Abstract
The Wnt/β-catenin signaling pathway is indispensable for embryonic development, maintenance of adult tissue homeostasis and repair of epithelial injury. Unsurprisingly, aberrations in this pathway occur frequently in many cancers and often result in increased nuclear β-catenin. While mutations in key pathway members, such as β-catenin and adenomatous polyposis coli, are early and frequent occurrences in most colorectal cancers (CRC), mutations in canonical pathway members are rare in pancreatic ductal adenocarcinoma (PDAC). Instead, in the majority of PDACs, indirect mechanisms such as promoter methylation, increased ligand secretion and decreased pathway inhibitor secretion work in concert to promote aberrant cytosolic/nuclear localization of β-catenin. Concomitant with alterations in β-catenin localization, changes in mucin expression and localization have been documented in multiple malignancies. Indeed, numerous studies over the years suggest an intricate and mutually regulatory relationship between mucins (MUCs) and β-catenin. In the current review, we summarize several studies that describe the relationship between mucins and β-catenin in gastrointestinal malignancies, with particular emphasis upon colorectal and pancreatic cancer.
Introduction
The Wnt signaling pathway is an important developmental regulatory pathway and plays critical roles in embryogenesis, including roles in regulating delineation of the body axis and in the formation of the germ layer (1). The binding of Wnt ligands, a group of secreted lipid-modified proteins, activate both the canonical and non-canonical Wnt-signaling pathways (2). The canonical Wnt pathway in particular, hinges upon the activity of β-catenin, a molecule important for both cell adhesion and signaling, both functions being indispensable for normal cellular processes.
There are two separate pools of β-catenin-cytosolic and membrane-localized (2–5). The membrane-localized fraction participates in cell adhesion, where it forms part of the adherens junction. Here, membrane-localized β-catenin links E-cadherin to the cytoskeleton via α-catenin. The cytosolic fraction is typically degraded through phosphorylation at the N-terminus by a destruction complex. This complex consists of glycogen synthase kinase β (GSK3-β), Axin1 and casein kinase 1 (CK1) (5). In the presence of a Wnt ligand, which binds to the Frizzled seven-pass transmembrane receptor and a co-receptor, the low density lipoprotein receptor-related protein (LRP), this complex is destroyed via a cascade of reactions triggered by the recruitment of dishevelled segment polarity protein 1 (DVL-1) to the receptor complex. Here, DVL-1 recruits Axin1 and GSK3-β to form part of the Wnt signalosome, thus destabilizing the destruction complex (2–5). Next, β-catenin is released from the destruction complex and enters the nucleus through direct contact with the nuclear pore complex (6). Nuclear β-catenin upregulates a host of tissue-specific target genes, typically partnering with the TCF/LEF family of transcription factors, which usually function as transcriptional repressors in the absence of nuclear β-catenin (2). Wnt ligands can also activate the non-canonical pathway, which is independent of β-catenin and comprises the planar cell polarity and the Wnt/Ca(2+) pathways (2).
The β-catenin molecule is remarkably well conserved, as evidenced by the presence of a β-catenin-like molecule in all metazoans. The Drosophila analogue of β-catenin, armadillo, was crucial in the discovery of the signaling function of β-catenin in a screen for mutations that affect segmentation of the embryo (7). Remarkably, an amoebozoan, Dictyostelium discoideum, expresses a β-catenin analogue Aardvark, which maintains cell–cell junctional polarity in multicellular aggregates that comprise the fruiting bodies of the normally single-celled organism (8). In the developing embryo, β-catenin is required for mesoderm formation, where the signaling function of the molecule plays a crucial role. The β-catenin molecule is also required for formation of the neuroepithelial structures and the endoderm. However, here the structural, junction-forming function of β-catenin takes precedence over the signaling function (9). While Wnt/β-catenin signaling is not as active in adult tissue as the embryo, the Wnt/β-catenin pathway is required for the maintenance of tissue homeostasis and cell renewal, in addition to maintenance of the cell–cell junctions (2).
Given the multifarious nature of β-catenin and far-ranging effects of the perturbations in this critical pathway, Wnt/β-catenin signaling plays an important role in both normal tissue homeostasis and tumorigenesis. This review summarizes the significance of the Wnt/β-catenin signaling pathway in gastrointestinal malignancies, with an emphasis upon PDAC and colorectal cancer (CRC). Also, this review describes the relationship between the Wnt/β-catenin pathway and mucins, which are glycoproteins that play important roles in various malignancies.
The role of Wnt/β-catenin signaling in cancer
The Wnt/β-catenin signaling plays important role in development as well as homeostasis of adult tissue. As expected, mutations in this pathway occur frequently in cancer, most commonly in CRC, where around 80% of the patient population possesses either inactivating mutations in adenomatous polyposis coli (APC) or activating mutations in β-catenin (5). However, aberrant activation of this pathway also occurs in pancreatic cancer (PC), breast cancer, multiple myelomas, melanoma, hepatocellular carcinoma and other malignancies (4,10–12). Both mutations in Axin 1/2 (13) and activating mutations in β-catenin (3) occur in hepatocellular carcinoma. Mutations that prevent the phosphorylation-mediated degradation of β-catenin also occur in medulloblastoma (3) and the pediatric renal cancer Wilm’s tumor (14). Activation of Wnt/β-catenin signaling may also be wrought by epigenetic mechanisms, as observed in CRC and PC, as well as medulloblastoma, where the promoters of Wnt inhibitors were found to be hypermethylated (15,16).
Activation of the Wnt/β-catenin pathway can be precipitated either by overt mutations in pathway components or indirect mechanisms, such as increased secretion of ligands or decreased secretion of inhibitors. These two mechanisms of Wnt/β-catenin activation are exemplified by CRC and PC, both gastrointestinal malignancies where the Wnt/β-catenin pathway plays a significant role in disease progression, albeit through distinct mechanisms.
Wnt/β-catenin in CRC
Aberrations in the Wnt/β-catenin pathway frequently occur in CRC. While mutations in several Wnt/β-catenin pathway have been recorded, an overwhelming majority of CRCs (70-80%) possess truncating mutations in APC (17). Individuals with familial adenomatous polyposis possess truncating mutations in APC, rendering them liable to the formation of hundreds of polyps in their colon, ultimately leading to CRC (1). The increase in cytosolic/nuclear β-catenin could also be due to mutations in the exon 3 of β-catenin, which render it resistant to degradation, seen in less than 5% of CRCs (17). Mutations in Transcription factor 7-like 2 (TCF7L2 or TCF4), the nuclear partner of β-catenin, have also been observed in 5% of CRCs (17). It must be noted, however, that distinct molecular subtypes of CRC exist, and that while mutations in the Wnt/β-catenin pathway are very frequent, not all CRCs are driven by aberrant Wnt/β-catenin signaling.
The majority of CRCs follow what is often referred to as the ‘suppressor’ pathway (18). Here, both precursor lesions; ‘traditional’ adenomas, as well as full-blown tumors are characterized by aberrantly localized β-catenin, typically a consequence of truncated APC mutations (1). Further, truncating mutations in APC are present in the earliest lesions, aberrant crypt foci, suggestive of a driving role for the Wnt/β-catenin pathway (1). In addition, around 50% of these tumors have a Kras mutation (19), which has been shown to aid in nuclear localization of β-catenin (20). The levels of nuclear β-catenin steadily increase during the progression of CRC, starting from adenomas to full-blown carcinomas (1). A subset of CRCs are characterized by frequent aberrations in the DNA mismatch repair machinery, often called the ‘mutator’ pathway (18). These tumors possess microsatellite instability (MSI) and are less likely to possess Wnt/β-catenin driver mutations (21,22). Yet another subtype, mucinous CRC, comprising roughly 10% of all CRCs (23) has also been observed. These tumors are characterized by excessive mucin production (chiefly MUC2) are also less likely to have aberrations in the Wnt/β-catenin as driving mutations since they also frequently possess MSI-high (MSI-H) status (24). Each of these CRC subtypes are further stratified by varying frequencies of BRAF, KRAS mutations as well as CIMP (CpG island methylator phenotype) (25). A detailed analysis of the various subtypes of CRC is, however, beyond the purview of this review.
