Highlight
The cysteine-rich receptor-like protein kinase CRK5 is a potentially positive regulator of ABA signaling in early seedling growth, stomatal movement and plant drought tolerance.
Keywords: ABI2, abscisic acid, CRK5, drought tolerance, receptor-like kinase, WRKY18, WRKY40, WRKY60.
Abstract
Receptor-like kinases (RLKs) have been reported to regulate many developmental and defense process, but only a few members have been functionally characterized. In the present study, our observations suggest that one of the RLKs, a membrane-localized cysteine-rich receptor-like protein kinase, CRK5, is involved in abscisic acid (ABA) signaling in Arabidopsis thaliana. Overexpression of CRK5 increases ABA sensitivity in ABA-induced early seedling growth arrest and promotion of stomatal closure and inhibition of stomatal opening. Interestingly, and importantly, overexpression of CRK5 enhances plant drought tolerance without affecting plant growth at the mature stages and plant productivity. Transgenic lines overexpressing a mutated form of CRK5, CRK5 K372E with the change of the 372nd conserved amino acid residue from lysine to glutamic acid in its kinase domain, result in wild-type ABA and drought responses, supporting the role of CRK5 in ABA signaling. The loss-of-function mutation of the CRK5 gene does not affect the ABA response, while overexpression of two homologs of CRK5, CRK4 and CRK19, confers ABA responses, suggesting that these CRK members function redundantly. We further showed that WRKY18, WRKY40 and WRKY60 transcription factors repress the expression of CRK5, and that CRK5 likely functions upstream of ABI2 in ABA signaling. These findings help in understanding the complex ABA signaling network.
Introduction
The phytohormone abscisic acid (ABA) plays an essential role in the regulation of plant growth and development, including inhibition of seed germination and seedling growth, promotion of seed dormancy, modulation of stomatal movement, and adaptive responses to various abiotic stresses (Finkelstein et al., 2002). Recent advances in ABA signaling deepen greatly our understanding of the functional mechanism of this phytohormone from primary events of signal perception to downstream gene expression. A group of the START domain proteins PYR/PYL/RCARs, which are identified as cytosolic ABA receptors, interact with protein phosphatase 2Cs (PP2Cs) such as ABA-INSENSITIVE1/2 (ABI1/2) and HYPERSENSITIVE TO ABA1 (HAB1) to release their dephosphorylation effects on SnRK2 protein kinases, which phosphorylate downstream transcription factors to induce ABA-responsive gene expression (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). The H subunit of Mg-chelatase (CHLH/putative ABA receptor ABAR), which is identified as a candidate receptor for ABA in chloroplasts/plastids, functions together with the chloroplast protein cochaperonin CPN20 and interacts with a group of WRKY transcription factors, WRKY18/40/60, to regulate the expression of downstream ABA-responsive transcription factor genes such as ABA INSENSITIVE4/5 (ABI4/ABI5) (Shen et al., 2006; Wu et al., 2009; Shang et al., 2010; Du et al., 2012; Liu et al., 2012, 2013; Yan et al., 2013; Zhang et al., 2013, 2014). A recent study showed that CHLH/ABAR cross-talks with PYR/PYL/RCARs to regulate SnRK2.6 in stomatal response to ABA (Liang et al., 2015). CHLH/ABAR also regulates a nucleocytosolic PPR-domain protein, SOAR1, which functions as a hub of ABA signaling to nuclear gene expression (Mei et al., 2014; Jiang et al., 2014, 2015; Wang and Zhang, 2014). Functioning at the cell surface, plasma membrane GPCR-type G proteins GTG1 and GTG2 perceive extracellular ABA signals to regulate seed germination and stomatal behavior (Pandey et al., 2009). It is widely believed, however, that ABA signal transduction involves highly complex signaling pathways, and many other components remain to be identified to fully understand the complex ABA signaling network.
Receptor-like kinases (RLKs) have been reported to regulate many developmental and defense process, such as root and shoot growth, cell differentiation regulation, self-incompatibility, brassinosteroid signaling and disease resistance (Morillo & Tax, 2006; De Smet et al., 2009; Osakabe et al., 2013). In Arabidopsis, RLKs are the largest membrane receptor family and belong to a large gene family with more than 610 members (Shiu & Bleecker, 2001; Morillo & Tax, 2006). Based on amino acid sequence and structure differences, RLKs are categorized into several subfamilies, including leucine-rich repeat RLKs (LRR-RLKs), cysteine-rich repeat (CRR) RLKs (CRKs), domain of unknown function 26 RLKs, S-domain RLKs, and others (Shiu & Bleecker, 2001). A typical RLK structure contains an extracellular domain, a transmembrane domain and a cytoplasmic kinase domain, whereas receptor-like cytosolic kinases (RLCKs) contain no apparent signal sequence or transmembrane domain (Shiu & Bleecker, 2001; Torii, 2004). Similar to animal receptor tyrosine kinases (RTKs), extracellular domains of RLKs bind to ligand specifically and the conserved intracellular kinase domains transduce signals to their downstream targets in the cytoplasm by catalytic processes of protein phosphorylation (Torii, 2004; Lemmon & Schlessinger, 2010).
Several RLKs have been reported to be involved in ABA signaling pathways and stress tolerance in Arabidopsis (Osakabe et al., 2005; Bai et al., 2009; Deng et al., 2009; Xin et al., 2009; Osakabe et al., 2010; Hua et al., 2012; Tanaka et al., 2012; Yu et al., 2012). Mutation of receptor-like kinase 1 (RPK1) decreases ABA sensitivity during the process of seed germination, seedling growth, and stomatal movement, whereas RPK1 overproduction increases plant tolerance to dehydration and oxidative stress (Osakabe et al., 2005; Osakabe et al., 2010). Impairment of proline-rich extensin-like receptor kinase 4 (PERK4) reduces ABA-inhibited root growth by decreasing cytosolic free calcium concentration and Ca2+ signaling (Bai et al., 2009). ABA INSENSITIVE3 (ABI3)-activated lectin receptor-like kinase LecRK-b2 positively regulates ABA signaling during seed germination, whereas the A4 subfamily of lectin receptor kinase members, LecRKA4.1, LecRKA4.2 and LecRKA4.3, play negative and redundant roles in ABA responses (Deng et al., 2009; Xin et al., 2009). GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1) participates in ABA- and H2O2-regulated activation of S-type anion currents in guard cells, which can be inhibited by ABI2 but not ABI1 (Hua et al., 2012). A positive regulator of auxin signaling, FERONIA (FER), interacts with guanine exchange factors GEF1, GEF4, and GEF10, which activate GTPase ROP11/ARAC10 and the activated ROP11 enhances the ABI2 phosphatase activity (Yu et al., 2012).
The RLK subfamily of cysteine-rich receptor-like protein kinases (CRKs) includes 46 members in Arabidopsis, which are defined by two copies of DUF26 domains each of which contain C–8X–C–2X–C motifs forming disulfide bonds for protein–protein interactions in the extracellular region (Wrzaczek et al., 2010). Over-production of CRK5 or CRK13 enhances plant resistance to Pseudomonas syringae (Chen et al., 2003; Acharya et al., 2007). Induced expression of the four structurally closely related CRKs, CRK4, CRK5, CRK19 and CRK20, leads to hypersensitive response-associated cell death in transgenic Arabidopsis (Chen et al., 2003, 2004). CRK7 has been reported to be involved in mediating the responses to extracellular ROS production (Idanheimo et al., 2014). The signaling pathways mediated by several CRKs, such as BR-insensitive 1 (BRI1) (Wang et al., 2001) and FLAGELLIN-SENSITIVE2 (FLS2) (Gomez-Gomez et al., 2001), have been well characterized in hormone perception and pathogen response. Two abiotic stress-inducible CRK members, CRK36 and a receptor-like cytosolic kinase (RLCK), ARCK1, interact with each other and negatively regulate ABA and osmotic stress signal transduction (Tanaka et al., 2012). However, the function of most of the CRK members remains unknown.
In this study, we showed that overexpression of a membrane-localized cysteine-rich repeat RLK-encoding gene, CRK5, enhances plant sensitivity to ABA and improves drought resistance, whereas overexpression of a mutant form of CRK5, CRK5 K372E, induces no significant ABA-related phenotypes. Transgenic lines of the two homologous genes of CRK5, CRK4 and CRK19, also exhibit ABA-hypersensitive phenotypes in early seedling growth. Overexpression of CRK4 also enhances ABA sensitivity of stomatal movement and drought tolerance. The expression of CRK5 is repressed by cooperation of the WRKY transcription factors WRKY18, WRKY40 and WRKY60. These data suggest that CRK5 is positively involved in ABA signaling. Additionally, genetic evidence suggests that CRK5 may function upstream of ABI2. These findings help in understanding the complex ABA signaling network.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Columbia (Col-0) was used as an Arabidopsis wild-type. The loss of function mutants crk5-1 (SALK_063519C with Col-0 ecotype as background) and crk5-2 (SALK_109339 with Col-0 ecotype as background), and the knock-down mutants crk19-1 (SALK_019639C with Col-0 ecotype as background) and crk19-2 (SALK_120859C with Col-0 ecotype as background) were purchased from the Arabidopsis Biological Resource Center (ABRC). The seeds of the aba2 mutant (CS156: aba2-1, with Col-0 ecotype as background) were also obtained from ABRC. The wrky single, double (wrky40 wrky18, wrky18 wrky60, and wrky40 wrky60), and triple (wrky40 wrky18 wrky60) mutants used in this study were identified as described previously (Shang et al., 2010; Liu et al., 2012). The primers for identification of these mutants are listed in Supplementary Table S1 at JXB online. For the generation of the CRK-overexpression lines, the open reading frame (ORF) sequences of CRK4, CRK5, CRK19, or CRK20 were amplified by PCR and cloned into the binary vector pCAMBIA-1300-221 (http://www.cambia.org) with a green fluorescent protein (GFP) tag driven by cauliflower mosaic virus (CaMV) 35S promoter. For the generation of the CRK5 promoter–β-glucuronidase (GUS) transgenic plants, the genomic DNA fragment from 1963bp to 1bp upstream of translation initiation site of CRK5 was introduced into pCAMBIA-1381 plasmid carrying GUS (http://www.cambia.org). The constructed plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into Arabidopsis (Col-0) plants by the floral infiltration method. Transgenic plants with single T-DNA insertion were screened by hygromycin resistance and confirmed by real-time PCR. The homozygous T3 generation seeds were used for further analysis. All the primer sequences used for generation of the transgenic plants are presented in Supplementary Table S1.
The full length sequence of CRK5 K372E, a mutated form of CRK5 (site-directed mutagenesis of a conserved active-site residue), was obtained by overlap extension PCR (OE-PCR), which introduces a point mutation into the CRK5 gene sequence by synthesizing a pair of mutated primers (K372E-Middle-F and K372E-Middle-R) that were designed with flanking sequences at the mutation site. Briefly, the procedure of OE-PCR includes two steps. For the first step, the PCR was performed with the wild-type CRK5 gene as template for amplification of the two mutated CRK5 K372E segments containing overlapped and mutated sequences using the following primer pairs: forward primer (CRK5-GFP-F) and mutated reverse primer (K372E-Middle-R) for cloning part of the CRK5 K372E segment with the mutation in its 3′ end; reverse primer (CRK5-GFP-R) and mutated forward primer (K372E-Middle-F) for cloning the other part of the CRK5 K372E segment with the mutation in its 5′ end. For the second step, the PCR was performed with the two overlapped and mutated CRK5 K372E segments obtained in the first step as templates for amplification of the full length of the CRK5 K372E gene using the following primer pair: forward primer (CRK5-GFP-F) and reverse primer (CRK5-GFP-R). The primers used for creating the mutation are listed in Supplementary Table S1. The CRK5 K372E fragment was cloned into the binary vector pCAMBIA-1300-221 and the methods for transformation of A. tumefaciens strain GV3101 and Arabidopsis (Col-0) plants were performed as described above.
