Abstract
Emerging evidence indicates that disruption of the gut microbial community (dysbiosis) impairs mental health. Germ-free mice and antibiotic-induced gut dysbiosis are two approaches to establish causality in gut microbiota-brain relationships. However, both models have limitations, as germ-free mice display alterations in blood-brain barrier and brain ultrastructure and antibiotics may act directly on the brain. We hypothesized that the concerns related to antibiotic-induced gut dysbiosis can only adequately be addressed if the effect of intragastric treatment of adult mice with multiple antibiotics on (i) gut microbial community, (ii) metabolite profile in the colon, (iii) circulating metabolites, (iv) expression of neuronal signaling molecules in distinct brain areas and (v) cognitive behavior is systematically investigated. Of the antibiotics used (ampicillin, bacitracin, meropenem, neomycin, vancomycin), ampicillin had some oral bioavailability but did not enter the brain. 16S rDNA sequencing confirmed antibiotic-induced microbial community disruption, and metabolomics revealed that gut dysbiosis was associated with depletion of bacteria-derived metabolites in the colon and alterations of lipid species and converted microbe-derived molecules in the plasma. Importantly, novel object recognition, but not spatial, memory was impaired in antibiotic-treated mice. This cognitive deficit was associated with brain region-specific changes in the expression of cognition-relevant signaling molecules, notably brain-derived neurotrophic factor, N-methyl-D-aspartate receptor subunit 2B, serotonin transporter and neuropeptide Y system. We conclude that circulating metabolites and the cerebral neuropeptide Y system play an important role in the cognitive impairment and dysregulation of cerebral signaling molecules due to antibiotic-induced gut dysbiosis.
Keywords: Cognition, Microbiome, Metabolome, Antibiotic, Brain, Gut, Dysbiosis, Serotonin transporter, Neuropeptide Y, GRIN2B
1. Introduction
The digestive tract is colonized by trillions of microbes, collectively termed gut microbiota. This extensive microbial community, comprising approximately 1012 colony-forming units/mL in the colon, influences gastrointestinal physiology as well as function of distant organs and susceptibility of the host to disease (Lozupone et al., 2012). Detailed analysis indicates that the gut microbiota regulates the development and function of the brain, and that disturbances of the microbial community may contribute to neuropsychiatric diseases (Collins et al., 2012; Cryan and Dinan, 2012; Bienenstock et al., 2015; De Palma et al., 2015; Sampson and Mazmanian, 2015). Conceptually, the effects of the gut microbiota on the brain may involve metabolic, immune, endocrine and neuronal pathways (Cryan and Dinan, 2012; Sharon et al., 2014; Liu et al., 2015), but gut microbiota-brain communication is still insufficiently understood.
Studies in germ-free (GF) mice have been crucial in establishing a causal role of the commensal gut microbiota on shaping brain function and behavior (Stilling et al., 2014a). GF mice, raised under sterile conditions, have been shown to display changes in anxiety-like, social and cognitive behavior (Gareau et al., 2011; Diaz Heijtz et al., 2011; Desbonnet et al., 2014; Stilling et al., 2014b). However, they represent a highly artificial model that is characterized by a leaky blood-brain barrier (Braniste et al., 2014) and alterations in brain structure and neurochemistry (Diaz Heijtz et al., 2011). In addition, compensatory processes are likely to dampen physiologic deficits caused by the life-long absence of microbiota. In view of these considerations, antibiotic-induced short-term disruption of the intestinal microbial community (dysbiosis) is thought to be a less intrusive model to probe causality in microbiota-dependent effects (Bercik et al., 2011; Farzi et al., 2012; Desbonnet et al., 2015). Possible drawbacks of antibiotic-induced gut dysbiosis are systemic or even central effects of the antibiotics themselves and alterations in ingestion if the antibiotics are administered via the drinking water.
Given that short-term antibiotic treatment impairs cognitive performance (Farzi et al., 2012), we hypothesized that this effect is due to gut dysbiosis, alterations in gut microbiota-brain communication via microbial metabolites, and/or dysregulation of neuronal signaling systems in the brain. To this end, we examined the effects of intragastric treatment of adult mice with multiple antibiotics on gut microbial community, metabolite profile in colon and circulation, expression of select neuronal signaling molecules in distinct brain areas, and cognitive behavior. With this systematic investigation and a pharmacokinetic analysis of the antibiotics under study we consider antibiotic-induced gut dysbiosis a valid approach to establish causality in gut microbiota-brain relationships, relative to GF mice. In addition, we suppose that circulating metabolites and distinct signaling systems in the brain, such as the neuropeptide (NPY) system, make an important contribution to gut dysbiosis-associated brain dysfunction.
2. Materials and Methods
2.1. Experimental animals
The experiments were carried out with adult male C57BL/6N mice obtained from Charles River Laboratories (Sulzfeld, Germany). Mice were kept in groups of two to three. Light conditions (12 hour light/dark cycle), temperature (set point 22°C) and relative air humidity (set point 50%) were tightly controlled. Throughout the experiments tap water and standard laboratory chow were provided ad libitum.
2.2. Ethics statement
The experimental procedures and number of animals used were approved by the ethical committee at the Federal Ministry of Science, Research and Economy of the Republic of Austria (BMWF-66.010/0026-WF/II/3b/2014) and conducted according to the Directive of the European Parliament and of the Council of 22 September 2010 (2010/63/EU). The experiments were designed in such a way that both the number of animals used and their suffering was minimized.
2.3. Experimental groups and timelines
To minimize environmental stress, mice were transferred to the behavioral test room (12 hour light/dark cycle, set points 22°C and 50% relative air humidity, maximal light intensity 100 lux) 2 days before the start of the antibiotic treatment and maintained in this room until sacrifice.
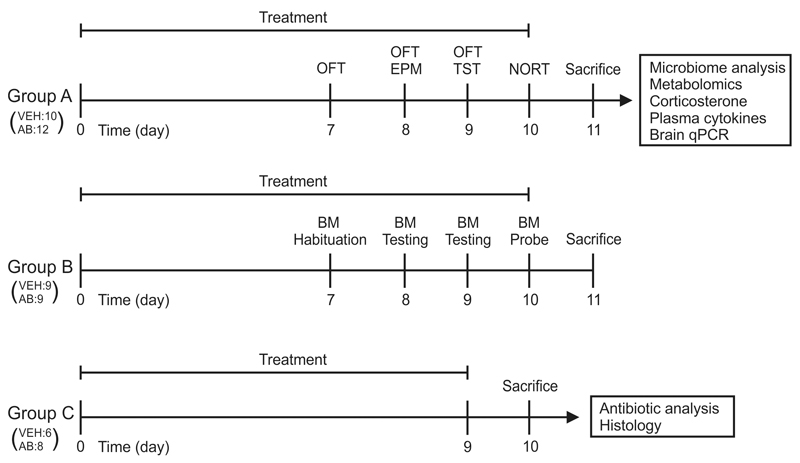
Animals were allocated to one of three groups (Fig. 1). Group A was subjected to a behavioral test battery consisting of the open field test on days 7, 8 and 9, elevated plus maze test on day 8, tail suspension test on day 9, and novel object recognition test (NORT) on day 10. Group B was habituated to the Barnes maze (BM) on day 7. BM training sessions were performed on days 8 and 9, and the probe session was conducted on day 10. Groups A and B were sacrificed on day 11. Plasma, colon, luminal colonic contents, and brain tissues collected from group A were used for molecular analysis. Group C was also treated with vehicle or the antibiotic mix but was not subjected to any behavioral tests; they were sacrificed on day 10 (the time point of cognitive testing) for analysis of antibiotic concentrations in blood and brain and for histological examination of the colon (Fig. 1). In all experiments, vehicle- and antibiotic-treated animals were run in parallel.
Fig. 1. Experimental groups and time lines.
Mice of group A were subjected to a test battery including OFT, EPM test, TST and NORT. After sacrifice, tissues of group A animals were analyzed as indicated. Mice of group B were habituated and trained in the BM. Animals of group C were not subjected to any behavioral tests but euthanized on the day 10 (the day of cognitive evaluation in group A and B) to measure antibiotic concentrations in plasma and brain and investigate the effect of antibiotics alone on colon histology. In all experiments vehicle (VEH)- and antibiotic (AB)-treated animals were run in parallel, the number of mice in each group being indicated in brackets. OFT, open field test; EPM, elevated plus maze; TST, tail suspension test; NORT, novel object recognition test; BM, Barnes maze; qPCR, quantitative real-time polymerase chain reaction.
2.4. Antibiotic mix
An antibiotic mix consisting of ampicillin (ampicillin sodium salt, catalogue number A9518, Sigma-Aldrich, Vienna, Austria), bacitracin (bacitracin from Bacillus licheniformis, catalogue number 11702, Sigma-Aldrich), meropenem (OptinemR, AstraZeneca Österreich GmbH, Vienna, Austria), neomycin (neomycin trisulfate salt hydrate, catalogue number N5285, Sigma-Aldrich) and vancomycin (vancomycin hydrochloride from Streptomyces orientalis, catalogue number 4747, Sigma-Aldrich) was used to deplete the gut microbiota.
2.5. Antibiotic treatment
At the age of 8-11 weeks mice were treated with a mix of five antibiotics (pH 6.98-7.14) or vehicle (distilled water) by oral gavage (10 mL/kg) for 11 days (Fig. 1). The antibiotics (108.0 mg bacitracin, 108.0 mg neomycin, 43.2 mg ampicillin, 21.6 mg meropenem, 6.48 mg vancomycin) were dissolved in 4.5 mL distilled water. Dosing of antibiotics was based on studies where ampicillin (Membrez et al., 2008; Khosravi et al., 2014; Desbonnet et al., 2015), bacitracin and neomycin (Bercik et al., 2011), meropenem (Moller et al., 2005; Gadjeva et al., 2010) and vancomycin (Lawley et al., 2012) were added to the drinking water. The antibiotic concentrations used in these studies were converted to equivalent gavage doses, calculated relative to the average daily water intake. Each day the mice were weighed before the first gavage, and the gavage volume was adjusted accordingly. Because of their coprophagic behavior all cage mates received the same treatment. For each gavage session 4.5 mL of the antibiotic mix was prepared and used within 18 h. From the first day of treatment (day 0) until day 6 the mice were gavaged twice daily (8:00 am, 5:00 pm). On day 7 the mice were gavaged shortly after the open field test (12:00 am) and before onset of the dark phase (5:00 pm). From day 8 onwards the mice were gavaged once daily at 3:00 pm (after the behavioral testing of the mice scheduled for the respective day had been completed). The mice received the last treatment at 3:00 pm on the day before sacrifice (day 11).