Wnt/β-catenin in PC
Unlike CRC, where mutations in the Wnt pathway are important driver mutations, PC does not usually display such mutations. Despite this, around 65% of PDACs show aberrant nuclear/cytosolic localization of β-catenin and active Wnt signaling (26). A significant fraction of PDAC patients also show elevated Axin2 expression, widely regarded as a universal marker of active Wnt/β-catenin signaling (27). Further, the Wnt pathway was found to be one of the 12 core signaling pathways most frequently dysregulated in PDAC (28).
The proposed causes for the increase in Wnt/β-catenin signaling in PDAC include epigenetic regulation of the Wnt pathway components, increased ligand secretion and decreased expression of pathway inhibitors. For example, the promoter of the Wnt-inhibitor SFRP1 was found to be hypermethylated in PDAC (16). Also, the canonical Wnt ligand, Wnt 7b, was found to be over-expressed in PC (29). Wnt 7b independently confers a poorer prognosis to patients. Furthermore, oncogenic Kras has been shown to induce expression of the ataxia telangiectasia group D complementing gene (ATDC) (30), which indirectly activates β-catenin signaling via stabilization of Dishevelled-2 (Dvl2), abolishing the destruction complex. In a mouse model that expressed transgenic ATDC and mutant Kras driven by a pancreas-specific promoter p48-Cre, ATDC was found to induce the epithelial-to-mesenchymal transition (EMT) and metastasis via β-catenin (30,31). It has also been reported that Wnt ligand agonists Sulfatase 1 (SULF-1) and SULF-2 are overexpressed in PDAC (32). Other developmental pathways, such as the Notch and Hedgehog pathways, have also been shown to cause increased nuclear/cytosolic β-catenin in PDAC (33).
A number of mouse models have also shed light on the role played by Wnt/β-catenin in PDAC progression. The KRAS mutation is considered the driving mutation in PDAC, present in around 90% of patients (31). Mice that express the Kras G12D mutation, driven by the expression of a pancreas-specific Cre (Pdx or p48), develop precursor lesions (mPanINs) that eventually form PDACs reminiscent of the majority of human lesions (34). In contrast, mice that express mutant, stabilized β-catenin (exon 3-deleted), driven by a pancreas-specific p48-Cre, develop solid pseudopapillary neoplasms, an extremely uncommon form of the disease (35). Mice that express both stabilized β-catenin and mutant Kras, driven by p48-Cre (p48-Cre; Cttnb1exon3/+; KrasG12D), develop tumors that resemble intraductal tubular neoplasms, yet another extremely rare disease (35). However, activation of canonical Wnt signaling is necessary for the formation of pancreatic intra-epithelial neoplasia (PanINs) and PDAC in the Ptf1a-Cre; KrasG12D (KC) mouse model (36), albeit at levels substantially lower than observed in CRC, as demonstrated by Zhang et al, who generated a Ptf1a-Cre; KrasG12D; β-cateninf/f mouse model. It was observed that the loss of β-catenin prevented the formation of mPanINs (mouse precursor lesions). Notably, β-catenin-depleted cells expressed lower levels of mucins in these mice, as determined by Periodic acid–Schiff (PAS) staining.
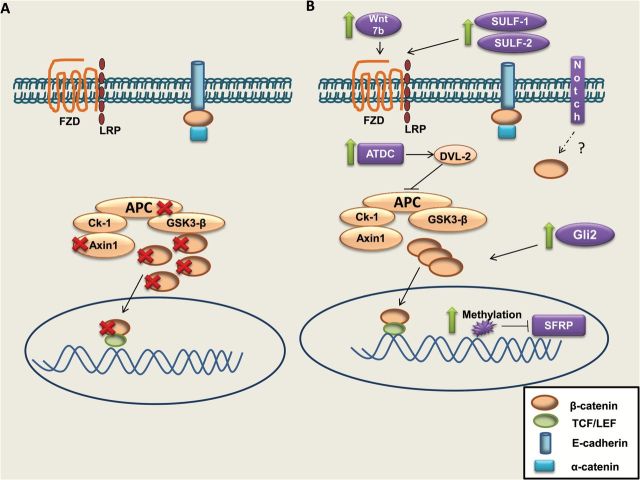
Thus, while β-catenin alone is unable to initiate pancreatic tumorigenesis, canonical Wnt/β-catenin signaling is required for PDAC progression and contributes to EMT and metastasis. The temporal regulation of Wnt/β-catenin pathway activation appears to be critical considering that β-catenin signaling has been reported to actively suppresses Kras-mediated tumorigenesis by acinar cell regeneration in a mouse model of pancreatitis (37). Figure 1 pictorially summarizes the mechanism by which the Wnt/β-catenin pathway is dysregulated in both PDAC and CRC.
Figure 1.
Mechanisms of Wnt/β-catenin upregulation in CRC and PDAC. (A) In CRC, mutations in Adenomatous polyposis coli (APC; most common), β-catenin and Axin1 contribute to increased nuclear β-catenin. (B) In pancreatic ductal adenocarcinoma (PDAC), no mutations in canonical pathway members are typically found. However, an increase in the WNT 7B ligand, SULF1 and SULF2, which enhance Wnt ligand binding and an increased expression of the ataxia telangiectasia group D complementing gene (ATDC), inhibit formation of the destruction complex and work together to precipitate cytosolic/nuclear β-catenin. In addition, other developmental pathways, such as Notch and Hedgehog, can contribute to the pool of nuclear β-catenin.
Mucins and their roles in cancer
Mucins are heavily O-glycosylated proteins that are normally expressed in the epithelial lining of the lungs, and gastrointestinal and reproductive tracts (38). Mucins can be broadly categorized as follows: (i) membrane-bound/trans-membrane mucins, which include MUC1, MUC3A/MUC3B, MUC4, MUC11, MUC12, MUC13, MUC15, MUC16, MUC17 and MUC21, (ii) secreted (gel-forming) mucins, which include MUC2, MUC5AC, MUC5B, MUC6 and MUC19, and (iii) soluble (non-gel-forming) mucins, which include MUC7, MUC8, MUC9 and MUC20 (38). The normal functions of mucins involve protection of the epithelial surfaces via entrapment of pathogens, which is primarily a function of secreted mucins (39). The transmembrane mucins can also be involved in cell signaling (39). Thus, mucins are critical in maintaining cellular functions, particularly those of epithelial surfaces.
While critical in maintaining and protecting the normal epithelium, decades of research have revealed that a number of mucins are aberrantly expressed in cancer. For instance, the transmembrane mucin MUC4, which is normally absent in the pancreas, is aberrantly overexpressed in PDAC (40). MUC4 has been shown to act as a binding partner for HER2 via its EGF-like domains, thereby playing a role in promoting metastasis, cell proliferation, and invasion (41–45). Furthermore, MUC4 can interact with secreted endothelial proteins such as galectin (46), thus aiding in the invasion and metastasis of PDAC cells. Likewise, mucins MUC1, MUC16 and MUC5AC are also overexpressed in PDAC (39).
