Highlight
Brassinosteroid-regulated transcription factor BES1 targets the BRL3 receptor gene and finely modulates its transcription in the vascular and stem cells of the Arabidopsis primary root.
Key words: BES, BRL3, brassinosteroid, quiescent center, root, vascular.
Abstract
Brassinosteroid (BR) hormones are important regulators of plant growth and development. Recent studies revealed the cell-specific role of BRs in vascular and stem cell development by the action of cell-specific BR receptor complexes and downstream signaling components in Arabidopsis thaliana. Despite the importance of spatiotemporal regulation of hormone signaling in the control of plant vascular development, the mechanisms that confer cellular specificity to BR receptors within the vascular cells are not yet understood. The present work shows that BRI1-like receptor genes 1 and 3 (BRL1 and BRL3) are differently regulated by BRs. By using promoter deletion constructs of BRL1 and BRL3 fused to GFP/GUS (green fluorescent protein/β-glucuronidase) reporters in Arabidopsis, analysis of their cell-specific expression and regulation by BRs in the root apex has been carried out. We found that BRL3 expression is finely modulated by BRs in different root cell types, whereas the location of BRL1 appears to be independent of this hormone. Physiological and genetic analysis show a BR-dependent expression of BRL3 in the root meristem. In particular, BRL3 expression requires active BES1, a central transcriptional effector within the BRI1 pathway. ChIP analysis showed that BES1 directly binds to the BRRE present in the BRL3 promoter region, modulating its transcription in different subsets of cells of the root apex. Overall our study reveals the existence of a cell-specific negative feedback loop from BRI1-mediated BES1 transcription factor to BRL3 in phloem cells, while contributing to a general understanding of the spatial control of steroid signaling in plant development.
Introduction
Brassinosteroids (BRs) are polyhydroxylated plant steroid hormones that were first identified by their ability to elongate plant stems when applied exogenously (Grove et al., 1979; Mandava, 1988). The identification of mutants affected in BR synthesis or signal transduction has revealed an essential role for this hormone in cell elongation and differentiation (Fukuda, 1997; Yamamoto et al., 1997). So far, BRs have been reported to be involved in the regulation of multiple developmental and physiological processes such as cell division, elongation, and also the differentiation of vascular and stem cells, among others (Fabregas and Caño-Delgado, 2014). The BRASSINOSTEROID INSENSITIVE 1 (BRI1) gene was identified by a genetic screening of a BR-insensitive loss-of-function mutant performed in the model plant Arabidopsis (Arabidopsis thaliana) (Clouse, 1996). This mutant exhibited severe dwarfism with characteristic dark green and epinastic leaves and a reduced apical dominance and fertility (Szekeres et al., 1996; Clouse, 1996; Kauschmann et al., 1996; Li et al., 1996; Azpiroz et al., 1998; Choe et al., 1999, 2001; Noguchi et al., 1999). The BRI1 gene encodes a membrane-localized leucine-rich repeat receptor-like kinase (LRR-RLK) comprising an extracellular LRR domain, a single transmembrane domain, a juxtamembrane domain, a cytoplasmic serine/threonine kinase domain, and a C-terminal regulatory region (Li and Chory, 1997). Genetic and biochemical assays have demonstrated that the BRI1 receptor complex directly bind BRs with high affinity (Wang et al., 2001). This occurs via direct binding of BRs to the extracellular domain of the LRR-RLK proteins at the cell membrane (Wang et al., 2001; Kinoshita et al., 2005).
Recent structural studies have confirmed that brassinolide (BL) binds to the BRI1 plasma membrane receptor through a 70 amino acid island domain located within the extracellular domain of BRI1, creating a surface pocket for ligand binding (Hothorn et al., 2011; She et al., 2011). Upon direct binding of BL to this extracellular domain (Kinoshita et al., 2005), BRI1 forms a heterodimer with its co-receptor BAK1 ASSOCIATED RECEPTOR KINASE 1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (BAK1/SERK3), yet another LRR-RLK (Li et al., 2002; Nam and Li, 2002; Russinova et al., 2004). Subsequently, the BR signal is modulated intracellularly in a phosphorylation- and dephosphorylation-dependent manner, ending in the de-phosphorylation of the BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1) genes (Li et al., 2002; Wang et al., 2002; Yin et al., 2002; Mora-García et al., 2004; Gampala et al., 2007 ). Both BES1 and BZR1 are members of a plant-specific family of basic-helix–loop–helix (bHLH) transcription factors that act as homo- or heterodimers (Yin et al., 2002; He et al., 2005; Yu et al., 2008; Li et al., 2009). The gain-of-function mutant bes1-D is known to be constitutively active mutant independent of BR and BRI1 signaling and able to suppress bri1 phenotypes. The accumulation of de-phosphorylated BES1 protein in the nucleus, where BES1 is activating its target genes, is higher in bes1-D lines than in the wild type (Wang et al., 2002; Yin et al., 2002).
Detailed analysis of promoter elements indicated that both BES1 and BZR1 are able to bind BR response elements (BRREs) as well as E-boxes (Sun et al., 2010; Yu et al., 2011). Binding of BES1 to BRREs was shown to be much stronger than to E-boxes, since efficient BES1 binding to the latter needs a partner (Yin et al., 2005). While BRREs are mostly enriched in BR-repressed genes, E-box elements are mostly enriched in BR-induced genes (Sun et al., 2010; Yu et al., 2011). Additionally, it was previously suggested that BZR1 acts as a transcriptional repressor (He et al., 2005) while BES1 acts as a transcriptional activator (Yin et al., 2005). However, recent genome-wide ChIP analysis showed that both BZR1 and BES1 function either as activators or repressors. BES1 and BZR1 regulation of downstream targets is most probably determined by additional promoter sequence elements and/or BES1- and BZR1-interacting proteins (Sun et al., 2010; Ye et al., 2011; Yu et al., 2011).
In addition to BRI1, three additional LRR-RLK proteins have been identified as BRI1 homologs named BRI1-LIKE RECEPTORS 1, 2, and 3 (BRL1, BRL2 and BRL3) (Caño-Delgado et al., 2004). Unlike BRL2, previously described as VASCULAR HIGHWAY 1 (VH1) (Clay and Nelson, 2002), BRL1 and BRL3 encode membrane-localized receptors able to bind BL with high affinity. The expression of the BRL1 and BRL3 genes under the BRI1 promoter reverts the phenotypic defects in the bri1 mutant, demonstrating that both BRL1 and BRL3 are functional BR receptor genes (Caño-Delgado et al., 2004; Zhou et al., 2004). In contrast to BRI1 that is widely expressed in plants, BRL1 and BRL3 exhibit an enriched expression in the vasculature (Caño-Delgado et al., 2004). Biochemical purification of the BRL3 complex has addressed the cellular specificity of BR receptor complexes in plants (Fàbregas et al., 2013), thus suggesting that the localization of these receptors accounts for specific cellular functions (Fàbregas et al., 2013). It has been proposed that the BRI1, the BRL1, and the BRL3 receptors signal together in BR-mediated root growth and quiescent center (QC) division dynamics, although whether this interaction occurs at the receptor level or by a downstream signaling component is not yet established.
