Abstract
Attention-deficit/hyperactivity disorder (ADHD) is a prevalent developmental disorder, associated with a range of long-term impairments. Variation in DNA methylation, an epigenetic mechanism, is implicated in both neurobiological functioning and psychiatric health. However, the potential role of DNA methylation in ADHD symptoms is currently unclear. In this study, we examined data from the Avon Longitudinal Study of Parents and Children (ALSPAC) – specifically the subsample forming the Accessible Resource for Integrated Epigenomics Studies (ARIES) – which includes (i) peripheral measures of DNA methylation (Illumina 450k) at birth (n=817, 49% male) and age 7 (n=892, 50% male) and (ii) trajectories of ADHD symptoms (7-15 yrs). We first employed a genome-wide analysis to test whether DNA methylation at birth associates with later ADHD trajectories; and then followed up at age 7 to investigate the stability of associations across early childhood. We found that DNA methylation at birth differentiated ADHD trajectories across multiple genomic locations, including probes annotated to SKI (involved in neural tube development), ZNF544 (previously implicated in ADHD), ST3GAL3 (linked to intellectual disability) and PEX2 (related to perixosomal processes). None of these probes maintained an association with ADHD trajectories at age 7. Findings lend novel insights into the epigenetic landscape of ADHD symptoms, highlighting the potential importance of DNA methylation variation in genes related to neurodevelopmental and peroxisomal processes, which play a key role in the maturation and stability of cortical circuits.
Keywords: DNA Methylation, methylome-wide, ADHD, longitudinal, ALSPAC
Introduction
Attention-Deficit Hyperactivity Disorder (ADHD) is one of the most prevalent and disabling psychiatric conditions in childhood and adolescence,1 often persisting into adulthood2 and associating with a range of long-term impairments.3–6 Like other psychiatric disorders, the aetiology of ADHD is complex and multifactorial. While epidemiological studies have identified numerous environmental risk factors,7,8 family-based studies have also documented the importance of genetic factors in the development of ADHD symptomatology,9–11 although genome-wide association studies have yet to find genetic variants robustly associated with ADHD.10,12,13 The mechanisms underlying the association between these risk factors and the development of ADHD symptoms remain to be elucidated. In recent years, epigenetic processes, such as DNA methylation, regulating gene expression have emerged as candidate mechanisms as they have been associated with environmental/genetic risk as well as neurobiological functioning and psychiatric wellbeing.14–17
Recent research has begun to demonstrate the potential of epigenetic research for understanding ADHD.18,19 However, current literature on the topic remains scant and presents a number of important limitations. First, the majority of existing studies have been based on candidate genes (e.g. dopaminergic genes), which precludes the identification of novel findings. A methylome-wide analysis, in contrast, is hypothesis-free with respect to which genes might be involved and hence has the potential to detect novel biological associations.20 Second, studies have relied primarily on cross-sectional designs featuring DNA methylation at a single time point. This has precluded the possibility of examining whether altered DNA methylation patterns are a risk factor for and/or consequence of ADHD, as well as establishing the stability of associations across time.
To our knowledge, only one study to date has conducted a methylome-wide analysis of ADHD, implicating several potential biological pathways related to inflammatory mechanisms, such as homocysteine and fatty acid oxidation.21 However, the study focussed on a relatively small sample (N = 105) of boys and relied in part on a priori information to identify differentially methylated sites. The cross-sectional design also meant that it was not possible to disentangle epigenetic predictors of ADHD from a posteriori markers of ADHD and/or associated characteristics (e.g. medication or stress resulting from ADHD symptoms). In the present study we aimed to address these gaps in the literature, by conducting the first methylome-wide study of ADHD symptomatology in a large population-based sample featuring repeated measures of DNA methylation and the use of a prospective design spanning birth to adolescence.
Methods
Participants
Participants were drawn from the Accessible Resource for Integrated Epigenomics Studies (ARIES, http://www.ariesepigenomics.org.uk),22 containing DNA methylation data for a subset of 1018 mother-offspring pairs and nested within the Avon Longitudinal Study of Parents and Children (ALSPAC). ALSPAC is an ongoing epidemiological study of children born from 14,541 pregnant women residing in Avon, UK, with an expected delivery date between April 1991 and December 1992 (85% of eligible population).23 Informed consent was obtained from all ALSPAC participants and ethical approval was obtained from the ALSPAC Law and Ethics Committee as well as Local Research Committees. The original ALSPAC sample is representative of the general population.24 Please note that the study website contains details of all the data that is available through a fully searchable data dictionary: http://www.bris.ac.uk/alspac/researchers/dataaccess/data-dictionary/.
For this study, we included youth from ARIES who had available data on ADHD symptomatology ratings (age 7-15) as well as epigenetic data at birth (n = 817, 49% male) and/or age 7 (n = 892, 50% male). The overlap was n = 783 participants with DNA methylation at birth and age 7 as well as ADHD ratings (see Supplementary section 1.1 for details).
Measures
ADHD
ADHD symptomatology was assessed via maternal ratings at ages 7, 10, 13 and 15 years, using the well-validated Development and Well-Being Assessment interview (DAWBA).25 The DAWBA was administered via computer, generating the following ‘probability bands’ (i.e. levels of prediction of the probability of disorder for a DSM-IV diagnosis of ADHD, ranging from 0 – very unlikely – to 5 – probable): 0: < 0.1% probability of children in this band having the disorder; 1: ~0.5%; 2: ~3%; 3: ~15%; 4: ~40%; 5: >70%, respectively. See Supplementary section 1.2 for more details.