For the generation of the CRK5/ABI2 double-overexpression line OE-1×ABI2OE, the over-expressed ABI2 gene was introduced into the CRK5-transgenic line OE-1 by crossing the ABI2OE line with the OE-1 plant. For the generation of the CRK5-overexpression plant in the aba2 mutant background, the ABA2 gene mutation was introduced into the CRK5-transgenic line OE-2 by crossing aba2 mutant with the OE-2 plant. The precise T-DNA insertion points in OE-1, OE-2 and ABI2OE plants were identified by thermal asymmetric interlaced PCR (TAIL-PCR).
Seeds of different genotypes sown on Murashige and Skoog (MS) medium (Sigma-Aldrich, St Louis, MO, USA) were placed for 72h at 4 °C for stratification and then transferred to a growth chamber at 20–21 °C with about 80 μmol photons m −2 s−1 or in compost soil with about 120 μmol photons m −2 s−1 light intensity using cool white fluorescent lamps under 16h of light–8h of dark and 60% relative humidity.
Real-time PCR analysis and TAIL-PCR
The rosette leaves from 4-week-old plants were used for determination of the transcription levels of CRK5, CRK5 K372E, CRK4, CRK19 and CRK20 in the wild-type Col-0 and their corresponding transgenic lines. Seedlings grown 4 days after stratification were sampled for detection of the expression level of ABA-responsive genes. Total RNA was extracted and purified using the total RNA Rapid Extraction Kit (BioTeke, Beijing, China) and RNA Purification Kit (BioTeke, Beijing, China), respectively, according to the manufacturer’s instructions. Single-strand cDNA was synthesized using 2 µg of total RNA with the Roche Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany). Real-time PCR was performed using the CFX96TM Real-Time System of C1000TM Thermal Cycler and SYBR Premix Ex Taq (TaKaRa, Dalian, China) with the program as follows: 5min at 94 °C and then 30 cycles of 5s at 94 °C, 30s at 60 °C. ACTIN2/8 gene was used as an internal control. All the real-time PCR assays were performed in triplicate and means of the three biological repeats were calculated to represent gene expression level. Primers for real-time PCR are listed in Supplementary Table S1. TAIL-PCR was performed essentially as described previously (Liu et al., 1995; Mei et al., 2014). Random primers and the specific left border primer of pCAMBIA-1300-221 are listed in Supplementary Table S1.
Phenotypic analysis
For the cotyledon greening assay, about 200 seeds of different genotypes were sterilized and planted on ABA-free or (±)ABA-containing MS medium that contained 3% sucrose and 0.7% agar (pH 5.8~6.0). The seeds were placed at a growth chamber after stratification for 72h, and green cotyledons were scored 5 days later.
Two methods were used to test ABA-mediated inhibition of post-germination growth. For the first method, the seeds were directly planted in ABA-free or ABA-containing MS medium, and seedling growth was recorded at the indicated times when the primary root length was measured using a ruler. For the second method, seeds were planted in ABA-free medium, subjected to a 3-d stratification, and 60-h-old germinating seeds/young seedlings were transferred to ABA-free (0 μM) or (±)ABA-containing MS medium and continued to grow for 10 d before investigation.
Stomatal aperture was determined essentially as described previously (Wu et al., 2009; Shang et al., 2010). Rosette leaves of 4-week-old plants were used. To assay ABA-induced stomatal closure, detached leaves of different genotype plants were immersed in solutions containing 50mM KCl and 10mM MES-KOH (pH 6.15) under a halogen cold light source (Colo-Parmer) for 3h before treatment with different concentrations of (±)ABA for 2h. Apertures were recorded on epidermal strips to estimate ABA-induced stomatal closure before and after ABA treatment. To assay ABA-inhibited stomatal opening, plants were placed in dark for 24h before leaves were immersed in the same buffer described above containing different concentration of ABA for 2h under the cold light, and the apertures were then determined.
For drought stress treatment, plants of different genotypes grown for 2 weeks on ABA-free MS medium under normal conditions were transferred into soil and placed in the greenhouse without irrigation. After 16 d of water deficiency, the plants’ growth status was photographed before and after re-watering for 24h followed by recording and calculating survival rates.
Yeast one-hybrid assays
Yeast one-hybrid assays were performed with a Matchmaker™ One-Hybrid Library Construction and Screening kit (Clontech, Mountain View, CA, USA) using the AH109 yeast strain according to the manufacturer’s instructions. The promoter fragment of CRK5 and the open reading frame of WRKY18/40/60 were cloned into the pHIS2 bait vector and pGADT7 prey vector, respectively. Yeast cells were co-transformed with pGADT7 prey vector containing WRKY18, WRKY40, or WRKY60 and pHIS2 bait vector containing the promoter of CRK5. The corresponding transformation with pGADT7 prey vector containing WRKY18, WRKY40, or WRKY60 and pHIS2 bait vector containing p53 promoter fragment were used as negative controls. Co-transformation of pHIS2-p53 with pGADT7-p53 was used as positive control, and co-transformation of pGADT7-p53 with empty pHIS2 was used as its own negative control. Transformed yeast cells with different combinations of plasmids were first grown in SD-2 medium (lacking Trp, Leu) at 30 °C for 4 days to ensure that the yeast cells were successfully co-transformed, and then co-transformed yeast cells were grown overnight in liquid SD-2 medium to an OD600 of 0.1 and diluted in a 10× dilution series. For each dilution, 10 μl yeast cells was spotted on SD-2 and SD-3 medium (lacking Trp, Leu, His) supplemented with 40mM 3-aminotriazole (3-AT; Sigma-Aldrich, USA) and then cultured at 30 °C for another 4 days. The primers used for constructing the related plasmids are listed in Supplementary Table S1.
Trans-inhibition of CRK5 promoter activity by WRKYs in tobacco leaves
This experiment was performed essentially according to the previously described procedures (Liu et al., 2012). The full-length ORF fragments of the WRKY genes were amplified by PCR and cloned into the pCAMBIA1300-Flag vectors under the promotion of CaMV 35S promoter, forming the effector constructs. The CRK5 promoter fused with the reporter construct, a modified form of pCAMBIA-1381 vector with the full length ORF of luciferase (LUC) gene, was inserted into the Sal1/Spe1 sites before the start codon of the GUS reporter gene. Primers used are listed in Supplementary Table S1. The constructs were transformed into A. tumefaciens strain GV3101. Bacterial suspensions were infiltrated into healthy and fully expanded leaves of 7-week-old Nicotiana benthamiana plants using a needleless syringe. The amounts of the infiltrated constructs must be kept the same among treatments and controls for each group of assays. After infiltration, plants were placed in darkness for 24h and then with 16h of light–8h of dark for another 24h, and the LUC activity (assessed by fluorescence intensity) was observed 48h after infiltration with CCD imaging apparatus (Andor iXon, Andor, UK). We used the ImageJ software (an image processing program developed by the National Institutes of Health, which can calculate area and pixel value statistics of user-defined areas and intensity-thresholded objects) to calculate the average optical density (OD, integrated density divided by area) of the fluorescence area and background area of the experimental groups and control groups, respectively. The values of the fluorescence intensity as shown in Fig. 10C–E are the result of the OD value of fluorescence area minus the corresponding OD value of background area.
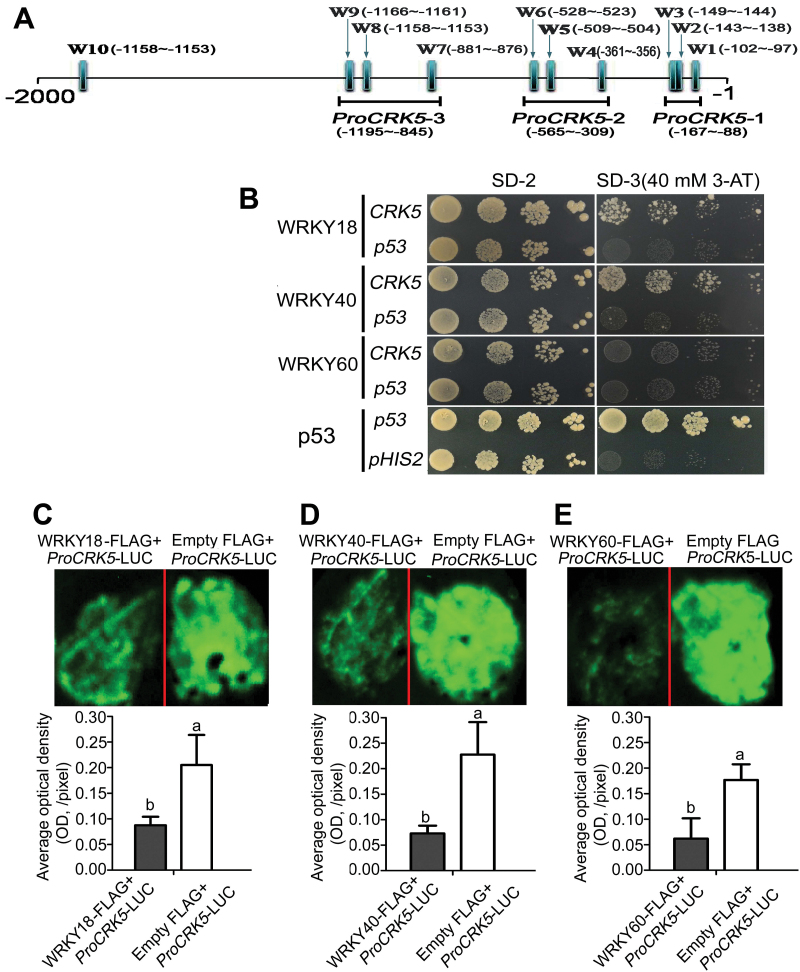
Fig. 10.
Test of the interaction of WRKY18, WRKY40 and WRKY60 with the promoter of the CRK5 gene. (A) Promoter diagram of the CRK5 gene. W1–W10 represent the W-box numbered from left to right with their location sites relative to the start codon (ATG). The segments marked with ProCRK5-1, ProCRK5-2 and ProCRK5-3 indicate the probe fragments used in the gel shift assays described in Fig. 11. (B) Yeast one-hybrid assays to test the interaction of WRKYs with the CRK5 promoter. Yeast cells were co-transformed with pGADT7 prey vector containing WRKY18, WRKY40, or WRKY60 and pHIS2 bait vector containing the promoter of CRK5. The corresponding transformation with pGADT7 prey vector containing WRKY18, WRKY40, or WRKY60 and pHIS2 bait vector containing p53 promoter fragment were used as negative controls. Co-transformation of pHIS2-p53 and pGADT7-p53 was used as positive control, and co-transformation of pGADT7-p53 and empty pHIS2 was used as its own negative control. Three 10-fold series dilutions were dropped vertically for each assay on SD-2 medium (synthetic dropout medium lacking Leu, Trp) and SD-3 medium (synthetic dropout medium lacking Trp, Leu, His) supplemented with 40mM 3-aminotriazole (3-AT). All the experiments were repeated five times with the same results. (C) WRKY18, (D) WRKY40 and (E) WRKY60 inhibit the transcription activity of the CRK5 promoter in tobacco system, assayed with luciferase (LUC) imaging. Tobacco leaves were co-transformed with the constructs ProCRK5-LUC plus WRKY18-FLAG or ProCRK5-LUC plus empty FLAG (C), or with the constructs ProCRK5-LUC plus WRKY40-FLAG or ProCRK5-LUC plus empty FLAG (D), or with the constructs ProCRK5-LUC plus WRKY60-FLAG or ProCRK5-LUC plus empty FLAG (E). Top panels in (C), (D) and (E): LUC fluorescence imaging. Bottom panels in (C), (D) and (E): optical densities calculated with the ImageJ software. The experiments were repeated three times with similar results. Each value for the columns in (C), (D) and (E) are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
Protein production and purification in E. coli
6×His-tagged full length proteins of WRKY18, WRKY40 and WRKY60 were produced in E. coli and purified essentially as described previously (Wu et al., 2009; Shang et al., 2010). The cDNA fragments encoding these proteins were cloned by PCR and the primers are listed in Supplementary Table S1. Full-length ORFs of WRKY18, WRKY40 and WRKY60 were cloned into the protein expression vector pET48b(+). The recombinant plasmids were transformed and expressed in E. coli BL21 (DE3) strains (Novagen, Darmstadt, Germany). The transformed E. coli were grown at 37 °C overnight in 1 liter of liquid Luria–Bertani (LB) medium containing 50 μg ml–1 kanamycin until the OD600 of the cultures was 0.6–0.8. Then, isopropyl-β-D-thiogalactopyranoside was added to the cultures to a final concentration of 0.5mM, and culture was continued at 16 °C at 150rpm for 16h. The WRKY18, WRKY40 and WRKY60 proteins were expressed in the inclusion body, and resolved by 8M urea after the collected cells were lysed, followed by protein purification using a Ni2+-chelating column (Novagen, San Diego, CA USA) as described in the manufacturer’s instructions for the Ni2+-chelating column. After that, the denatured WRKY proteins were treated with slow dialysis in a dialysis buffer containing 25mM Tris (pH 8.0), 150mM NaCl, and 1×Protease Inhibitor Cocktail (Roche, Mannheim, Germany) with a gradually decreased amount of urea (6, 4, 2, 0M) for about 24h until renaturation of recombinant proteins. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was then conducted to detect the quality of purified proteins.
