb.2014.0107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godfrey SS. 2013. Networks and the ecology of parasite transmission: a framework for wildlife parasitology. Int. J. Parasitol. 2, 235–245. ( 10.1016/j.ijppaw.2013.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White L, Forester J, Craft M. 2015. Using contact networks to explore mechanisms of parasite transmission in wildlife. Camb Philos Soc. ( 10.1111/brv.12236) [DOI] [PubMed] [Google Scholar]
- 4.Böhm M, Hutchings MR, White PCL. 2009. Contact networks in a wildlife–livestock host community: identifying high-risk individuals in the transmission of bovine TB among badgers and cattle. PLoS ONE 4, e5016, 1–12. ( 10.1371/journal.pone.0005016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal S, Grenfell BT, Meyers LA. 2007. When individual behaviour matters: homogeneous and network models in epidemiology. J. R. Soc. Interface 4, 879–891. ( 10.1098/rsif.2007.1100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeling MJ, Eames KTD. 2005. Networks and epidemic models. J. R. Soc. Interface 2, 295–307. ( 10.1098/rsif.2005.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ames GM, George DB, Hampson CP, Kanerek AR, McBee CD, Lockwood DR, Achter JD, Webb CT. 2011. Using network properties to predict disease dynamics on human contact networks. Proc. R. Soc. B 278, 3544–3550. ( 10.1098/rspb.2011.0290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature 438, 355–359. ( 10.1038/nature04153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimbach R, Bisanzio D, Galvis N, Link A, di Fiore A, Gillispie TR. 2015. Brown spider monkeys (Ateles hybridus): a model for differentiating the role of social networks and physical contact on parasite transmission dynamics. Phil. Trans. R. Soc. B 370, 20140110 ( 10.1098/rstb.2014.0110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanderWaal KL, Atwill ER, Hooper S, Buckle K, McCowan B. 2013. Network structure and prevalence of Cryptosporidium in Belding's ground squirrels. Behav. Ecol. Sociobiol. 67, 1951–1959. ( 10.1007/s00265-013-1602-x) [DOI] [Google Scholar]
- 11.Godfrey SS, Moore JA, Nelson NJ, Bull CM. 2010. Social network structure and parasite infection patterns in a territorial reptile, the tuatara (Sphenodon punctatus). Int. J. Parasitol. 40, 1575–1585. ( 10.1016/j.ijpara.2010.06.002) [DOI] [PubMed] [Google Scholar]
- 12.Drewe JA, Eames KTD, Madden JR, Pearce GP. 2011. Integrating contact network structure into tuberculosis epidemiology in meerkats in South Africa: implications for control. Prev. Vet. Med. 101, 113–120. ( 10.1016/j.prevetmed.2011.05.006) [DOI] [PubMed] [Google Scholar]
- 13.Otterstatter MC, Thomson JD. 2007. Contact networks and transmission of an intestinal pathogen in bumble bee (Bombus impatiens) colonies. Oecologia 154, 411–421. ( 10.1007/s00442-007-0834-8) [DOI] [PubMed] [Google Scholar]
- 14.Corner LAL, Pfeiffer DU, Morris RS. 2003. Social-network analysis of Mycobacterium bovis transmission among captive brushtail possums (Trichosurus vulpecula). Prev. Vet. Med. 59, 147–167. ( 10.1016/S0167-5877(03)00075-8) [DOI] [PubMed] [Google Scholar]
- 15.VanderWaal KL, Atwill ER, Isbell LA, McCowan B. 2014. Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa camelopardalis). J. Anim. Ecol. 86, 406–414. ( 10.1111/1365-2656.12137) [DOI] [PubMed] [Google Scholar]
- 16.Blyton MDJ, Banks SC, Peakall R, Lindenmayer DB, Gordon DM. 2014. Not all types of host contacts are equal when it comes to E. coli transmission. Ecol. Lett. 17, 970–978. ( 10.1111/ele.12300) [DOI] [PubMed] [Google Scholar]
- 17.Hoppitt W, Boogert NJ, Laland KN. 2010. Detecting social transmission in networks. J. Theor. Biol. 263, 544–555. ( 10.1016/j.jtbi.2010.01.004) [DOI] [PubMed] [Google Scholar]
- 18.Ribeiro-Lima J, Enns EA, Thompson B, Craft ME, Wells SJ. 2015. From network analysis to risk analysis—an approach to risk-based surveillance for bovine tuberculosis in Minnesota, US. Prev. Vet. Med. 118, 328–340. ( 10.1016/j.prevetmed.2014.12.007) [DOI] [PubMed] [Google Scholar]