DNA methylation data
500ng genomic DNA from blood (cord at birth; whole at age 7) was bisulfite-converted using the EZ-DNA methylation kit (Zymo Research, Orange, CA, USA). DNA methylation was quantified using the Illumina HumanMethylation450 BeadChip (HM450k; Illumina, USA) with arrays scanned using an Illumina iScan (software version 3.3.28). Samples (nbirth = 25; nage 7 = 8) or probes (nbirth = 7873; nage 7 = 4861) that failed quality control (>1% probes/samples with background detection p-value >= 0.05) were excluded from further analysis. Sex checks were performed using X/Y chromosome methylation. Genotype probes on the HM450k were compared between samples from the same individual and against SNP-chip data to identify and remove any sample mismatches. Samples were quantile normalised using the dasen function within the wateRmelon package (version 1.4.0) in R. Normalization performance was evaluated using all three testing metrics in wateRmelon (genki assessing SNP-related probes, dmrse assessing imprinted probes and seabi, assessing gender differences). Methylation levels were then indexed by beta values (corresponding to the ratio of methylated signal divided by the sum of the methylated and unmethylated signal). Probes known to be cross-reactive or polymorphic26,27 and SNP (i.e. “rs”) probes were removed (n = 72,068). We also removed participants with non-caucasian or missing ethnicity (based on self-reports; n = 61), leaving a total of 828 (cord) and 903 samples (age 7) after quality control. Cell type proportions (CD8 T lymphocytes, CD4 T lymphocytes, natural killer cells, B lymphocytes, monocytes and granulocytes) for each participant were estimated using the reference-based approach detailed in Houseman et al.28 As a final step, we regressed out chip and cell type to remove potentially confounding effects. For more information, see Supplementary section 1.3.
Analyses
ADHD trajectories
Trajectories of hyperactivity/inattention across 7 to 15 years were estimated using k-means for longitudinal data (Package KmL).29–31 This non-parametric procedure classifies participants into developmental trajectories, i.e. homogenous subgroups following similar developmental patterns. Each participant is first assigned arbitrarily to one initial trajectory. Next, the center (mean) of each trajectory is calculated and each participant is reassigned to the closest trajectory. The operation is repeated until convergence (i.e. until no further mean adjustment change occurs in the trajectories). The process from assignment to convergence is then repeated (500 times in the present study) to make sure that the solution is not dependent on the initial assignment. The best solution is determined by a criterion that maximizes a ratio computed by dividing the trace of the between-variance by the trace of the within-variance (i.e. maximizing the differences between trajectories and maximizing the homogeneity within trajectories). Additional details on the procedure and the choice of the best solution are provided in Supplementary sections 1.4 and 1.5. The final sample consisted of nbirth = 777 and nage7 = 842 in the low trajectory group against nbirth = 40 and nage7 = 50 in the high trajectory group (see results section for further details).
Methylome-wide analysis
Methylome-wide association analysis between DNA methylation (407,462 probes, cell type and batch-corrected) and ADHD trajectories were performed at birth, adjusting for sex, using a general linear model. Differentially methylated probes (DMPs) surviving a False Discovery Rate (FDR) correction of q < 0.05 were visually inspected to assess equal variance between trajectories and analysed further to determine whether they were also significant at age 7 (i.e. follow-forward approach, FDR-corrected q<0.05) and whether the direction of association was consistent across time points. All analyses were performed in R (version 3.0.2) using package IMA.32
Code availability
Computer code used in our analyses is available from the authors upon request.
Network analysis
To further analyse underlying genetic networks of ADHD trajectory-associated DMPs, we imported and analysed all genes related to FDR-corrected DMPs at birth using GeneMANIA (http://www.genemania.org) bioinformatics software with default parameters. For more information on GeneMANIA methods, see Supplementary section 2.2 and references.33,34
Results
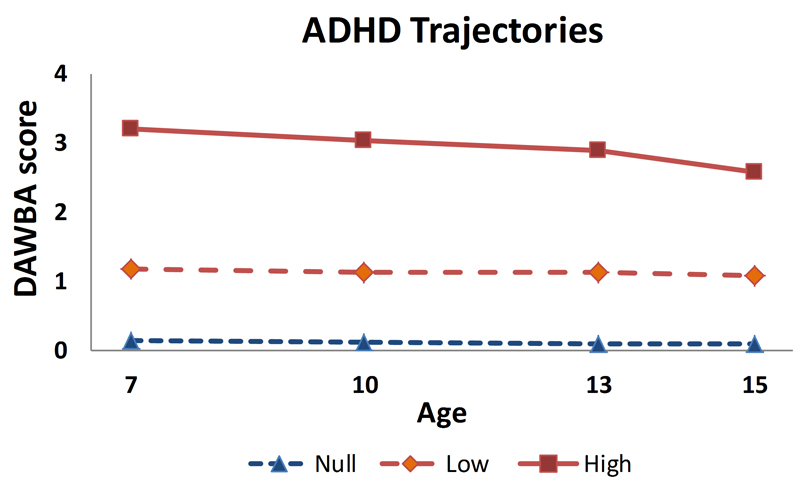
Trajectory analyses of the DAWBA ADHD scores yielded a 3 trajectory solution (Figure 1 and Supplementary section 1.5). Participants in the high trajectory (6.1%) had DAWBA band scores of around 3 across time points. Conversely, participants in the null trajectory (67.4%) and in the low trajectory (26.5%), had stable scores of around 0 and 1 respectively, corresponding to a close to zero probability of being ADHD cases. In the methylome-wide analyses, we therefore used a binary variable grouping the null and the low trajectories (called low thereafter) (nbirth = 777 and nage7 = 842) against the high trajectory (nbirth = 40 and nage7 = 50).