2.6. Novel object recognition test
Mice were habituated to the open field box (as described in the open field test in the Supplementary Data) each day during three consecutive days (days 7-9). On day 10, the mice were placed in the open field apparatus and given 5 min to explore two objects that were placed at adjacent edges of the central area of the field. One hour later the animals were re-exposed to one familiar object (old object) together with a novel object (new object) for 5 min. The exploratory behavior directed at each object was recorded in both sessions. The time of object exploration was measured with the VideoMot2 software (TSE Systems, Bad Homburg, Germany) and the performance of each mouse was expressed by the memory index (MI) according to the formula: MI = (tn – to)/(tn + to). The time exploring the new object is represented by tn, whereas to represents the time exploring the old object (Redrobe et al., 2004). Mice that explored the objects for less than 5 s in total were excluded from the results. To avoid spatial and object bias, the position of objects was alternated between trials, and the choice of familiar versus novel object was changed from mouse to mouse. After each test session the objects were cleaned with 70% ethanol.
2.7. Barnes maze
The BM test protocol was adapted from that used by Attar et al (Attar et al., 2013), with minor modifications. In short, the BM apparatus consisted of a circular white polyvinyl chloride slab with a diameter of 91 cm elevated 61.5 cm above the floor. At a distance of 2.5 cm from the edge, 20 holes with a diameter of 5 cm were evenly distributed along the perimeter. Below the surface of the maze a small escape cage was affixed underneath one of the 20 holes (target hole). The surface of the BM was illuminated by 35 lux. Four visual cues (colored circles and squares) were mounted around the room for spatial orientation on the slab. The animals were subjected to the BM test on three consecutive days. On day 7 the mice were habituated to the BM, and on days 8 and 9 the mice were trained to quickly find and enter the escape hole. This short training phase consisted of two training trials on day 8 and three training trials on day 9. The probe session during which no escape cage was affixed to the BM apparatus was scheduled on day 10. The movements of the mice on the training and probe days were tracked for 2 min with a video camera and evaluated with the VideoMot2 software. Spatial learning manifested itself in a shortening of the latency to identify the target hole (target latency) during consecutive training trials. Spatial memory was assessed by the time the mice spent in the target area (quadrant of BM with target hole in the center) on the probe day. To avoid spatial or visual cue bias, the location of the escape cage was alternated after every three mice. After each test session the BM and escape cage were cleaned with 70% ethanol. Mice which did not learn the correct position of the target hole, or did not voluntarily enter the escape cage, were excluded from the evaluation.
2.8. Blood and tissue harvesting for biochemical analysis
Animals were anesthetized with pentobarbital (150 mg/kg i.p.). Blood was drawn via cardiac puncture with a syringe that was filled with 100 µL of 3.8% sodium citrate as anticoagulant. The average blood vs sodium citrate ratio was 4:1. After 15 min of centrifugation at 4°C and 7000 rpm, blood plasma was collected and stored at -70°C. Following blood collection, brains were collected and immediately frozen in 2-methylbutane (Sigma-Aldrich) on dry ice for 5 s. Afterwards the brains were wrapped in aluminum foil and kept at -70°C. For microbiome and metabolome analyses the large intestine including the luminal contents was removed and stored at -70°C.
2.9. Microbiome analysis
Frozen colon tissue including the luminal contents was homogenized on a MagNA Lyser Instrument MagNA Lyser Green Beads (Roche Diagnostics GmbH, Mannheim, Germany). Bacterial DNA was extracted with the Power Lyzer® Power Soil® DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. Subsequently, the DNA concentration was determined, and bacterial 16S rRNA was amplified by PCR with the Rotor-Gene SYBR Green PCR Kit (Qiagen, Hilden, Germany) using 20 ng DNA as template. To this end, the 16S primers F27—AGAGTTTGATCCTGGCTCAG and R357—CTGCTGCCTYCCGTA were used as fusion primers containing Ion Torrent sequencing adapters. In order to assess contaminations imported during the microbiome analysis workflow, a sample devoid of any tissue and colon content (blank) was included in the PCR run. Afterwards PCR products were gel-purified and the amplicon DNA concentration was determined. Sequencing of pooled amplicons was performed with the Ion PGM Sequencer and an Ion Sequencing 400 Kit (both from Life Technologies, Carlsbad, CA, USA). Contaminating non-bacterial sequences were removed and Acacia error correction was applied on all reads using standard parameters (Bragg et al., 2012). Chimeras were identified by Usearch algorithm and removed. The resulting bam file was introduced into QIIME (v1.8.0) 16S workflow (www.qiime.org) (Caporaso et al., 2010). Differences in microbial communities between groups were investigated using the phylogeny-based weighted UniFrac distance metric. Alpha diversity and ADONIS calculations were performed with the respective QIIME scripts (alpha_diversity.py and compare_categories.py).
2.10. Brain microdissection
The working area and dissection instruments were cleaned with RNeasy AWAY (Carl Roth, Karlsruhe, Germany). The microdissection was performed by a trained researcher on a cold plate (Weinkauf Medizintechnik, Forchheim, Germany) set at -20°C as described previously (Brunner et al., 2014). The dissected brain tissues (medial prefrontal cortex, hypothalamus, amygdala, hippocampus) were collected in micro packaging vials filled with some Precellys beads (Peqlab, Erlangen, Germany) and stored at -70°C until RNA extraction.
2.11. Reverse transcriptase polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qPCR)
Brain tissues were homogenized with the Precellys 24 homogenizer (Peqlab). RNA extraction was performed according to the manufacturer’s instructions using the RNeasy lipid tissue mini kit (Qiagen). The RNA concentration was measured and 2 µg of RNA was reverse-transcribed in the Mastercycler Gradient (Eppendorf, Hamburg, Germany), using the high capacity cDNA reverse transcription kit (Life Technologies) according to the manufacturer’s instructions. A control without reverse transcriptase for each group and area was always included. For relative quantification of mRNA levels, qPCR was performed on a LightCycler®480 System with TaqMan inventoried gene expression assays (listed in Supplementary Data) and the TaqMan gene expression master mix (Life Technologies). All samples were measured as triplicates. ACTB, GAPDH and PPIL3 were used as reference (endogenous housekeeping) genes for quantification of target gene expression. Quantitative measurements of target gene levels relative to controls were performed with the 2-ΔΔCt method using the mean value of the vehicle-treated group as the calibrator (Livak and Schmittgen, 2001). Group differences were expressed as fold changes.
2.12. Colonic content metabolomics
Metabolites in the colonic content of mice were analyzed by untargeted 1H NMR analysis at the Institute of Biology of Leiden University (Netherlands). Colonic content extracts were prepared by mixing 20 mg of frozen luminal colonic content material with 1 mL of saline phosphate buffer that consisted of 1.9 mM Na2HPO4, 8.1 mM NaH2PO4, 150 mM NaCl and 1 mM sodium 3-(trimethylsilyl)-propionate-D4 (TSP) in D2O. After mixing thoroughly, samples were centrifuged at 17,000 x g for 5 min. Each supernatant was filtered through a 0.2 µm membrane filter, and 300 µL filtrate was transferred to a 3 mm NMR tube for analysis. High resolution 1H NMR spectra were recorded using a Bruker AV 600 spectrometer (Bruker, Karlsruhe, Germany). Details of the spectrometric analysis are given in Supplementary Data.
2.13. Plasma metabolomics
Metabolites in the plasma of mice were analyzed by targeted LC-HRMS metabolomics according to Bajad et al. (Bajad et al., 2006) by hydrophilic interaction liquid chromatography (HILIC) at the HEALTH Institute for Biomedicine and Health Sciences, JOANNEUM RESEARCH Forschungsgesellschaft mbH (Graz, Austria). The plasma samples were processed according to Yuan et al. (Yuan et al., 2012). Details of the LC-HRMS procedure are given in Supplementary Data.
Raw data were converted into mzXML by msConvert (ProteoWizard Toolkit v3.0.5) (Chambers et al., 2012), and metabolites were targeted-searched by the in-house developed tool PeakScout (as detailed in Supplementary Data), with a reference list containing accurate mass and retention times in agreement to standards outlined by Sumner et al. (Sumner et al., 2007). Molecular masses for all substances were taken from the literature and available online databases (HMDB, KEGG, Metlin) (Kanehisa and Goto, 2000; Smith et al., 2005; Wishart et al., 2007; Wishart et al., 2013; Kanehisa et al., 2014). Additionally, pure substances of all analytes, except lipids, were run on the same system to obtain reference retention times and fragmentation spectra.
To correct for dilution differences resulting from different blood volumes (100-560 µL) median normalization was performed. Each metabolite was scaled 0 to 1 as AUC0-1scaled = (AUC-minimum(AUC))/(range(AUC)), for each sample the samplemedian was calculated as the median of all AUC0-1scaled in the sample, and AUC values were normalized as median normalized AUC = AUC/samplemedian. Finally, median normalized AUC values were log10 transformed to achieve sufficiently normal distribution and homoscedasticity in the data set.
2.14. Antibiotic analysis
A sensitive and selective analytical method for the quantitative analysis of antibiotics in plasma and brain was developed. Plasma samples were thawed to room temperature. A 50 µL aliquot was diluted with 100 µL of 0.1% acetic acid (Sigma-Aldrich), spiked with 10 ng amoxicillin (Sigma-Aldrich) as internal standard and mixed. A 30 mg strata-x solid phase extraction cartridge (30 mg; 1 cc; Phenomenex, Aschaffenburg, Germany) was activated (1 mL methanol (Sigma-Aldrich) followed by 1 mL 0.1% acetic acid), after which the diluted sample was loaded on the cartridge and washed with 1 mL of 0.1% acetic acid. Analytes were eluted with 1 mL methanol. The elution solvent was evaporated to dryness and the residual sample was resolved in 100 µL of 0.1% formic acid (Sigma-Aldrich).
For brain analysis, half a brain was placed into 2 mL ZR Bashing Bead Lysis Tubes (VWR, Vienna, Austria), spiked with 10 ng amoxicillin (internal standard) and 1 mL water. After homogenization the vial was centrifuged at 10,000 rpm for 10 min and the supernatant was used for solid phase extraction. The supernatant was diluted with 1 mL of 0.1% acetic acid and mixed. A 100 mg strata-x solid phase extraction cartridge (100 mg, 3 cc; Phenomenex) was activated (3 mL methanol followed by 3 mL 0.1% acetic acid), and the diluted sample was loaded on the cartridge and washed with 3 mL of 0.1% acetic acid. Analytes were eluted with 3 mL methanol. The elution solvent was evaporated to dryness and the residual sample was resolved in 200 µL of 0.1% formic acid.