In this study, the 5' intergenic region of BRL1 (ProBRL1) and BRL3 (ProBRL3) has been analyzed, to identify cis-acting elements required for QC and vascular-specific expression patterns. The ProBRL1 expression seemed to be BR independent, whereas the expression of ProBRL3 showed a BR dose-dependent spatial expression pattern. This analysis reveals that binding of the BR-activated transcription factor BES1 to a cis-acting BRRE located at base pair −1441 of the BRL3 promoter controls the spatial localization of the receptor in plants. Overall this study advances the idea that BRI1 and BRL3 receptor signals are interconnected by BES1, which provide the cellular specificity for BRL3 transcription in specific cells.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana Columbia ecotype (Col-0) was used to generate all the ProBRL3::GUS and ProBRL1::GUS transgenic plants.The bes1-D mutant introgressed into the Col-0 ecotype was used in this study (Ibanes et al., 2009). Seeds were surface sterilized in 35% sodium hypochlorite, vernalized for 72h at 4 °C in the dark, and grown on plates containing 1× Murashige and Skoog (MS) salt mixture, 1% sucrose, and 0.8% agar in the absence or presence of different concentrations of BL (C28H48O6; Wako, Osaka, Japan). In the case of the 35S::bes1-D:GR transgenic plants, the agar was supplemented with 1 µM dexamethasone. Plants were grown under fluorescent light (12h light/12h dark cycles) for 6 d prior to analysis.
In silico analysis of the promoters
The search for pre-determined regulatory promoter elements was done using the program DNA-pattern (Thomas-Chollier et al., 2008).
Generation of promoter constructs
To generate the various BRL3 and BRL1 promoter fusion constructs, Invitrogen’s Gateway technology was used. In the first reaction step, pDONR221 or pDONR207 was used to generate entry clones. In the second step, the destination vectors pHGWFS7 [(green fluorescent protein/β-glucuronidase) GFP/GUS] and pGWB635 (firefly luciferase) (Nakamura et al., 2010) were used to generate the expression clones. Transgenic plants (GFP/GUS) from 10 independent T4 homozygous lines were selected by hygromycin resistance and homozygous plants were used for expression pattern analysis. In addition, the constructs 35S::bes1-D:GFP (Vilarrasa-Blasi et al., 2014) and 35S::bes1-D:GR were generated by using the destination vector pB7m34GW (Karimi et al., 2007). Primers used in the cloning of the above constructs are listed in Supplementary Table S1 at JXB online).
Histology and microscopy
For GUS detection, 6-day-old seedlings were immersed in ice-cold 90% (v/v) acetone, incubated for 20min on ice, rinsed twice in dH2O, infiltrated with GUS [100mM sodium phosphate buffer (pH 7.2), 10mM sodium EDTA, 0.1% Triton X-100, 1mg ml–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Xgluc; Duchefa, Haarlem, The Netherlands), 10mM potassium ferrocyanide and potassium ferricyanide] and incubated at 37 °C for 15h in the dark. Samples were rinsed three times in dH2O and treated with 70% ethanol. Stained roots were visualized with an AxioPhot (Zeiss, Jena, Germany) microscope. For a cell type-specific expression analysis of ProBRL1::GUS and ProBRL3::GUS within the root meristem, GUS-stained seedlings were subsequently immersed in 10% acetic acid supplemented with 50% MetOH solution and stained using a modified Pseudo-Schiff (mPS)-propidium iodide (PI) staining method (adapted from Truernit et al., 2008).
To analyze the GFP localization in ProBRL3::GFP lines, 6-day-old roots were stained in 10 μg ml–1 PI and visualized after excitation by a Kr/Ar 488nm laser line. PI and GFP were detected with a 570–670nm and a 500–545nm band-pass filter, respectively. An FV 1000 confocal microscope (Olympus, Tokyo, Japan) was used throughout the study.
Luciferase expression assays
Arabidopsis protoplasts were isolated as previously described (Sheen, 2002) and transfected with different ProBRL3::LUC promoter fusions (pGWB635 vector), 35S::bes1-D:GFP or 35S::GFP and 35S::Renilla (PHTT672, from Pioneer) (Morohashi et al., 2012). For the expression assay per se, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used. The bioluminescent signal was measured using a luminometer Centre LB 960 (Berthold). The data were normalized for Renilla activity. After normalization, the fold change was calculated as the ratio between each particular treatment and the treatment with the promoter constructs without transcription factor (Schagat et al., 2007). For each experiment, three technical and three biological replicates were used.
ChIP assays
35S::bes1-D:GFP (Vilarrasa-Blasi et al., 2014) and Col-0 plants were grown in 1/2 MS (12h light/12h dark cycles) for 6 d. Seedlings were fixed with 1% formaldehyde and nuclei were extracted according to Deal and Henikoff (2011). ChIP experiments using anti-GFP antibodies were performed according to Gendrel et al. (2005). Detection of PCR products was performed using Absolute qPCR SYBR Green mix (Thermo Scientific) in a Biorad thermocycler. Two different biological replicas were performed for each region of interest. The ChIP-quantitative PCR (qPCR) data were analyzed using the Percent Input Method (Nagaki et al., 2003). With this method, signals obtained from the ChIP are divided by signals from an input sample. This input sample represents the amount of chromatin used in the ChIP. Primers used for qPCR are listed in Supplementary Table S1.
Results
Promoter deletion analysis of BRL3 and BRL1 receptor genes
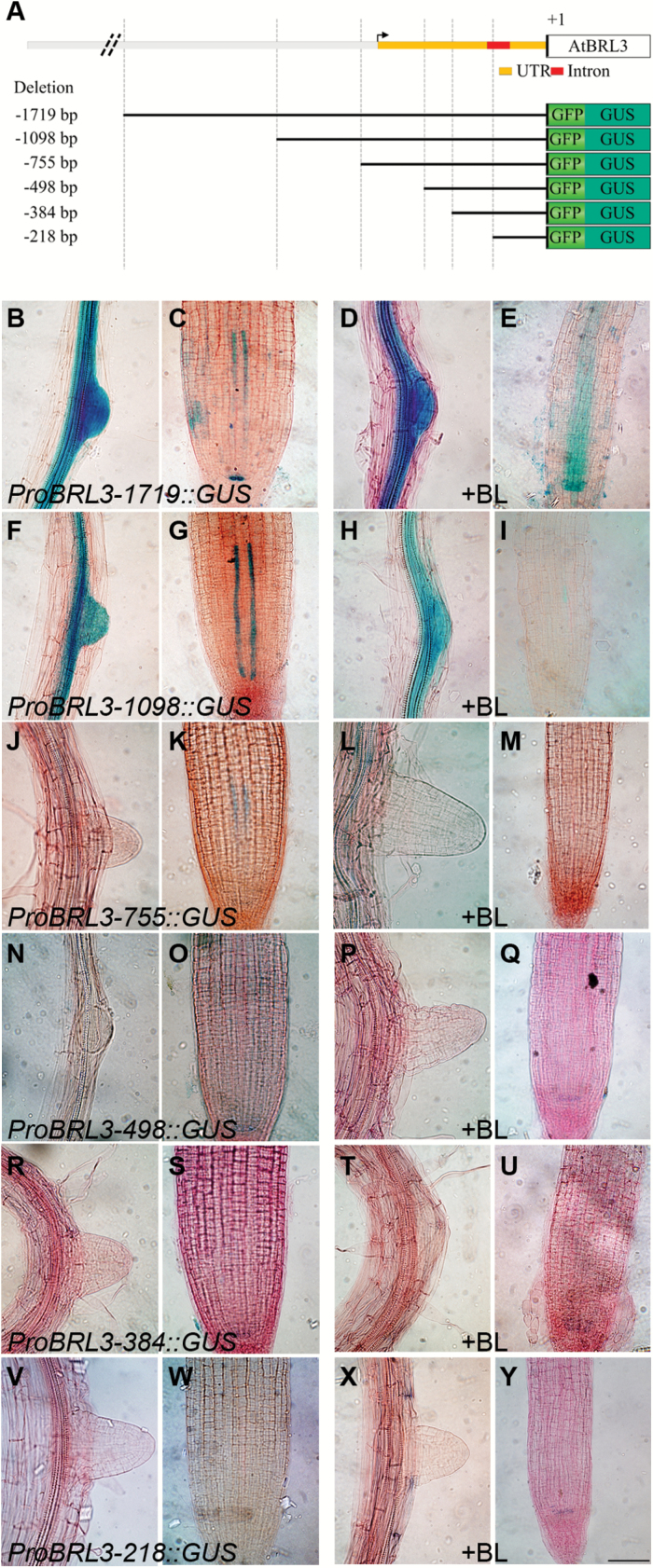
Previous GUS reporter gene assays in transgenic Arabidopsis lines using a 750bp BRL3 promoter fragment and a 1.72kb BRL1 promoter fragment, respectively, showed an overlapping expression pattern for both genes in the plant vascular tissue (Caño-Delgado et al., 2004). To identify cis-elements important in the regulation of BRL3 and BRL1, promoter::GUS truncations were generated and used to analyze their expression (Fig. 1A; Supplementary Fig. S1A).