Figure 1.
Trajectory analyses of the DAWBA ADHD scores yielded a 3 trajectory solution. Trajectories were estimated using k-means for longitudinal data, a non-parametric procedure, which classifies participants into homogenous subgroups following similar developmental patterns (trajectories). See method and results section as well as Supplementary sections 1.4 and 1.5 for statistical details.
Methylome-wide analysis of ADHD trajectories
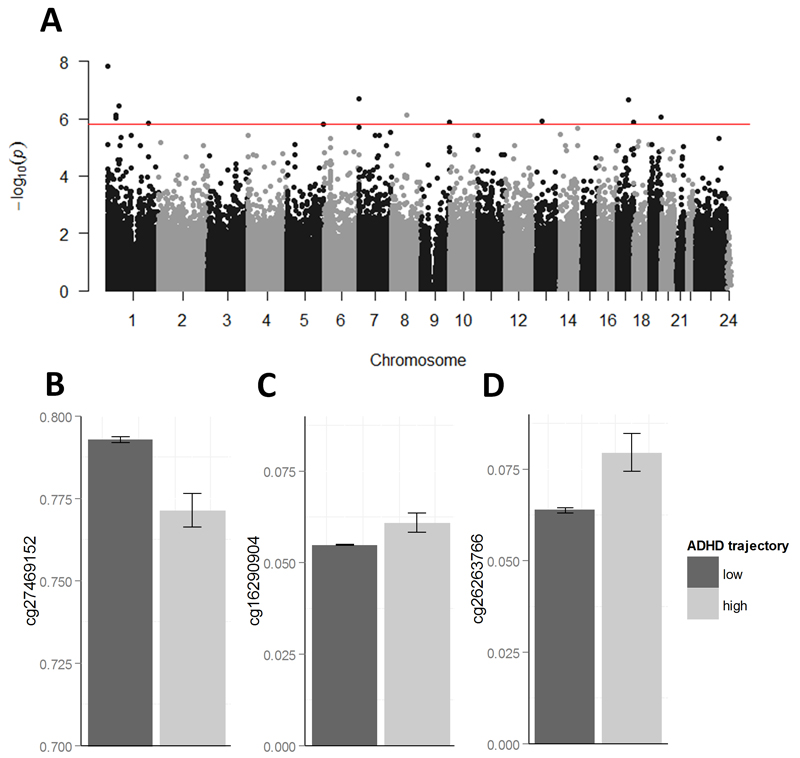
At birth, 13 probes were differentially methylated between ADHD trajectories after FDR correction (q < 0.05; Table 1 and Figure 2A). Inspection of the QQ-plot (Supplementary Figure 3) and a lambda statistic of 1.056 provided little evidence of inflation of test statistics. Additionally, visual inspection of boxplots gave no strong indication for a violation of equal variance assumption between trajectories for any of these 13 probes (see Supplementary section 2.1). Cg24481594, the most significant DMP (βstdn = -0.198; p = 1.51*10-8; q = 0.006), was hypo-methylated in the high trajectory and is annotated to SKI, a gene related to Transforming Growth Factor-beta (TGF-beta) signalling and neural tube development.35,36 Other DMPs of interest were located in genes such as (i) EPX (cg27469152; βstdn = -0.181; p = 2.26*10-7; q = 0.031; Figure 2B), a member of the peroxidase gene family, and PEX2 (cg16290904; βstdn = 0.173; p = 7.35*10-7; q = 0.048; Figure 2C), a peroxisomal membrane protein gene involved in myelin production and fatty-acid metabolism; (ii) ST3GAL3 (cg09989037; βstdn = -0.172; p = 9.46*10-7; q = 0.048), a gene linked to mental retardation;37 (iii) FBXW5 (cg13714586; βstdn = 0.170; p = 1.30*10-6; q = 0.048), associated with interleukin-1B signalling; (iv) ELF3 (cg05653018; βstdn = 0.169; p = 1.43*10-6; q = 0.048), involved in preimplantation development; and (v) ZNF544 (cg26263766; βstdn = 0.173; p = 8.72*10-7; q = 0.048; Figure 2D), implicated in transcriptional regulation and previously shown to associate with ADHD.38 Absolute mean percent methylation difference between the high and low trajectory group for the 13 DMPs passing FDR-correction was 2.3% (range 0.6 – 4.8).
Table 1.
FDR-corrected probes that associate with ADHD trajectory. Estimates given for birth and age 7, ranked by birth p-values. Chr, chromosome; StdB, standardized regression beta (negative values indicate hypomethylation in the high trajectory group); p, uncorrected p-value; q, FDR-corrected value; s.d., standard deviation.