Analysis was performed with an Agilent 6460 triple quadrupole mass spectrometer equipped with an electrospray ionization source coupled to a 1290 binary UHPLC system (Agilent, Waldbronn, Germany). For chromatographic separation a Phenomenex Kinetex C18 (100x2.1mm; 2.6 µm) column (Phenomenex) was used.
Gradient elution with a binary mobile phase system of channel A (0.1% formic acid in water) and channel B (0.1% formic acid in acetonitrile (Sigma-Aldrich)) was performed at 30°C column temperature and a 0.4 mL /min flow rate. The gradient profile was 3% channel B for 0.5 min, linearly increased to 24% channel B over 1.5 min and ramped to 90% channel B over 0.1 min. The run time was set to 3.5 min following a post time of 1.5 min. The autosampler was maintained at 4°C and the injection volume was set at 5 µL. Mass spectrometry detection was conducted in positive mode, using dynamic multiple reaction monitoring. The cycle time was set at 250 ms. Two precursor/product ion transitions were used as quantifier and qualifier for each analyte, respectively. Quantitation was performed using the ion transitions m/z 366.1 → m/z 349;114 (amoxicillin; internal standard), m/z 350.1 → m/z 114;106 (ampicillin) and m/z 724.8 → m/z 144;100.1 (vancomycin). According to bioanalytical validation guidelines the method was tested for the following parameters: selectivity, linearity, lower limit of quantification (LLOQ), accuracy and precision, extraction efficiency, matrix effect, and autosampler stability. Calibration curves were constructed using linear regression with 1/x2 weighting based on a minimum of 8 calibrator peak area ratios, excluding the blank. Calibration levels were 0.1, 0.2, 0.5, 1.0, 2.0, 5.0, 10, 20, 50, 100 and 200 ng/mL for ampicillin and 1.0, 2.0, 5.0, 10, 20, 50, 100 and 200 ng/mL for vancomycin. Calibrators and triplicate quality control samples at low, medium and high concentrations were analyzed in each set of specimens. Samples in which no antibiotic was detected were labeled with the respective LLOQ value for numerical evaluation and statistical comparison.
2.15. Statistics
Results were statistically evaluated either with SPSS 22 (SPSS Inc., Chicago, IL, USA) or with R (R Development Core Team, 2011) (v3.2.1, packages stats, missMDA, nlme) using Tibco® Spotfire® (v7.0.0). Principal component analysis (PCA) was performed centered and scaled to unit variance (R function prcomp). Missing values of NMR metabolites were imputed by zero, missing values of LC-MS metabolites by a regularized expectation-maximization (function imputePCA and estim_ncpPCA). For all data except LC-MS metabolomics differences between two independent groups were analyzed with the independent samples t-test in SPSS. The homogeneity of variances was assessed with the Levene test. In case of a non-parametric distribution of data the Mann–Whitney U test was used. For repeated measurements repeated measures analysis of variance (ANOVA) was performed. For multiple comparisons, p-values were adjusted with the false discovery rate. Probability values of p ≤ 0.05 were regarded as statistically significant. Pearson’s correlation coefficient was used to determine correlations between variables.
For microbiome analysis the ADONIS test of weighted UniFrac distances was conducted with the QIIME compare_categories script. For LC-MS metabolomics the normal data distribution was found to be sufficient according to Shapiro-Wilk (91% normally distributed) and Kolmogorov-Smirnov (99% normally distributed) after log10 transformation of median normalized AUC. Scedasticity was found to be sufficient according to Bartlett (86% homoscedastic) and Levene (81% homoscedastic) after log10 transformation of median normalized AUC. Differences between two independent groups were analyzed by a simple ANOVA (R function aov) model with the categorically fixed factor treatment. p-Values were adjusted according to Benjamini-Hochberg (R function p.adjust).
3. Results
3.1. Brain levels of ampicillin and vancomycin are below the lower limit of quantification at the time point of cognitive testing
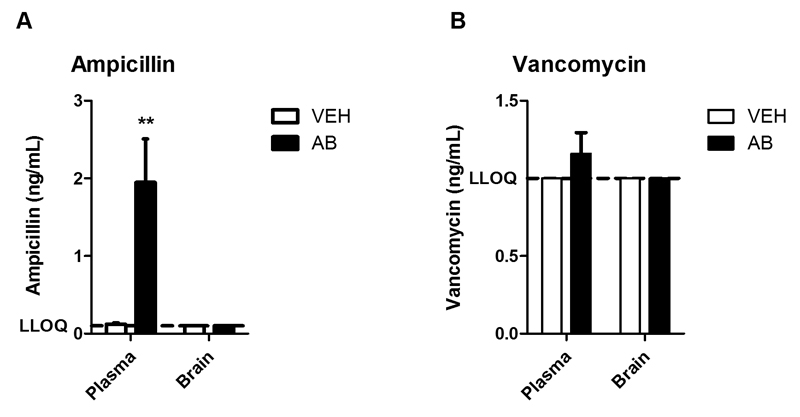
Multiple antibiotics with little or no oral bioavailability were used to ablate a wide range of bacteria specifically in the intestine. Of the antibiotics chosen (ampicillin, bacitracin, meropenem, neomycin, vancomycin), only ampicillin is absorbed to some extent from the human gut (Craig and Stitzel, 2004; Lafforgue et al., 2008). In order to examine whether ampicillin enters murine circulation and brain, plasma and brain levels of ampicillin were compared with those of vancomycin, a non-absorbable antibiotic, at a time point when cognitive performance was assessed (day 10). The plasma levels of ampicillin were around 2 ng/mL in antibiotic-treated mice (Fig. 2A) while the plasma concentrations of vancomycin were, except for two samples, below the LLOQ of 1 ng/mL (Fig. 2B). In the brain the concentrations of both ampicillin (Fig. 2A) and vancomycin (Fig. 2B) were below the respective LLOQ.
Fig. 2. Brain concentrations of ampicillin and vancomycin are below the respective lower limit of quantification.
The graphs depict concentrations of ampicillin (A) and vancomycin (B) in plasma and brain after 10 days of antibiotic (AB) treatment. The bars represent means + SEM, n=6-8; **p ≤ 0.01 compared to vehicle (VEH)-treated mice, Mann-Whitney U test.
3.2. Antibiotic treatment strongly disrupts microbial composition in the colon
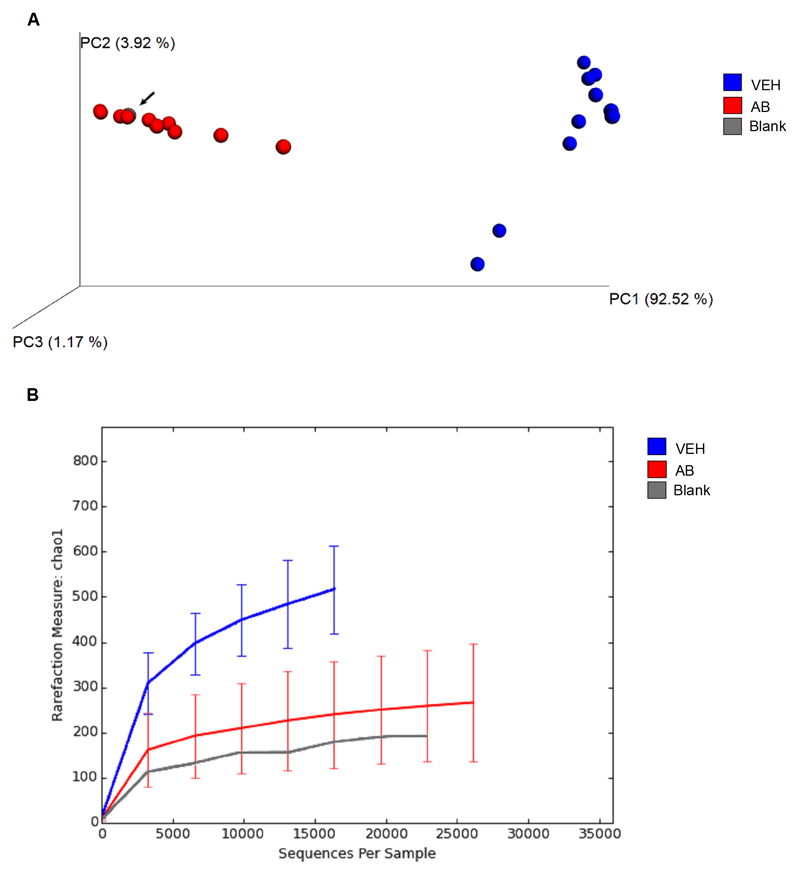
Antibiotic treatment vigorously changed the microbiome, reducing bacterial load (Fig. 3A) and diversity of bacteria (Fig. 3B). Principal coordinate analysis (PCoA) showed that antibiotic-treated mice had a significantly different (p = 0.001 by ADONIS test) microbial community than vehicle-treated mice (Fig. 3A).The profile of the microbial composition of antibiotic-treated mice clustered more closely to the blank than to the profile of vehicle-treated mice, indicating that most of the commensal bacteria were eradicated by antibiotic treatment (Fig. 3A). The residual operational taxonomic units (OTU) in antibiotic-treated mice represent mainly bacterial species known to be common contaminations of DNA extraction and PCR reagents and solutions (Salter et al., 2014) (see Supplementary Table 1).
Fig. 3. Antibiotic treatment strongly disrupts and diminishes the microbial community in the colon.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. (A) Principal coordinate analysis (PCoA) plot based on weighted UniFrac distance between samples. The microbiome analysis blank is depicted as a dot highlighted by an arrow. Two individual dots of antibiotic-treated mice are not visible because they are overlaid by other dots. (B) Alpha-rarefaction curves using the chao1 index. Values in B represent median ± SD, n=10-12.