Gel shift assay
Gel shift assay (GSA) was performed with a Light-Shift Chemiluminescent EMSA Kit (Thermo Scientific, Waltham, MA, USA) using the recombinant 6×His-WRKY18, 6×His-WRKY40 and 6×His-WRKY60 fusion proteins purified from E. coli according to the manufacturer’s instructions. The promoter fragments used for the GSA were synthesized using the following primer pairs: forward primer 5′-TTGATGTTACTCGTCTAGTTGACCTTGACTTGCAAG ATATTGTATTATTTTACAAAAACCAAAATTTGACT GGCTTGGCT-3′ and reverse primer 5′-AGCCAAGCCAGTCAAA TTTTGGTTTTTGTAAAATAATACAATATCTTGCAAGTC AAGGTCAACTAGACGAGTAACATCAA-3′ for the CRK5 promoter fragment ProCRK5-1; forward primer 5′-TTGATGTTACTCGTCTAGTTGACCTTGACTTGCAA GATATTGTATTATTTTACAAAAACCAAAATTT AAATGGCTTGGCT-3′ and reverse primer 5′-AGCCAAGCCATT TAAATTTTGGTTTTTGTAAAATAATACAA TATCTTGCAAGTCAAGGTCAACTAGACG AGTAACATCAA-3′ for the W-box mutation of W1 in the fragment ProCRK5-1; forward primer 5′-TTGATGTTACTCGTCTA GTTAAACTTAAATTGCAAGATATTGTATTATT TTACAAAAACCAAAATTTGACTGGCTTG GCT-3′ and reverse primer 5′-AGCCAAGCCAGTCAAATT TTGGTTTTTGTAAAATAATACAATATCTTGCAATTTAA GTTTAACTAGACGAGTAACATCAA-3′ for the double W-box mutations of W2 and W3 in the fragment ProCRK5-1. The fragment of ProCRK5-1 and its mutant form mW1 and mW2/W3 were synthesized directly by annealing of the above described forward primers and reverse primers, and each of the primers were synthesized with the biotin labeling in the 5′ end for biotin-labeled fragment or synthesized without labeling for competitor fragments. Forward primer 5′-AGTTGTAAAGTTCAGAAGGAAAAGTACTAA-3′ and reverse primer 5′-GGATATTTAATAGGTTTGTGATTATTCAG-3′ were used for PCR amplification of the fragment ProCRK5-2. Forward primer 5′-AGTATAAGATGGGTTGTGGTGACTATAAGA-3′ and reverse primer 5′-TGGAAGTAATTTAACTAAGAA AAATCGAAG-3′ were used for PCR amplification of the fragment ProCRK5-3. Biotin labeled fragments were obtained by PCR amplification using the above described primer pairs with the first base in the 5′ end labeled with biotin. Unlabeled fragments of the same sequences were used as competitors. Binding reactions were performed in a binding buffer containing 25mM Hepes (pH 8.0), 40mM KCl, 5mM MgCl2, 1mM DTT, 1mM EDTA and 8% glycerol with 50ng recombinant 6×His-WRKY fusion protein and 20fmol probes for each of the biotin-labelled promoter fragments. Competition binding experiments were performed using a 50- and 200-fold molar excess of unlabeled fragments and the mixture was cultured for 30min at 28 °C followed by PAGE without SDS.
Protein targeting and histochemical analysis
Roots of 1-week-old transgenic plants expressing CRK5-GFP or empty GFP driven by CaMV 35S promoter were used to detect the subcellular localization of CRK5 or GFP, respectively, using a confocal laser scanning microscope (LSM780, Carl Zeiss, Germany). The chemical reagent N-(3-triethylammomiumpropyl)-4-(p-diethylaminophenylhexatrienyl) (FM4-64; Invitrogen, Carlsbad, CA, USA) is widely used as an endocytic tracer and plasma membrane stain that offers red fluorescence (Fischer-Parton et al., 2000; Lu et al., 2016). For the FM4-64 staining, roots were immersed in 20ng ml–1 FM4-64 solution for 2min before investigation under a confocal microscope. GFP fluorescence was detected using an emission filter at 505–530nm with excitation at 488nm, and the red signal of FM4-64 staining was collected using an emission filter at 585–615nm with excitation at 543nm.
For the GUS staining, whole plants or tissues of the transgenic lines expressing CRK5-promoter-GUS were immersed in a reaction buffer containing 1mM 5-bromo-4-chloro-3-indolyl-β-GlcUA (X-gluc; Sigma-Aldrich, USA), 100mM sodium phosphate (pH 7.0), 2mM EDTA, 0.05mM ferricyanide, 0.05mM ferrocyanide and 0.1% (v/v) Triton X-100 for 12h at 37 °C. Chlorophyll was removed from the tissues with a mixture of 30% acetic acid and 70% ethanol.
Results
Overexpression of CRK5, but not its mutated form CRK5 K372E, results in ABA hypersensitivity in post-germination growth
Our preliminary experiment suggested that expression of CRK5 is likely to be regulated by the ABA-responsive WRKY transcription factors WRKY18/40/60, which are negatively involved in ABA signaling (Shang et al., 2010; Liu et al., 2012, 2013; Yan et al., 2013; Zhang et al., 2013, 2014). To test whether CRK5 is involved in ABA signaling, we created transgenic plants overexpressing CRK5 or its mutated form CRK5 K372E. It is known that the mutation of the conserved lysine to glutamic acid abolishes the activities and functions of RLKs (Chen et al., 2003, 2004; Torii, 2004; Wang et al., 2008; Lemmon and Schlessinger, 2010; Osakabe et al., 2010; Tanaka et al., 2012). An amino acid sequence alignment of the conserved cytoplasmic kinase domain of the Arabidopsis receptor-like protein kinases CRK5, CRK36, ARCK1, BAK1 and RPK1 indicated that the 372nd amino acid is the conserved lysine in the kinase domain of CRK5 protein (see Supplementary Fig. S1), and thus the site of the conserved lysine 372 was chosen for point mutagenesis. The CRK5K372E mutation, which involves the change of the 372nd lysine to glutamic acid residue in the kinase domain of CRK5, may lead to loss-of-function of this protein kinase.
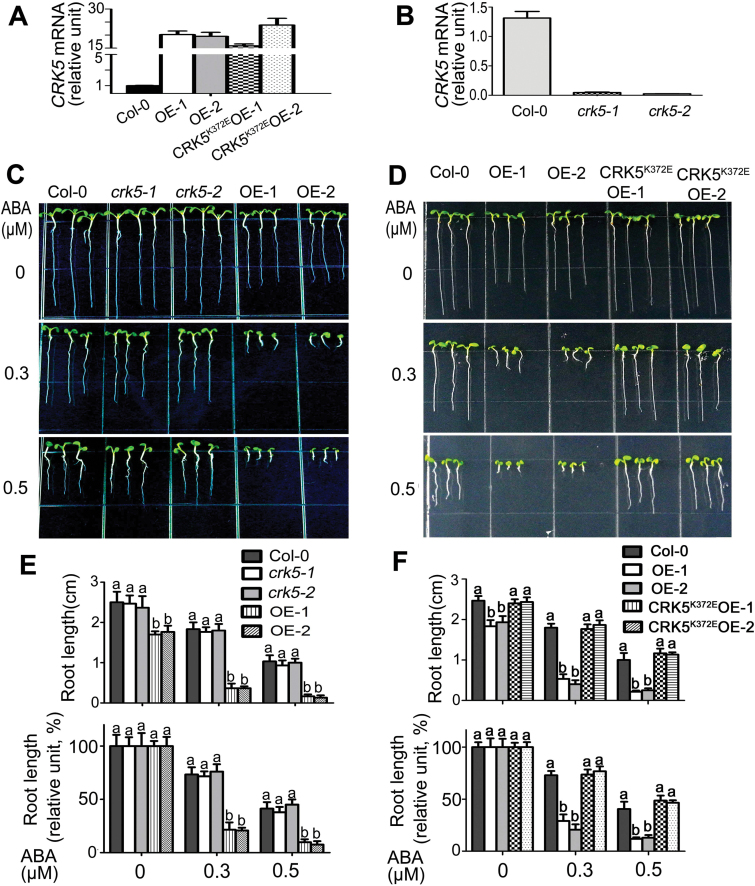
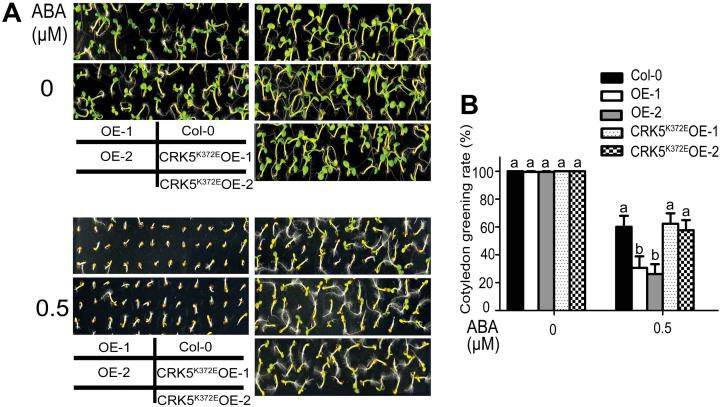
We selected five CRK5-overexpression lines (OE-1, OE-2, OE-5, OE-6 and OE-7) and five CRK5 K372E-overexpression lines (CRK5K372EOE-1, CRK5K372EOE-2, CRK5K372EOE-3, CRK5K372EOE-5 and CRK5K372EOE-7) as representatives used in this study (Figs 1–5; Supplementary Figs S2 and S3). Two loss-of-function T-DNA insertion mutant alleles of the CRK5 gene, crk5-1 (Salk_063519C) and crk5-2 (SALK_109339), were also obtained and identified (Fig. 1B). We assayed ABA sensitivity of the different genotypes by directly sowing seeds in ABA-free or ABA-containing medium, and observed that the CRK5-overexpression lines OE-1 and OE-2 were significantly hypersensitive to ABA in ABA-induced post-germination growth arrest, estimated by both root length and cotyledon greening rate (Figs 1C–F and 2A, B). The GFP-transgenic plants showed wild-type ABA response (see Supplementary Fig. S4), excluding the possible disturbance of the GFP tag of CRK5 in the experiments. It is noteworthy that the CRK5-transgenic lines OE-1 and OE-2 exhibited shorter roots compared with wild-type plants in ABA-free medium, which may be due to a complex, currently unknown, function of CRK5 to reduce root growth, while the root length of the OE-1 and OE-2 lines was significantly much more reduced than that of wild-type plants in the presence of ABA treatment (Fig. 1E, F). However, the loss-of-function mutants crk5-1and crk5-2, as well as CRK5 K372E-overexpression lines CRK5K372EOE-1 and CRK5K372EOE-2, showed wild-type ABA responses in ABA-induced post-germination growth arrest (Figs 1C–F and 2A, B).