| CpG | Gene | Chr | Position | Birth | Age 7 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| StdB | p | q | mean±s.d. low trajectory group | mean±s.d. high trajectory group | StdB | p | q | mean±s.d. low trajectory group | mean±s.d. high trajectory group | ||||
| cg24481594 | SKI | 1 | 2190850 | -0.198 | 1.51E-08 | 0.006 | 0.828 (0.026) | 0.805 (0.034) | 0.005 | 0.876 | 0.963 | 0.777 (0.025) | 0.777 (0.024) |
| cg03905179 | MAFK | 7 | 1582588 | -0.182 | 2.00E-07 | 0.031 | 0.761 (0.040) | 0.727 (0.076) | -0.002 | 0.951 | 0.963 | 0.753 (0.034) | 0.752 (0.028) |
| cg27469152 | EPX | 17 | 56282313 | -0.181 | 2.26E-07 | 0.031 | 0.793 (0.024) | 0.771 (0.032) | -0.090 | 0.008 | 0.104 | 0.772 (0.032) | 0.758 (0.030) |
| cg15096815 | JUN | 1 | 59249838 | -0.178 | 3.53E-07 | 0.036 | 0.105 (0.010) | 0.097 (0.009) | 0.002 | 0.963 | 0.963 | 0.104 (0.010) | 0.104 (0.010) |
| cg01324543 | CCDC30 | 1 | 42999439 | -0.174 | 7.21E-07 | 0.048 | 0.871 (0.025) | 0.851 (0.036) | 0.023 | 0.497 | 0.881 | 0.876 (0.020) | 0.877 (0.022) |
| cg16290904 | PEX2 | 8 | 77912348 | 0.173 | 7.35E-07 | 0.048 | 0.055 (0.006) | 0.061 (0.017) | -0.077 | 0.022 | 0.143 | 0.061 (0.011) | 0.058 (0.013) |
| cg26263766 | ZNF544 | 19 | 58739734 | 0.173 | 8.72E-07 | 0.048 | 0.064 (0.019) | 0.079 (0.032) | 0.012 | 0.729 | 0.948 | 0.063 (0.020) | 0.064 (0.025) |
| cg09989037 | ST3GAL3 | 1 | 44300942 | -0.172 | 9.46E-07 | 0.048 | 0.443 (0.055) | 0.402 (0.069) | -0.056 | 0.099 | 0.322 | 0.463 (0.044) | 0.454 (0.048) |
| cg18587973 | CDADC1 | 13 | 49822535 | 0.170 | 1.20E-06 | 0.048 | 0.076 (0.034) | 0.108 (0.092) | -0.015 | 0.656 | 0.948 | 0.086 (0.029) | 0.085 (0.039) |
| cg22193912 | MAFG | 17 | 79881523 | 0.169 | 1.28E-06 | 0.048 | 0.171 (0.056) | 0.218 (0.082) | -0.041 | 0.224 | 0.570 | 0.311 (0.053) | 0.303 (0.063) |
| cg13714586 | FBXW5 | 9 | 139838358 | 0.170 | 1.30E-06 | 0.048 | 0.049 (0.005) | 0.055 (0.026) | -0.038 | 0.263 | 0.570 | 0.050 (0.007) | 0.049 (0.005) |
| cg05653018 | ELF3 | 1 | 201979533 | 0.169 | 1.43E-06 | 0.048 | 0.799 (0.045) | 0.833 (0.035) | 0.059 | 0.082 | 0.322 | 0.841 (0.027) | 0.847 (0.021) |
| cg24843380 | ZNF454 | 5 | 178367827 | 0.168 | 1.56E-06 | 0.049 | 0.075 (0.010) | 0.087 (0.053) | 0.021 | 0.542 | 0.881 | 0.093 (0.021) | 0.096 (0.015) |
Figure 2.
Manhattan plot and bar graphs of methylome-wide results at birth. A) CpG chromosome positions are plotted against -log10 p-values. The red line indicates FDR-corrected significance threshold. Results were derived using a general linear model between DNA methylation (407,462 probes at birth, cell type and batch-corrected) and ADHD trajectories, adjusting for sex. See method section for further statistical details. B-D) Bar graphs (mean ± s.e.m.) of three DMPs associated with ADHD trajectories.
Network analysis of FDR-corrected probes
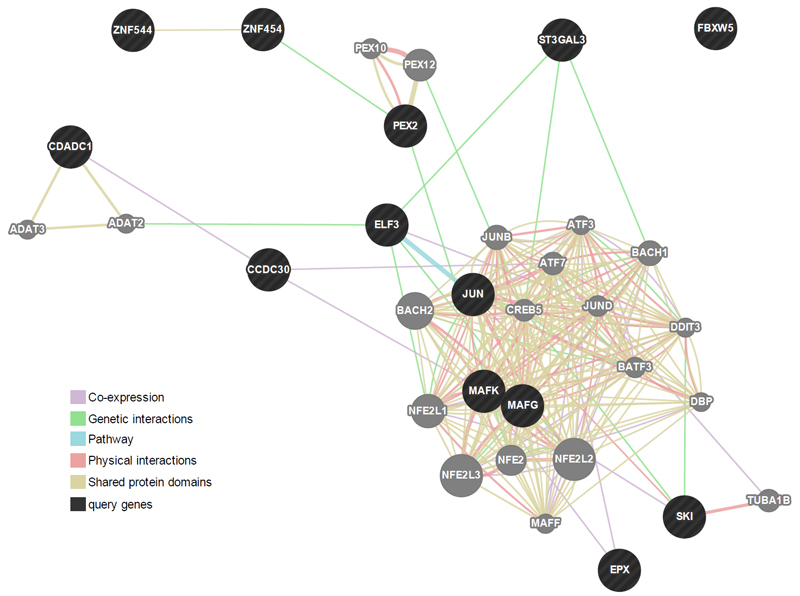
All genes annotated to FDR-corrected DMPs (n = 13) were entered into a network analysis using GeneMANIA, which is based on known genetic and physical interactions, shared pathways and protein domains as well as protein co-expression data. The analysis showed that these genes form a compact cluster network (see Figure 3). The most enriched biological functions related to peroxisomal processes (involving the genes PEX2, PEX10 and PEX12; pFDR ranging from 0.008 - 0.012; Supplementary Table 3), followed by functions related to transcription activity (pFDR = 0.019). However, it should be noted that only PEX2 was directly measured, while the involvement of PEX10 and PEX12 was indirectly inferred based on shared protein domains and physical interactions.