3.3. Antibiotic treatment strongly decreases microbial metabolite levels in the colonic luminal contents
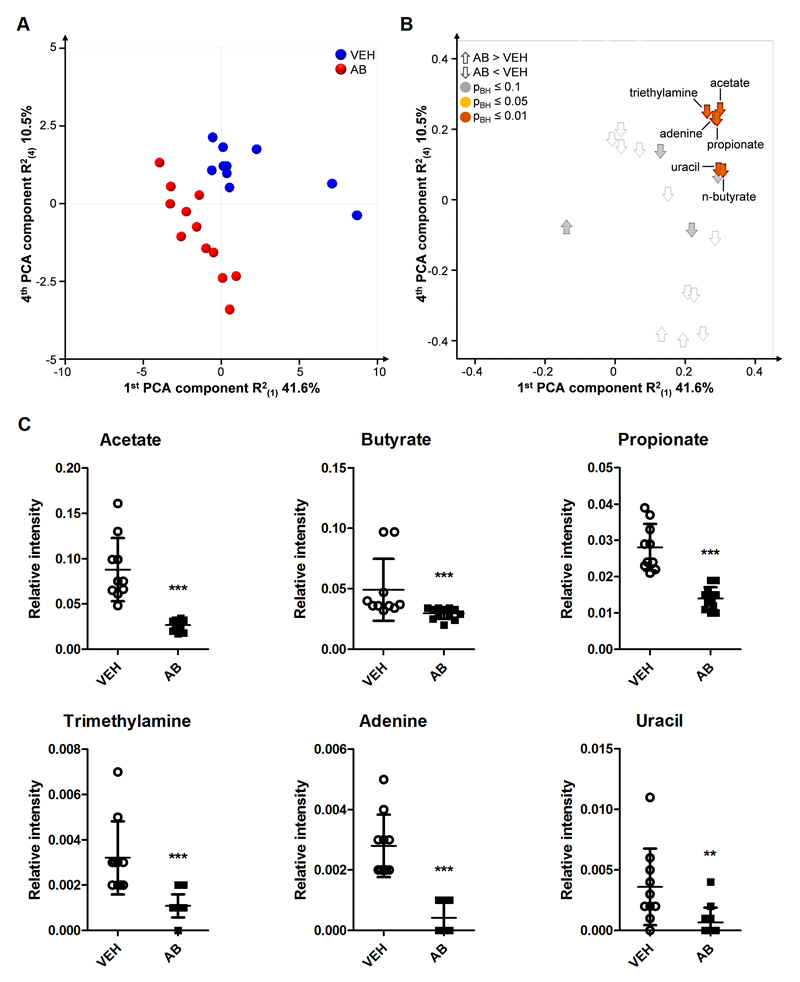
Given that the gut microbiota contributes to digestion (Russell et al., 2013), antibiotic-induced changes in the microbial community profile are likely to alter the colonic metabolite profiles. Untargeted 1H NMR analysis revealed a separation of the treatment groups, which indicates a considerable difference in colonic metabolic composition (Fig. 4A, B). Many metabolites known to be produced by the colonic microbiota were decreased in the luminal contents of antibiotic-treated mice. In particular, the levels of the short-chain fatty acids (SCFA) acetate, butyrate and propionate as well as of trimethylamine, adenine and uracil were significantly diminished by antibiotic treatment (Fig. 4C).
Fig. 4. Distinct microbial metabolite levels in the colonic contents are markedly decreased by antibiotic treatment.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. (A) Scores plot of the principal component analysis (PCA) with 21 identified colonic metabolites analyzed by 1H NMR, showing group separation in the first and fourth components. (B) Corresponding loadings plot, showing the contribution of metabolites to group separation. The metabolites are colored according to p-values calculated with the Mann-Whitney U test. (C) Graphs of significantly different metabolites. Vertical bars represent the group mean, whiskers SD, n=10-12; **p ≤ 0.01, ***p ≤ 0.001 compared to VEH-treated mice, Mann-Whitney U test.
3.4. Antibiotic treatment alters the profile of circulating metabolites, but not cytokines
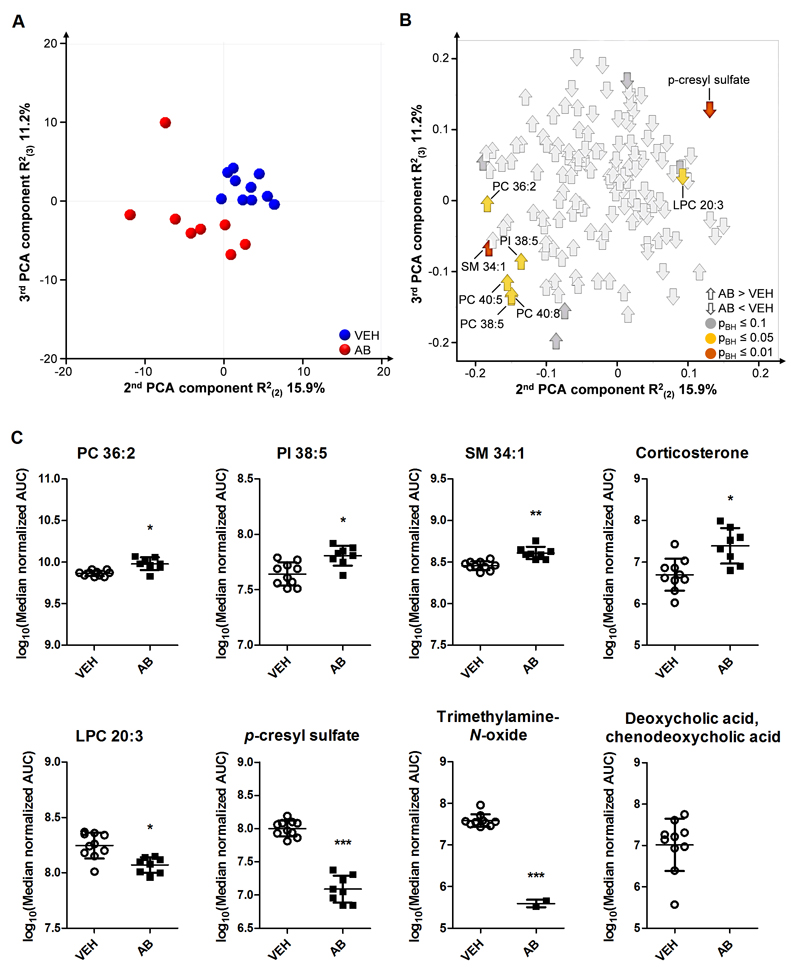
As microbial colonization impacts on circulating metabolites (Wikoff et al., 2009), the plasma metabolic profile was assessed with targeted LC-MS metabolomics. Of the 170 identified metabolites, 142 were suitable for multiple analyses. PCA showed that the treatment groups separated to a significant extent, indicating a marked difference in overall plasma metabolic composition (Fig. 5A, B). In particular, 11 metabolites were present in significantly different amounts in antibiotic-treated mice compared to vehicle-treated animals (Fig. 5B). The levels of corticosterone, sphingomyelin 34:1, phosphatidylinositol 38:5, phosphatidylcholine (PC) 36:2, PC 38:5, PC 40:5 and PC 40:8 were significantly increased in antibiotic-treated mice whereas the levels of lysophosphatidylcholine (LPC) 20:3 and p-cresyl sulfate were reduced (Fig. 5C). The relative levels of corticosterone measured by metabolomic analysis corresponded well with those measured immunochemically (see Supplementary Fig. 1). Moreover, the antibiotic-induced reduction of circulating trimethylamine-N-oxide levels (Fig. 5C) concorded with the reduction of this metabolite in the colonic contents (Fig. 4C). The levels of deoxycholic acid/chenodeoxycholic acid were below the limit of detection in antibiotic-treated mice whereas in vehicle-treated mice the metabolite was clearly present (Fig. 5C).
Fig. 5. Circulating metabolite levels are markedly altered by antibiotic treatment.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. (A) Scores plot of the principal component analysis (PCA) with 148 identified circulating metabolites analyzed by LC-MS, showing group separation in the second and third components. (B) Corresponding loadings plot, showing the contribution of metabolites to group separation. Metabolites are colored according to Benjamini-Hochberg adjusted p-values. (C) Graphs of selected metabolites that were significantly different. Vertical bars represent the group mean, whiskers SD, n=8-10; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 compared to VEH-treated mice, Benjamini-Hochberg adjusted p-values. AUC, area under the curve; PC, phosphatidylcholine; PI, phosphatidylinositol; LPC, lysophosphatidylcholine; SM, sphingomyelin.
The plasma concentrations of interleukin (IL)-1β, IL-6, IL-10, tumor necrosis factor (TNF)-α and interferon (IFN)-γ did not differ between vehicle- and antibiotic-treated mice (see Supplementary Fig. 1).
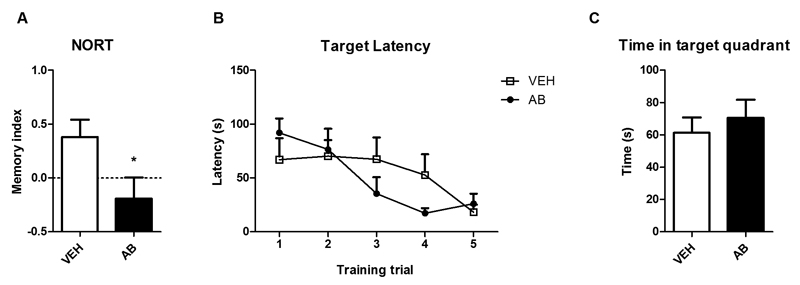
3.5. Novel object recognition memory is impaired in antibiotic-treated mice whereas spatial learning and memory remains intact
Since memory is impaired in GF mice (Gareau et al., 2011), we evaluated non-spatial memory with the NORT on day 10 of the antibiotic treatment and, in a separate group of animals, spatial learning and memory with the BM test on days 7-10 of the antibiotic treatment (Fig. 6A-C). Antibiotic-treated mice had a significantly lower memory index than vehicle-treated mice (Fig. 6A), which indicates a disruption of novel object recognition memory. The BM test revealed that spatial learning (Fig. 6B) and memory (Fig. 6C) were not changed by the antibiotic treatment. Repeated measures ANOVA revealed that only the number of trials significantly influenced target latency (F(4, 52) = 7.561; p ≤ 0.001).
Fig. 6. Object recognition memory is impaired in antibiotic-treated mice whereas spatial learning and memory remain intact.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. In the NORT (A) a positive memory index (MI) indicates that the mice spent more time exploring the novel object than the known object. In the BM test (B,C) panel B shows the decrease in target latency with consecutive training trials, and panel C shows memorization of the target hole in the target quadrant on the probe day. Values represent means + SEM, n=6-8 (A), n=7-8 (B), n=6-7 (C); *p ≤ 0.05 compared to VEH-treated mice, t-test.