Fig. 1.
Overexpression of CRK5, but not its mutated form CRK5 K372E, results in an ABA-hypersensitive phenotype in early seedling growth. (A) Real-time PCR analysis of the transgenic lines overexpressing CRK5 (OE-1 and OE-2) or a mutated form of CRK5 encoding CRK5K372E with a point mutation at its kinase domain (CRK5K372E OE-1 and OE-2). Expression level of CRK5 or CRK5 K372E was normalized to that of Actin2/8, and the expression level of CRK5 in Col-0 was set to 1. Values are the mean±SE of three independent biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test). (B) Real-time PCR analysis of the CRK5 expression level in wild-type Col-0, and crk5-1 and crk5-2 T-DNA insertion mutant plants. Values are the mean±SE of three independent biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test). (C, D) Root growth of wild-type Col-0, crk5-1, crk5-2, OE-1, OE-2 (C) or Col-0, OE-1, OE-2, CRK5K372EOE-1 and CRK5K372EOE-2 (D) growing on ABA-free (0 μM) or (±)ABA-containing (0.3 and 0.5 μM) MS medium. Seeds were directly planted in the medium for a 72-h stratification and germinating seeds/young seedlings continued to grow for 10 d before investigation. The experiments were repeated three times with similar results. (E, F) Statistical analysis of absolute (top) and relative values (bottom) of root length of different genotypes described in (C) and (D), respectively. Relative values of the root length of each genotype grown on MS medium containing 0.3 and 0.5 μM (±)ABA are normalized relative to the value of the corresponding genotype at 0 μΜ (±)ABA, which is taken as 100%. Values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
Fig. 2.
Overexpression of CRK5, but not its mutated form CRK5 K372E, results in an ABA-hypersensitive phenotype in ABA-induced inhibition of cotyledon greening. (A) Cotyledon greening of wild-type Col-0, CRK5-transgenic lines OE-1 and OE-2, and CRK5 K372E-transgenic lines CRK5K372EOE-1 and CRK5K372EOE-2 in ABA-free (0 μM, top) or (±)ABA-containing (0.5 μM, bottom) MS medium. (B) Percentages of green cotyledons of the different genotypes as described in (A). Green cotyledons were scored 5 days after stratification. Values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
We used a different technique to further test the ABA response of early seedling growth of these different genotypes. Seeds were planted in ABA-free medium, subjected to a 3-d stratification, and 60-h-old germinating seeds/young seedlings were transferred to ABA-containing medium and continued to grow for 10 d before investigation. The same ABA-hypersensitive phenotypes of the CRK5-overexpression lines OE-1 and OE-2 were observed, while other genotypes, including crk5-1, crk5-2, CRK5K372EOE-1 and CRK5K372EOE-2, showed wild-type ABA responses (see Supplementary Fig. S2). These findings confirmed the observations mentioned above. Together, these findings suggest that CRK5 is positively involved in ABA signaling as a functional protein kinase and that a functional redundancy occurs in the CRK-mediated ABA signaling.
Moreover, we observed that the ABA sensitivity of the different CRK5-transgenic lines OE-1, OE-5, OE-6 and OE-7 with a gradient of the CRK5 expression levels was positively correlated with the CRK5 expression levels (see Supplementary Fig. S3A, C, E), whereas the CRK5 K372E expression did not modify the ABA response regardless of its expression levels (Supplementary Fig. S3B, D, F). These findings further suggest that CRK5 is positively involved in ABA signaling.
Overexpression of CRK5, but not CRK5 K372E, results in ABA hypersensitivity in stomatal movement and increases plant drought tolerance without affecting plant productivity under non-stressful conditions
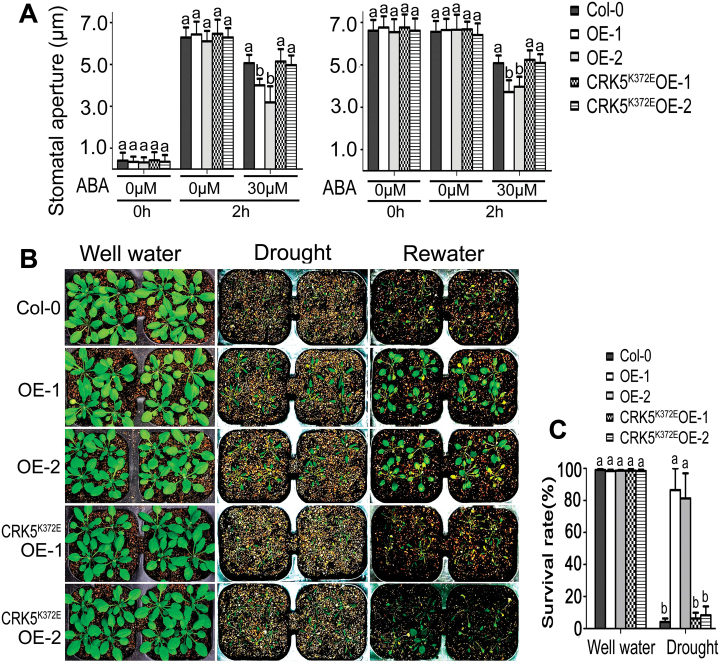
The CRK5-overexpression lines OE-1 and OE-2 showed significant ABA-hypersensitive phenotypes, whereas the CRK5 K372E-transgenic lines CRK5K372EOE-1 and CRK5K372EOE-2 exhibited wild-type ABA responses, in ABA-induced promotion of stomatal closure and inhibition of stomatal opening (Fig. 3A). We further assayed dehydration tolerance of these genotypes, and observed that the CRK5-overexpression lines OE-1 and OE-2 showed significantly higher tolerance to drought than the CRK5 K372E-transgenic lines CRK5K372EOE-1 and CRK5K372EOE-2 that exhibited drought-sensitive as wild-type plants (Fig. 3B, C). These findings are consistent with the ABA-hypersensitive phenotypes of the CRK5-overexpression lines in stomatal movement.
Fig. 3.
Overexpression of CRK5, but not CRK5 K372E, results in ABA hypersensitivity in stomatal movement and increases plant drought tolerance. (A) ABA-induced inhibition of stomatal opening (left) and promotion of stomatal closure (right) in wild-type Col-0 plants, CRK5-transgenic lines OE-1 and OE-2, and CRK5 K372E-transgenic lines CRK5K372EOE-1 and CRK5K372EOE-2. The experiments were repeated five times with similar results. The values are the mean±SE from 60 stomata for each time point, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test). (B) Test of drought tolerance of the different genotypes described above. Plants were well watered (control, ‘Well water’) or drought stressed by withholding water (‘Drought’) for 16 d (D) and then re-watered (‘Rewater’). The experiments were repeated five times, and at least 30 plants per individual line were used for each experiment. (C) Survival rates of the plants described in (B). The values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
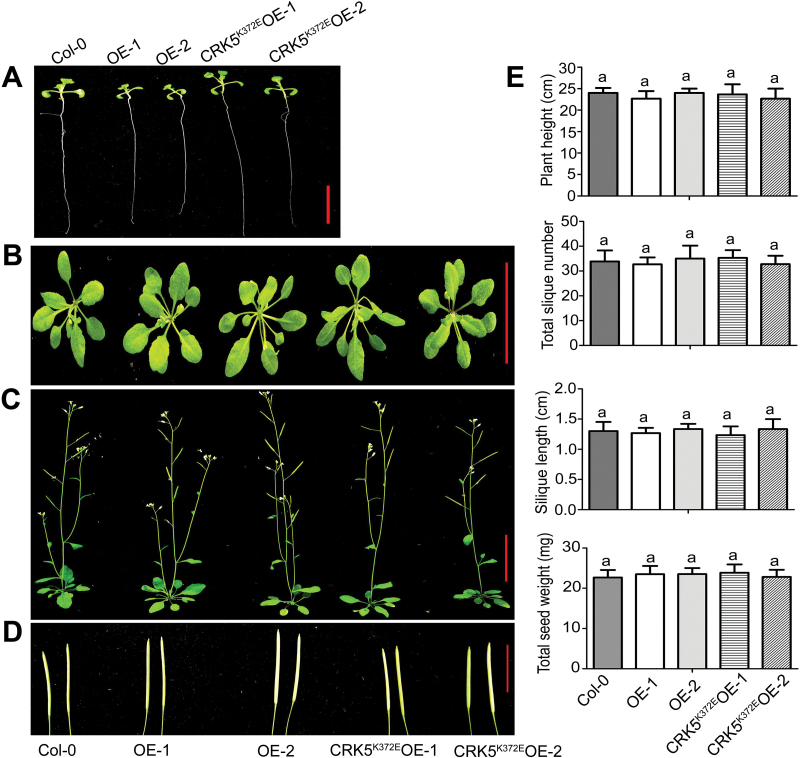
As plant drought tolerance is often associated with reduction of plant development, and the CRK5-transgenic plants displayed shorter roots compared with the wild-type plants in the early growth stage, we assayed the plant growth and productivity of wild-type Col-0, the CRK5-overexpression lines OE-1 and OE-2, and the CRK5 K372E-transgenic lines CRK5K372EOE-1 and CRK5K372EOE-2 at the different mature stages under non-stressful conditions. We found no significant differences in the aspects of rosette leaves, plant height, total silique number per plant, silique length and total seed weight (dry weight) per plant among the different genotypes (Fig. 4A–E).
Fig. 4.
Effects of over-expression of CRK5 on plant growth and productivity. Two-week-old (A; bar=1cm), 4-week-old (B; bar=3cm), and 6-week-old seedlings (C; bar=4cm), and siliques (D; bar=1cm) are shown for wild-type Col-0 plants, CRK5-transgenic lines OE-1 and OE-2, and CRK5 K372E-transgenic lines CRK5K372EOE-1 and CRK5K372EOE-2. (E) Statistics of the plant height, total silique number per plant, silique length and total seed weight (dry weight) per plant of the different genotypes as described above. Each value is the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
Functional interaction of CRK5 with ABI2 and ABA2
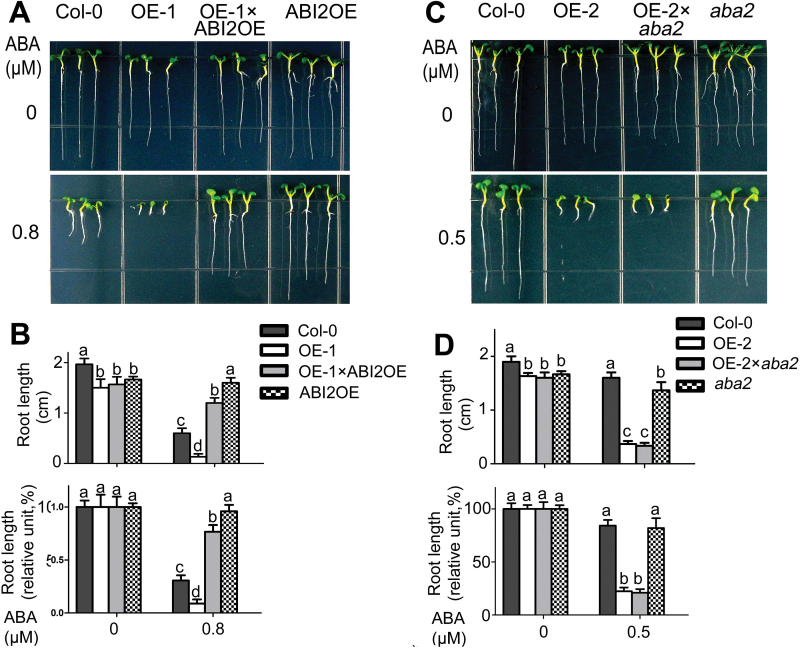
We created CRK5/ABI2-double-overexpression line OE-1×ABI2OE line (Fig. 5; Supplementary Fig. S5) and the CRK5-overexpressing line OE-2 in the aba2 mutant background (OE-2×aba2; Fig. 5; Supplementary Fig. S5). The OE-1×ABI2OE line showed an ABA-hyposensitive phenotype in early seedling growth like the ABI2-overexpression line ABI2OE, which suppresses the ABA-hypersensitive phenotype of the CRK5-overexpressing line OE-1 (Fig. 5A, B). These findings reveal that ABI2 is genetically epistatic to CRK5, suggesting that CRK5 may function upstream of ABI2 in ABA signaling. To the contrary, the OE-2×aba2 plants displayed an ABA-hypersensitive phenotype in early seedling growth like the CRK5-overexpression line OE-2, so loss-of-function of ABA2 (aba2) did not affect the ABA-hypersensitive response resulting from overexpression of CRK5 (Fig. 5C, D). Given that ABA2 is a key, rate-limiting enzyme for ABA biosynthesis (Nambara & Marion-Poll, 2005; Taylor et al., 2005), these data reveal that CRK5 regulates ABA signaling independently of ABA biosynthesis. However, we further found that CRK5-overexpression could partly restored the drought-sensitive phenotype of the aba2 mutant (see Supplementary Fig. S6A, B), suggesting that overexpression of CRK5 stimulates drought response of this mutant by promoting cell signaling in response to the low level of ABA in the aba2 mutant.