Figure 3.
Gene network analysis using GeneMANIA. Darker circles represent genes associated with the 13 probes found to be related to ADHD trajectories in the methylome-wide analysis at birth. Lighted circles represent additional genes predicted by GeneMANIA based on genetic and physical interactions, shared pathways and protein domains as well as protein co-expression data. The gene network analysis demonstrates that, rather than being isolated (e.g. FBXW5), these genes clustered into a complex interconnected network. For more information on GeneMANIA methods, see Supplementary section 2.2.
Follow-forward analysis at age 7
As a last step, we investigated whether any of the 13 DMPs that differentiated ADHD trajectories at birth also did so at age 7. While two of the 13 DMPs at birth also showed nominally significant effects at age 7 (cg27469152 located in the gene EPX; βstdn = -0.090; p = 0.008; and cg16290904 located in the gene PEX2; βstdn = -0.077;p = 0.022; Table 1), no probe remained significant after FDR-correction for 13 test. The direction of effect was consistent for cg27469152 (decreased methylation in the high trajectory group, both at birth and at age 7), but not for cg16290904.
Additional analyses
Given that a minority (see Supplementary Figure 1) of participants had DNA methylation data only at birth or age 7, we repeated the analyses in a subsample with complete DNA methylation data at both time points (n = 783). In agreement with our previous findings, effect sizes at birth were consistent in direction and size. Mean percent variation compared to the original effect sizes was 5.5% (standard error ± 2.6). Similar to the main analysis, no marker replicated at a FDR-corrected significance level at age 7 years (Supplementary section 2.3).
Discussion
In this study, we employed a methylome-wide prospective analysis (birth, age 7) with trajectories of ADHD symptoms (7-15 years). We identified 13 probes at birth that were differentially methylated between high and low trajectories of ADHD symptoms, none of which continued to be differentially methylated at age 7. Detected probes were located in the vicinity of genes implicated in peroxisomal processes, neural tube development and mental retardation, as well as one gene previously associated with ADHD. We first discuss our findings in light of the few previous epigenetic studies on ADHD, before turning to potentially relevant biological mechanisms suggested by our findings. We also discuss differences in results between birth and age 7 years from a developmental perspective.
Our findings both contrast and support previous epigenetic studies of ADHD. With regard to contrasting results, we found no evidence of differential methylation in VIPR2 (a gene linked to mood disorders and circadian rhythm regulation), which was identified as a top hit by the only other published methylome-wide study on ADHD.21 We also found no evidence of differential methylation in DRD4, which was reported as significantly associated with ADHD in two previous candidate gene studies.18,19 A number of factors may explain these discrepancies, including differences in: (i) samples (population versus clinical); (ii) assessment (diagnosis versus continuous ratings); (iii) analytical methods (candidate genes vs. methylome-wide); (iv) design (prospective/longitudinal vs cross-sectional); and (v) developmental period examined (birth vs childhood). As a result, consideration of these differences should be applied when interpreting the findings of the present study.
Conversely, our findings regarding peroxisomal processes lend support to the involvement of mechanisms related to fatty acid oxidation in ADHD, as previously reported by Wilmot et al.21 Peroxisomes are cell components that play a key role in the metabolism of essential fatty acids from the omega-3 family. In particular, dietary alpha-linolenic acids are transformed into docosahexaenoic acid (DHA) through a final β-oxidation reaction that takes place in the peroxisomes.40 DHA is particularly relevant as it accumulates in brain tissue at a rapid rate during the third trimester of pregnancy and continues to do so throughout early childhood and adolescence, playing an essential role in the maturation and stability of cortical circuits as well as in several other processes (e.g. implication in neurotransmitter systems including dopamine and serotonin).42 Deficits in DHA have been related to several psychiatric disorders, including ADHD.42,43 For instance, a recent meta-analysis of case control studies has demonstrated that ADHD is associated with robust blood DHA deficits.44 Furthermore, meta-analyses of randomized controlled trials in ADHD patients have shown a small but statistically significant effect of omega-3 fatty acid supplementation on ADHD symptoms43 as well as some effects on cognition in people with low dietary omega-3 intake.45 Consequently, our findings, together with those of previous research,21 suggest that early life methylation patterns in the peroxisomal network contribute to ADHD symptomatology in childhood and adolescence, possibly through disruptive effects on DHA synthesis. If the association between DNA methylation patterns in this network and ADHD is indeed mediated by peroxisome abnormalities leading to DHA deficits, then future studies should investigate whether dietary intake of preformed DHA, which bypasses peroxisome biosynthesis, mitigates the effect of altered DNA methylation patterns.