Fig. 1.
Promoter deletion analysis of ProBRL3 in the root. (A) Schematic diagram of the 5'-flanking regions of ProBRL3. The figure represents part of the 5'-flanking region of ProBRL3 including the 5'-UTR and an intron, labeled in orange and red, respectively. The lower part represents the deletion constructs generated fused to the reporter genes GFP and GUS. (B–Y) Histochemical GUS assay in the root differentiation and meristematic zone of 6-day-old ProBRL3 transgenics with and without 4nM BL treatment for 48h. Scale bar=125 µm. (B, C) ProBRL3-1719::GUS showed expression in regions where lateral roots emerge, in the differentiation zone, in the two protophloem cell files, and in the QC within the meristem. (F, G) ProBRL3-1098::GUS showed expression in regions where lateral roots emerge, in the differentiation zone, and in the two protophloem cell files. (J, K) ProBRL3-755::GUS showed quite similar expression to ProBRL3-1098::GUS although the expression was weaker, especially in the two protophloem cell files and in the differentiation zone. (N, O) ProBRL3-498::GUS, (R, S) ProBRL3-384::GUS, and (V, W) ProBRL3-218::GUS showed no expression in the root. After treatment with BL (D, E) ProBRL3-1719::GUS showed expression in regions where lateral roots emerge, in the differentiation zone and in the QC. (E) Expression of the two protophloem cell layers is expanded and showed expression in the stele. (H, I) ProBRL3-1098::GUS showed expression in regions where lateral roots emerge and in the differentiation zone, and (I) a significantly repressed expression pattern in the two protophloem cell files. (L, M) ProBRL3-755::GUS showed quite similar expression to ProBRL3-1098::GUS and also exhibited (M) significantly reduced expression in the two protophloem cell files. (P, Q) ProBRL3-498::GUS and (T, U) ProBRL3-384::GUS did not show any difference from the untreated lines analyzed. (X, Y) ProBRL3-218::GUS was not expressed in any tissue analyzed.
The ProBRL3::GUS expression in the root was analyzed in 6-day-old seedlings of two representative independent T4 homozygous lines generated for each construct. The ProBRL3-1719::GUS construct showed GUS expression in the root differentiation zone, in lateral root primordia, in the two protophloem cell files, and in the QC within the root meristematic zone (Fig. 1B, C). Removal of a 621bp region (ProBRL3-1098::GUS) eliminated expression in the QC, while expression in the root differentiation zone, in lateral root primordia, and in the two protophloem cell files was still visible (Fig. 1F, G). In the next shorter promoter deletion construct, ProBRL3-755::GUS, reduced expression was detected in the two protophloem cell files and in the differentiation zone, whereas in lateral root primordia GUS staining was lost (Fig. 1J, K). Subsequent analysis of the ProBRL3-498::GUS, ProBRL3-384::GUS, and ProBRL3-218::GUS lines revealed that BRL3 expression in the root was completely lost (Fig. 1N, O, R, S, V, W). These results indicate that the BRL3 promoter requires a minimal promoter length of 755bp for proper BRL3 expression in the root vascular tissue, although the expression pattern differs slightly from the one detected in ProBRL3-1719::GUS transgenic lines. The loss of BRL3 expression in emerging lateral roots in ProBRL3-755::GUS transgenics suggests that additional relevant elements may exist within the region betrween base pairs 1098 and 755. Finally, elements within the 5'-flanking region between base pairs −1719 and −1098 are controlling BRL3 expression in the QC.