Interestingly, while some mucins are aberrantly overexpressed in cancer, expression of other mucins decreases in certain malignancies. For instance, expression of the secreted-mucin MUC2, which comprises most of the secreted mucus layer in the colon, is markedly reduced in most CRCs, with certain notable exceptions, such as mucinous CRCs (47,48). The loss of MUC2 is more frequently associated with non-mucinous, non-MSI CRCs and epigenetic factors as well as other regulatory mechanisms have been shown to be responsible for this loss (49,50). The role played by MUC4 in CRC is, however, unclear (51,52). While some reports suggest that MUC4 is lost as CRC progresses (51), another report indicates that MUC4 expression is enhanced in a subset of patients where it confers a poorer prognosis (52). No correlation between high MUC4 expression and MSI or mucinous status of CRC has been observed (23). MUC1 is expressed in both normal and cancerous colons, but its expression increases in CRC and strongly confers with disease progression (53). As with MUC4, MUC1 expression does not correlate with MSI status (23). Other mucins, such as MUC5AC, which is not normally expressed in the colon, are also aberrantly overexpressed in CRC (54). The de novo expression of MUC5AC is more frequently observed in mucinous and MSI-high tumors (55,56). Altered mucin glycosylation has also been shown to contribute to metastasis (57). Thus, despite varying expression levels in disparate malignancies, the importance of mucins in disease progression is evident, either due to loss of protective mucins such as MUC2 in non-mucinous CRCs or the gain of aberrantly glycosylated/abnormally abundant mucins.
Given their role in promoting cancer progression, mucins have been proposed to be important diagnostic and prognostic markers. Consequently, the mechanisms by which these molecules promote and/or suppress the progression of cancer have been subject of intense investigation. In this regard, a number of studies have focused on the interaction of β-catenin and mucins. Most prominently, the cytoplasmic tail of the transmembrane mucin MUC1 has been shown to interact with β-catenin in various malignancies and aid in the nuclear localization of the molecule (58). Other mucins, like MUC16 and MUC4, have also been shown to influence the localization and/or stabilization of β-catenin (59,60).
While many studies have shown the regulation of β-catenin by mucins, the Wnt/β-catenin pathway has also been shown to regulate mucin expression. In CRC, where the β-catenin pathway is a driving force, β-catenin has been shown to suppress mucin expression. A siRNA targeting β-catenin resulted in the loss of mucin expression, as measured by alcian blue staining, in the CRC cell line LS174T (61). Activation of β-catenin has been shown to result in the loss of colonocyte differentiation, resulting in a crypt progenitor phenotype in CRC (62). The loss of mucin expression is among the myriad of changes associated with this de-differentiated phenotype. Mucin-depleted foci, first identified in rats treated with the carcinogen azoxymethane, are pre-cancerous lesions in CRC characterized by both aberrant β-catenin signaling and loss of mucins (63). Mucin-depleted foci have also been described in human colon tissue samples (64). Thus, a number of studies have implied that the loss of certain mucins is a consequence of Wnt/β-catenin pathway activation in CRC. The following sections of the review summarize the diverse roles played by mucins and β-catenin in cancer.
The relationship between β-catenin and membrane-bound mucins in cancer (MUC1, MUC4 and MUC16)
MUC1.
The relationship between MUC1 and β-catenin has been extensively studied in various malignancies. The MUC1 cytoplasmic domain (CD) has been shown to possess a serine-rich motif (SXXXXXSSL) required for the binding of β-catenin (65), which is used to bind the Armadillo repeat domain of β-catenin, thus preventing the phosphorylation induced degradation of β-catenin by GSK3β. Cleavage of the MUC1 CD has been demonstrated to occur through γ-secretase, thus untethering the cytoplasmic tail from the membrane (66). In addition, the MUC1 CD possesses a binding site for GSK3β, at the STDRSPYEKV site (67). Phosphorylation by GSK3β of the serine residue next to the proline at this site inhibits the MUC1–β-catenin interaction and stimulates the formation of the β-catenin–E-cadherin complex at the membrane. The MUC1 CD also possesses a phosphorylation site for the epidermal growth factor receptor (EGFR), which phosphorylates the tyrosine residue in the YEKV motif on the MUC1 CD, thus priming the tail for binding by the c-Src tyrosine kinase, which leads to increased β-catenin–MUC1 CD interaction (68,69). Protein kinase-C δ phosphorylates the tail at the TDR site, also leading to increased β-catenin binding to the MUC1 CD (68–70). The MUC1 CD–β-catenin complex can enter the nucleus, where it partners with TCF4 to upregulate β-catenin target genes, such as cyclin D1 (71,72). Not only does MUC1 stabilize β-catenin, it also binds the nuclear co-factor TCF4, preventing binding of the repressive C-terminal binding proteins to TCF4 and recruiting transcriptional co-activators such as p300 on the cyclin D1 promoter (73). The expression of MUC1 has been linked to the Wnt target gene Cyclin D1 in a number of cancers, such as breast cancer (73), H.pylori-induced gastric cancer (71), and PDAC (72,74).
The expression of MUC1 and aberrant β-catenin at the invasive front in gastric cancer and CRC has been shown to be independent predictors of poorer prognosis (75,76). The MUC1–β-catenin interaction is implicated in inducing invasion and EMT in breast, renal, gastric and PCs (73,77,78). In PC, the seven tyrosine residues present in the MUC1 cytoplasmic tail were found to be critical for its interaction with β-catenin and mediation of EMT (78). In mouse NIH3T3 fibroblast cells, the interaction between Galectin-3 and the N-terminal domain of MUC1 was found to trigger recruitment of β-catenin to the C-terminus of MUC1 (79). In renal carcinoma, the MUC1–β-catenin complex has been found to directly bind the Zinc finger protein SNAI1 (SNAIL) promoter, thus triggering EMT and invasion (77,80). Additionally, the KL6 variant of MUC1 has been found to exacerbate metastasis of PDAC through interactions with β-catenin (81). Further, the MUC1 cytoplasmic tail has been shown to interact with APC in some breast cancer cell lines and in human metastatic breast cancer tissue (82). Moreover, MUC1 was found to aid in the nuclear localization of β-catenin in CRC (83). Thus, multiple lines of evidence show a direct relationship between MUC1 and EMT, metastasis and progression of various cancers.
In contrast to the aforementioned findings, in HEK293 cells, MUC1 has been implicated in suppressing the proliferation of cancer by preventing nuclear localization of β-catenin (84), thus contradicting a number of studies. The MUC1–β-catenin interaction may also promote cancer progression without necessitating the nuclear localization of β-catenin. For example, in breast cancer, the deletion of MUC1 in MMTV-Wnt-1 transgenic mice prolonged the time required for tumor formation (85). However, the MUC1–β-catenin complex was observed in the membrane and cytosol of wild-type mice, as opposed to the nucleus. Further, the MUC1–β-catenin complex was present at the invading edge of the cell membrane connecting to the collagenous matrix; this complex co-localized with the focal adhesion proteins fascin and vinculin, thus presumably aiding in invasion and metastasis despite preventing the nuclear localization of β-catenin. Accordingly, MUC1–β-catenin interactions were found to be greatly enriched in metastatic tumors (85).
In summary, MUC1–β-catenin interactions may either (i) promote nuclear localization of β-catenin, thereby upregulating numerous EMT-, metastasis- and proliferation-related genes or (ii) prevent nuclear localization of β-catenin by sequestering it at the membrane/cytoskeleton. It has been suggested that the relative abundance of these two proteins may determine which path is followed (84). A recent study of PDAC observed that the MUC1–β-catenin regulation of cyclin D1 requires the presence of p120 catenin, which sequesters the transcriptional repressor Kaiso. This observation suggests that the relative abundance of various p120 isoforms determines the ability of MUC1–β-catenin to activate gene transcription (72). Thus, while the MUC1–β-catenin interaction possesses the ability to promote EMT and metastasis, several variables such as the relative abundance of MUC1/β-catenin and the presence of requisite isoforms of as p120 catenin influence the MUC1–β-catenin dynamic.
MUC4.