In addition to peroxisomal processes, significant probes pointed towards several other processes associated with the development of the central nervous system. In particular, the most significant probe at birth was linked to SKI proto-oncogene, which functions as a suppressor of transforming growth factor-beta signalling, and is implicated in neural tube development and myelination.35,36,46 A study by Atanasoski et al.46 showed that overexpression of Ski in cultured Schwann cells – the main glial cells in the peripheral nervous system - causes upregulation of myelin protein genes, indicating that Ski is an essential regulator that controls Schwann cell myelination. Interestingly, children with neurofibromatosis type 1 – a genetic disorder that can result in neurofibromas, which primarily affect Schwann cells47 – are three times as likely to meet DSM-IV diagnostic criteria for ADHD compared to their unaffected siblings.48 An additional probe, which we identified, was linked to ST3GAL3, a gene encoding a membrane protein involved in cellular communication located on chr1p34.1. This region (and ST3GAL3 itself) had been previously associated with intellectual disability using linkage analysis, chromosome sorting and next-generation sequencing.37,49,50 Furthermore, we identified two probes that are linked to ZNF544 and ZNF454, respectively. Both genes belong to the same zinc finger family (C2H2-type) and are involved in gene transcription. Strikingly, ZNF544 - along with several other ZNF genes - was previously flagged up in GWAS analysis of ADHD.38 Using data from 376 family trios and a composite quantitative phenotype of ADHD symptoms based on DSM-IV criteria, the authors of that study identified, among others, a SNP linked to ZNF544. This marker is around 30,000 basepairs away from the CpG site identified in this study, which is associated with the same gene.
Among the 13 probes identified at birth, none were still associated at age 7 years with ADHD trajectories after FDR-correction. A number of non-exclusive factors may drive this non-replication. First, a number of the 13 probes detected at birth may have been false positives. Although we had a much larger sample than any previous study, false positives are still possible. Second, early life methylation patterns may be particularly important for the development of ADHD. For example, one probe was linked to a gene involved in preimplantation development whose differential methylation may be only detectable at birth. In addition, it has been shown that DHA accrual in the brain is particularly important in the third trimester of pregnancy and the first year of life.42 These and other processes that are more salient during early development may lead to enduring individual differences (e.g. in brain structure) without the epigenetic association being maintained. Third, there is mounting evidence that DNA methylation patterns change considerably across development.51,52 Consequently, a large proportion of epigenetic effects may be specific to certain developmental epochs. In the present case, differences in methylation patterns between birth and age 7 may reflect differences in environmental exposures at both ages and/or developmental genetic influences that have been reported for ADHD symptoms.11 Importantly, given the current lack of similar prospective epigenetic studies in ADHD, it is not yet clear what should be expected in terms of continuity vs. discontinuity in epigenetic patterns. Therefore, the above considerations remain inevitably speculative and necessitate further investigation.
Limitations
The present findings should be interpreted in light of a number of limitations. Because of the use of a population-based sample, the proportion of youth showing severe ADHD symptomatology across time was relatively small and necessitates replication using larger clinical samples. Furthermore, although the DAWBA is a well-validated and extensively used measure based on DSM-IV criteria, it is not, per se, a clinical diagnostic tool. As such, it will be important in future to replicate findings in clinical populations were diagnostic assessments are available. As psychostimulants can influence DNA methylation,53 the use of clinical samples would also facilitate analyses investigating the effect of psychostimulant treatment on DNA methylation, which we were unable to do in the present sample. The use of a larger number of cases vs controls will also make it possible to examine potential sex differences in the association between DNA methylation and ADHD symptoms.
Findings were based on DNA methylation from blood samples. Given that methylation patterns can be tissue-specific, the extent to which these changes reflect changes in the brain will need to be established. This is particularly relevant given that markers identified were related to genes involved in neural function and development. The analysis of transcriptomic data will also be important for assessing the functional significance of DNA methylation changes to gene expression levels.
The markers identified in this study were not validated using alternative methods such as bisulfite-pyrosequencing. While we note that our previous work with bisulfite-pyrosequencing has demonstrated that the Illumina HM450 is a robust and sensitive platform for the detection of DNA methylation differences,54,55 future work is needed to validate our findings further.
Conclusion
In conclusion, the present findings suggest that epigenetic mechanisms related to specific neurodevelopmental processes, such as neural tube development and peroxisomal mechanisms, are implicated in ADHD symptomatology. These results lend novel insights into longitudinal epigenetic risk markers for ADHD, pinpointing specific targets for further interrogation.
Supplementary Material
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. With regard to the ALSPAC DNA methylation, we thank all involved, particularly the laboratory scientists and bioinformaticians who contributed considerable time and expertise to the data in this paper. The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731) and the University of Bristol provide core support for ALSPAC. This work was funded by the National Institute of Child and Human Development grant (R01HD068437). ARIES was funded by the BBSRC (BBI025751/1 and BB/I025263/1). ARIES is maintained under the auspices of the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_12013/2 and MC_UU_12013/8). Dr Walton is supported by the German Research Foundation. Dr Pingault is supported by a European Commission Marie Curie Intra-European Fellowship [N° 330699]. Dr Cecil is supported by the Economic and Social Research Council (grant ref: ES/N001273/1).