- 19.Wey T, Blumstein DT, Shen W, Jordán F. 2008. Social network analysis of animal behaviour: a promising tool for the study of sociality. Anim. Behav. 75, 333–344. ( 10.1016/j.anbehav.2007.06.020) [DOI] [Google Scholar]
- 20.Croft DP, James R, Krause J. 2008. Exploring animal social networks. Princeton, NJ: Princeton University Press. [Google Scholar]
- 21.Wasserman S, Faust K. 1994. Social network analysis: methods and applications. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 22.Godfrey SS, Bull CM, James R, Murray K. 2009. Network structure and parasite transmission in a group living lizard, gidgee skink, Egernia stokesii. Behav. Ecol. Sociobiol. 63, 1045–1056. ( 10.1007/s00265-009-0730-9) [DOI] [Google Scholar]
- 23.Rushmore J, Caillaud D, Matamba L, Stumpf RM, Borgatti SP, Altizer S. 2013. Social network analysis of wild chimpanzees provides insights for predicting infectious disease risk. J. Anim. Ecol. 82, 976–986. ( 10.1111/1365-2656.12088) [DOI] [PubMed] [Google Scholar]
- 24.Habig B, Archie EA. 2015. Social status, immune response and parasitism in males: a meta-analysis. Phil. Trans. R. Soc. B 370, 20140109 ( 10.1098/rstb.2014.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VanderWaal KL, Ezenwa VO. 2016. Heterogeniety in pathogen transmission: mechanisms and methodology. Funct. Ecol. ( 10.1111/1365-2435.12645) [DOI] [Google Scholar]
- 26.Franz M, Nunn CL. 2009. Network-based diffusion analysis: a new method for detecting social learning. Proc. R. Soc. B 276, 1829–1836. ( 10.1098/rspb.2008.1824) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boogert NJ, Reader SM, Hoppitt W, Laland KN. 2008. The origin and spread of innovations in starlings. Anim. Behav. 75, 1509–1518. ( 10.1016/j.anbehav.2007.09.033) [DOI] [Google Scholar]
- 28.Farine DR, Aplin LM, Sheldon BC, Hoppitt W. 2015. Interspecific social networks promote information transmission in wild songbirds. Proc. R. Soc. B 282, 20142804 ( 10.1098/rspb.2014.2804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobaiter C, Poisot T, Zuberbühler K, Hoppitt W, Gruber T. 2014. Social network analysis shows direct evidence for social transmission of tool use in wild chimpanzees. PLoS ONE 12, e1001960 ( 10.1371/journal.pbio.1001960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert A, et al. 2013. Deciphering serology to undestand the ecology of infectious diseases in wildlife. EcoHealth 10, 298–313. ( 10.1007/s10393-013-0856-0) [DOI] [PubMed] [Google Scholar]
- 31.Smith RL, Schukken YH, Lu Z, Mitchell RM, Grohn YT. 2013. Development of a model to simulate infection dynamics on Mycobacterium bovis in cattle herds in the United States. J. Am. Vet. Med. Assoc. 243, 411–423. ( 10.2460/javma.243.3.411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eames K, Bansal S, Frost S, RIley S. 2015. Six challenges in measuring contact networks for use in modelling. Epidemics 10, 72–77. ( 10.1016/j.epidem.2014.08.006) [DOI] [PubMed] [Google Scholar]
- 33.Wakefield JC, Kelsall JE, Morris SE. 2001. Clustering, cluster detection, and spatial variation in risk. In Spatial epidemiology: methods and applications (eds Elliot P, Wakefield JC, Best NG, Briggs DJ), pp. 128–152. Oxford, UK: Oxford University Press. [Google Scholar]
- 34.VanderWaal KL, Atwill ER, Isbell LA, McCowan B. 2014. Quantifying microbe transmission networks for wild and domestic ungulates in Kenya. Biol. Conserv. 169, 136–146. ( 10.1016/j.biocon.2013.11.008) [DOI] [Google Scholar]
- 35.MacIntosh AJJ, Jacobs A, Garcia C, Shimizu K, Mouri K, Huffman MA, Hernandez AD. 2012. Monkeys in the middle: parasite transmission through the social network of a wild primate. PLoS ONE 7, e51144 ( 10.1371/journal.pone.0051144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJounral, Complex Systems 1695. [Google Scholar]
- 37.Erdos P, Renyi A. 1959. On random graphs. Publ. Math. 6, 290–297. [Google Scholar]
- 38.Newman MEJ, Girvan M. 2004. Finding and evaluating community structure in networks. Phys. Rev. E 69, 1–16. [DOI] [PubMed] [Google Scholar]