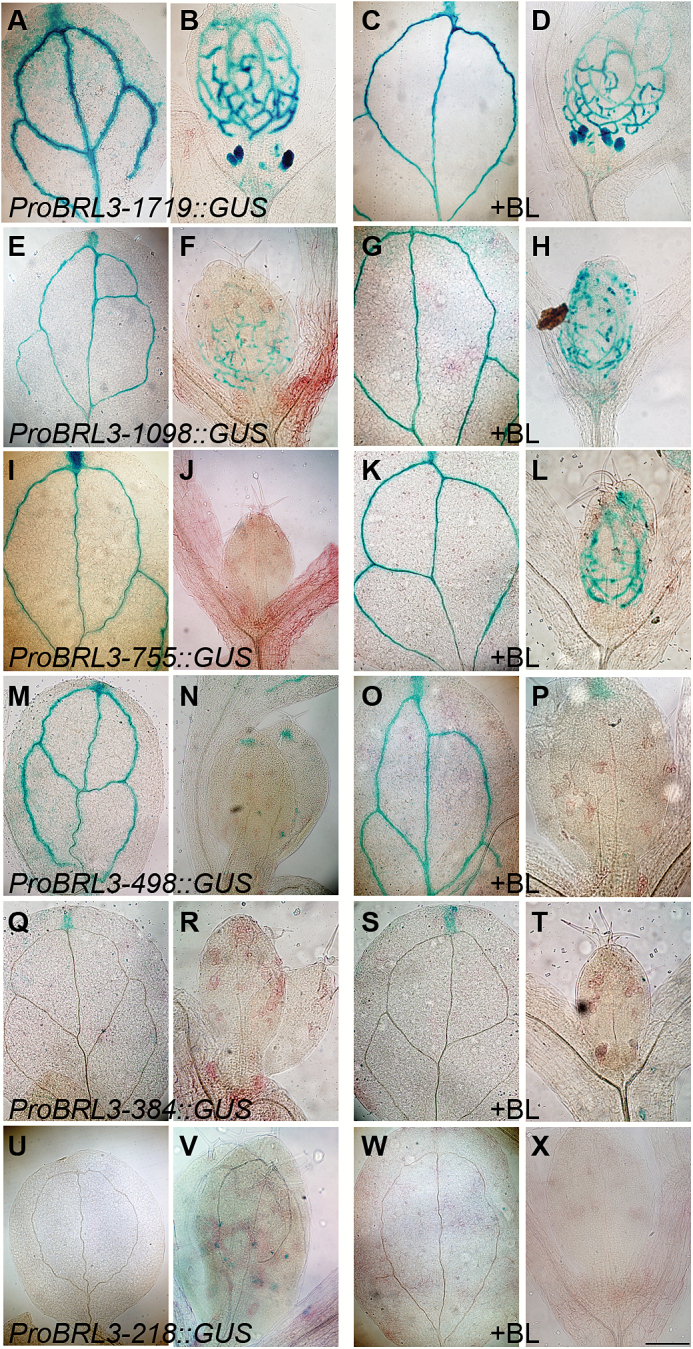
In addition, ProBRL3-1719::GUS constructs displayed BRL3 expression in the vascular tissue, the tip of the cotyledons, and in the shoot apex (Fig. 2A, B). In ProBRL3-1098::GUS,ProBRL3-755::GUS, and ProBRL3-498::GUS transgenic lines, BRL3 expression was lost in the shoot apex, but still visible in the vascular tissue and also in the tip of the cotyledons (Fig. 2E, F, I, J, M, N). The ProBRL3-384::GUS plants only showed expression at the tip of the cotyledon (Fig.2Q, R). However, the shortest construct generated, ProBRL3-218::GUS, shows a complete loss of the GUS reporter activity (Fig.2U, V). Thus, ProBRL3::GUS expression is spatially repressed as the construct of ProBRL3 was reduced in size, starting from a loss in the QC (ProBRL3-1098::GUS) to a complete loss in the roots (ProBRL3-498::GUS). In the shorter constructs, BRL3 continues to be present in the leaf vascular tissue (ProBRL3-384::GUS) and ends in a complete abolishment of BRL3 expression (ProBRL3-218::GUS).
Fig. 2.
Promoter deletion analysis of BRL3. (A–X) Histochemical GUS assay in cotyledons and the shoot apical meristem (SAM) of 6-day-old ProBRL3 transgenics with and without 4nM BL treatment for 48h. Scale bar=125µm. (A, B) ProBRL3-1719::GUS showed expression in the veins and the tip of the cotyledons and in the SAM. (E, F) ProBRL3-1098::GUS, (I, J) ProBRL3-755::GUS, and (M, N) ProBRL3-498::GUS showed expression in the veins and the tip of the cotyledons. (Q, R) ProBRL3-384::GUS was expressed only in the tip of the cotyledon. (U, V) ProBRL3-218::GUS did not show any expression in the tissues analyzed. After treatment with BL, an alteration in the expression pattern of BRL3 in the cotyledons and in the SAM was not observed. (C, D) ProBRL3-1719::GUS showed expression in the veins and the tip of the cotyledons and in the SAM. (G, H) ProBRL3-1098::GUS, (K, L) ProBRL3-755::GUS, and (O, P) ProBRL3-498::GUS showed expression in the veins and the tip of the cotyledons. (S, T) ProBRL3-384::GUS showed only expression in the tip of the cotyledons. (W, X) ProBRL3-218::GUS was not expressed in any tissue analyzed.
As for ProBRL3::GUS, the expression pattern of ProBRL1::GUS was examined in 6-day-old seedlings of two representative and independent T4 homozygous lines for each construct generated. The longest truncated construct analyzed (ProBRL1-1641::GUS) showed GUS expression in the differentiation zone and in the tip of lateral roots, while no GUS expression was detected in the root meristem (Supplementary Fig. S1B, C). Progressively shorter promoter constructs, ProBRL1-978::GUS and ProBRL1-790::GUS, resulted in a loss of GUS expression in the tip of lateral roots, whereas in the differentiation zone GUS expression was still present (Supplementary Fig. S1F, G, J, K). In the ProBRL1−479bp::GUS and ProBRL1-334::GUS deletions, that are missing parts of the 5'-untranslated region (UTR), the expression in the root was completely abolished (Supplementary Fig. S1N, O, R, S). These results suggest that the region between base pairs −790 and −479 contains regulatory elements needed for BRL1 expression.
Brassinosteroids control the expression of BRL3 but not BRL1
The expression levels of BRL3 are known to be repressed by BRs (Mussig et al., 2002; Vert et al., 2005; Sun et al., 2010; Yu et al., 2011). Therefore, the effects of BRs on the expression pattern of BRL3 were investigated. The transgenic lines described above were treated with 4nM BL for 48h. Previous publications already reported that treatments for 24h and 48h, using physiological BL concentrations (below the K d of the receptors), showed effects on root expression even when there were no dramatic morphological effects present (González-García et al., 2011). Analysis of BL-treated ProBRL3-1719::GUS roots showed a shifted and diffuse expression in the stele and in the two protophloem cell files (Fig. 1E), whereas the expression in the root differentiation zone, in lateral roots, and in the QC (Fig. 1D, E) was similar to that in the untreated plants (Fig. 1B, C). In the shorter promoter deletion lines (ProBRL3-1098::GUS, ProBRL3-755::GUS, ProBRL3-498::GUS ProBRL3-384::GUS, and ProBRL3-218::GUS), no significant differences were observed in BRL3 expression when treated with BL (Fig. 1H, I, L, M, P, Q, T, U, X, Y). However, GUS expression in the two protophloem cell files of BL-treated ProBRL3-1098::GUS and ProBRL3-755::GUS was significantly down-regulated (Fig. 1I, M). This region between base pairs −1719 and −1098 contains a BRRE at base pair −1441, capable of binding BES1 and BZR1 transcription factors, suggesting a specific regulatory role for BRs in the expression of BRL3. Thus, these results indicate that BR modulates the expression of BRL3 in the root, since BRL3 expression in other vascular parts remained unchanged (Fig. 2). Conversely, no significant changes in the expression of ProBRL1::GUS upon BL treatment were observed in any of the transgenic lines generated (Supplementary Figs S1, S2).