Fig. 5.
Test of genetic interaction of CRK5 with ABI2 involved in ABA signaling or with ABA2 involved in ABA biosynthesis. (A) ABI2 is genetically epistatic to CRK5. Seeds of wild-type Col-0, CRK5-overexpression line OE-1, ABI2-overexpression line ABI2OE and CRK5/ABI2-double-overexpression line OE1×ABI2OE were directly planted on the ABA-free (0 μM) or 0.8 μM-ABA-containing MS medium, and the growth status was recorded 10 d after stratification. The experiments were repeated three times with similar results. (B) Statistical analysis of absolute (top) and relative values (bottom) of root length of different genotypes described in (A). Relative values of the root length of each genotype grown on ABA-containing medium are normalized relative to the value of the corresponding genotype at 0 μM ABA, which is taken as 100%. Values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test). (C) Loss-of-function of ABA2 (aba2) does not affect ABA-hypersensitive response of the CRK5-overexpression line OE-2. Seeds of wild-type Col-0, aba2 mutant, CRK5-overexpression line OE-2 and CRK5-overexpression line OE2 in the aba2 mutant background (OE-2×aba2) were directly planted on the ABA-free (0 μM) or 0.5 μM-ABA-containing MS medium, and the growth status was recorded 10 d after stratification. The experiments were repeated three times with similar results. (D) Statistical analysis of absolute (top) and relative values (bottom) of root length of different genotypes described in (C). Relative values of the root length of each genotype grown on ABA-containing medium are normalized relative to the value of the corresponding genotype at 0 μM ABA, which is taken as 100%. Values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
CRK5 protein is localized to plasma membrane, and CRK5 gene is expressed preferentially in roots and leaves
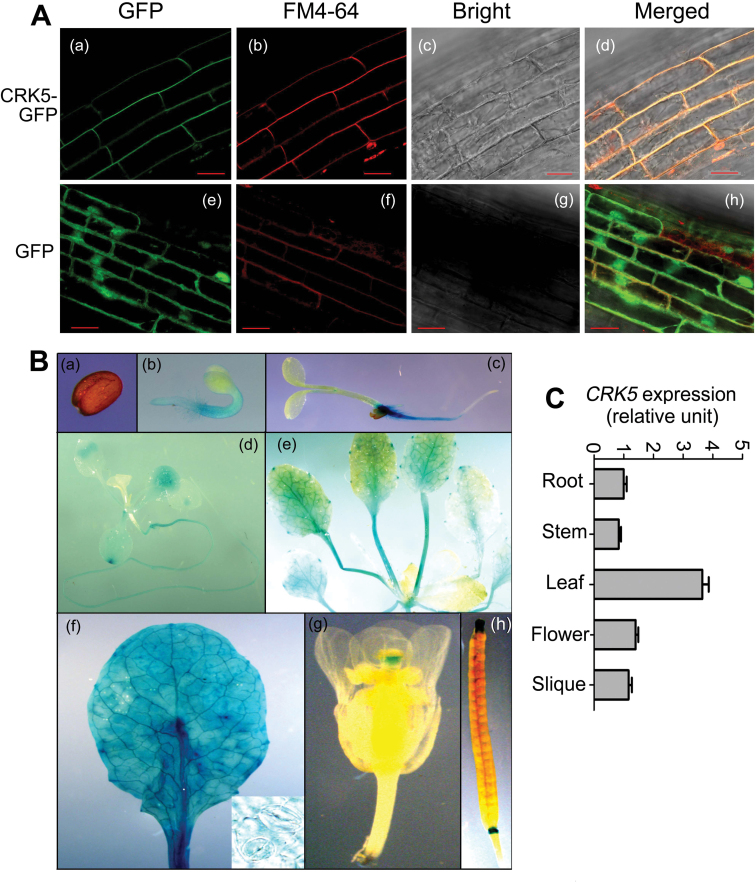
We used the homozygous transgenic plants expressing the CRK5 protein fused with GFP (CRK5–GFP) to investigate the subcellular localization of CRK5. Confocal imaging showed that CRK5–GFP fusion protein was localized in the plasma membrane of the roots of the transgenic plants (Fig. 6A), and that the GFP fluorescence of CRK5–GFP protein merged well with the red fluorescence of the FM4-64 dye that stains the plasma membrane (Fig. 6A). It is noteworthy that FM4-64 is a lipophilic probe used as an endocytic tracer to study the vesicle trafficking of the plasma membrane, and so can be used as a transient plasma membrane stain (within 10min after staining) (Fischer-Parton et al., 2000; Lu et al., 2016). In this experiment, we observed that the fluorescence of FM4-64 was slightly moved from the plasma membrane to cytoplasmic space (Fig. 6A, b, bottom), which may be a phenomenon of endocytosis. As a control, the GFP fluorescence of transgenic plants expressing GFP protein alone were found in the nucleus, cytoplasm and membranes, and only the green signal in the membrane merged with the red fluorescence of FM4-64 (Fig. 6A). Consistently, a prediction with the ‘DAS’ Transmembrane Prediction server and TMHMM algorithm suggests that CRK5 is associated with cell membranes (see Supplementary Fig. S7). Together, these data showed that CRK5 is a plasma membrane-localized protein.
Fig. 6.
Subcellular localization of CRK5 protein and expression profile of CRK5 gene. (A) CRK5 is localized to plasma membrane. The Col-0 plants were transformed with the construct carrying CRK5-GFP or empty GFP, respectively, driven by CaMV 35S promoter, and the roots of transgenic plants were investigated by a confocal laser scanning microscope. (a) CRK5–GFP localization in the mature root zone. (b) FM4-64 staining of the CRK5-GFP-transgenic plant in the mature root zone. (c) The corresponding bright field of (a) and (b). (d) Merged imagine of (a), (b) and (c). (e) Empty GFP localization in the mature root zone. (f) FM4-64 staining of GFP-transgenic plants in the mature root zone. (g) The corresponding bright field (e) and (f). (h) Merged imagine of (e), (f) and (g). Bars=20 μm. (B) Expression of the CRK5-promoter–GUS in transgenic lines. (a) Dry seed. (b) Young seedling 48h after stratification. (c) Young seedling 72h after stratification. (d) Young seedling 14 d after stratification. (e) Young seedling 21 d after stratification. (f) Rosette leaves and stomata (shown at bottom, right). (g) Flower. (h) Silique. (C) Relative expression levels of CRK5 in different tissues/organs determined by real-time PCR analysis.
We created the CRK5-promoter–GUS transgenic lines to investigate the spatial expression pattern of CRK5, and observed that CRK5 was ubiquitously expressed in all the organs/tissues except for seeds (Fig. 6B). The GUS-expression level appeared higher in roots and leaves, but almost no GUS staining was detected in the seeds, including dry seeds, imbibed seeds and mature seeds residing in siliques (Fig. 6B). Similarly, the real-time PCR data showed that the CRK5 gene was expressed in different organs/tissues and had a higher expression level in leaves than in other tissues (Fig. 6C).
Overexpression of CRK5, but not its mutated form CRK5 K372E, alters expression of a subset of ABA-responsive or ABA-signaling-related genes
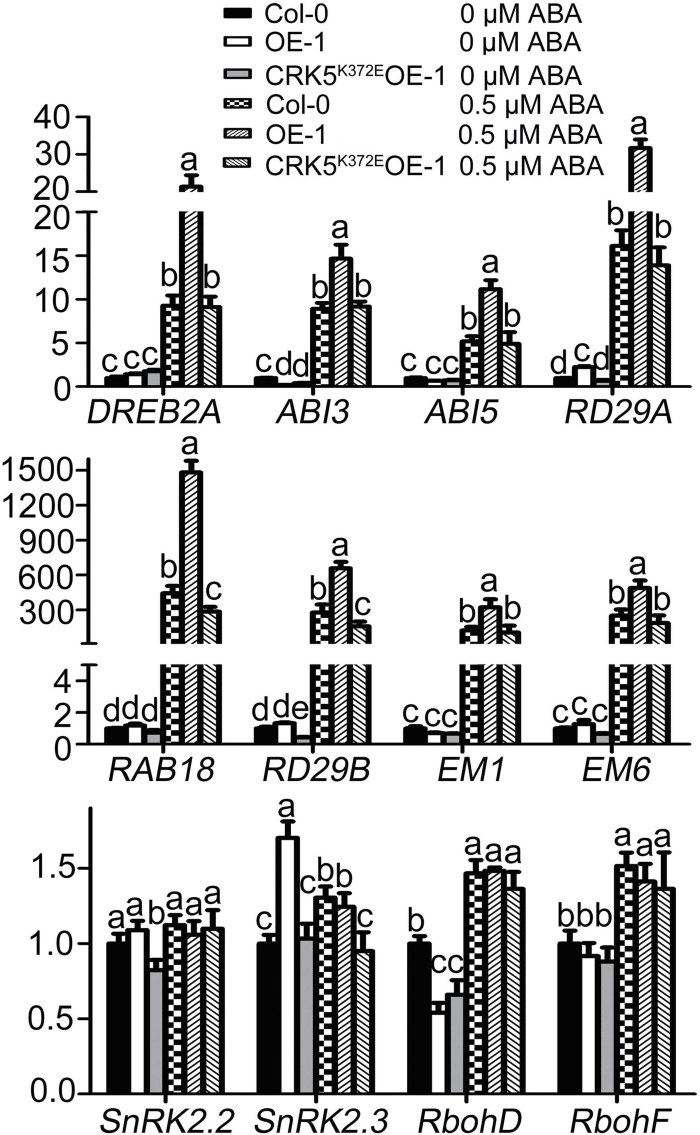
We assayed the expression levels of a subset of ABA-responsive or ABA-signaling-related genes in CRK5 transgenic line OE-1 and CRK5 K372E-transgenic line CRK5K372EOE-1. The assayed ABA-responsive or ABA-signaling-related genes include RD29A and RD29B (Yamaguchi-Shinozaki & Shinozaki, 1994), RAB18 (Lang & Palva, 1992), DREB2A (Liu et al., 1998), ABI3 (Giraudat et al., 1992), ABI5 (Finkelstein & Lynch, 2000), EM1 and EM6 (Gaubier et al., 1993; Devic et al., 1996), SnRK2.2, SnRK2.3 (Fujii & Zhu, 2009), RbohD and RbohF (Kwak et al., 2003). In the absence of ABA, expression levels of these genes in Col-0, OE-1 and CRK5K372EOE-1 showed no marked difference except SnRK2.3 with a higher level in OE-1 and RbohD with a lower level in both OE-1 and CRK5K372EOE-1 (Fig. 7). In the presence of ABA, the expression levels of RD29A, RD29B, RAB18, DREB2A, ABI3, ABI5, EM1 and EM6 were significantly and markedly increased in the CRK5 overexpression line, whereas the expression levels of these genes in the CRK5K372EOE-1 line were not altered significantly except for RAB18 and RD29B of which the expression slightly decreased in the CRK5K372EOE-1 line (Fig. 7). No significant difference was detected for the expression of other ABA-signaling regulator-encoding genes including SnRK2.2, RbohD and RbohF in the OE-1 and the CRK5K372EOE-1 lines compared with the wild-type plants in the presence of ABA (Fig. 7). These data of the expression of the ABA-responsive genes are essentially consistent with the ABA-related phenotypes of the CRK5 transgenic lines and wild-type responses of the CRK5 K372E-transgenic lines.