While the relationship between MUC1 and β-catenin has been extensively studied, the potential relationship between β-catenin and other membrane-bound mucins, such as MUC4, is less known. In PDAC, MUC4 induces the dissociation of β-catenin from E-cadherin, by triggering lysosomal degradation of E-cadherin via HER2/Src/FAK signaling, and thereby causes nuclear localization of β-catenin (59). Gao et al. proposed that MUC4 can also inhibit nuclear localization of β-catenin in lung cancer, where MUC4 plays a protective role (86). MUC4 may also be governed by β-catenin. For example, a recent study, using genetically ablated β-catenin by zinc finger nucleases in the PDAC cell line BXPC3, performed a subsequent microarray to demonstrate that MUC4 was one of the most significantly downregulated transcripts upon the depletion of β-catenin (Supplementary Table 3 of paper by Olson et al.) (87). Moreover, whole-exome sequencing of a case of osteosarcoma showed that the Wnt/β-catenin pathway is an important disease driver and that MUC4 was upregulated, hinting at a regulatory relationship between β-catenin and MUC4 (88).
A number of other studies also support the existence of a β-catenin–MUC4 regulatory relationship. A study by Hashimoto et al., which examined the role of β-catenin in developing lungs, used a lung-specific rCCSP-Cre recombinase in mice and found that when a constitutively active (exon 3-deleted) β-catenin was overexpressed, MUC4 transcript levels were significantly increased in the bronchial epithelium (89). Our MUC4-promoter analysis using the MatInspector (Genomatix) software showed the presence of 3 putative TCF/LEF binding sites (one in the proximal promoter and two in the distal promoter) in the MUC4 promoter. Additional studies conducted in our laboratory further suggest that, in PDAC, MUC4 is a direct transcriptional target of β-catenin (90).
Overall, most studies thus far indicate that the Wnt/β-catenin pathway likely regulates the expression of MUC4. Furthermore, MUC4 may regulate nuclear localization of β-catenin through its interactions with HER2, which then triggers a cascade of signaling events that culminate in the Src-mediated phosphorylation of E-cadherin and result in the release of β-catenin from E-cadherin. However, MUC4 has also been shown to prevent the nuclear localization of β-catenin in lung cancer through a mechanism that has not yet been delineated (86), thus contributing to a certain degree of uncertainty in the field.
MUC16.
MUC16 is a membrane-bound mucin that is upregulated in various cancers, including ovarian, pancreatic and breast cancers (91). The first report suggesting an interaction between MUC16 and β-catenin was published by Comamala et al. in 2011; here, MUC16 was shown to interact with β-catenin in the ovarian carcinoma cell line OVCAR3 (92). Comamala et al. proposed that MUC16 interacts with E-cadherin and β-catenin, thus ensuring their membrane localization and preventing EMT. This co-related with the loss of MUC16 in late-stage, metastatic ovarian cancer. A study by Akita et al. focused on the interaction between the cytoplasmic tail of MUC16 and β-catenin in a colon cancer cell line, HCT116. It was determined that overexpression of the MUC16 cytoplasmic tail resulted in reduced expression of membranous E-cadherin and β-catenin. This reduced expression was attributed to the increased recruitment of Src family kinases to the membrane, which in turn caused degradation of E-cadherin and dissociation of β-catenin from E-cadherin (93).
In 2014, Giannakouros et al. proposed that the MUC16–β-catenin complex promotes the formation of multicellular aggregates. These aggregates precede dissemination of ovarian cancer cells and the MUC16–β-catenin interaction tethers β-catenin at the membrane, preventing phosphorylation-mediated degradation of β-catenin by GSK-3β (60). Thus, while most studies indicate that the MUC16 cytoplasmic tail does indeed interact with β-catenin, the role played by the MUC16–β-catenin complex appears to be context-dependent, and either causes increased cytosolic/nuclear localization of β-catenin or prevents the nuclear localization and enhances the E-cadherin binding propensities of β-catenin.
The relationship between β-catenin in cancer and secreted-mucins MUC2, MUC5AC and MUC6
MUC2 is the most abundantly secreted mucin in the intestines, produced primarily by goblet cells (51). Notably, unlike most colorectal carcinomas in humans, Muc2−/− mice develop colorectal carcinomas and colitis in the absence of any other mutations (94,95) and without aberrantly localized β-catenin. However, when Muc2−/− mice were crossed with Apc1638N/+ or ApcMin/+ mice in a study by Yang et al., increased tumor formation occurred in the distal colon (96). This is unlike mice where the Apc gene alone has been mutated or lost, and tumor lesions are located primarily in the small intestine (96). Importantly, these tumors in the distal colon showed an increase in aberrant Wnt/β-catenin signaling, which suggests that the loss of MUC2 acts in concert with Wnt/ β-catenin signaling to cause CRC (95,96). Because these mice also showed signs of an inflammatory response in their tumors, it was suggested that loss of MUC2 results in tumors through an inflammatory mechanism that complements activation of the Wnt pathway (96). Interestingly, in colon carcinomas, β-catenin has also been shown to negatively regulate MUC2 expression (97). This negative regulation of MUC2 was found to be driven by Sox9, which in turn is upregulated by β-catenin (98). Another mechanism of down-regulation of MUC2 by β-catenin involves Hath-1, a transcription factor that is proteasomally degraded by active Wnt/β-catenin signaling (99). Hath-1 up-regulates MUC2 expression (97) and is repressed by the Wnt/β-catenin pathway (99). Other mechanisms such as MUC2 promoter methylation have also been shown to contribute to the MUC2 mucin loss in CRC (100). In conclusion, the loss of MUC2 plays an important role in CRC progression and the Wnt/β-catenin pathway aids in precipitating the loss of MUC2 expression. The loss of MUC2 may promote inflammatory responses which exacerbate the severity of the disease.
The gastric, gel-forming, secreted-mucin MUC5AC is aberrantly overexpressed in colorectal carcinoma (51), PDAC (39) and in certain infections, including the infection caused by the bacterial pathogen Shigella dysenteriae (101). Some studies suggest that β-catenin can regulate MUC5AC. It has been shown that S.dysenteriae stimulates secretion of interleukin-1β, which in turn causes Trefoil factor 3 to stimulate the Akt pathway by phosphorylating the EGF receptor (101). The Akt pathway then potentiates the nuclear localization of β-catenin, which in turn upregulates MUC5AC (101). In rats treated with 1, 2-dimethylhydrazine, a carcinogen, it was seen that crypts that contained aberrant localization of β-catenin showed progressively increasing levels of MUC5AC (33% immunopositivity at 8 weeks and 90% immunopositivity at 36 weeks) concurrent with progressively decreasing levels of MUC2 (102). A study by Mucenski et al. used transgenic mice that constitutively overexpressed transcriptionally active, exon 3-deleted β-catenin, which was achieved using doxycycline-regulated Cre recombinase regulated by the lung-specific rat Clara cell secretory protein (rCCSP) promoter. Here, it was observed that mice displayed goblet cell dysplasia and increased MUC5AC expression (103). All the aforementioned studies suggest that MUC5AC expression is likely governed by β-catenin, although the precise manner in which β-catenin regulates MUC5AC has not yet been studied.
Concurrent with findings that suggest a regulatory relationship between β-catenin and MUC5AC, it has also been found that MUC5AC can increase nuclear accumulation of β-catenin. Specifically, a study by Inaguma et al. determined that MUC5AC can prevent membranous accumulation of E-cadherin, and therefore untether β-catenin from the adherens junction complex and stimulate nuclear accumulation of β-catenin in PDAC (104). Thus, MUC5AC expression has been reported to be governed by Wnt/β-catenin signaling in S.dysenteriae infections in the colon, and possibly also in colon cancer and in the developing lungs. On the other hand, MU5AC has also been shown to regulate β-catenin localization in PC.