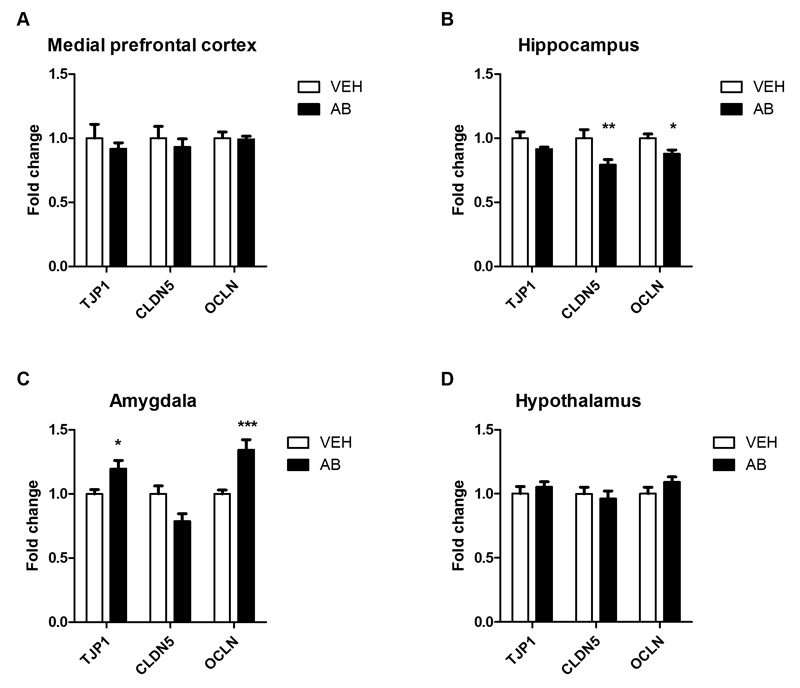
3.6. Antibiotic treatment differentially alters tight junction protein and cytokine mRNA expression in the amygdala and hippocampus
As tight junction protein expression in frontal cortex, striatum and hippocampus is lowered in GF mice (Braniste et al., 2014), we measured mRNA expression of three tight junction proteins in four brain regions of antibiotic-treated mice (Fig. 7A-D). Brain regions were chosen based on their importance in cognition and memory (amygdala, medial prefrontal cortex, hippocampus), and the established impact of the gut microbiota on the hypothalamic-pituitary-adrenal (HPA) axis (hypothalamus) (Sudo et al., 2004). In the medial prefrontal cortex (Fig. 7A) and hypothalamus (Fig. 7D) the expression of the tight junction proteins claudin 5 (CLDN5), tight junction protein 1 (TJP1) and occludin (OCLN) was similar in vehicle- and antibiotic-treated animals. In the hippocampus, however, antibiotic treatment significantly reduced the expression of CLDN5 and OCLN mRNA (Fig. 7B). The expression of TJP1 and OCLN mRNA in the amygdala was increased in antibiotic-treated mice relative to vehicle controls (Fig. 7C).
Fig. 7. Antibiotic treatment alters tight junction protein mRNA expression in amygdala and hippocampus.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. The panels show the expression of tight junction protein 1 (TJP1), claudin 5 (CLDN5) and occludin (OCLN) mRNA in the medial prefrontal cortex (A), hippocampus (B), amygdala (C), and hypothalamus (D). mRNA expression is expressed as fold change relative to VEH-treated mice. Values represent means + SEM, n=10-12; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 compared to VEH-treated mice, t-test.
Antibiotic treatment failed to enhance the expression of IL-1β, IL-6, TNF-α, IFN-γ and chemokine (C-C motif) ligand 2 (CCL2) mRNA in any of the four brain regions examined (see Supplementary Fig. 2).
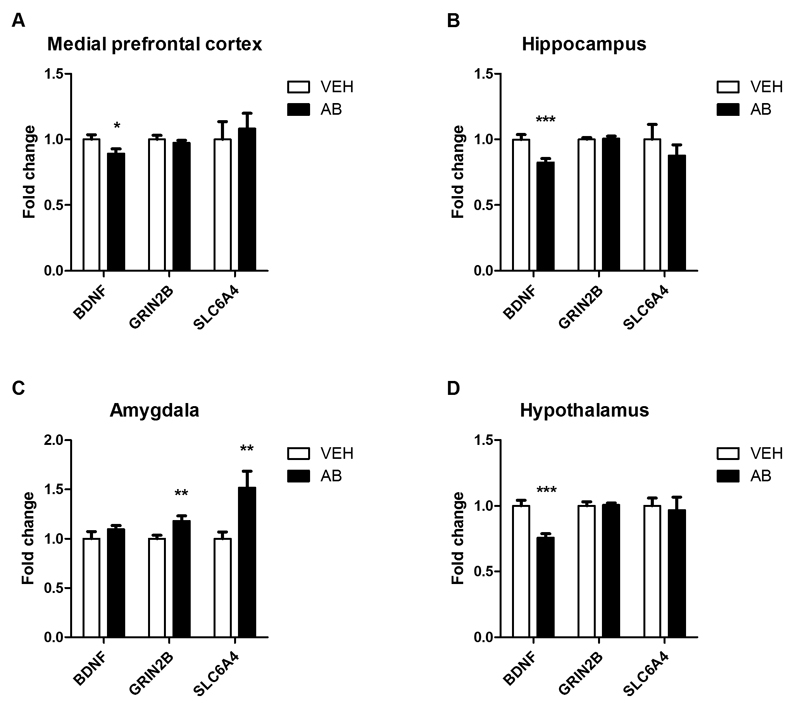
3.7. Antibiotic treatment alters expression patterns of neural signaling-related molecules in the brain
Given that cognitive performance is impaired in antibiotic-treated mice, we examined the cerebral expression of neural signaling-related molecules that are related to learning and memory. Particular focus was put on brain-derived neurotrophic factor (BDNF), N-methyl-D-aspartate receptor subunit GRIN2B (glutamate receptor, ionotropic, NMDA2B, epsilon 2) and serotonin transporter SLC6A4 (solute carrier family 6, neurotransmitter transporter, member 4), all of which may be involved in the cognitive deficit of GF mice (Sudo et al., 2004; Neufeld et al., 2011; Gareau et al., 2011; Clarke et al., 2013). In the medial prefrontal cortex (Fig. 8A), hippocampus (Fig. 8B) and hypothalamus (Fig. 8D) BDNF mRNA expression was significantly diminished in antibiotic-treated mice. In contrast, the expression of GRIN2B and SLC6A4 was left unaltered by antibiotic treatment in these three brain regions. In the amygdala, however, the expression of GRIN2B and SLC6A4 mRNA was significantly enhanced while that of BDNF mRNA was not affected (Fig. 8C).
Fig. 8. Antibiotic treatment alters the expression of neural signaling-related molecules in a brain region-specific manner.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. The panels show the expression of brain-derived neurotrophic factor (BDNF), N-methyl-D-aspartate receptor subunit 2B (GRIN2B) and serotonin transporter (SLC6A4) mRNA in the medial prefrontal cortex (A), hippocampus (B), amygdala (C), and hypothalamus (D). mRNA expression is expressed as fold change relative to VEH-treated mice. Values represent means + SEM, n=10-12; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 compared to VEH-treated mice, t-test.
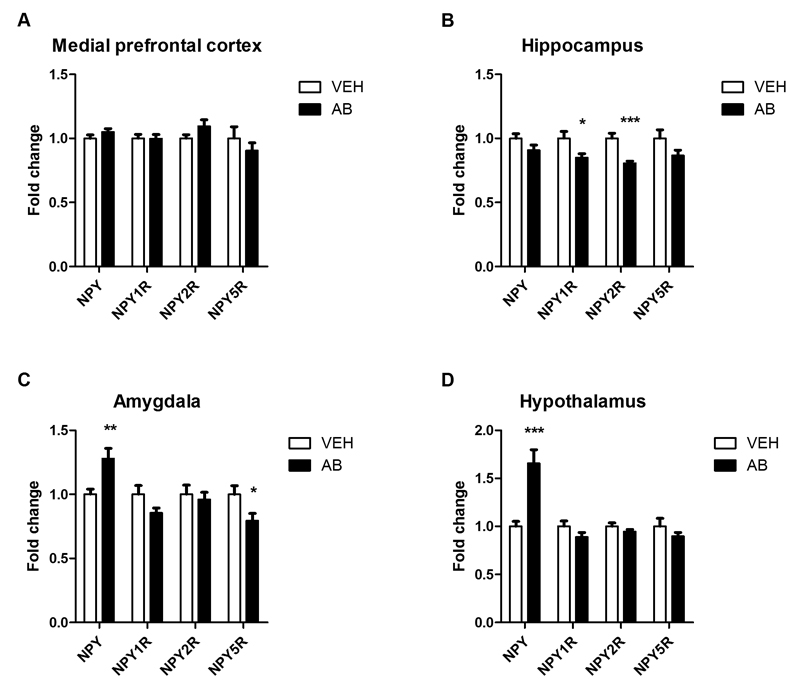
Since NPY is involved in the regulation of cognitive processes (Lach and de Lima, 2013; Reichmann and Holzer, 2015; Tasan et al., 2015) and its mRNA expression is increased in GF mice (Schele et al., 2013), we examined the expression of NPY and three of its receptors (Y1, NPY1R; Y2, NPY2R; Y5, NPY5R) in four brain regions of antibiotic-treated mice (Fig. 9A-D). In the medial prefrontal cortex no differences in the expression of NPY, NPY1R, NPY2R and NPY5R mRNA between vehicle- and antibiotic-treated mice were found (Fig. 9A). Antibiotic treatment led to a significant attenuation of NPY1R and NPY2R mRNA expression in the hippocampus (Fig. 9B), but NPY and NPY5R expression remained unaltered. Conversely, in the amygdala of antibiotic-treated mice NPY mRNA expression was significantly elevated and NPY5R expression decreased whereas NPY1R and NPY2R mRNA expression stayed unchanged (Fig. 9C). In the hypothalamus antibiotic treatment led to a prominent increase in NPY mRNA expression but did not affect the expression of any NPY receptor under study (Fig. 9D).
Fig. 9. Antibiotic treatment alters the expression of neuropeptide Y and its receptors in a region-specific manner.
Mice were treated with antibiotic (AB) mix or vehicle (VEH) by gavage for 11 days. The panels show the expression of neuropeptide Y (NPY) and the NPY receptors NPY1R, NPY2R and NPY5R in the medial prefrontal cortex (A), hippocampus (B), amygdala (C), and hypothalamus (D). mRNA expression is expressed as fold change relative to VEH-treated mice. Values represent means + SEM, n=10-12; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 compared to VEH-treated mice, t-test.
4. Discussion
The present study shows that intragastric treatment of mice with an antibiotic mix impairs novel object recognition, but not spatial memory. This behavioral change is associated with a disruption of the microbial community in the colon, distinct alterations of the colonic and circulating metabolite profile and particular changes of neurochemical brain activity. Although ampicillin was absorbed to some extent into the circulation, it remained undetectable in the brain. Neurotoxic effects may only occur if very high doses of ampicillin are given parenterally, the blood-brain barrier is leaky, and ampicillin’s concentration in the cerebrospinal fluid is elevated (Browning and Tune, 1983; Grill and Maganti, 2011). We therefore conclude that it is gut dysbiosis rather than a systemic antibiotic response that causes cognitive impairment. Furthermore, we propose that short-term treatment with antibiotics, which do not reach the brain, is a valid model to probe causality in microbiota-dependent changes of brain function and behavior. In substantiating this conclusion we address the multiple actions of antibiotic treatment along the gut microbiota-brain axis and compare them with the relevant phenotype of GF mice.