Fig. 7.
Expression of some ABA-responsive genes in CRK5-transgenic line OE-1 and CRK5 K372E-transgenic line CRK5K372EOE-1. The seeds were germinated and grown on ABA-free (–ABA) or 0.5 μM-ABA-containing (+ABA) MS medium for 4 days before sampling for RNA extraction. Transcription levels of these genes were assayed by real-time PCR. Expression level of each gene is normalized to that of Actin2/8, and the relative expression level of each gene is normalized relative to the level of this gene of the wild-type Col grown in ABA-free medium, which is taken as 1. Values are the mean±SE of three independent biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
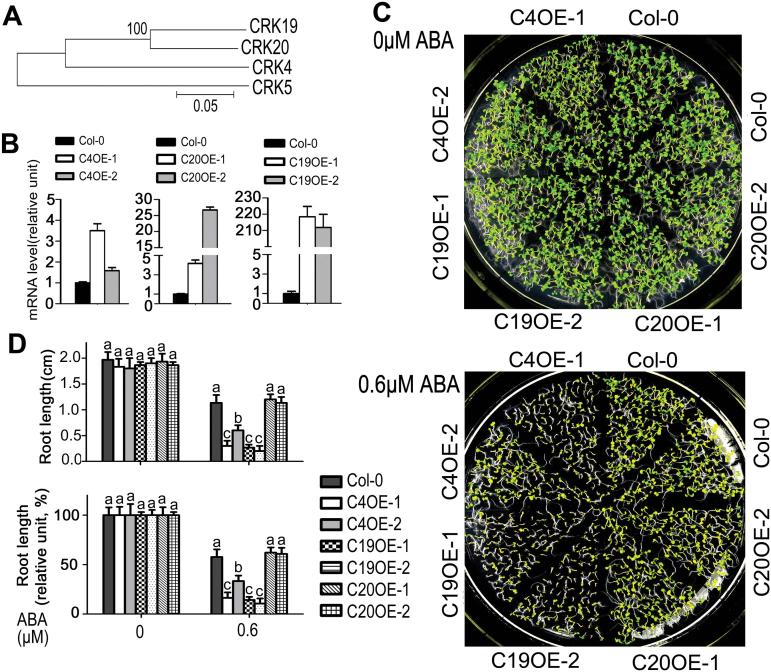
Overexpression of homologs of CRK5, CRK4 and CRK19, but not CRK20, results in ABA hypersensitivity in post-germination growth
CRK4, CRK19 and CRK20 are homologous proteins of CRK5 (Fig. 8A; Supplementary Fig. S8). We observed that the CRK4- and CRK19-overexpression lines, like the CRK5-overexpressing lines, displayed an ABA-hypersensitive phenotype in early seedling growth (Fig. 8B–D). However, CRK20-overexpression lines exhibited a wild-type ABA response in early seedling growth (Fig. 8B–D). Further experiments showed that two knock-down mutants of CRK19, crk19-1 and crk19-2, showed no ABA-related phenotype in early seedling growth (see Supplementary Fig. S9A–C). These data suggest that CRK4 and CRK19, together with CRK5, redundantly regulate ABA signaling, whereas CRK20 is not involved in ABA-mediated early seedling growth arrest.
Fig. 8.
Phenotypes of the transgenic lines overexpressing CRK4, CRK19 or CRK20 homologous to CRK5 in ABA-induced early seedling growth arrest. (A) Phylogenic analysis of Arabidopsis CRK4, CRK5, CRK19 and CRK20 using the neighbor-joining method with MEGA version 4 by alignment of the amino acid sequences with ClustalW. (B) Real-time PCR analysis of the transgenic lines. C4OE-1 and C4OE-2 denotes CRK4-overexpression lines; C19OE-1 and C19OE-2, CRK19-overexpression lines; C20OE-1 and C20OE-2, CRK20-overexpression lines. Values are the mean±SE of three independent biological determinations. (C) Phenotypes of ABA-induced inhibition of early seedling growth in different transgenic lines as described in (B). Seeds were directly planted in ABA-free (0 μM ABA, top) or 0.6 μM-ABA-containing MS medium, and the growth status was recorded 10 d after stratification. The experiments were repeated three times with similar results. (D) Statistical analysis of absolute (top) and relative values (bottom) of root length of different genotypes described in (C). Relative values of the root length of each genotype grown on ABA-containing medium are normalized relative to the value of the corresponding genotype at 0 μM ABA, which is taken as 100%. Values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
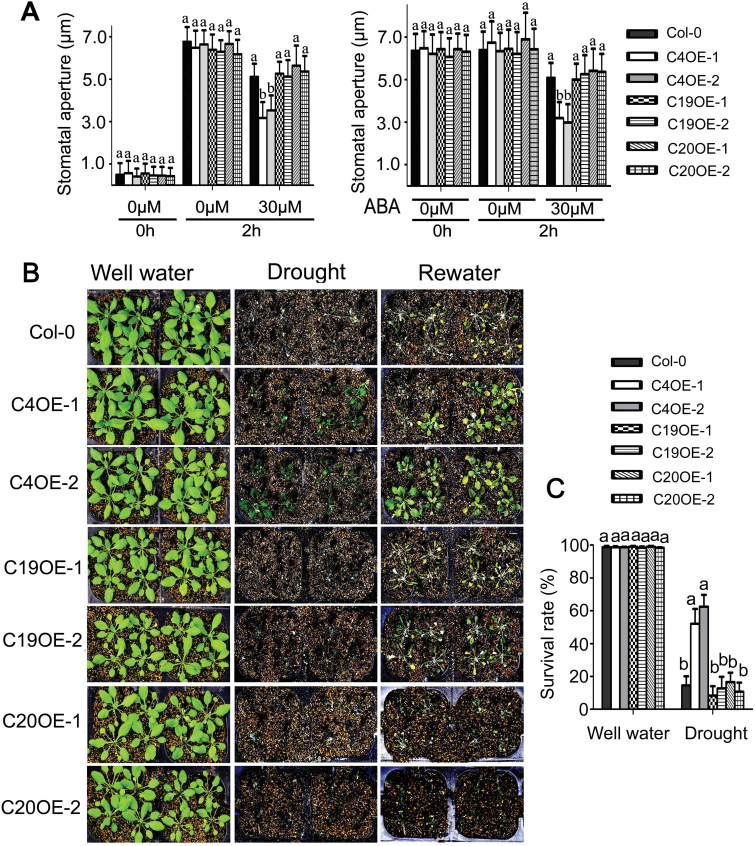
Overexpression of CRK4 results in ABA hypersensitivity in stomatal movement and enhances drought tolerance
We further found that the CRK4-overexpression lines, like the CRK5-overexpressing lines, showed ABA-hypersensitive phenotype in stomatal movement (Fig. 9A) and enhanced drought tolerance (Fig. 9B, C), but neither CRK19- nor CRK20-overexpression lines showed significantly different phenotypes in stomatal movement in response to ABA and in drought response compared with the wild-type plants (Fig. 9A–C). These data suggest that CRK4, but not CRK19, cooperates with CRK5 to regulate ABA response of guard cells as well as drought response, and that the function of CRK19 is only involved in the CRK5-mediated ABA response in early seedling growth.
Fig. 9.
Overexpression of CRK4, but not CRK19 or CRK20, results in ABA hypersensitivity in stomatal movement and increases plant drought tolerance. (A) ABA-induced inhibition of stomatal opening (left) and promotion of stomatal closure (right) in wild-type Col-0 plants, CRK4 (C4OE1, C4OE2), CRK19 (C19OE1, C19OE2) and CRK20 (C20OE1, C20OE2) transgenic lines. The experiments were repeated five times with similar results. The values are the mean±SE from 60 stomata for each time point, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test). (B) Test of drought tolerance of the different genotypes described above. Plants were well watered (control, ‘Well water’) or drought stressed by withholding water (‘Drought’) for 16 d and then re-watered (‘Rewater’). The experiments were repeated five times, and at least 30 plants per individual line were used for each experiment. (C) Survival rates of the plants described in (B). The values are the mean±SE of three biological determinations, and different letters represent significant differences at P<0.05 (Duncan’s multiple range test).
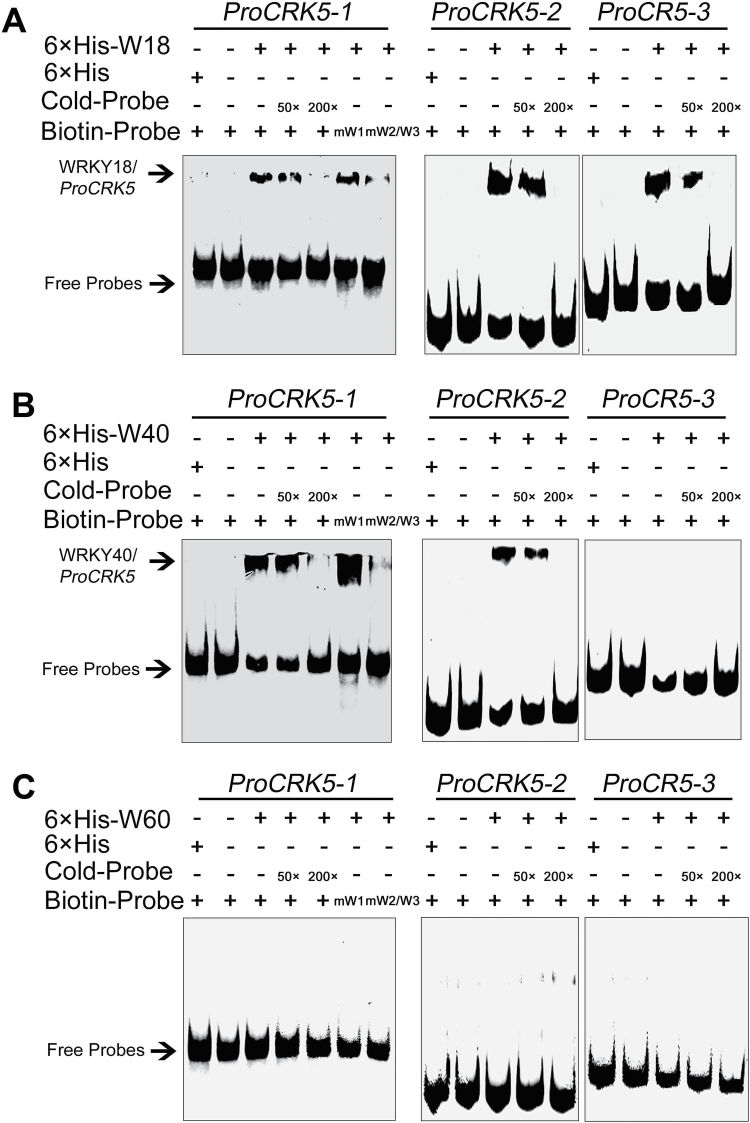
WRKY18 and WRKY40, but not WRKY60, bind to the promoter of the CRK5 gene, while all the three WRKYs inhibit promoter activity of this gene
Sequence analysis shows that there are ten W-boxes in the putative promoter region of the CRK5 gene (Fig. 10A), and we tested whether the three closely related ABA-responsive transcription factors (Shang et al., 2010; Liu et al., 2012, 2013; Yan et al., 2013; Geilen and Böhmer, 2015), WRKY18/40/60, regulate CRK5 expression. In the yeast one-hybrid system, yeast cells co-transformed with pGADT7 prey vector containing WRKY18 or WRKY40 and pHIS2 bait vector containing the promoter of CRK5 grew well in SD-3 medium (lacking Trp, Leu, and His) supplemented with 40mM 3-AT (Fig. 10B), indicating a possible interaction between WRKY18/40 and the CRK5 promoter. However, yeast cells co-transformed with pGADT7 prey vector containing WRKY60 and pHIS2 bait vector containing the promoter of CRK5 could not grow in the SD-3 medium supplemented with 40mM 3-AT (Fig. 10B), suggesting that WRKY60 do not bind to the promoter of CRK5.