Cell-specific and dose-dependent control of BRL3 transcription by BRs
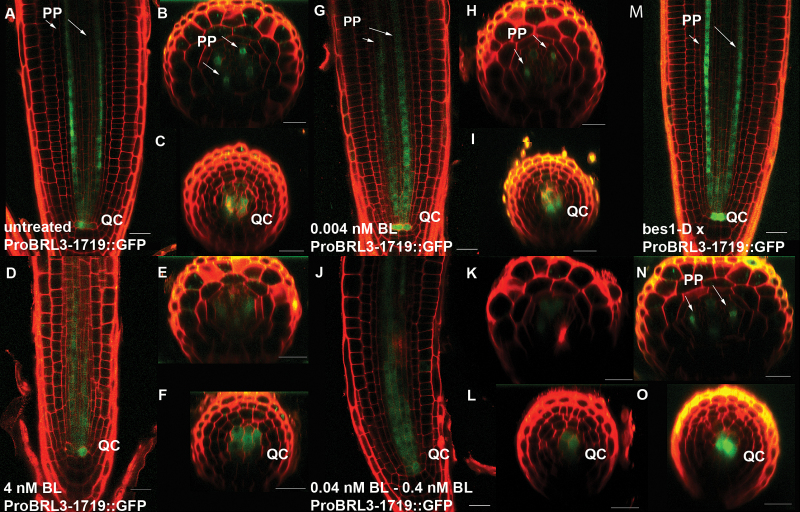
To understand how BRs affect the expression pattern of BRL3 within the root meristem in more detail, confocal visualization of GFP in the ProBRL3::GFP deletion were counterstained with PI to label the cell walls. Investigating ProBRL3-1719:: GFP in the meristem using confocal microscopy revealed specific expression of BRL3 in the protophloem cell files at the transition zone where undifferentiated protophloem starts to differentiate, as well as in the QC (Fig. 3A–C). Upon 48h of BL treatment of the transgenic line ProBRL3-1719::GFP, a shift in expression from the protophloem cell files into the stele and towards the QC was observed (Fig. 3D–F). Due to a spatial shift in expression pattern of ProBRL3-1719::GFP in the stele, its specific expression at the transition zone, where protophloem starts to differentiate, could not be detected.
Fig. 3.
BRL3 expression pattern in the root meristem is BR dose dependent. (A–F) Confocal images of primary roots expressing ProBRL3-1719::GFP in the root differentiation and meristematic zone of 6-day-old ProBRL3-1719::GFP transgenics with and without 4nM BL treatment for 48h. PP, protophloem; QC, quiescent center. (A) Untreated lines showed expression in the protophloem cell files at the transition zone where protophloem differentiates, and in the QC. (B, C) Transversal images of the meristem and the QC of untreated lines. (D) Lines treated with 4nM showed expression in the QC and in a diffuse pattern in the stele. (E, F) Transversal images of the meristem in 4nM BL-treated lines. (G–L) ProBRL3 transgenics treated with increasing concentrations of BL (0.004–4nM continuous treatment) (G) ProBRL3-1719::GFP treated with 0.004nM BL showed an increased and expanded expression in the protophloem cell files towards the QC, where the GFP reporter is also expressed. (H, I) Transversal images of the meristem and the QC of ProBRL3-1719::GUS after treatment with 0.004nM BL. (J) ProBRL3-1719::GFP treated with 0.04–0.4nM BL showed a reduced and misplaced expression in the stele and repression in the QC. (K, L) Transversal images of the meristem and the QC after treatment with 0.04–0.4nM BL. (M–O) ProBRL3-1719::GFP transgenics crossed to bes1-D lines showed increased expression in the protophloem cell files towards the QC and in the QC, similar to transgenics treated with 0.004nm BL. (N, O) Transversal images of the meristem and the QC of ProBRL3-1719::GUS crossed to bes1-D. Scale bar=20 µm.
Next a dose-dependent control of BRL3 expression by BRs in the primary root was investigated by treating ProBRL3-1719 reporter lines with different concentrations of BL. At growth-promoting concentrations of BRs (0.004nM) (González-García et al., 2011), the expression of BRL3 was also enhanced (Fig. 3G–I). The ProBRL3::GFP was extended towards the root transition zone and along the protophloem cell files towards the QC (Fig. 3G). A moderate shift of ProBRL3::GUS expression into the stele was observed (Fig. 3G–I). In contrast, the root growth-inhibitory BL concentrations (>0.004nM) repressed the BRL3 expression in the two protophloem cell files while BRL3 expression was spatially shifted towards the stele and the QC (Fig. 3J–L). The fact that low BL concentrations promote BRL3 in the protophloem cell files, while it is strongly repressed at higher BL concentrations, indicates that BRL3 regulation by BR follows the same trend as the effect in BRs in root growth. Furthermore, our data reveal that BRL3 transcription levels in the root apex are tightly regulated by BRs.
BES1 directly targets and drives the expression of BRL3 in specific cell types in the root
The presence of a BRRE (base pair −1441) and/or an E-box (base pair −892) in the 5'-flanking region of BRL3 prompted us to investigate whether BRs might regulate BRL3 transcription via direct binding of BES1 and/or BZR1 transcription factors. It could be hypothesized that BES1 is able to regulate the specific expression of BRL3 based on the observation that high levels of BRs repress and/or misexpress BRL3 in protophloem cells and vascular cells, respectively, whereas low levels promote its expression. The differential role of BES1 over BRZ1 in the control of QC function in the root (Vilarrasa-Blasi et al., 2014) and the fact that the binding of active BES1 to BRREs appeared to be much stronger than to E-boxes (Yin et al., 2005) point to BES1 as the most suitable factor regulating BRL3 receptors.
To investigate the functional role of BES1 in the BR-regulated BRL3 expression, ProBRL3-1719::GFP was analyzed by genetic crosses with the BR-activated bes1-D mutants and an inducible bes1-D line (35S::Bes1-D:GR). In agreement with physiological data (0.004nM of BR treatment for 6 d) in ProBRL3-1719::GFP plants, the ProBRL3-1719::GFP×bes1-D plants exhibited a similar expression pattern of ProBRL3::GFP in the meristem. BRL3 expression showed an increased and continued expression in the protophloem cell files and in the QC, and a spatial shift to the stele (Fig. 3G–I, M–O). In contrast, the expression in ProBRL3-1719::GUS plants in the background of overexpressing bes1-D-inducible lines resembled the ProBRL3::GFP expression when treated with high BR levels, showing a spatial shift of BRL3 towards the stele and the QC (Fig. 3D, J; Supplementary Fig. S4). These results support the idea of a dose-dependent BR-regulated BRL3 expression pattern in the root meristem mainly based on active BES1 protein levels.
BES1 binds to the BRRE present in the promoter of BRL3
Next, the BRRE at base pair −1441 present in the 5'-flanking region of BRL3 seemed to be an important regulatory element in response to BL, and previous results already demonstrated that ProBRL3-1719::GFP expression is modulated by BES1. It might be obvious that BES1 regulates BRL3 via binding to the BREEs. However, an identified E-box at base pair −892 might also play an additional role in BRL3 regulation. Additionally, BES1 is known to bind both the BRRE and E-box (Yu et al., 2011). Therefore, we further investigated whether BES1 binds to both the BRRE and E-box.