4.1. Intragastrically administered non-absorbable antibiotics are unlikely to reach the brain
Intragastric treatment of mice with non-absorbable antibiotics was used to specifically deplete the gut of a wide range of bacteria, including gram-positive as well as gram-negative anaerobic taxa. Of the antibiotics employed here (ampicillin, bacitracin, meropenem, neomycin, vancomycin), only ampicillin is known to have some oral bioavailability in humans (Craig and Stitzel, 2004; Lafforgue et al., 2008). Since a limited amount of ampicillin was found in murine plasma, but not brain tissue, following gastric gavage we conclude that the antibiotics under study were not or negligibly present in the brain at the time of behavioral and neurochemical analysis. Although systemic actions of circulating ampicillin cannot be excluded, intact spatial memory and a lack of cytokine upregulation (see Supplementary Fig. 2) negate a direct effect on the brain. In keeping with this contention, global vitality markers such as body weight, feeding and drinking (see Supplementary Fig. 3) likewise stayed unchanged. In addition, histology of the small and large intestine failed to unveil any signs of inflammation (see Supplementary Fig. 4).
4.2. Multiple antibiotic treatment causes dysbiosis and metabolite depletion in the gut
Antibiotic treatment profoundly disturbed the colonic microbial community and gut dysbiosis manifested itself in changes of several metabolite concentrations in the colonic luminal contents. The depletion of the SCFAs acetate, n-butyrate and propionate, which are products of microbial fermentation of dietary fiber, is in agreement with other studies (Yap et al., 2008; Romick-Rosendale et al., 2009; Swann et al., 2011a). Dietary choline is either metabolized to glycine by a mammalian pathway or processed to trimethylamine by gut bacterial enzymes (Zeisel et al., 1985). The depletion of trimethylamine in our study might indicate that choline metabolism was shifted towards the glycine pathway. Reduced bacterial load may also be the cause for the fall of colonic adenine and uracil contents in antibiotic-treated mice, since the colonic levels of nucleobases (adenine, cytosine, guanine and uracil) are likewise reduced in GF mice (Matsumoto et al., 2012). It should be noted, however, that the biochemical effects of antibiotic treatment in the colon were largely restricted to bacteria-derived metabolites (see Supplementary Table 2).
4.3. Circulating metabolites may be messengers in the communication between gut dysbiosis and brain
In contrast to circulating cytokines which remained unaltered (see Supplementary Fig. 1), the levels of several plasma metabolites (lipid species and converted bacteria-derived molecules) were significantly altered in antibiotic-treated mice. These changes are not fully congruent with those seen in GF mice (Wikoff et al., 2009; Velagapudi et al., 2010; Swann et al., 2011b). Whereas many PCs and sphingomyelins are decreased in the serum of fasted GF mice, PC 36:2 is increased in serum and adipose tissue (Velagapudi et al., 2010). While this is in agreement with the present findings in antibiotic-treated mice, the serum levels of PC 40:5, PC 40:8 and sphingomyelin 34:1 were increased in antibiotic-treated mice but left unaltered in fasted GF mice (Velagapudi et al., 2010). Given that the metabolism of PC and LPC is interconnected (Richmond and Smith, 2011), the increase in PC and concomitant decrease in LPC levels in antibiotic-treated mice points to a decrease in PC lipolysis. Unlike the LPC levels, the plasma concentration of phosphatidylinositol 38:5 was elevated in antibiotic-treated mice. As PCs are relevant to biological membrane structure and membrane-mediated cell signaling (Exton, 1990), a change in their circulating concentrations may have an impact on many organs including the brain. It remains to be elucidated in which way the gut microbiota modifies dietary absorption and hepatic biosynthesis of these important lipid species (Velagapudi et al., 2010).
The circulating levels of p-cresyl sulfate were markedly decreased in antibiotic-treated mice, which is in line with the absence of p-cresol sulfate in the serum of GF mice (Wikoff et al., 2009). Given that bacterial fermentation of dietary tyrosine generates p-cresol sulfate which is then absorbed and further metabolized to p-cresyl sulfate by the host (Meijers and Evenepoel, 2011), decreased levels of p-cresyl sulfate reflect a lack of gut bacterial enzymes. A similar explanation applies to the decrease in the plasma levels of trimethylamine-N-oxide, an oxidation product of trimethylamine (Rebouche and Chenard, 1991). The unconjugated secondary bile acid deoxycholic acid, which is produced by intestinal bacteria (Ridlon et al., 2006) and which could not be differentiated from the primary bile acid chenodeoxycholic acid due to isomerism, was completely depleted from the plasma of antibiotic-treated mice. As the levels of unconjugated bile acids are also decreased in plasma, heart, liver and kidney tissue of GF mice (Swann et al., 2011b), it is obvious that disruption of the gut microbial community has a strong impact on bile acid metabolism (Ridlon et al., 2006). The observation that nucleobase concentrations were not changed in the plasma of antibiotic-treated mice (see Supplementary Table 3) may indicate that the nucleobase depletion in the colonic contents reflects reduced bacterial load rather than changes in endogenous production.
Taken all metabolite data together, it is evident that gut dysbiosis-related changes in the concentrations of several circulating molecules may be a means to signal to the brain and modify its function. Cytokines are unlikely to be involved in this communication as deduced from the present study. These data add to the important search for specialized metabolites whereby the gut microbiota impacts on the brain in health and disease (Sharon et al., 2014).
4.4. Antibiotic-induced dysbiosis impairs cognitive performance
GF mice exhibit a deficit in non-spatial and working memory as assessed with the NORT and T-maze, respectively (Gareau et al., 2011). The current findings thus add to the emerging concept that factors emanating from the gut microbiota impact on cognitive performance (Gareau et al., 2011; Neufeld et al., 2011; Farzi et al., 2012; Davari et al., 2013; Gareau, 2014; Desbonnet et al., 2015).
The specific disturbance of this type of learning and memory also supports the conclusion that antibiotic-induced dysbiosis as studied under the current experimental conditions does not have a generalized, or neurotoxic, effect on brain function. The finding that in none of the cerebral areal areas under study (medial prefrontal cortex, hippocampus, amygdala, hypothalamus) the expression of cytokines was enhanced (see Supplementary Fig. 2) affirms this contention and indicates that neuroinflammatory processes were not induced by gut dysbiosis.
4.5. Cognitive impairment due to gut dysbiosis is associated with changes in the expression of tight junction proteins, brain-derived neurotrophic factor, N-methyl-D-aspartate receptor subunit 2B, serotonin transporter, NPY system and corticosterone
In analyzing neurochemical alterations that may underlie the cognitive deficit evoked by gut dysbiosis we focused on the medial prefrontal cortex, hippocampus and amygdala, all of which make distinct contributions to object recognition (Moses et al., 2005; Balderas et al., 2008; Broadbent et al., 2009; Barker and Warburton, 2011; Antunes and Biala, 2012; Beldjoud et al., 2015). In addition, the hypothalamus was included in view of the established impact of the gut microbiota on HPA axis activity (Sudo et al., 2004) and the role of corticosteroids in cognitive function (McEwen, 2007). The molecular analysis focused on mRNA quantitation, because this parameter provides a better reflection of the dynamics of intervention-induced gene expression in neuronal somata than peptide or protein levels that may mirror homeostatic balance between translation, storage capacity, axonal transport and neuronal release (Larsen et al., 1993; Ji et al., 1994).
The expression of tight junction proteins at the blood-brain barrier is regulated by the gut microbiota (Braniste et al., 2014), and a decrease in tight junction protein expression is thought to reflect an increase in blood-brain barrier permeability (Tietz and Engelhardt, 2015). Antibiotic-induced gut dysbiosis was associated with a downregulation of OCLN and CLDN5 in the hippocampus, as also seen in GF mice (Braniste et al., 2014), whereas OCLN and TJP1 were upregulated in the amygdala. Although the underlying mechanisms are not yet understood, our data add to the contention that the gut microbiota has an influence on the blood-brain barrier, the functionality of which modulates information transfer between the gut and brain. The change in cerebral tight junction expression in antibiotic-treated mice did not, however, translate into neuroinflammatory processes as would be reflected by increased cytokine expression in the brain.
There is ample evidence that BDNF plays a role in synaptic plasticity and cognition (Rattiner et al., 2005; Lu et al., 2014), and BDNF expression has been found to be altered both in GF mice (Sudo et al., 2004; Diaz Heijtz et al., 2011; Clarke et al., 2013; Schele et al., 2013) and mice with antibiotic-induced gut dysbiosis (Bercik et al., 2011; Desbonnet et al., 2015). We found a positive correlation between hippocampal BDNF expression and the memory index of the NORT in antibiotic-treated mice (Pearson’s correlation; r = 0.775, p = 0.024), which attests to a strong relationship between the deficit of hippocampal BDNF and the disturbance of novel object recognition memory.
Enhanced forebrain expression of GRIN2B, which encodes one subunit of the N-methyl D-aspartate glutamate receptor, is associated with improved performance in the NORT (Tang et al., 1999; Niimi et al., 2008). In the present study the expression of GRIN2B remained unaltered in the medial prefrontal cortex, but increased in the amygdala, which is at variance with the decreased formation of GRIN2B in the central amygdala of GF mice (Neufeld et al., 2011). In view of the established role of the amygdala in the regulation of anxiety it remains to be explored whether this disparity is of relevance to the observation that anxiety-like behavior is attenuated in GF mice but left unaltered in the antibiotic-treated mice of the current study (see Supplementary Fig. 5).
There is ample evidence that the metabolism and activity of tryptophan-derived mediators such as serotonin and kynurenine are under the influence of the gut microbiota (Diaz Heijtz et al., 2011; Desbonnet et al., 2015; O'Mahony et al., 2015; Yano et al., 2015). Serotonin-mediated neurotransmission is regulated by reuptake of this indoleamine into the presynaptic nerve terminals via a serotonin transporter encoded by the gene SLC6A4. Inhibition of serotonin reuptake by fluoxetine has been found to improve novel object recognition memory in stressed mice (El Hage et al., 2004; Urani et al., 2011) but to impair it in non-stressed mice (Carlini et al., 2012). A potential implication of serotonin reuptake in gut dysbiosis-mediated alterations in brain function can be deduced from the enhanced expression of SLC6A4 specifically in the amygdala of antibiotic-treated mice. There was a negative correlation between SLC6A4 expression in the amygdala and the NORT memory index (Pearson’s correlation; r = -0.711, p = 0.048) as well as a negative correlation between BDNF expression in the hippocampus and SLC6A4 expression in the amygdala (Pearson’s correlation; r = -0.679, p = 0.015). These relationships suggest that the serotonin system in the amygdala is in some way involved in the object recognition deficit of antibiotic-treated mice.