The secreted-mucin MUC6, in conjunction with MUC1 and MUC2, is frequently associated with the presence of nuclear β-catenin in gastrointestinal type gastric cancer and has been proposed for a prognostic marker by Aihara et al. (105). A separate study by Silva et al. found that patients with gastric cancer who were younger (less than or equal to 40 years old) were more likely to express MUC6, MUC5AC and MUC2, as well as β-catenin, compared to older patients (above 40 years old) (106). However, the prognostic value of these observations remains unclear.
Conclusions and future directions
Despite the proven importance of the Wnt/β-catenin pathway and mucins in regulating neoplastic transformation and malignant growth, a number of questions remain unanswered. While the MUC1–β-catenin relationship is the most well delineated of all mucin–β-catenin relationships, the determinants or context that governs whether the MUC1–β-catenin complex enters the nucleus or remains bound to members of the cytoskeleton have not yet been identified. Notably, another transmembrane mucin, MUC16, has been shown to interact with β-catenin. While the cleavage and nuclear localization of the MUC16 cytoplasmic tail has been demonstrated (107), it is not known whether the β-catenin-bound MUC16 cytoplasmic tail can also enter the nucleus. Also, the role played by the MUC16-CT–β-catenin complex in ovarian cancer, where it prevents EMT by sequestering β-catenin at the membrane, and in colon and breast cancers, where it enhances EMT by enabling cytosolic/nuclear localization of β-catenin, is lacking sufficient explanation. A similar pattern has been observed for MUC4, where MUC4 prevents β-catenin nuclear localization in lung cancer but enhances nuclear localization in PC.
Another stream of research focuses on the regulation of mucin expression by β-catenin. This field is replete with findings reflective of the disparate roles played by the collusion of mucins and β-catenin in various malignancies. For example, the depletion of β-catenin in the Ptf1a-Cre; KrasG12D; β-cateninf/f, as well as inhibition of β-catenin in the Ptf1a-Cre; LSL-KrasG12D; Rosa26rtTa/+; TetO-Dkk1 mouse models of PDAC resulted in reduced overall mucin staining compared to KC (Ptf1a-Cre;KrasG12D) mice, hinting at a regulatory β-catenin–mucin relationship (36). While this is not conclusive proof of a direct β-catenin–mucin relationship, a study that performed a microarray on β-catenin-null PDAC cells found that MUC4 was one of the top significantly down-regulated transcripts, implying that MUC4 is regulated by β-catenin (87). Studies from our laboratory support direct β-catenin-mediated upregulation of the MUC4 transcript in PC (90). In CRC, however, β-catenin has been shown to repress mucin expression. The β-catenin-mediated repression of MUC2 in CRC occurs via Sox9 up-regulation. Additional studies from our laboratory (unpublished data) indicate that β-catenin can repress the MUC4 transcript in CRC. The reasons for this seemingly contradictory relationship could stem from differing status of MUC4 promoter methylation in these two cancers. Studies by Yamada et al. showed using the CaCo2 CRC cell line negative for MUC4 mRNA, that while the MUC4 promoter is methylated at key positions (CpG sites 108-112 in the proximal promoter), these sites are unmethylated in PDAC cell lines expressing MUC4 (108). A repressive histone code including deacetylated Histone 3 and trimethylated K27H3 in the MUC4 5′ UTR is also present in MUC4-negative cell lines (109). One possibility is that, given the wide disparity in the degree of Wnt/β-catenin pathway activity in PDAC and CRC (50–80% nuclear β-catenin in CRC versus 4–11% nuclear β-catenin in PDAC (5)), a different set of target genes are activated. A study by Hlubek et al. supports this possiblity. Here, it was observed that even within CRC tumors, varying degrees of nuclear β-catenin activate different target genes in the tumor center as opposed to the invasive front (110). Another factor to consider is that while aberrant β-catenin is the driving force in CRC progression, dysregulation of β-catenin occurs at a later stage in the natural history of PDAC. Thus, a number of other mutations are at play, possibly influencing methylation of the mucin promoter and other variables that affect the β-catenin–mucin relationship. Furthermore, β-catenin regulates a number of micro-RNAs (mi-RNAs) in CRC (111). Interestingly, MUC4 has also been shown to be regulated by mi-RNAs such as miR-200c, miR-219-1-3p and miR-150 (112–114), suggesting the possibility that β-catenin may indirectly regulate MUC4 via mi-RNAs. Further studies to address the potentially disparate roles played by β-catenin vis-à-vis MUC4 in these two cancers can delineate the precise relationship between β-catenin and MUC4. Like the apparent differential regulation of MUC4 by β-catenin in CRC and PDAC, other mucins such as MUC5AC are possibly differentially regulated by Wnt/β-catenin, highlighting the context-dependent nature of interactions between β-catenin and mucins.
The apparent discordance between mucin–β-catenin relationships in PDAC and CRC could be due to the distinct functions of secretory and membrane bound mucins in normal and cancerous conditions. The normal adult gastrointestinal system is characterized by specific levels of secretory and transmembrane mucins in every tract/organ. For instance, MUC2 is the predominant secretory mucin in the intestine, while MUC5AC is highly preponderant in the stomach (115). The outer, loose layer of MUC2 harbors the intestinal microflora, while the dense inner MUC2 layer protects the colonic epithelium from microbial assault (115). Transmembrane mucins such as MUC1, MUC3, MUC17, MUC12 and MUC4 form the protective glycocalyx while also participating in cell signaling events, although their functions in normal conditions are not as well studied (115). The variable lengths of the extracellular portions of each mucin form a multi-tiered barrier to pathogen invasion (116). This delicate balance between the levels and functions of transmembrane and secreted mucins is disrupted in cancer. In conjunction with varying degrees of aberrant β-catenin signaling, it is likely that each transmembrane and secretory mucin plays a highly context-dependent role in cancer, based on its expression and role in the normal tissue.
Figure 2 and Table 1 summarize the relationship between the Wnt/β-catenin pathway and mucins. Notably, the cytoplasmic tails of MUC1 and MUC16 have been shown to interact with β-catenin. In both instances, the mucin–β-catenin complexes can either promote or repress tumor progression. The MUC1 cytoplasmic tail–β-catenin complex, in particular, plays an important role in various cancers. Likewise, in PDAC and lung cancer, MUC4 has been shown to affect β-catenin localization. In colorectal and PC, the Wnt/β-catenin pathway, on the other hand, can also regulate expression of MUC5AC, MUC2 and MUC4. Thus, mucins and the Wnt/β-catenin pathway have an intricate, mutually regulatory relationship that often culminates in cancer progression. As mentioned earlier, further studies are needed to determine the factors governing β-catenin–MUC1 mucin nuclear localization as well as the individual roles played by β-catenin–mucin complexes in different cancers. While it is apparent that β-catenin can govern mucin expression, both indirectly (i.e. MUC2 in CRC) and directly (i.e. MUC4 in PC), future studies are needed to explain the seemingly conflicting nature of the β-catenin–mucin relationship in various malignancies.
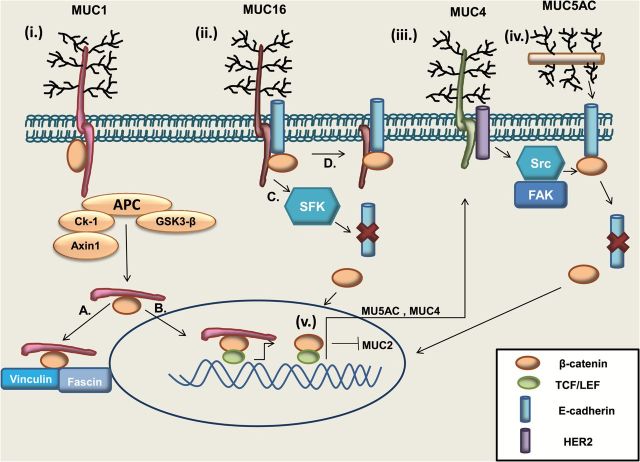
Figure 2.