- 39.Barabasi A, Albert R. 1999. Emergence of scaling in random networks. Science 286, 509–512. ( 10.1126/science.286.5439.509) [DOI] [PubMed] [Google Scholar]
- 40.Watts DJ, Strogatz SH. 1998. Collective dynamics of 'small world' networks. Nature 393, 440–442. ( 10.1038/30918) [DOI] [PubMed] [Google Scholar]
- 41.VanderWaal KL, Picasso C, Enns EA, Craft ME, Alvarez J, Fernandez F, Gil A, Perez A, Wells S. 2016. Network analysis of cattle movements in Uruguay: quantifying heterogeneity for risk-based disease surveillance and control. Prev. Vet. Med. 123, 12–22. ( 10.1016/j.prevetmed.2015.12.003) [DOI] [PubMed] [Google Scholar]
- 42.Picasso C, Alvarez J, VanderWaal KL, Fernandez F, Gil A, Wells SJ, Perez AM. Submitted. Epidemiological investigation of bovine tuberculosis outbreaks in Uruguay (2011–2013). [DOI] [PubMed]
- 43.Humblet M-F, Boschiroli ML, Saegerman C. 2009. Classification of worldwide bovine tuberuclosis risk factors in cattle: a stratified approach. Vet. Res. 40, 50 ( 10.1051/vetres/2009033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks-Pollock E, Roberts GO, Keeling MJ. 2014. A dynamic model of bovine tuberculosis spread and control in Great Britain. Nature 511, 228–231. ( 10.1038/nature13529) [DOI] [PubMed] [Google Scholar]
- 45.RoelkeParker ME, et al. 1996. A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature 379, 441–445. ( 10.1038/379441a0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craft ME, Volz E, Packer C, Meyers A. 2009. Distinguishing epidemic waves from disease spillover in a wildlife population. Proc. R. Soc. B 276, 1777–1785. ( 10.1098/rspb.2008.1636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craft ME, Hawthorne PL, Packer C, Dobson AP. 2008. Dynamics of a multihost pathogen in a carnivore community. J. Anim. Ecol. 77, 1257–1264. ( 10.1111/j.1365-2656.2008.01410.x) [DOI] [PubMed] [Google Scholar]
- 48.Worton BJ. 1989. Kernel methods for estimating the utilization distribution in home-range studies. Ecology 70, 164–168. ( 10.2307/1938423) [DOI] [Google Scholar]
- 49.Harris S, Cresswell WJ, Forde PG, Trewhella WJ, Woollard T, Wray S. 1999. Home-range analysis using radio-tracking data—a review of problems and techniques particularly as applied to the study of mammals. Mamm. Rev. 29, 97–123. [Google Scholar]
- 50.VanderWaal KL, Mosser A, Packer C. 2009. Optimal group size, dispersal decisions and postdispersal relationships in female African lions. Anim. Behav. 77, 949–954. ( 10.1016/j.anbehav.2008.12.028) [DOI] [Google Scholar]
- 51.Craft ME, Volz E, Packer C, Meyers LA. 2010. Disease transmission in territorial populations: the small-world network of Serengeti lions. J. R. Soc. Interface 8, 776–786. ( 10.1098/rsif.2010.0511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thakur KK, Revie CW, Hurnik D, Poljak Z, Sanchez J. 2016. Analysis of swine movement in four Canadian regions: network structure and implications. Transboundary Emerging Dis. 63, 14–26. [DOI] [PubMed] [Google Scholar]
- 53.Robinson SE, Everett MG, Christley RM. 2007. Recent network evolution increases the potential for large epidemics in the British cattle population. J. R. Soc. Interface 4, 669–674. ( 10.1098/rsif.2007.0214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.May RM. 2006. Network structure and the biology of populations. Trends Ecol. Evol. 21, 394–399. ( 10.1016/j.tree.2006.03.013) [DOI] [PubMed] [Google Scholar]
- 55.Farine D, Standburg-Peshkin A. 2015. Estimating uncertainty and reliability of social network data using Bayesian inference. R. Soc. open sci. 2, 150367 ( 10.1098/rsos.150367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blonder B, Wey TW, Dornhaus A, James R, Sih A. 2012. Temporal dynamics and network analysis. Methods Ecol. Evol. 3, 958–972. ( 10.1111/j.2041-210X.2012.00236.x) [DOI] [Google Scholar]
- 57.Vernon MC, Keeling MJ. 2009. Representing the UK's cattle herd as static and dynamic networks. Proc. R. Soc. B 276, 469–476. ( 10.1098/rspb.2008.1009) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the lion CDV case study have been uploaded as part of the electronic supplementary material.