Another messenger with a potential implication in the cognitive impairment due to gut dysbiosis is NPY, given that this peptide is involved in various physiologic functions including cognition (Farzi et al., 2015; Reichmann and Holzer, 2015). NPY expression was markedly enhanced in the amygdala and hypothalamus of antibiotic-treated mice, a change that has also been reported for GF mice (Schele et al., 2013). Since antibiotic treatment did not increase food intake (see Supplementary Fig. 3), we hypothesize that elevated NPY expression in the hypothalamus and amygdala is related to the elevated plasma level of corticosterone (Lee et al., 2009; Gelfo et al., 2012; Farzi et al., 2015). Increased NPY synthesis in the amygdala and hypothalamus may also be the reason why anxiety- and depression-like behavior remained unchanged despite a rise in plasma corticosterone (see Supplementary Fig. 1 and Fig. 5), which otherwise is known to enhance anxiety- and depression-like behavior (Lee et al., 2009; Kutiyanawalla et al., 2011; Baldock et al., 2014; Farzi et al., 2015). In addition, increased hypothalamic NPY formation may have contributed to the decreased BDNF expression in the hypothalamus (Gelfo et al., 2012).
The antibiotic-induced upregulation of NPY in the amygdala and hypothalamus was matched by a downregulation of NPY1R and NPY2R in the hippocampus. In line with their preferential post- and presynaptic location, respectively, NPY1R and NPY2R often mediate opposing effects of NPY in neurotransmission and behavior (Farzi et al., 2015). While NPY1R plays an important role in maintaining stress resilience and emotional-affective homeostasis, NPY2R has also been implicated in cognition and schizophrenia (Farzi et al., 2015). It is particularly worth noting that NPY2R knockout mice exhibit a similar deficit in novel object recognition memory (Redrobe et al., 2004; Painsipp et al., 2008) as the antibiotic-treated mice in the current study. Decreased NPY2R expression in the hippocampus could hence be a crucial factor in the cognitive impairment due to gut dysbiosis. The antibiotic-induced increase of NPY expression in the amygdala occurred alongside a decrease in NPY5R expression. The functional relevance of this finding in the context of the current study remains to be analyzed as the implication of NPY5R in cognition is still little understood (Farzi et al., 2015).
The plasma concentration of corticosterone was enhanced in antibiotic-treated mice, which is reminiscent of the exaggerated corticosterone response of GF mice to stress (Sudo et al., 2004). Enhanced circulating corticosterone could have a bearing on the cognitive deficit seen in antibiotic-treated mice, since chronic stress-induced enhancement of glucocorticoid levels has a negative impact on cognitive function (McEwen, 2007). In addition, the rise of plasma corticosterone in antibiotic-treated mice may, in analogy to other findings (Dwivedi et al., 2006; Naert et al., 2015), contribute to the downregulation of BDNF seen in the hypothalamus.
4.6. Conclusions
Taking all data together, we show that a short-term intragastric treatment of adult mice with an antibiotic mixture disrupts the bacterial community in the colon, specifically impairs novel object recognition memory and causes distinct alterations in circulating metabolite levels and in the expression of molecules relevant to cerebral function. Profound alterations in the metabolite profile of the colon and circulation may be relevant to the communication between gut and brain. The cognitive impairment due to gut dysbiosis is related to changes in the expression of tight junction proteins, BDNF, GRIN2B, the serotonin transporter and the NPY system and HPA axis activity. Several of the behavioral and biochemical alterations brought about by short-term antibiotic treatment are similar to those found in GF mice although distinct differences have also been noted. Our findings add to the understanding of the microbiota-gut-brain axis and highlight the potential and limitation of antibiotic-induced gut dysbiosis as model system to probe causality in the interaction between gut microbiota and brain.
Appendix A. Supplementary Data
Supplementary Data
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Acknowledgements
This work was supported by the Austrian Science Fund (FWF grants P25912-B23 and W1241-B18) and the Medical University of Graz (Doctoral College “Molecular Fundamentals of Inflammation – MOLIN”). Part of this work has been carried out with the K1 COMET Competence Center CBmed, which is funded by the Federal Ministry of Transport, Innovation and Technology (BMVIT); the Federal Ministry of Science, Research and Economy (BMWFW); Land Steiermark (Department 12, Business and Innovation); the Styrian Business Promotion Agency (SFG); and the Vienna Business Agency. The COMET program is executed by the FFG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The results contained in this work form part of the PhD thesis of Esther E. Fröhlich. The authors thank Dr. Young Hae Choi and Dr. Hye Kyong Kim (University of Leiden) for performing the NMR analysis of intestinal metabolites and Isolde Besseling van der Vaart (Winclove, Amsterdam) for managing this cooperation. The technical assistance of Claudia Meindl, Markus Absenger-Novak, Martina Hatz and Theresa Maierhofer (Center for Medical Research, Medical University of Graz) is greatly appreciated. In addition, the authors are grateful to Edgar Gander (JOANNEUM RESEARCH Forschungsgesellschaft mbH) for performing the LC-MS metabolomics analysis and to Gunnar Libiseller (JOANNEUM RESEARCH Forschungsgesellschaft mbH) for writing the PeakScout tool.
Abbreviations
- ANOVA
analysis of variance
- AUC
area under the curve
- BDNF
brain-derived neurotrophic factor
- BM
Barnes maze
- CCL2
chemokine (C-C motif) ligand 2
- CLDN5
claudin 5
- GF
germ-free
- GRIN2B
glutamate receptor, ionotropic, NMDA2B (epsilon 2); N-methyl-D-aspartate receptor subunit 2B
- HPA
hypothalamic-pituitary-adrenal
- IFN
interferon
- IL
interleukin
- LLOQ
lower limit of quantification
- LPC
lysophosphatidylcholine
- NORT
novel object recognition test
- NPY
neuropeptide Y
- NPY1R
neuropeptide Y receptor Y1
- NPY2R
neuropeptide Y receptor Y2
- NPY5R
neuropeptide Y receptor Y5
- OCLN
occludin
- PC
phosphatidylcholine
- PCA
principal component analysis
- SCFA
short-chain fatty acids
- SLC6A4
solute carrier family 6 (neurotransmitter transporter), member 4; serotonin transporter
- TJP1
tight junction protein 1
- TNF
tumor necrosis factor
References
- Antunes M, Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process. 2012;13:93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar A, Liu T, Chan WT, Hayes J, Nejad M, Lei K, Bitan G. A shortened Barnes maze protocol reveals memory deficits at 4-months of age in the triple-transgenic mouse model of Alzheimer's disease. PLoS One. 2013;8:e80355. doi: 10.1371/journal.pone.0080355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. S0021-9673(06)00993-9 [pii] [DOI] [PubMed] [Google Scholar]
- Balderas I, Rodriguez-Ortiz CJ, Salgado-Tonda P, Chavez-Hurtado J, McGaugh JL, Bermudez-Rattoni F. The consolidation of object and context recognition memory involve different regions of the temporal lobe. Learn Mem. 2008;15:618–624. doi: 10.1101/lm.1028008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldock PA, Lin S, Zhang L, Karl T, Shi Y, Driessler F, Zengin A, Hormer B, Lee NJ, Wong IP, Lin EJ, et al. Neuropeptide y attenuates stress-induced bone loss through suppression of noradrenaline circuits. J Bone Miner Res. 2014;29:2238–2249. doi: 10.1002/jbmr.2205. [DOI] [PubMed] [Google Scholar]
- Barker GR, Warburton EC. When is the hippocampus involved in recognition memory? J Neurosci. 2011;31:10721–10731. doi: 10.1523/JNEUROSCI.6413-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldjoud H, Barsegyan A, Roozendaal B. Noradrenergic activation of the basolateral amygdala enhances object recognition memory and induces chromatin remodeling in the insular cortex. Front Behav Neurosci. 2015;9:108. doi: 10.3389/fnbeh.2015.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. 609.e1-3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- Bienenstock J, Kunze W, Forsythe P. Microbiota and the gut-brain axis. Nutr Rev. 2015;73(Suppl 1):28–31. doi: 10.1093/nutrit/nuv019. [DOI] [PubMed] [Google Scholar]
- Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods. 2012;9:425–426. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, Korecka A, Bakocevic N, Ng LG, Kundu P, Gulyas B, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent NJ, Gaskin S, Squire LR, Clark RE. Object recognition memory and the rodent hippocampus. Learn Mem. 2009;17:5–11. doi: 10.1101/lm.1650110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning MC, Tune BM. Toxicology of beta-lactam antibiotics. Handb Exp Pharmacol. 1983;67/II:371–397. [Google Scholar]
- Brunner SM, Farzi A, Locker F, Holub BS, Drexel M, Reichmann F, Lang AA, Mayr JA, Vilches JJ, Navarro X, Lang R, et al. GAL3 receptor KO mice exhibit an anxiety-like phenotype. Proc Natl Acad Sci USA. 2014;111:7138–7143. doi: 10.1073/pnas.1318066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini VP, Poretti MB, Rask-Andersen M, Chavan RA, Ponzio MF, Sawant RS, de Barioglio SR, Schioth HB, de Cuneo MF. Differential effects of fluoxetine and venlafaxine on memory recognition: possible mechanisms of action. Prog Neuropsychopharmacol Biol Psychiatry. 2012;38:159–167. doi: 10.1016/j.pnpbp.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- Craig CR, Stitzel RE. Modern Pharmacology with Clinical Applications. 6th edn. Lippincott Williams & Wilkins; Philadelphia: 2004. [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Davari S, Talaei SA, Alaei H, Salami M. Probiotics treatment improves diabetes-induced impairment of synaptic activity and cognitive function: behavioral and electrophysiological proofs for microbiome-gut-brain axis. Neuroscience. 2013;240:287–296. doi: 10.1016/j.neuroscience.2013.02.055. [DOI] [PubMed] [Google Scholar]