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Willcutt EG. The prevalence of DSM-IV attention-deficit/hyperactivity disorder: a meta-analytic review. Neurotherapeutics. 2012;9:490–499. doi: 10.1007/s13311-012-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franke B, Faraone SV, Asherson P, Buitelaar J, Bau CHD, Ramos-Quiroga JA, et al. The genetics of Attention Deficit/Hyperactivity Disorder in adults, a review. Mol Psychiatry. 2012;17:960–987. doi: 10.1038/mp.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mannuzza S, Klein RG, Moulton JL., III Lifetime criminality among boys with attention deficit hyperactivity disorder: A prospective follow-up study into adulthood using official arrest records. Psychiatry Res. 2008;160:237–246. doi: 10.1016/j.psychres.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polderman TJC, Boomsma DI, Bartels M, Verhulst FC, Huizink AC. A systematic review of prospective studies on attention problems and academic achievement. Acta Psychiatr Scand. 2010;122:271–284. doi: 10.1111/j.1600-0447.2010.01568.x. [DOI] [PubMed] [Google Scholar]
- 5.Pingault J-B, Tremblay RE, Vitaro F, Carbonneau R, Genolini C, Falissard B, et al. Childhood trajectories of inattention and hyperactivity and prediction of educational attainment in early adulthood: a 16-year longitudinal population-based study. Am J Psychiatry. 2011;168:1164–1170. doi: 10.1176/appi.ajp.2011.10121732. [DOI] [PubMed] [Google Scholar]
- 6.Pingault J-B, Côté S, Galéra C, Genolini C, Falissard B, Vitaro F, et al. Childhood trajectories of inattention, hyperactivity and oppositional behaviors and prediction of substance abuse/dependence: a 15-year longitudinal population-based study. Mol Psychiatry. 2013;18:806–812. doi: 10.1038/mp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foulon S, Pingault J-B, Larroque B, Melchior M, Falissard B, Côté SM. Developmental predictors of inattention-hyperactivity from pregnancy to early childhood. PLoS ONE. 2015;10:e0125996. doi: 10.1371/journal.pone.0125996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galéra C, Côté SM, Bouvard MP, Pingault J-B, Melchior M, Michel G, et al. Early risk factors for hyperactivity-impulsivity and inattention trajectories from age 17 months to 8 years. Arch Gen Psychiatry. 2011;68:1267–1275. doi: 10.1001/archgenpsychiatry.2011.138. [DOI] [PubMed] [Google Scholar]
- 9.Chang Z, Lichtenstein P, Asherson PJ, Larsson H. Developmental twin study of attention problems: high heritabilities throughout development. JAMA Psychiatry. 2013;70:311–318. doi: 10.1001/jamapsychiatry.2013.287. [DOI] [PubMed] [Google Scholar]
- 10.Merwood A, Asherson P. Attention Deficit Hyperactivity Disorder: a lifespan genetic perspective. Adv Ment Health Intellect Disabil. 2011;5:33–46. [Google Scholar]
- 11.Pingault J-B, Viding E, Galéra C, Greven CU, Zheng Y, Plomin R, et al. Genetic and Environmental Influences on the Developmental Course of Attention-Deficit/Hyperactivity Disorder Symptoms From Childhood to Adolescence. JAMA Psychiatry. 2015;72:651–658. doi: 10.1001/jamapsychiatry.2015.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch K-P, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Neale BM, Liu L, Lee SH, Wray NR, Ji N, et al. Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome-wide association study of both common and rare variants. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:419–430. doi: 10.1002/ajmg.b.32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cecil CaM, Lysenko LJ, Jaffee SR, Pingault J-B, Smith RG, Relton CL, et al. Environmental risk, Oxytocin Receptor Gene (OXTR) methylation and youth callous-unemotional traits: a 13-year longitudinal study. Mol Psychiatry. 2014;19:1071–1077. doi: 10.1038/mp.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mill J, Petronis A. Pre- and peri-natal environmental risks for attention-deficit hyperactivity disorder (ADHD): the potential role of epigenetic processes in mediating susceptibility. J Child Psychol Psychiatry. 2008;49:1020–1030. doi: 10.1111/j.1469-7610.2008.01909.x. [DOI] [PubMed] [Google Scholar]
- 16.Lutz P-E, Turecki G. DNA methylation and childhood maltreatment: From animal models to human studies. Neuroscience. 2014;264:142–156. doi: 10.1016/j.neuroscience.2013.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014;15:1–10. doi: 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Mil NH, Steegers-Theunissen RPM, Bouwland-Both MI, Verbiest MMPJ, Rijlaarsdam J, Hofman A, et al. DNA methylation profiles at birth and child ADHD symptoms. J Psychiatr Res. 2014;49:51–59. doi: 10.1016/j.jpsychires.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Chen X-T, Luo M, Tang Y, Zhang G, Wu D, et al. Multiple epigenetic factors predict the attention deficit/hyperactivity disorder among the Chinese Han children. J Psychiatr Res. 2015;64:40–50. doi: 10.1016/j.jpsychires.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan PF. The Psychiatric GWAS Consortium: Big Science Comes to Psychiatry. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilmot B, Fry R, Smeester L, Musser ED, Mill J, Nigg JT. Methylomic analysis of salivary DNA in childhood ADHD identifies altered DNA methylation in VIPR2. J Child Psychol Psychiatry. 2015 doi: 10.1111/jcpp.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Relton CL, Gaunt T, McArdle W, Ho K, Duggirala A, Shihab H, et al. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES) Int J Epidemiol. 2015:dyv072. doi: 10.1093/ije/dyv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Cohort Profile: The Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort Profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman A, Heiervang E, Collishaw S, Goodman R. The ‘DAWBA bands’ as an ordered-categorical measure of child mental health: description and validation in British and Norwegian samples. Soc Psychiatry Psychiatr Epidemiol. 2011;46:521–532. doi: 10.1007/s00127-010-0219-x. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price EM, Cotton AM, Lam LL, Farré P, Emberly E, Brown CJ, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics & Chromatin. 2013;6:4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genolini C, Falissard B. KmL: a package to cluster longitudinal data. Comput Methods Programs Biomed. 2011;104:e112–121. doi: 10.1016/j.cmpb.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. Journal of the Royal Statistical Society Series C (Applied Statistics) 1979;28:100–108. [Google Scholar]