Schematic representation of β-catenin–mucin dynamics. (i) MUC1: The cytoplasmic tail of MUC1 binds to β-catenin at the serine-rich domain. Formation of the MUC1–β-catenin complex stabilizes β-catenin by preventing its phosphorylation-mediated degradation. Following cleavage of the cytoplasmic tail, the MUC1–β-catenin complex can either, (A) localize adjacent to the membrane and bind cytoskeleton members Fascin and Vinculin, or (B) enter the nucleus, where MUC1 can also bind Transcription factor 7-like 2 (TCF7L2/TCF4) and aid in transcription of β-catenin target genes such as Cyclin D1. (ii) The MUC16 cytoplasmic tail can bind both β-catenin and E-cadherin. This may either cause (C), the recruitment of Src family kinases (SFKs) resulting in the degradation of E-cadherin and release of β-catenin in the cytosol or (D), the stabilization of the β-catenin–E-cadherin complex. (iii) MUC4 can act as a binding partner for HER2, triggering the activation of Src and FAK, which cause the lysosomal degradation of E-cadherin and release of β-catenin into the cytosol. (iv) MUC5AC in the extracellular space can hinder the membranous localization of E-cadherin and stimulate cytosolic and nuclear β-catenin. (v) β-catenin has been shown to transcriptionally upregulate MUC5AC and MUC4, and transcriptionally repress MUC2.
Table 1.
The relationship between β-catenin and mucins in cancer
| Mucin | Type | Cancer | Relationship with β-catenin | References |
|---|---|---|---|---|
| MUC1 | Transmembrane | PDAC | The cytoplasmic tail binds β-catenin, enters the nucleus, and upregulates Cyclin D1, which induces EMT | (72,74) |
| CRC | The cytoplasmic tail binds β-catenin, enters nucleus and colocalizes with β-catenin at invasive front | (76,83) | ||
| Breast cancer | The cytoplasmic tail binds β-catenin, enters nucleus, co-localizes with β-catenin at the invasive front | (73) | ||
| MUC16 | Transmembrane | Ovarian cancer | The cytoplasmic tail binds β-catenin, suppresses EMT by preventing cytosolic localization and helps form multicellular aggregates that precede metastasis by tethering β-catenin to membrane | (60,92) |
| CRC | The cytoplasmic tail binds β-catenin and recruits Src family kinases, thus triggering degradation of E-cadherin and enhancing cytosolic β-catenin | (93) | ||
| MUC4 | Transmembrane | PDAC | MUC4 causes dissociation of β-catenin from E-cadherin by HER2/Src/ FAK signaling, enhances nuclear β-catenin, and is transcriptionally up-regulated by β-catenin | (59,90) |
| Lung cancer | MUC4 prevents nuclear localization of β-catenin | (86) | ||
| MUC5AC | Secreted, gel forming | PDAC | Enhances nuclear β-catenin by disrupting E-cadherin at the membrane | (104) |
| CRC | β-catenin upregulates MUC5AC in HT29 CRC cell line; nuclear β-catenin is associated with enhanced MUC5AC expression | (101,102) | ||
| MUC2 | Secreted, gel forming | CRC | β-catenin represses MUC2; loss of MUC2 co-operates with β-catenin signaling to aid in cancer progression | (96–99) |
| MUC6 | Secreted, gel forming | Gastric cancer | Expression frequently associated with nuclear β-catenin; co-expression is prognostic marker | (105,106) |
Funding
National Institutes of Health, NIH (EDRN UO1 CA111294, SPORE P50 CA127297, TMEN U54 CA163120 and RO1 CA183459).
Acknowledgements
We would like to thank Ms. Melody Montgomery, Editor, for professionally editing this manuscript.
Conflict of interest statement: None declared.
Glossary
Abbreviations
- APC
adenomatous polyposis coli
- CRC
colorectal cancer
- EMT
epithelial-to-mesenchymal transition
- MUCs
mucins
- MSI
microsatellite instability
- GSK3-β
glycogen synthase kinase β
- PC
pancreatic cancer
- PDAC
pancreatic ductal adenocarcinoma
References
- 1. Fodde R., et al. (2001) APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer, 1, 55–67. [DOI] [PubMed] [Google Scholar]
- 2. Valenta T., et al. (2012) The many faces and functions of beta-catenin. EMBO J., 31, 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polakis P. (2007) The many ways of Wnt in cancer. Curr. Opin. Genet. Dev., 17, 45–51. [DOI] [PubMed] [Google Scholar]
- 4. Polakis P. (2012) Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol., 4, a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. White B.D., et al. (2012) Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology, 142, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jamieson C., et al. (2014) Targeting the β-catenin nuclear transport pathway in cancer. Semin. Cancer Biol., 27, 20–29. [DOI] [PubMed] [Google Scholar]
- 7. Nüsslein-Volhard C., et al. (1980) Mutations affecting segment number and polarity in Drosophila. Nature, 287, 795–801. [DOI] [PubMed] [Google Scholar]
- 8. Coates J.C., et al. (2002) Loss of the beta-catenin homologue aardvark causes ectopic stalk formation in Dictyostelium. Mech. Dev., 116, 117–127. [DOI] [PubMed] [Google Scholar]
- 9. Lyashenko N., et al. (2011) Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat. Cell Biol., 13, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmeel L.C., et al. (2013) Targeting the Wnt/beta-catenin pathway in multiple myeloma. Anticancer Res., 33, 4719–4726. [PubMed] [Google Scholar]
- 11. Geyer F.C., et al. (2011) β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod. Pathol., 24, 209–231. [DOI] [PubMed] [Google Scholar]
- 12. Damsky W.E., et al. (2011) β-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell, 20, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salahshor S., et al. (2005) The links between axin and carcinogenesis. J. Clin. Pathol., 58, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Su M.C., et al. (2008) Beta-catenin expression and mutation in adult and pediatric Wilms’ tumors. APMIS, 116, 771–778. [DOI] [PubMed] [Google Scholar]
- 15. Kongkham P.N., et al. (2010) The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene, 29, 3017–3024. [DOI] [PubMed] [Google Scholar]
- 16. Vincent A., et al. (2011) Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin. Cancer Res., 17, 4341–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fearon E.R. (2011) Molecular genetics of colorectal cancer. Annu. Rev. Pathol., 6, 479–507. [DOI] [PubMed] [Google Scholar]
- 18. Pino M.S., et al. (2010) The chromosomal instability pathway in colon cancer. Gastroenterology, 138, 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson P.M., et al. (2010) Molecular markers in the treatment of metastatic colorectal cancer. Cancer J., 16, 262–272. [DOI] [PubMed] [Google Scholar]
- 20. Janssen K.P., et al. (2006) APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology, 131, 1096–1109. [DOI] [PubMed] [Google Scholar]
- 21. Jass J.R., et al. (2003) APC mutation and tumour budding in colorectal cancer. J. Clin. Pathol., 56, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Løvig T., et al. (2002) APC and CTNNB1 mutations in a large series of sporadic colorectal carcinomas stratified by the microsatellite instability status. Scand. J. Gastroenterol., 37, 1184–1193. [DOI] [PubMed] [Google Scholar]
- 23. Biemer-Hüttmann A.E., et al. (2000) Mucin core protein expression in colorectal cancers with high levels of microsatellite instability indicates a novel pathway of morphogenesis. Clin. Cancer Res., 6, 1909–1916. [PubMed] [Google Scholar]
- 24. Debunne H., et al. (2013) Mucinous differentiation in colorectal cancer: molecular, histological and clinical aspects. Acta Chir. Belg., 113, 385–390. [PubMed] [Google Scholar]