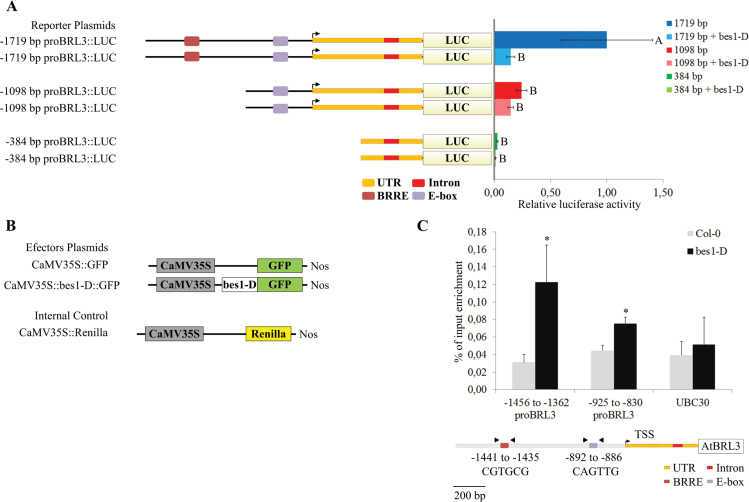
To elucidate whether BES1 regulates the expression of BRL3, a reporter gene assay and ChIP experiments were performed. In the former, Arabidopsis protoplasts were co-transformed with both 35S::bes1-D:GFP (effector gene) and different BRL3 promoter deletion constructs (ProBRL3-1719, ProBRL3-1098, and ProBRL3-384) controlling the luciferase gene (reporter gene). The construct with 35S::GFP was used as a control. Co-transfected protoplasts using ProBRL3-1719::LUC and 35S::bes1-D:GFP showed a strong reduction in the luciferase activity compared with the co-transfected combination using the control. The ProBRL3-1098::LUC or ProBRL3-384::LUC constructs did not show significant luciferase activity, neither in co-transfection with overexpressed BES1 nor in co-transfection with the control (Fig. 4A, B).
Fig. 4.
BES1 regulates the BRL3 expression pattern in the root meristem through binding to the BRRE. (A, B) BES1 represses BRL3 expression through binding the BRRE. In a luciferase reporter gene assay, protoplasts of Arabidopsis have been co-transfected with either ProBRL3-1719::LUC, ProBRL3-1098::LUC, or ProBRL3-384::LUC and either 35S::bes1-D:GFP or the empty vector 35S:GFP (control), and additionally with 35S::Renilla. The transient transactivation assays were done in biological triplicates and data were normalized for Renilla activity (Grotewold et al., 2000). The ratio was calculated as the ratio of each treatment and the treatment of the longest construct used without the repressor. The plotted diagram shows the arithmetic means and the SEM, demonstrating a strong repression of BRL3 due to the binding of BES1 to the BRRE in ProBRL3-1719::LUC. Letters indicate significant differences in ProBRL3-driven luciferase intensity using a one-way ANOVA followed by a Tukey’s test (P<0.05; Tukey’s least significant difference). (C) The 5'-flanking region of BRL3 containing the BRRE was enriched in the ChIP experiment using antibodies against GFP. BES1 ChIP assays showed strong enrichment at the BRL3 promoter region containing the BRRE (base pairs −1441 to −1435) and a low enrichment in the region containing an E-box (base pairs −892 to −886). The position and the sequence of the BRRE and E-box elements present in the BRL3 promoter are shown on the bottom of the scheme. The arrows indicate the primers’ annealing positions. Results are represented as percentage input; the error bars indicate the SD of the data obtained from three technical replicates. As a negative control, UBC30 has been used. Statistical analysis of differences between fragments of the ProBRL3 promoter and internal negative controls (UBC30) was performed using Student’s t-test. Asterisks refer to a significant difference of *P<0.05. Two independent biological replicates gave the same result (Supplementary Fig. S3).
The ChIP experiment was performed in 35S::bes1-D:GFP and wild-type plants using an anti-GFP antibody. The BES1-D ChIP was performed similarly to previous reported work (Vilarrasa-Blasi et al., 2014). In the area of the BRRE (base pair −1441), an enrichment of BES1 was detected, whereas in the region between base pairs −1098 and −755, containing the E-box (base pair −892), only a slight enrichment was observed (Fig. 4C; Supplementary Fig. S3). This indicates that BES1 primarily regulates BRL3 in vivo, binding the BRRE at position −1441; although additional regulatory effects exhibited by BES1 bound to the E-box cannot be completely ruled out. In summary, these results address the BR-regulated expression pattern of BRL3 in the root meristem, which is basically based on BES1 binding to the BRRE at position −1441.
Discussion
The BRL3 and BRL1 genes have been described as BRI1 homologs (serine/threonine kinase receptors) capable of binding BRs with high affinity and are specifically expressed in the plant vasculature (Caño-Delgado et al., 2004). Histological analysis of a 750bp promoter fragment of BRL3 fused to a GUS reporter revealed expression in the two protophloem cell files of the Arabidopsis primary root (Caño-Delgado et al., 2004). The recent demonstration of the expression in the vascular tissue and their close homology to the BRI1 receptor led to the proposition that BRL3 as well as BRL1 might also have a functional role in vascular development (Caño-Delgado et al., 2004; Fàbregas et al., 2013). Interestingly, the root length analysis of 6-day-old seedlings showed that bak1-3 roots are significantly shorter than those of wild-type Col-0 plants (Nam and Li, 2002; Albrecht et al., 2008; Fàbregas et al., 2013), and brl1 brl3 bak1-3 triple mutants enhanced the bak1-3 short root phenotype (Fàbregas et al., 2013). In addition, the triple mutant brl1 brl3 bak1-3 also exhibited a wider stele than Col-0 wild-type and bak1-3 plants under normal conditions. This points to an involvement of BRL3 and/or BRL1 in vascular root development and supports the importance of BRL3 for BR-mediated root growth.
In addition, recent studies showed that BRL3 is a BZR1 putative target repressed by BR (Sun et al., 2010) and a down-regulated BR putative target of BES1 (Yu et al., 2011), whereas BRL1 is only detected as a non-BR-regulated BZR1 putative target (Sun et al., 2010). Nevertheless, the specific mechanisms of the spatial and temporal regulation of BRL3 and BRL1 expression patterns within the root vascular tissue remained unknown. Further identification of regulatory elements, factors driving the expression of BRL3 and BRL1, and crosstalk with other signaling pathways is fundamental to understanding vascular development. In this study, a detailed expression analysis for BRL3 and BRL1 and their 5' regulatory regions essential for proper gene expression has been carried out. In addition, this result confirm that the expression of BRL3 in the root vasculature underlies a dose-dependent hormone-regulatory mechanism.
Tissue-specific expression of BRL3 and BRL1
This study demonstrates a highly specific expression pattern for BRL3 and BRL1 throughout the plant vascular tissue. BRL3 showed expression in the vascular tissues and the tip of the cotyledons, in the shoot apex, in lateral root primordia, in the differentiation zone, in the two protophloem cell files, and in the QC within the meristematic zone. The BRL3 promoter domain ranging from base pair −1719 to −1098 as well as the region from base pair −1098 to −755 5' of the translational start codon has been demonstrated to contain essential regulatory elements to drive BRL3 in different vascular tissues. The minimal promoter length for BRL3 expression in the root has been confirmed to be 755bp. In transgenic plants carrying promoter constructs shorter than −755bp, the expression in the root was completely abolished, and in deletions generated close to the translational start codon the expression was completely lost in seedlings, indicating the presence of fundamental regions within the 5'-UTR.