- 31.Genolini C, Falissard B. KmL: k-means for longitudinal data. Computational Statistics. 2010;25:317–328. [Google Scholar]
- 32.Wang D, Yan L, Hu Q, Sucheston LE, Higgins MJ, Ambrosone CB, et al. IMA: An R package for high-throughput analysis of Illumina’s 450K Infinium methylation data. Bioinformatics. 2012:bts013. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amaravadi LS, Neff AW, Sleeman JP, Smith RC. Autonomous neural axis formation by ectopic expression of the protooncogene c-ski. Dev Biol. 1997;192:392–404. doi: 10.1006/dbio.1997.8780. [DOI] [PubMed] [Google Scholar]
- 36.Chen D, Lin Q, Box N, Roop D, Ishii S, Matsuzaki K, et al. SKI knockdown inhibits human melanoma tumor growth in vivo. Pigment Cell Melanoma Res. 2009;22:761–772. doi: 10.1111/j.1755-148X.2009.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H, Eggers K, Chen W, Garshasbi M, Motazacker MM, Wrogemann K, et al. ST3GAL3 Mutations Impair the Development of Higher Cognitive Functions. The American Journal of Human Genetics. 2011;89:407–414. doi: 10.1016/j.ajhg.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasky-Su J, Neale BM, Franke B, Anney RJL, Zhou K, Maller JB, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet. 2008;147B:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 39.Cecil CaM, Walton E, Viding E. Epigenetics of addiction: Current knowledge, challenges and future directions. J Stud Alcohol Drugs. 2016 doi: 10.15288/jsad.2016.77.688. In Press. [DOI] [PubMed] [Google Scholar]
- 40.Masters C. Omega-3 fatty acids and the peroxisome. Mol Cell Biochem. 1996;165:83–93. doi: 10.1007/BF00229469. [DOI] [PubMed] [Google Scholar]
- 41.Farooqui AA, Horrocks LA. Plasmalogens, phospholipase A2, and docosahexaenoic acid turnover in brain tissue. J Mol Neurosci. 2001;16:263–272. doi: 10.1385/jmn:16:2-3:263. discussion 279-284. [DOI] [PubMed] [Google Scholar]
- 42.McNamara RK, Vannest JJ, Valentine CJ. Role of perinatal long-chain omega-3 fatty acids in cortical circuit maturation: Mechanisms and implications for psychopathology. World J Psychiatry. 2015;5:15–34. doi: 10.5498/wjp.v5.i1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bloch MH, Qawasmi A. Omega-3 fatty acid supplementation for the treatment of children with attention-deficit/hyperactivity disorder symptomatology: systematic review and meta-analysis. J Am Acad Child Adolesc Psychiatry. 2011;50:991–1000. doi: 10.1016/j.jaac.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkey E, Nigg JT. Omega-3 fatty acid and ADHD: blood level analysis and meta-analytic extension of supplementation trials. Clin Psychol Rev. 2014;34:496–505. doi: 10.1016/j.cpr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper RE, Tye C, Kuntsi J, Vassos E, Asherson P. Omega-3 polyunsaturated fatty acid supplementation and cognition: A systematic review and meta-analysis. J Psychopharmacol (Oxford) 2015;29:753–763. doi: 10.1177/0269881115587958. [DOI] [PubMed] [Google Scholar]
- 46.Atanasoski S, Notterpek L, Lee H-Y, Castagner F, Young P, Ehrengruber MU, et al. The protooncogene Ski controls Schwann cell proliferation and myelination. Neuron. 2004;43:499–511. doi: 10.1016/j.neuron.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Williams VC, Lucas J, Babcock MA, Gutmann DH, Korf B, Maria BL. Neurofibromatosis Type 1 Revisited. Pediatrics. 2009;123:124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 48.Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–1044. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- 49.Najmabadi H, Motazacker MM, Garshasbi M, Kahrizi K, Tzschach A, Chen W, et al. Homozygosity mapping in consanguineous families reveals extreme heterogeneity of non-syndromic autosomal recessive mental retardation and identifies 8 novel gene loci. Hum Genet. 2007;121:43–48. doi: 10.1007/s00439-006-0292-0. [DOI] [PubMed] [Google Scholar]
- 50.Kuss AW, Garshasbi M, Kahrizi K, Tzschach A, Behjati F, Darvish H, et al. Autosomal recessive mental retardation: homozygosity mapping identifies 27 single linkage intervals, at least 14 novel loci and several mutation hotspots. Hum Genet. 2011;129:141–148. doi: 10.1007/s00439-010-0907-3. [DOI] [PubMed] [Google Scholar]
- 51.Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiers H, Hannon E, Schalkwyk LC, Smith R, Wong CCY, O’Donovan MC, et al. Methylomic trajectories across human fetal brain development. Genome Res. 2015;25:338–352. doi: 10.1101/gr.180273.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCowan T, Dhasarathy A, Carvelli L. The Epigenetic Mechanisms of Amphetamine. [(accessed 18 Feb2016)];J Addiction Prevention. 2015 doi: 10.13188/2330-2178.S100001. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.681.8437&rep=rep1&type=pdf. [DOI] [PMC free article] [PubMed]
- 54.Pidsley R, Viana J, Hannon E, Spiers H, Troakes C, Al-Saraj S, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dempster EL, Wong CCY, Lester KJ, Burrage J, Gregory AM, Mill J, et al. Genome-wide Methylomic Analysis of Monozygotic Twins Discordant for Adolescent Depression. Biol Psychiatry. 2014;76:977–983. doi: 10.1016/j.biopsych.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.