- De Palma G, Blennerhassett P, Lu J, Deng Y, Park AJ, Green W, Denou E, Silva MA, Santacruz A, Sanz Y, Surette MG, et al. Microbiota and host determinants of behavioural phenotype in maternally separated mice. Nat Commun. 2015;6:7735. doi: 10.1038/ncomms8735. [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Traplin A, O'Sullivan O, Crispie F, Moloney RD, Cotter PD, Dinan TG, Cryan JF. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav Immun. 2015;48:165–173. doi: 10.1016/j.bbi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Pandey GN. Antidepressants reverse corticosterone-mediated decrease in brain-derived neurotrophic factor expression: differential regulation of specific exons by antidepressants and corticosterone. Neuroscience. 2006;139:1017–1029. doi: 10.1016/j.neuroscience.2005.12.058. S0306-4522(06)00059-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage W, Peronny S, Griebel G, Belzung C. Impaired memory following predatory stress in mice is improved by fluoxetine. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:123–128. doi: 10.1016/j.pnpbp.2003.09.028. S0278-5846(03)00241-0 [pii] [DOI] [PubMed] [Google Scholar]
- Exton JH. Signaling through phosphatidylcholine breakdown. J Biol Chem. 1990;265:1–4. [PubMed] [Google Scholar]
- Farzi A, Gorkiewicz G, Holzer P. Non-absorbable oral antibiotic treatment in mice affects multiple levels of the microbiota-gut-brain axis. Neurogastroenterol Motil. 2012;24:78–78. [Google Scholar]
- Farzi A, Reichmann F, Holzer P. The homeostatic role of neuropeptide Y in immune function and its impact on mood and behaviour. Acta Physiol (Oxf) 2015;213:603–627. doi: 10.1111/apha.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjeva M, Paradis-Bleau C, Priebe GP, Fichorova R, Pier GB. Caveolin-1 modifies the immunity to Pseudomonas aeruginosa. J Immunol. 2010;184:296–302. doi: 10.4049/jimmunol.0900604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau MG. Microbiota-gut-brain axis and cognitive function. Adv Exp Med Biol. 2014;817:357–371. doi: 10.1007/978-1-4939-0897-4_16. [DOI] [PubMed] [Google Scholar]
- Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, Macqueen G, Sherman PM. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- Gelfo F, Tirassa P, De Bartolo P, Croce N, Bernardini S, Caltagirone C, Petrosini L, Angelucci F. NPY intraperitoneal injections produce antidepressant-like effects and downregulate BDNF in the rat hypothalamus. CNS Neurosci Ther. 2012;18:487–492. doi: 10.1111/j.1755-5949.2012.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill MF, Maganti RK. Neurotoxic effects associated with antibiotic use: management considerations. Br J Clin Pharmacol. 2011;72:381–393. doi: 10.1111/j.1365-2125.2011.03991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Zhang X, Wiesenfeld-Hallin Z, Hokfelt T. Expression of neuropeptide Y and neuropeptide Y (Y1) receptor mRNA in rat spinal cord and dorsal root ganglia following peripheral tissue inflammation. J Neurosci. 1994;14:6423–6434. doi: 10.1523/JNEUROSCI.14-11-06423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. gkd027 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutiyanawalla A, Terry AV, Jr, Pillai A. Cysteamine attenuates the decreases in TrkB protein levels and the anxiety/depression-like behaviors in mice induced by corticosterone treatment. PLoS One. 2011;6:e26153. doi: 10.1371/journal.pone.0026153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lach G, de Lima TC. Role of NPY Y1 receptor on acquisition, consolidation and extinction on contextual fear conditioning: dissociation between anxiety, locomotion and non-emotional memory behavior. Neurobiol Learn Mem. 2013;103:26–33. doi: 10.1016/j.nlm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Lafforgue G, Arellano C, Vachoux C, Woodley J, Philibert C, Dupouy V, Bousquet-Melou A, Gandia P, Houin G. Oral absorption of ampicillin: role of paracellular route vs. PepT1 transporter. Fundam Clin Pharmacol. 2008;22:189–201. doi: 10.1111/j.1472-8206.2008.00572.x. [DOI] [PubMed] [Google Scholar]
- Larsen PJ, Mikkelsen JD, Jessop DS, Lightman SL, Chowdrey HS. Neuropeptide Y mRNA and immunoreactivity in hypothalamic neuroendocrine neurons: effects of adrenalectomy and chronic osmotic stimulation. J Neurosci. 1993;13:1138–1147. doi: 10.1523/JNEUROSCI.13-03-01138.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Shim I, Lee HJ, Yang Y, Hahm DH. Effects of acupuncture on chronic corticosterone-induced depression-like behavior and expression of neuropeptide Y in the rats. Neurosci Lett. 2009;453:151–156. doi: 10.1016/j.neulet.2009.01.076. [DOI] [PubMed] [Google Scholar]
- Liu X, Cao S, Zhang X. Modulation of Gut Microbiota-Brain Axis by Probiotics, Prebiotics, and Diet. J Agric Food Chem. 2015;63:7885–7895. doi: 10.1021/acs.jafc.5b02404. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Nagappan G, Lu Y. BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb Exp Pharmacol. 2014;220:223–250. doi: 10.1007/978-3-642-45106-5_9. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. 87/3/873 [pii] [DOI] [PubMed] [Google Scholar]
- Meijers BK, Evenepoel P. The gut-kidney axis: indoxyl sulfate, p-cresyl sulfate and CKD progression. Nephrol Dial Transplant. 2011;26:759–761. doi: 10.1093/ndt/gfq818. [DOI] [PubMed] [Google Scholar]
- Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Mace K, Chou CJ. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- Moller PL, Paerregaard A, Gad M, Kristensen NN, Claesson MH. Colitic scid mice fed Lactobacillus spp. show an ameliorated gut histopathology and an altered cytokine profile by local T cells. Inflamm Bowel Dis. 2005;11:814–819. doi: 10.1097/01.mib.0000175906.77340.15. 00054725-200509000-00005 [pii] [DOI] [PubMed] [Google Scholar]
- Moses SN, Cole C, Driscoll I, Ryan JD. Differential contributions of hippocampus, amygdala and perirhinal cortex to recognition of novel objects, contextual stimuli and stimulus relationships. Brain Res Bull. 2005;67:62–76. doi: 10.1016/j.brainresbull.2005.05.026. S0361-9230(05)00235-2 [pii] [DOI] [PubMed] [Google Scholar]
- Naert G, Zussy C, Tran Van Ba C, Chevallier N, Tang YP, Maurice T, Givalois L. Involvement of endogenous brain-derived neurotrophic factor in hypothalamic-pituitary-adrenal axis activity. J Neuroendocrinol. 2015 doi: 10.1111/jne.12324. [DOI] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–64. e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- Niimi K, Takahashi E, Itakura C. Improved short-term memory and increased expression of NR2B observed in senescence-accelerated mouse (SAM) P6. Exp Gerontol. 2008;43:847–852. doi: 10.1016/j.exger.2008.06.010. [DOI] [PubMed] [Google Scholar]
- O'Mahony SM, Clarke G, Dinan TG, Cryan JF. Early-life adversity and brain development: Is the microbiome a missing piece of the puzzle? Neuroscience. 2015 doi: 10.1016/j.neuroscience.2015.09.068. S0306-4522(15)00895-7 [pii] [DOI] [PubMed] [Google Scholar]
- Painsipp E, Wultsch T, Edelsbrunner ME, Tasan RO, Singewald N, Herzog H, Holzer P. Reduced anxiety-like and depression-related behavior in neuropeptide Y Y4 receptor knockout mice. Genes Brain Behav. 2008;7:532–542. doi: 10.1111/j.1601-183X.2008.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2011. [Google Scholar]
- Rattiner LM, Davis M, Ressler KJ. Brain-derived neurotrophic factor in amygdala-dependent learning. Neuroscientist. 2005;11:323–333. doi: 10.1177/1073858404272255. 11/4/323 [pii] [DOI] [PubMed] [Google Scholar]
- Rebouche CJ, Chenard CA. Metabolic fate of dietary carnitine in human adults: identification and quantification of urinary and fecal metabolites. J Nutr. 1991;121:539–546. doi: 10.1093/jn/121.4.539. [DOI] [PubMed] [Google Scholar]
- Redrobe JP, Dumont Y, Herzog H, Quirion R. Characterization of neuropeptide Y, Y(2) receptor knockout mice in two animal models of learning and memory processing. J Mol Neurosci. 2004;22:159–166. doi: 10.1385/JMN:22:3:159. JMN:22:3:159 [pii] [DOI] [PubMed] [Google Scholar]
- Reichmann F, Holzer P. Neuropeptide Y: A stressful review. Neuropeptides. 2015 doi: 10.1016/j.npep.2015.09.008. S0143-4179(15)00097-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond GS, Smith TK. Phospholipases A(1) Int J Mol Sci. 2011;12:588–612. doi: 10.3390/ijms12010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. R500013-JLR200 [pii] [DOI] [PubMed] [Google Scholar]
- Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, Chona DL, Kennedy MA. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn Reson Chem. 2009;47(Suppl 1):S36–46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- Russell WR, Hoyles L, Flint HJ, Dumas ME. Colonic bacterial metabolites and human health. Curr Opin Microbiol. 2013;16:246–254. doi: 10.1016/j.mib.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12 doi: 10.1186/s12915-014-0087-z. 87-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schele E, Grahnemo L, Anesten F, Hallen A, Backhed F, Jansson JO. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology. 2013;154:3643–3651. doi: 10.1210/en.2012-2151. [DOI] [PubMed] [Google Scholar]
- Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014;20:719–730. doi: 10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Stilling RM, Bordenstein SR, Dinan TG, Cryan JF. Friends with social benefits: host-microbe interactions as a driver of brain evolution and development? Front Cell Infect Microbiol. 2014a;4:147. doi: 10.3389/fcimb.2014.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Dinan TG, Cryan JF. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014b;13:69–86. doi: 10.1111/gbb.12109. [DOI] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JR, Tuohy KM, Lindfors P, Brown DT, Gibson GR, Wilson ID, Sidaway J, Nicholson JK, Holmes E. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res. 2011a;10:3590–3603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. 2011b;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, Tsien JZ. Genetic enhancement of learning and memory in mice. Nature. 1999;401:63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- Tasan RO, Verma D, Wood J, Lach G, Hormer B, de Lima TC, Herzog H, Sperk G. The role of Neuropeptide Y in fear conditioning and extinction. Neuropeptides. 2015 doi: 10.1016/j.npep.2015.09.007. S0143-4179(15)00096-7 [pii] [DOI] [PubMed] [Google Scholar]
- Tietz S, Engelhardt B. Brain barriers: Crosstalk between complex tight junctions and adherens junctions. J Cell Biol. 2015;209:493–506. doi: 10.1083/jcb.201412147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urani A, Philbert J, Cohen C, Griebel G. The corticotropin-releasing factor 1 receptor antagonist, SSR125543, and the vasopressin 1b receptor antagonist, SSR149415, prevent stress-induced cognitive impairment in mice. Pharmacol Biochem Behav. 2011;98:425–431. doi: 10.1016/j.pbb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Boren J, Oresic M, Backhed F. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–6. doi: 10.1093/nar/gkl923. 35/suppl_1/D521 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel SH, DaCosta KA, Fox JG. Endogenous formation of dimethylamine. Biochem J. 1985;232:403–408. doi: 10.1042/bj2320403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.