BR-regulated expression of BRL3 in the root vasculature is dose dependent
The characterization of ProBRL3-1719::GUS lines in response to 4nM BL for 48h revealed a shift in the expression pattern in the root meristem. Whereas native BRL3 was expressed at the transition zone between undifferentiated and differentiated protophloem in the protophloem cell files and the QC, BL treatment expanded the expression from the protophloem cell files to encompass other stele cell types. However, the same treatment in ProBRL3-1098::GUS and ProBRL3-755::GUS lines did not drive the expression into the stele, but showed a significant reduction of BRL3 expression in the protophloem cell files. Interestingly, the 5' region between base pairs −1719 and −1098 and between −1098 and −755 contains a BRRE and an E-box, respectively, elements that can be bound by BES1 and/or BZR1 proteins. It is worth mentioning that BRs regulate vascular differentiation, in particular promoting xylem and repressing the formation of phloem (Ibanes et al., 2009).
These results show: (i) that BRs regulate the expression of BRL3 receptor gene transcripts; and (ii) that this regulation is BR dose dependent. The expression pattern of BRL3 differs significantly when subjected to different levels of BL; likewise the root meristem needs an equilibrated BR signaling to maintain its length (González-García et al., 2011). For instance, very low levels of BL (0.004nM for 6 d) in ProBRL3-1719::GUS increased the expression in the protophloem cell files at the transition zone and an expansion towards the QC. This concentration has recently been demonstrated to promote both root epidermal cell number and size of the Arabidopsis primary root meristem (González-García et al., 2011). In addition, BRs are known to promote cell differentiation (Iwasaki and Shibaoka, 1991; Yamamoto et al., 1997). Thus, it can be assumed that low levels of BRs not only increase the epidermal cell number and size of the meristem but also promote the differentiation of phloem. This observation is opposite to the reported function for BRs in repressing phloem differentiation (Fukuda, 1997). However, recent studies indicate opposing effects of BR signaling in terms of root growth, depending on the tissue on which BR is acting (Vragovic et al., 2015). In the transgenic promoter deletion lines with the GUS/GFP reporter system, ProBRL3 −1719::GFP, −1098::GFP, and −755::GFP, treatment with high levels of BL (≥0.04nM) reduced the marker expression in the protophloem cell files, which could be a consequence of a direct repression caused by BRs and/or a repressed number of differentiated phloem cells. In addition, increasing BR concentrations significantly reduced the root length of wild-type plants (González-García et al., 2011) while brl1brl3bak1-3 triple mutant plants showed resistance to BR-mediated root shortening (Fàbregas et al., 2013).
Interestingly, and despite the similarity of BRL1 and BRL3 receptors (Caño-Delgado et al., 2004; Fàbregas et al., 2013), these results indicate that BRL1 transcription appeared not to be regulated by BRs. Moreover, an additional study already reported that BRL1 was detected as a non-BR regulated BZR1 putative target (Sun et al., 2010). Further biochemical studies involving the dephosphorylated and phosphorylated forms of BES1 will be necessary to understand the different regulatory mechanisms for these two functionally homologous vascular receptors during plant growth and development.
BRs regulate BRL3 expression through binding of BES1 to a BRRE
These findings reveal a role for BES1 as an important factor regulating the expression pattern of BRL3 in the root meristem. Interestingly, checking the expression pattern of BRL3 and BES1 in the eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) showed a correlation between low transcriptional levels of BES1 and high levels of BRL3, and between very high levels of BES1 and low levels of BRL3 (Brady et al., 2007) (Supplementsry Fig. S5).
Recently, Chaiwanon and Wang (2015) reported that the BZR1–yellow fluorescent protein (YFP) fusion protein expressed from either the BZR1 promoter or the constitutive 35S promoter accumulated at a low level in the nuclei of the stem cell region but at a higher level in the nuclei of epidermal cells in the transition and elongation zone as well as in the phloem. In addition, BES1–GFP under the endogenous promoter showed a similar pattern to BZR1–YFP (Chaiwanon and Wang, 2015). This correlates with the role of BRs in vascular differentiation and also supports the hypothesis of this study that in the case of BRL3, BES1 seems to act as an activator of BRL3 at low levels whereas at elevated levels, BES1 acts as a repressor of BRL3. Moreover, Jiang et al. (2015) recently identified a novel long isoform of BES1, called BES1-L. The BES1-L–GFP line presented in their study showed an expression pattern in the root meristem similar to that of the ProBRL3-1719::GUS line analyzed in this study.
In vivo experiments, such as ChIP and a luciferase reporter gene assay, confirmed the direct interaction of BES1 and the BRRE present in BRL3. ChIP assays showed that BES1 binds to the BRL3 promoter in a region comprising a BRRE cis-element. In the region containing an E-box element, only a slight enrichment of BES1 has been observed, indicating that the E-box plays a minor, perhaps additive, role in BRL3 regulation. In addition, it has already been reported that the binding affinity of BES1 for a BRRE is stronger than for E-boxes (Yin et al., 2005).
In the luciferase reporter gene assay, BES1 showed a significant reduction in the expression of the luciferase reporter in protoplasts co-transfected with ProBRL3-1719::LUC and 35S::bes1-D:GFP when compared with co-transfections of ProBRL3-1719::LUC and 35S::GFP alone. In summary, this study reveals that BRL3 expression in the root meristem is BES1 dose dependent and mediated mainly by the binding of BES1 to the BRRE present in the promoter of BRL3.
A role for BES1 in spatiotemporal control of BRL3 receptors in the stele
This study reveals that the levels of active BR-regulated BES1 control the spatiotemporal transcription of the BRL3 receptors in the root meristem. While the BES1 and BZR1 transcription factors can bind to the BRRE and the E-box, resulting in either activation or repression of the expression of their target genes (Sun et al., 2010; Yu et al., 2011), the mechanisms by which BES1/BZR1 mediate the BR-repressed gene expression are not well understood. It is well known that BES1 and BZR1 inhibit many genes involved in BR biosynthesis and signaling, probably as a feedback inhibition mechanism. Activation of BR signaling inhibits BR biosynthesis and perception through direct repression of DWF4, CPD, BRI1, and other genes by BES1 and BZR1 (Mathur et al., 1998; Noguchi et al., 1999; Choe et al., 2002; Mora-García et al., 2004; Sun et al., 2010; Clouse, 2011; Ye et al., 2011; Yu et al., 2011). BR-mediated gene regulation requires BES1/BZR1 interaction with other partners such as transcription factors, histone-modifying enzymes, and transcription elongation factors (Yin et al., 2005; Yu et al., 2008; Li et al., 2009, 2010; Zhang et al., 2014). Based on these results, a feedback mechanism for BRL3 regulation via BR signaling mediated by BES1 protein can be proposed.
The key factor in BRL3 regulation is the level of BES1 present in active BR signaling. When the level of the nuclear-localized BR-activated transcription factor BES1 is low, binding to the BRRE box in the BRL3 promoter results in an increase of BRL3 expression in the root meristem. However, at high BES1 levels, binding of the BRRE box suppresses the expression in the two protophloem cell files. Thus the spatiotemporal BRL3 expression is dependent on BES1 levels.
In addition, it has recently been proposed that the dose-dependent opposite effects of exogenous BR are due to a requirement for different BR levels in different developmental zones (Chaiwanon and Wang, 2015). Interestingly, BZR1 is activated by endogenous BR in a graded pattern along developmental zones and BRs act antagonistically with auxin on BZR1 nuclear localization, transcriptomic response, and cell elongation in a developmental zone-specific manner (Chaiwanon and Wang, 2015). Collectively, it can be argued that the repression of BRL3 in the two protophloem cell files functions in co-ordinating BR-mediated root development, especially during the differentiation of phloem cell files. Thus the integration of cell type-specific signaling events in response to environmental stimuli is important to understand plant growth and development completely.