- 25. Phipps A.I., et al. (2015) Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology, 148, 77–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeng G., et al. (2006) Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia, 8, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lowe A.W., et al. (2007) Gene expression patterns in pancreatic tumors, cells and tissues. PLoS One, 2, e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones S., et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science, 321, 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arensman M.D., et al. (2014) WNT7B mediates autocrine Wnt/β-catenin signaling and anchorage-independent growth in pancreatic adenocarcinoma. Oncogene, 33, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L., et al. (2015) ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev., 29, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collins M.A., et al. (2013) Kras as a key oncogene and therapeutic target in pancreatic cancer. Front. Physiol., 4, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nawroth R., et al. (2007) Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS One, 2, e392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morris J.P.4thet al. (2010) KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer, 10, 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hingorani S.R., et al. (2003) Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell, 4, 437–450. [DOI] [PubMed] [Google Scholar]
- 35. Heiser P.W., et al. (2008) Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology, 135, 1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., et al. (2013) Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res., 73, 4909–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morris J.P.4thet al. (2010) Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest., 120, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Andrianifahanana M., et al. (2006) Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim. Biophys. Acta, 1765, 189–222. [DOI] [PubMed] [Google Scholar]
- 39. Kaur S., et al. (2013) Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol., 10, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andrianifahanana M., et al. (2001) Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin. Cancer Res., 7, 4033–4040. [PubMed] [Google Scholar]
- 41. Rachagani S., et al. (2012) MUC4 potentiates invasion and metastasis of pancreatic cancer cells through stabilization of fibroblast growth factor receptor 1. Carcinogenesis, 33, 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Senapati S., et al. (2012) Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene, 31, 3346–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh A.P., et al. (2004) Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res., 64, 622–630. [DOI] [PubMed] [Google Scholar]
- 44. Jonckheere N., et al. (2012) The mucin MUC4 and its membrane partner ErbB2 regulate biological properties of human CAPAN-2 pancreatic cancer cells via different signalling pathways. PLoS One, 7, e32232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jonckheere N., et al. (2013) Membrane-bound mucin modular domains: from structure to function. Biochimie, 95, 1077–1086. [DOI] [PubMed] [Google Scholar]
- 46. Senapati S., et al. (2011) Novel INTeraction of MUC4 and galectin: potential pathobiological implications for metastasis in lethal pancreatic cancer. Clin. Cancer Res., 17, 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Imai Y., et al. (2013) Differential mucin phenotypes and their significance in a variation of colorectal carcinoma. World J. Gastroenterol., 19, 3957–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogata S., et al. (1992) Mucin gene expression in colonic tissues and cell lines. Cancer Res., 52, 5971–5978. [PubMed] [Google Scholar]
- 49. Van Seuningen I., et al. (2009) Mucins: a new family of epigenetic biomarkers in epithelial cancers. Expert Opin. Med. Diagn., 3, 411–427. [DOI] [PubMed] [Google Scholar]
- 50. Jonckheere N., et al. (2010) The membrane-bound mucins: From cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie, 92, 1–11. [DOI] [PubMed] [Google Scholar]
- 51. Biemer-Hüttmann A.E., et al. (1999) Immunohistochemical staining patterns of MUC1, MUC2, MUC4, and MUC5AC mucins in hyperplastic polyps, serrated adenomas, and traditional adenomas of the colorectum. J. Histochem. Cytochem., 47, 1039–1048. [DOI] [PubMed] [Google Scholar]
- 52. Shanmugam C., et al. (2010) Prognostic value of mucin 4 expression in colorectal adenocarcinomas. Cancer, 116, 3577–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kesari M.V., et al. (2015) Immunohistochemical study of MUC1, MUC2 and MUC5AC in colorectal carcinoma and review of literature. Indian J. Gastroenterol., 34, 63–67. [DOI] [PubMed] [Google Scholar]
- 54. Kocer B., et al. (2002) Expression of MUC5AC in colorectal carcinoma and relationship with prognosis. Pathol. Int., 52, 470–477. [DOI] [PubMed] [Google Scholar]
- 55. Renaud F., et al. (2015) The serrated neoplasia pathway of colorectal tumors: Identification of MUC5AC hypomethylation as an early marker of polyps with malignant potential. Int J Cancer., 138, 1472–1481. [DOI] [PubMed] [Google Scholar]
- 56. Renaud F., et al. (2015) MUC5AC hypomethylation is a predictor of microsatellite instability independently of clinical factors associated with colorectal cancer. Int. J. Cancer, 136, 2811–2821. [DOI] [PubMed] [Google Scholar]
- 57. Häuselmann I., et al. (2014) Altered tumor-cell glycosylation promotes metastasis. Front. Oncol., 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kufe D.W. (2013) MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene, 32, 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhi X., et al. (2014) MUC4-induced nuclear translocation of β-catenin: a novel mechanism for growth, metastasis and angiogenesis in pancreatic cancer. Cancer Lett., 346, 104–113. [DOI] [PubMed] [Google Scholar]
- 60. Giannakouros P., et al. (2015) MUC16 mucin (CA125) regulates the formation of multicellular aggregates by altering β-catenin signaling. Am. J. Cancer Res., 5, 219–230. [PMC free article] [PubMed] [Google Scholar]
- 61. van de Wetering M., et al. (2003) Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep., 4, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van de Wetering M., et al. (2002) The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell, 111, 241–250. [DOI] [PubMed] [Google Scholar]
- 63. Caderni G., et al. (2003) Identification of mucin-depleted foci in the unsectioned colon of azoxymethane-treated rats: correlation with carcinogenesis. Cancer Res., 63, 2388–2392. [PubMed] [Google Scholar]
- 64. Sakai E., et al. (2012) Identification of preneoplastic lesions as mucin-depleted foci in patients with sporadic colorectal cancer. Cancer Sci., 103, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamamoto M., et al. (1997) Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion. J. Biol. Chem., 272, 12492–12494. [DOI] [PubMed] [Google Scholar]
- 66. Julian J., et al. (2009) MUC1 is a substrate for gamma-secretase. J. Cell Biochem., 108, 802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li Y., et al. (1998) Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin. Mol. Cell Biol., 18, 7216–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Y., et al. (2001) The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin. J. Biol. Chem., 276, 6061–6064. [DOI] [PubMed] [Google Scholar]
- 69. Li Y., et al. (2001) The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin. J. Biol. Chem., 276, 35239–35242. [DOI] [PubMed] [Google Scholar]
- 70. Ren J., et al. (2002) Protein kinase C delta regulates function of the DF3/MUC1 carcinoma antigen in beta-catenin signaling. J. Biol. Chem., 277, 17616–17622. [DOI] [PubMed] [Google Scholar]
- 71. Udhayakumar G., et al. (2007) Interaction of MUC1 with beta-catenin modulates the Wnt target gene cyclinD1 in H. pylori-induced gastric cancer. Mol. Carcinog., 46, 807–817. [DOI] [PubMed] [Google Scholar]
- 72. Liu X., et al. (2014) MUC1 regulates cyclin D1 gene expression through p120 catenin and β-catenin. Oncogenesis, 3, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rajabi H., et al. (2012) MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J. Biol. Chem., 287, 10703–10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wen Y., et al. (2003) Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J. Biol. Chem., 278, 38029–38039. [DOI] [PubMed] [Google Scholar]