We assayed possible effects of the three WRKYs on the promoter activities of the CRK5 gene by co-transforming tobacco leaves with the CRK5 promoter linked to a LUC reporter gene together with WRKY18, WRKY40, or WRKY60 gene. We observed that all the three WRKYs showed inhibitory effects on the activity of the CRK5 promoter, as shown by LUC fluorescence intensity (Fig. 10C–E).
We performed the gel shift assays (GSA) in which three domains of the CRK5 promoter were used (ProCRK5-1, ProCRK5-2, ProCRK5-3) (Fig. 10A). 6×His tagged recombinant WRKY proteins were expressed and purified in E. coli (see Supplementary Fig. S10A–C). We showed that WRKY18 bound to all the three domains and WRKY40 bound to the ProCRK5-1 and ProCRK5-2 domains, while the binding was reduced by adding increasing amounts of unlabeled competitors (Fig. 11A, B). When a mutation of W-box 1 was introduced into the ProCRK5-1 domain, WRKY18 and WRKY40 still bound to the mutant form of the fragment (Fig. 11A, B). However, when double mutation of W-box 2 and W-box 3 was introduced into the ProCRK5-1 domain, WRKY18 and WRKY40 could scarcely bind to this domain (Fig. 11A, B), suggesting that W-box 2 and W-box 3 are two core cis-regulatory elements to which WRKY 18 and WRKY40 bind. WRKY60 showed no binding affinity to any of the three fragments (Fig. 11C). As negative controls, we purified empty 6×His protein (Supplementary Fig. S10D) and observed no shift bands in the control assays with 6×His protein (Fig. 11A–C). Together, these data indicate that WRKY18 and WRKY40, but not WRKY60, bind to the CRK5 promoter, which is consistent with data from the yeast-one hybrid, but not completely consistent with the data from assays in tobacco leaves where WRKY60, like WRKY18 and WRKY40, showed an inhibitory effect on the activity of CRK5 promoter. This may be due to a complex cooperation among these three homologous WRKYs, which interact functionally in a manner of redundancy, antagonistic, and distinct roles (Xu et al., 2006; Liu et al., 2012; Yan et al., 2013).
Fig. 11.
Gel shift assays to test interaction of WRKY18, WRKY40 or WRKY60 with the promoter of CRK5 gene. (A) WRKY18 binds to the ProCRK5-1, ProCRK5-2 and ProCRK5-3 fragments. 6×His-W18 indicates the purified 6×His-WRKY18 fusion protein; 6×His, the 6×His tag peptide served as negative control; Biotin-Probe, the biotin labeled CRK5 promoter fragments ProCRK5-1, ProCRK5-2, ProCRK5-3; mW1 and mW2/W3, the two mutant forms in W-boxes (W1 single mutation and W2/W3 double mutation) of biotin labeled ProCRK5-1 with W-box mutation at W1: TTGACT→TTAAAT, and W-box mutation at W2/W3: TTGACCTTGACT→TTAAACTTAAAT; Cold-Probe, the three unlabeled CRK5 promoter fragments; 50× and 200×, 50-fold or 200-fold cold-probe relative to the labeled probes added, respectively, for binding competition; Free-Probes, the labeled probes that do not bind the WRKY protein; WRKY18/ProCRK5, the shift bands of the complex of WRKY18 protein with corresponding ProCRK5 fragments. Each experiment was repeated three times with the same results. (B) WRKY40 binds to the ProCRK5-1, ProCRK5-2 fragments, but does not bind to the ProCRK5-3 sequence. 6×His-W40 denotes the purified 6×His-WRKY40 fusion protein; WRKY40/ProCRK5, the shift bands of the complex of WRKY40 protein with the corresponding ProCRK5 fragments. Other symbols are the same as described above in (A). Each experiment was repeated three times with the same results. (C) WRKY60 does not bind to any of the three promoter segments of CRK5 gene. 6×His-W60 denotes the purified 6×His-WRKY60 fusion protein, and other symbols are the same as described above in (A). Each experiment was repeated three times with the same results.
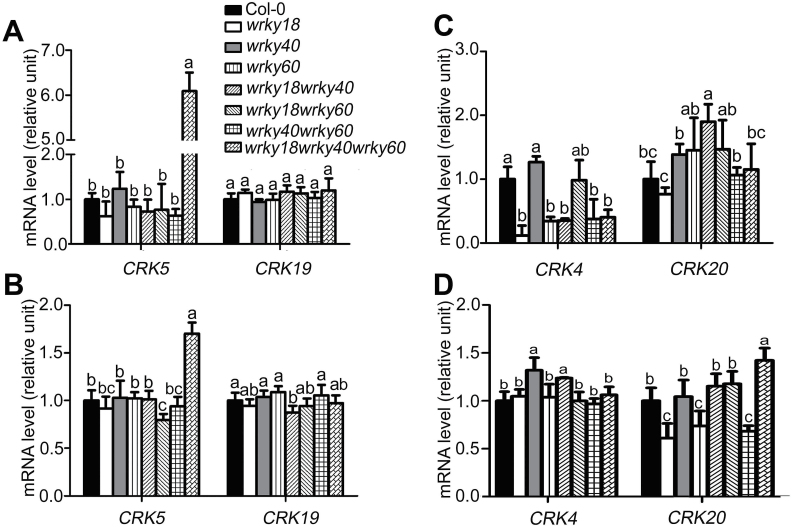
Triple loss-of-function mutation of WRKY18/40/60 enhances expression level of the CRK5 gene
We tested the mRNA level of CRK5 in wrky single, double (wrky40 wrky18, wrky18 wrky60, and wrky40 wrky60), and triple (wrky40 wrky18 wrky60) mutants, and observed that neither WRKY single nor double mutations affected the CRK5 transcription level, which, however, was significantly enhanced in the wrky40 wrky18 wrky60 triple mutant (Fig. 12A, B). This suggests that the three WRKYs act cooperatively to inhibit CRK5 expression. We also assayed the expression levels of three homologous genes of CRK5, CRK4, CRK19 and CRK20, and found that expression of CRK4 was down-regulated in several wrky mutants in 2-week-old young seedlings, contrarily to CRK5 (Fig. 12C), suggesting a positive regulation, but this effect was lost in the mature plants (4 weeks old) (Fig. 12D). These differences between CRK4 and CRK5 genes in the transcriptional regulation by the WRKYs suggest that a complex mechanism may be involved in the processes to maintain homeostasis of the CRK protein amounts to balance ABA signaling. Globally, however, there was no marked alteration of the expression levels of the three homologous CRK genes with loss-of-function of these WRKYs even in the wrky40 wrky18 wrky60 triple mutant (Fig. 12A–D).
Fig. 12.
Expression of CRK4, CRK5, CRK19 and CRK20 genes in wrky loss-of-function mutants. (A–D) Two-week-old seedlings grown on MS medium (A, C) or rosette leaves of 4-week-old seedlings (B, D) were sampled for RNA extraction. The transcription levels of CRK4, CRK5, CRK19 and CRK20 were assayed in the wrky single, double (wrky40 wrky18, wrky18 wrky60, and wrky40 wrky60), and triple (wrky40 wrky18 wrky60) mutants by real-time PCR. Actin2/8 was used as internal control. Each value is the mean±SE of three independent experiments, and the letters indicate significant differences at P<0.05 (Duncan’s multiple range test).
Given that WRKY18/40/60 are supposed to be regulated by ABA (Shang et al., 2010; Geilen and Böhmer, 2015), we tested whether expression of CRK5 is induced by ABA, and found that the expression levels of CRK5 were not significantly affected by ABA treatment (see Supplementary Fig. S11). This suggests that a complex feed-back and feed-forward mechanism may function to regulate homeostasis of CRK5 expression in response to ABA.
Discussion
CRK5 is a potentially positive regulator of ABA signaling
In this study, we observed that the CRK5-overexpression lines displayed ABA-hypersensitive phenotypes in ABA-induced post-germination growth arrest, promotion of stomatal closure and inhibition of stomatal opening (Figs 1–3; Supplementary Figs. S2 and S3). Consistent with the hypersensitivity of guard cells in response to ABA, the CRK5-overexpression lines showed drought tolerance (Fig. 3). Given that plant drought tolerance has been shown to be generally linked to reduction of growth and productivity, we observed, interestingly, that these CRK5-overexpression lines showed normal growth and productivity during mature stages under non-stressful conditions, thought the early seedlings showed shorter roots (Figs 1–4; Supplementary Figs S2 and S3), which suggests that CRK5 is likely to be useful in agriculture to improve crop tolerance to drought by transgenic manipulation. The enhancement of drought tolerance of the CRK5-overexpression lines may be attributed partly to an ABA-hypersensitive response of stomatal movement, which can minimize water transpiration from leaves under drought conditions that induce ABA accumulation (Leung and Giraudat 1998; Zhu, 2002; Kwak et al., 2008). However, ABA regulates plant adaptation to drought by regulating both water balance and osmotic stress/cellular dehydration tolerance, which is associated with both guard cell regulation and the induction of dehydration tolerance genes in nearly all cells (Zhu, 2002). Therefore, up-regulation of a set of ABA- and drought-responsive genes in the CRK5-overexpression lines (Fig. 7), which potentially induces cellular dehydration tolerance, additionally explains the mechanism of enhanced drought tolerance resulting from CRK5 overexpression.
Transgenic lines overexpressing the mutant form of CRK5 K372E showed wild-type ABA responses and drought sensitivity (Figs 1–3; Supplementary Figs S2 and S3). These observations reveal that CRK5 mediates ABA signaling through catalyzing phosphorylation of downstream targets by the cytoplasmic kinase domain in which the 372nd amino acid plays an essential role, and on the other hand, these data provide substantial, supporting evidence for involvement of CRK5 in ABA signaling.
Additionally, transgenic lines overexpressing the CRK5 homologous genes, CRK4 and CRK19, also exhibited ABA-hypersensitive phenotypes, whereas overexpression lines of CRK20 showed wild-type ABA responses in ABA-induced inhibition of early seedling growth, suggesting that CRK4 and CRK19, but not CRK20, function redundantly together with CRK5 in an overlapping manner in ABA signaling (Fig. 8). Interestingly, overexpression of CRK4 also results in ABA hypersensitivity in stomatal movement and enhances drought tolerance (Fig. 9), suggesting that CRK5 works, together with CRK4, to regulate guard cell response to ABA and to drought. We failed to obtain double or triple mutants of these CRK genes because CRK5 and CRK19 localize to the same chromosome with a relative close genetic distance and there is currently no crk4 or crk20 loss of function mutant in the public biological resources. Nevertheless, the possible functional redundancy of CRK4, CRK5 and CRK19 explains, at least partly, the wild-type ABA responses in the crk5 loss-of-function mutants and crk19 knockdown mutants (Fig. 1; Supplementary Fig. S9). Taken all together, in the present experiment, we identified a cysteine-rich receptor-like protein kinase, CRK5, as a potentially positive regulator of ABA signaling in early seedling growth and stomatal movement.