From the biotechnological point of view, identification of the cis-acting regulatory elements is gaining great importance because of the emergence of tools for genome editing such as the zinc-finger nucleases (ZFNs), the transcriptional activator-like effector nucleases (TALLENs), and the clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/CAS) system. Any modification of the promoter architecture of BR receptors via insertions or deletions in the E-box and/or BRRE can enable the modification of signaling events within the BR pathway. It is very well known that BRs play an important role in plant development including plant architecture, vascular differentiation, and flowering, and in the physiological responses such as tolerance to biotic and abiotic stress. In addition, the present work reporting the identification of promoter regions important for vascular expression may open the door to the identification and validation of new cis-regulatory elements important for plant vascular development.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Promoter deletion analysis of BRL1 in the root.
Figure S2. Promoter deletion analysis of BRL1.
Figure S3. BES1 was enriched in the 5'-flanking region of BRL3 containing the BRRE.
Figure S4. BRL3 expression pattern in the root meristem is BES1 dose dependent.
Figure S5. Relative and comparative expression patterns of expression for BRL3 and BES1.
Table S1. Primer sequences
Acknowledgements
We thank Dr Tsuyoshi Nakagawa (Shimane University) for providing Gateway binary vectors that contain the bar gene, which was identified by Meiji Seika Kaisha, Ltd. We thank Mary-Paz González-García and Norma Fàbregas for comments on the manuscript. RL was funded by the FWF Erwin Schrödinger Stipendum from Austria (J 3019-B16), and JES-H was supported by the Department of Innovation, Universities and Enterprise of the Generalitat de Catalunya, the European Social Fund FI Fellowship (AGAUR: FI-2006, Resolució EDU/3600/2006; FI-2008, Resolució IUE/2658/2007, and BE-DGR2010) in the Laboratory of AC-D. We acknowledge financial support from the Spanish Ministry of Economy and Competitiveness, through the ‘Severo Ochoa Programme for Centres of Excellence in R&D’ 2016–2019 (SEV‐2015‐0533). AIC-D is a recipient of a BIO2013-43873 grant from the Spanish Ministry of Economy and Competitiveness an European Research Council, ERC Consolidator Grant (ERC-2015-CoG – 683163).
References
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. 2008. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiology 148, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. 1998. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. The Plant Cell 10, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J. 2004. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY. 2015. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Current Biology 25, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. 2001. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. The Plant Journal 26, 573–582. [DOI] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, et al. 1999. The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. The Plant Cell 11, 207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE. 2002. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiology 130, 1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Nelson T. 2002. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. The Plant Cell 14, 2707–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. 1996. Plant hormones: brassinosteroids in the spotlight. Current Biology 6, 658–661. [DOI] [PubMed] [Google Scholar]
- Clouse SD. 2011. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. The Plant Cell 23, 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. 2011. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nature Protocols 6, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregas N, Cano-Delgado AI. 2014. Turning on the microscope turret: a new view for the study of brassinosteroid signaling in plant development. Physiologia Plantarum 151, 172–183. [DOI] [PubMed] [Google Scholar]
- Fàbregas N, Li N, Boeren S, Nash TE, Goshe MB, Clouse SD, de Vries S, Caño-Delgado AI. 2013. The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. The Plant Cell 25, 3377–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H. 1997. Tracheary element differentiation. The Plant Cell 9, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, et al. 2007. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Developmental Cell 13, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. 2005. Profiling histone modification patterns in plants using genomic tiling microarrays. Nature Protocols 2, 213–218. [DOI] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Cano-Delgado AI. 2011. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. 2000. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proceedings of the National Academy of Sciences, USA 97, 13579–13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthern JD, Jr, Steffens GL, Flippen-Anderson JL, Carter Cook J. 1979. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281, 216–217. [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. 2005. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. 2011. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanes M, Fàbregas N, Chory J, Caño-Delgado AI. 2009. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proceedings of the National Academy of Sciences, USA 106, 13630–13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T, Shibaoka H. 1991. Brassinosteroids act as regulators of tracheary-element differentiation in isolated Zinnia mesophyll cells. Plant and Cell Physiology 32, 1007–1014. [Google Scholar]
- Jiang J, Zhang C, Wang X. 2015. A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. The Plant Cell 27, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. 2007. Building blocks for plant gene assembly. Plant Physiology 145, 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. 1996. Genetic evidence for an essential role of brassinosteroids in plant development. The Plant Journal 9, 701–713. [Google Scholar]
- Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. 2005. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. 1997. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. 1996. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y. 2010. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proceedings of the National Academy of Sciences, USA 107, 3918–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. 2009. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. The Plant Journal 58, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava NB. 1988. Plant growth-promoting brassinosteroids. Annual Review of Plant Physiology and Plant Molecular Biology. 39, 23–52. [Google Scholar]
- Mathur J, Molnar G, Fujioka S, T et al. 1998. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. The Plant Journal 14, 593–602. [DOI] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J. 2004. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes and Development 18, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Falcone Ferreyra ML, et al. 2012. A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. The Plant Cell 24, 2745–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussig C, Fischer S, Altmann T. 2002. Brassinosteroid-regulated gene expression. Plant Physiology 129, 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J. 2003. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Mano S, Tanaka Y, et al. 2010. Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Bioscience, Biotechnology, and Biochemistry 74, 1315–1319. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. 1999. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiology 121, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC. 2004. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). The Plant Cell 16, 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagat T, Paguio A, Kopish K. 2007. Normalizing genetic reporter assays: approaches and considerations for increasing consistency and statistical significance. Cell Notes 17, 9–12. [Google Scholar]
- She J, Han Z, Kim TW, et al. 2011. Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. 2002. A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/.
- Sun Y, Fan XY, Cao DM, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell 19, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. 1996. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. 2008. RSAT: regulatory sequence analysis tools. Nucleic Acids Research 36, W119–W127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, Palauqui JC. 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. The Plant Cell 20, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. 2005. Molecular mechanisms of steroid hormone signaling in plants. Annual Review of Cell and Developmental Biology 21, 177–201. [DOI] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, Gonzalez-Garcia MP, Frigola D, et al. 2014. Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Developmental Cell 30, 36–47. [DOI] [PubMed] [Google Scholar]
- Vragovic K, Sela A, Friedlander-Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi-Goldstein S. 2015. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proceedings of the National Academy of Sciences, USA 112, 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, et al. 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. 2001. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Demura T, Fukuda H. 1997. Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant and Cell Physiology 38, 980–983. [DOI] [PubMed] [Google Scholar]
- Ye H, Li L, Yin Y. 2011. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. Journal of Integrative Plant Biology 53, 455–468. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. 2005. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-García S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y. 2008. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 105, 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, Rodermel S, Yin Y. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. The Plant Journal 65, 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang D, Ye H, Guo H, Johnson A, Lin H, Yin Y. 2014. Transcription factors involved in brassinosteroid repressed gene expression and their regulation by BIN2 kinase. Plant Signaling and Behavior 9, e27849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J. 2004. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. The Plant Journal 40, 399–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.