- 75. Retterspitz M.F., et al. (2010) Expression of {beta}-catenin, MUC1 and c-met in diffuse-type gastric carcinomas: correlations with tumour progression and prognosis. Anticancer Res., 30, 4635–4641. [PubMed] [Google Scholar]
- 76. Baldus S.E., et al. (2004) MUC1 and nuclear beta-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clin. Cancer Res., 10, 2790–2796. [DOI] [PubMed] [Google Scholar]
- 77. Gnemmi V., et al. (2014) MUC1 drives epithelial-mesenchymal transition in renal carcinoma through Wnt/β-catenin pathway and interaction with SNAIL promoter. Cancer Lett., 346, 225–236. [DOI] [PubMed] [Google Scholar]
- 78. Roy L.D., et al. (2011) MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene, 30, 1449–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tanida S., et al. (2014) Galectin-3 binds to MUC1-N-terminal domain and triggers recruitment of β-catenin in MUC1-expressing mouse 3T3 cells. Biochim. Biophys. Acta, 1840, 1790–1797. [DOI] [PubMed] [Google Scholar]
- 80. Bouillez A., et al. (2014) MUC1-C nuclear localization drives invasiveness of renal cancer cells through a sheddase/gamma secretase dependent pathway. Oncotarget, 5, 754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu H., et al. (2011) Expression of KL-6/MUC1 in pancreatic cancer tissues and its potential involvement in tumor metastasis. Oncol. Rep., 26, 371–376. [DOI] [PubMed] [Google Scholar]
- 82. Hattrup C.L., et al. (2004) MUC1 can interact with adenomatous polyposis coli in breast cancer. Biochem. Biophys. Res. Commun., 316, 364–369. [DOI] [PubMed] [Google Scholar]
- 83. Wang Z., et al. (2014) Interference of mucin 1 inhibits progression of colon carcinoma by repression of Wnt/β-catenin signaling. DNA Cell Biol., 33, 162–170. [DOI] [PubMed] [Google Scholar]
- 84. Lillehoj E.P., et al. (2007) MUC1 inhibits cell proliferation by a beta-catenin-dependent mechanism. Biochim. Biophys. Acta, 1773, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schroeder J.A., et al. (2003) MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion. Oncogene, 22, 1324–1332. [DOI] [PubMed] [Google Scholar]
- 86. Gao L., et al. (2014) Functional MUC4 suppress epithelial-mesenchymal transition in lung adenocarcinoma metastasis. Tumour Biol., 35, 1335–1341. [DOI] [PubMed] [Google Scholar]
- 87. Olsen P.A., et al. (2014) Implications of targeted genomic disruption of β-catenin in BxPC-3 pancreatic adenocarcinoma cells. PLoS One, 9, e115496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Reimann E., et al. (2014) Whole exome sequencing of a single osteosarcoma case–integrative analysis with whole transcriptome RNA-seq data. Hum. Genomics, 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hashimoto S., et al. (2012) beta-Catenin-SOX2 signaling regulates the fate of developing airway epithelium. J Cell Sci., 125, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pai P., et al. (2015) The canonical Wnt pathway regulates the metastasis promoting mucin MUC4 in pancreatic ductal adenocarcinoma. Mol. Oncol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Haridas D., et al. (2014) MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J., 28, 4183–4199. [DOI] [PubMed] [Google Scholar]
- 92. Comamala M., et al. (2011) Downregulation of cell surface CA125/MUC16 induces epithelial-to-mesenchymal transition and restores EGFR signalling in NIH:OVCAR3 ovarian carcinoma cells. Br. J. Cancer, 104, 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Akita K., et al. (2013) CA125/MUC16 interacts with Src family kinases, and over-expression of its C-terminal fragment in human epithelial cancer cells reduces cell-cell adhesion. Eur. J. Cell Biol., 92, 257–263. [DOI] [PubMed] [Google Scholar]
- 94. Velcich A., et al. (2002) Colorectal cancer in mice genetically deficient in the mucin Muc2. Science, 295, 1726–1729. [DOI] [PubMed] [Google Scholar]
- 95. Van der Sluis M., et al. (2006) Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology, 131, 117–129. [DOI] [PubMed] [Google Scholar]
- 96. Yang K., et al. (2008) Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res., 68, 7313–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Leow C.C., et al. (2004) Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res., 64, 6050–6057. [DOI] [PubMed] [Google Scholar]
- 98. Blache P., et al. (2004) SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol., 166, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tsuchiya K., et al. (2007) Reciprocal targeting of Hath1 and beta-catenin by Wnt glycogen synthase kinase 3beta in human colon cancer. Gastroenterology, 132, 208–220. [DOI] [PubMed] [Google Scholar]
- 100. Vincent A., et al. (2007) Epigenetic regulation (DNA methylation, histone modifications) of the 11p15 mucin genes (MUC2, MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene, 26, 6566–6576. [DOI] [PubMed] [Google Scholar]
- 101. Raja S.B., et al. (2012) Differential expression of gastric MUC5AC in colonic epithelial cells: TFF3-wired IL1 β/Akt crosstalk-induced mucosal immune response against Shigella dysenteriae infection. J. Cell Sci., 125(Pt 3), 703–713. [DOI] [PubMed] [Google Scholar]
- 102. Zoghbi S., et al. (2007) Intestinal MUC2 and gastric M1/MUC5AC in preneoplastic lesions induced by 1,2-dimethylhydrazine in rat: a sequential analysis. Int. J. Oncol., 30, 489–497. [PubMed] [Google Scholar]
- 103. Mucenski M.L., et al. (2005) Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol., 289, L971–L979. [DOI] [PubMed] [Google Scholar]
- 104. Inaguma S., et al. (2011) GLI1 facilitates the migration and invasion of pancreatic cancer cells through MUC5AC-mediated attenuation of E-cadherin. Oncogene, 30, 714–723. [DOI] [PubMed] [Google Scholar]
- 105. Aihara R., et al. (2005) Clinical significance of mucin phenotype, beta-catenin and matrix metalloproteinase 7 in early undifferentiated gastric carcinoma. Br. J. Surg., 92, 454–462. [DOI] [PubMed] [Google Scholar]
- 106. Silva E.M., et al. (2008) Cadherin-catenin adhesion system and mucin expression: a comparison between young and older patients with gastric carcinoma. Gastric Cancer, 11, 149–159. [DOI] [PubMed] [Google Scholar]
- 107. Das S., et al. (2015) Membrane proximal ectodomain cleavage of MUC16 occurs in the acidifying Golgi/post-Golgi compartments. Sci. Rep., 5, 9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yamada N., et al. (2009) Promoter CpG methylation in cancer cells contributes to the regulation of MUC4. Br. J. Cancer, 100, 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Vincent A., et al. (2008) Epigenetic regulation of the human mucin gene MUC4 in epithelial cancer cell lines involves both DNA methylation and histone modifications mediated by DNA methyltransferases and histone deacetylases. FASEB J., 22, 3035–3045. [DOI] [PubMed] [Google Scholar]
- 110. Hlubek F., et al. (2007) Heterogeneous expression of Wnt/beta-catenin target genes within colorectal cancer. Int. J. Cancer, 121, 1941–1948. [DOI] [PubMed] [Google Scholar]
- 111. Schepeler T., et al. (2012) Attenuation of the beta-catenin/TCF4 complex in colorectal cancer cells induces several growth-suppressive microRNAs that target cancer promoting genes. Oncogene, 31, 2750–2760. [DOI] [PubMed] [Google Scholar]
- 112. Lahdaoui F., et al. (2015) miR-219-1-3p is a negative regulator of the mucin MUC4 expression and is a tumor suppressor in pancreatic cancer. Oncogene, 34, 780–788. [DOI] [PubMed] [Google Scholar]
- 113. Radhakrishnan P., et al. (2013) MicroRNA-200c modulates the expression of MUC4 and MUC16 by directly targeting their coding sequences in human pancreatic cancer. PLoS One, 8, e73356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Srivastava S.K., et al. (2011) MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis, 32, 1832–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Johansson M.E., et al. (2013) The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol., 10, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pelaseyed T., et al. (2014) The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev., 260, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]