How may CRK5 function in ABA signaling?
Previous studies have reported that the three closely related WRKYs function as negative regulators of ABA signaling by repressing expression of a subset of ABA-responsive genes such as ABI4 and ABI5, in which WRKY60 plays a role in balancing the binding activities of WRKY18 and WRKY40 to the ABI4 and ABI5 promoters (Shang et al., 2010; Liu et al., 2012). Studies in plant response to pathogens showed that WRKY60 may function through interaction with WRKY18 and WRKY40 to promote or decrease their binding affinities to the promoters of their target genes (Xu et al., 2006; Shen et al., 2007; Wenke et al., 2012). Consistently, in this study, we showed that WRKY18, WRKY40 and WRKY60 transcription factors cooperatively repress CRK5 gene expression (Figs 10–12). WRKY18 and WRKY40, but not WRKY60, directly bind to the CRK5 promoter (Figs 10 and 11), but all the three WRKYs can repress promoter activity of CRK5 in a system of tobacco leaves (Fig. 10), suggesting that WRKY60 may interact, directly or indirectly, with other transcription factor(s) to act on the CRK5 promoter. The CRK5 gene is markedly relieved from inhibition only in the wrky18 wrky40 wrky60 triple mutant, but not in wrky18, wrky40 or wrky60 single or any double mutants (Fig. 12), indicating that a complex cooperation mechanism of the three WRKYs occurs in the repression of CRK5 expression, which is likely to function upstream of the CRK5-mediated ABA signaling.
CRK5 is a typical RLK member with an extracellular domain, a transmembrane domain, and a cytoplasmic protein kinase domain (see Supplementary Figs S7 and S8). In the present study, we showed that the cytoplasmic kinase domain may be of importance for ABA signaling (Figs 1–3; Supplementary Figs S2 and S3). Therefore, identification of the substrates of CRK5 is important for understanding of the CRK5-mediated ABA signaling. The present experiment provided genetic evidence that CRK5 may function upstream of ABI2 in ABA signaling (Fig. 5), but whether ABI2 is a direct downstream regulator of CRK5 remains unknown. It has been known that the protein phosphatase ABI2, functioning downstream of the cytosolic ABA receptors PYR/PYL/RCARs, is one of the central players in this PYR/PYL/RCAR-mediated, core ABA signaling pathway (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). So, further studies are needed to determine whether and how CRK5 is involved in the PYR/PYL/RCAR-mediated ABA signaling pathway and to understand the complex ABA signaling network.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Alignment of the conserved cytoplasmic kinase domain of the Arabidopsis receptor-like protein kinases CRK5, CRK36, ARCK1, BAK1 and RPK1.
Figure S2. Overexpression of CRK5, but not its mutated form CRK5 K372E, results in ABA hypersensitive phenotype in early seedling growth.
Figure S3. ABA-induced inhibition of seedling growth is negatively correlated with CRK5 expression levels.
Figure S4. Transgenic line expressing GFP tag alone shows wild-type ABA response in early seedling growth.
Figure S5. The precise T-DNA insertion site of the CRK5 and ABI2 transgenic lines.
Figure S6. Overexpression of CRK5 in aba2 mutant background partially restored drought tolerance of aba2 mutant.
Figure S7. Prediction of the potential transmembrane domains in CRK5 protein.
Figure S8. Alignment of the amino acids of the Arabidopsis CRK4, CRK5, CRK19 and CRK20.
Figure S9. Two knock-down mutants of CRK19, crk19-1 and crk19-2, showed no ABA-related phenotype in early seedling growth.
Figure S10. Identification of the recombined proteins used in this study.
Figure S11. Test of the effects of ABA treatment on CRK5 gene expression.
Table S1. PCR primers used in this study.
Acknowledgements
This research was supported by the National Key Basic Research Program of China (2012CB114300-002), National Natural Science Foundation of China (grant 31570275), and the Ministry of Agriculture of China (grant 2014ZX08009003).
References
- Acharya BR, Raina S, Maqbool SB, Jagadeeswaran G, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R. 2007. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. The Plant Journal 50, 488–499. [DOI] [PubMed] [Google Scholar]
- Bai L, Zhang GZ, Zhou Y, Zhang Z, Wang W, Du YY, Wu ZY, Song CP. 2009. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana . The Plant Journal 60, 314–327. [DOI] [PubMed] [Google Scholar]
- Chen KG, Du LQ, Chen ZX. 2003. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis . Plant Molecular Biology 53, 61–74. [DOI] [PubMed] [Google Scholar]
- Chen KG, Fan B, Du L, Chen ZX. 2004. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis . Plant Molecular Biology 56, 271–283. [DOI] [PubMed] [Google Scholar]
- Deng KQ, Wang QM, Zeng JX, Guo XH, Zhao XY, Tang DY, Liu XM. 2009. A lectin receptor kinase positively regulates aba response during seed germination and is involved in salt and osmotic stress response. Journal of Plant Biology 52, 493–500. [Google Scholar]
- De Smet I, Voss U, Jurgens G, Beeckman T. 2009. Receptor-like kinases shape the plant. Nature Cell Biology 11, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Devic M, Albert S, Delseny M. 1996. Induction and expression of seed-specific promoters in Arabidopsis embryo-defective mutants. The Plant Journal 9, 205–215. [DOI] [PubMed] [Google Scholar]
- Du SY, Zhang XF, Lu Z, Xin Q, Wu Z, Jiang T, Lu YF, Wang XF, Zhang DP. 2012. Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Molecular Biology 80, 519–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. 2002. Abscisic acid signaling in seeds and seedlings. The Plant Cell 14, Suppl 1, S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinson HA, Read ND. 2000. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. Journal of Microscopy 198, 246–259. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceeding of the National Academy of Sciences of the United States of America 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubier P, Raynal M, Hull G, Huestis GM, Grellet F, Arenas C, Pages M, Delseny M. 1993. Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Molecular and General Genetics 238, 409–418. [DOI] [PubMed] [Google Scholar]
- Geilen K, Böhmer M. 2015. Dynamic subnuclear relocalisation of WRKY40 in response to abscisic acid in Arabidopsis thaliana . Scientific Reports 5, 13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Bauer Z, Boller T. 2001. Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis . The Plant Cell 13, 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Hua DP, Wang C, He JN, Liao H, Duan Y, Zhu ZQ, Guo Y, Chen ZZ, Gong ZZ. 2012. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis . The Plant Cell 24, 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idanheimo N, Gauthier A, Salojarvi J, Siligato R, Brosche M, Kollist H, Mahonen AP, Kangasjarvi J, Wrzaczek M. 2014. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochemical and Biophysical Research Communications 445, 457–462. [DOI] [PubMed] [Google Scholar]
- Jiang SC, Mei C, Liang S, Yu YT, Lu K, Wu Z, Wang XF, Zhang DP. 2015. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Molecular Biology 88, 369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SC, Mei C, Wang XF, Zhang DP. 2014. A hub for ABA signaling to the nucleus: significance of a cytosolic and nuclear dual-localized PPR protein SOAR1 acting downstream of Mg-chelatase H subunit. Plant Signaling and Behavior 9, e972899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mäser P, Schroeder JH. 2008. The clickable guard cell, version II: interactive model of guard cell signal transduction mechanisms and pathways. The Arabidopsis Book 6, e0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis . The EMBO Journal 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Palva ET. 1992. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh . Plant Molecular Biology 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J. 1998. Abscisic acid signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Liang S, Lu K, Wu Z, Jiang SC, Yu YT, Bi C, Xin Q, Wang XF, Zhang DP. 2015. A link between magnesium-chelatase H subunit and sucrose nonfermenting 1 (SNF1)-related protein kinase SnRK2.6/OST1 in Arabidopsis guard cell signalling in response to abscisic acid. Journal of Experimental Botany 66, 6355–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis . The Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Xu YH, Jiang SC, et al. 2013. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. Journal of Experimental Botany 64, 5443–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP. 2012. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis . Journal of Experimental Botany 63, 6371–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xia Z, Wang W. 2016. The dynamic structure of Spitzenkörpers of Trichosporon asahii examined by the fluorescent probe FM4-64. Brazilian Journal of Microbiology 47, 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mei C, Jiang SC, Lu YF, et al. 2014. Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling. Journal of Experimental Botany 65, 5317–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SA, Tax FE. 2006. Functional analysis of receptor-like kinases in monocots and dicots. Current Opinion in Plant Biology 9, 460–469. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K. 2005. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis . The Plant Cell 17, 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Mizuno S, Tanaka H, Maruyama K, Osakabe K, Todaka D, Fujita Y, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. 2010. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis . Journal of Biological Chemistry 285, 9190–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. 2013. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. Journal of Experimental Botany 64, 445–458. [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. 2009. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis . Cell 136, 136–148. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. 2010. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell 22, 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. 2007. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, et al. 2006. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. 2001. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceeding of the National Academy of Sciences of the United States of America 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2012. Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis . The Plant Journal 70, 599–613. [DOI] [PubMed] [Google Scholar]
- Taylor I, Sonneveld T, Bugg TH, Thompson A. 2005. Regulation and manipulation of the biosynthesis of abscisic acid, including the supply of xanthophyll precursors. Journal of Plant Growth Regulation 24, 253–273. [Google Scholar]
- Torii KU. 2004. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. International Review of Cytology 234, 1–46. [DOI] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. 2008. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Developmental Cell 15, 220–235. [DOI] [PubMed] [Google Scholar]
- Wang XF, Zhang DP. 2014. ABA signal perception and ABA receptors. In: Zhang DP, ed. Abscisic Acid: Metabolism, Transport and Signaling.Heidelberg: Springer, 89–116. [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. 2001. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. [DOI] [PubMed] [Google Scholar]
- Wenke K, Wanke D, Kilian J, Berendzen K, Harter K, Piechulla B. 2012. Volatiles of two growth-inhibiting rhizobacteria commonly engage AtWRKY18 function. The Plant Journal 70, 445–459. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosche M, Salojarvi J, Kangasjarvi S, Idanheimo N, Mersmann S, Robatzek S, Karpinski S, Karpinska B, Kangasjarvi J. 2010. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis . BMC Plant Biology 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, et al. 2009. The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis . Plant Physiology 150, 1940–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin ZY, Wang AY, Yang GY, Gao P, Zheng ZL. 2009. The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiology 149, 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Chen CG, Fan BF, Chen ZX. 2006. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. The Plant Cell 18, 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Liu ZQ, Xu YH, Lu K, Wang XF, Zhang DP. 2013. Auto- and cross-repression of three Arabidopsis WRKY transcription factors WRKY18, WRKY40, and WRKY60 negatively involved in ABA signaling. Journal of Plant Growth Regulation 32, 399–416. [Google Scholar]
- Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li XQ, Lu CQ, Li H, Hou CC, Li L, Buchanan BB, Chen L, Cheung AY, Li D, Luan S. 2012. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proceeding of the National Academy of Sciences of the United States of America 109, 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Jiang T, Wu Z, Du SY, Yu YT, Jiang SC, Lu K, Feng XJ, Wang XF, Zhang DP. 2013. Cochaperonin CPN20 negatively regulates abscisic acid signaling in Arabidopsis . Plant Molecular Biology 83, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Jiang T, Yu YT, et al. 2014. Arabidopsis co-chaperonin CPN20 antagonizes Mg-chelatase H subunit to derepress ABA-responsive WRKY40 transcription repressor. Science China Life Sciences 57, 11–21. [DOI] [PubMed] [Google Scholar]
- Zhu JK. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.