Highlight
Of twenty-three TCP genes identified from the genome of Phalaenopsis orchid, PePCF10 and PeCIN8 play important roles in orchid ovule development by modulating cell division.
Key words: Cell proliferation, functional characterization, genome-wide identification, ovule development, Phalaenopsis equestris, TEOSINTE-BRANCHED/CYCLOIDEA/PCF (TCP) proteins.
Abstract
TEOSINTE-BRANCHED/CYCLOIDEA/PCF (TCP) proteins are plant-specific transcription factors known to have a role in multiple aspects of plant growth and development at the cellular, organ and tissue levels. However, there has been no related study of TCPs in orchids. Here we identified 23 TCP genes from the genome sequence of Phalaenopsis equestris. Phylogenetic analysis distinguished two homology classes of PeTCP transcription factor families: classes I and II. Class II was further divided into two subclasses, CIN and CYC/TB1. Spatial and temporal expression analysis showed that PePCF10 was predominantly expressed in ovules at early developmental stages and PeCIN8 had high expression at late developmental stages in ovules, with overlapping expression at day 16 after pollination. Subcellular localization and protein–protein interaction analyses revealed that PePCF10 and PeCIN8 could form homodimers and localize in the nucleus. However, PePCF10 and PeCIN8 could not form heterodimers. In transgenic Arabidopsis thaliana plants (overexpression and SRDX, a super repression motif derived from the EAR-motif of the repression domain of tobacco ETHYLENE-RESPONSIVE ELEMENT-BINDING FACTOR 3 and SUPERMAN, dominantly repressed), the two genes helped regulate cell proliferation. Together, these results suggest that PePCF10 and PeCIN8 play important roles in orchid ovule development by modulating cell division.
Introduction
Plant architecture and organ shape depends on complex coordination of cell proliferation and cell differentiation in response to genetic and environmental cues (Ingram and Waite, 2006; Kieffer et al., 2011). The final shape of shoot lateral organs, leaves and flowers is determined by coordinated growth after the initiation of primordia from shoot meristems in seed plants. This coordination involves the complex behavior of many transcription factors (TFs).
The TCP gene family was first described in 1999 and named after the first three characterized members, TEOSINTE BRANCHED1 (TB1) in maize, CYCLOIDEA (CYC) in snapdragon, and Proliferating Cell Factor (PCF) in rice (Cubas et al., 1999). They are ancient plant-specific TFs that originated near the base of streptophytes about 650–800 million years ago (Mondragon-Palomino and Trontin, 2011). From amino acid sequence similarity of the TCP domain, TCP proteins can be divided into two major classes: class I (also known as the PCF or TCP-P class) and class II (also known as the TCP-C class) (Martín-Trillo and Cubas, 2010; Manassero et al., 2013). TCP class II is further subdivided into the CINCINNATA (CIN) and CYC/TB1 subclades. In addition, several class II TCP members share an arginine-rich R domain and an ECE motif (a glutamic acid–cysteine–glutamic acid stretch), both of unknown biological function (Martín-Trillo and Cubas, 2010).
The TCP family of plant-specific TFs controls multiple traits in diverse plant species by controlling cell proliferation and differentiation. This control includes regulation of lateral branching, leaf development and senescence, flower development, gametophyte development, embryo growth, seed germination, circadian rhythm, hormone pathways and mitochondrial biogenesis (Aguilar-Martinez et al., 2007; Koyama et al., 2007; Martín-Trillo and Cubas, 2010; Aguilar-Martinez and Sinha, 2013; Balsemão-Pires et al., 2013; Manassero et al., 2013). Class I genes are suggested to promote plant growth and proliferation. The Arabidopsis multiple mutants (AtTCP8, 15, 21, 22, and 23) have fewer but larger leaves (Aguilar-Martinez and Sinha, 2013). However, most class I single mutants analysed have mild or no phenotypic defects, probably due to genetic redundancy (Martín-Trillo and Cubas, 2010). Class II genes are involved in lateral organ development, including CIN in snapdragon and CYC/TB1 clade genes controlling axillary meristem development, such as TB1 in maize (Lopez et al., 2015). In Arabidospsis, a group of CIN-type genes (AtTCP3, 4, 5, 10, and 13) functions redundantly to control shoot lateral organ morphology by negative regulation of boundary-specific genes (Koyama et al., 2010).
Orchidaceae is one of the largest families among the angiosperms, consisting of about 850 genera with 25 000 species (Dressler, 1993). They are well known for their broad geographic distribution, tremendous diversity in plant forms and growth habits, extraordinary diversity in flower morphology within the framework of a relatively conserved structure, initiation of ovule development precisely regulated by pollination, and high value of hybrids as a floricultural commodity. Despite the economic and biological significance of orchids, and the importance of TCP TFs in controlling floral architecture, the relationship between the function of TCP genes, flower morphology and reproductive development remains poorly understood in this family.
Recently, the whole genome of the orchid Phalaenopsis equestris was sequenced (Cai et al., 2015). The information provides a great opportunity to identify and characterize TCP TFs in orchid. In this study, we identified 23 non-redundant genes encoding TCP TFs in the genome of P. equestris. Systematic analysis including phylogenetic analysis and expression profiling was performed. Furthermore, a class I PCF-like gene, PePCF10, and a class II CIN-like gene, PeCIN8, were functionally characterized by cellular localization, protein–protein interaction and, in transgenic Arabidopsis, by gain-of-function and loss-of-function constructs. Our findings indicate that the two genes play important roles in orchid ovule development by modulating cell division.
Materials and methods
Accession numbers
The nucleotide sequences reported in this paper have been submitted to GenBank under the following accession numbers: PePCF1, KT258871; PePCF2, KT258872; PePCF3, KT258873; PePCF4, KT258874; PePCF5, KT258875; PePCF6, KT258876; PePCF7, KT258877; PePCF8, KT228878; PePCF9, KT258879; PePCF10, KT258880; PePCF11, KT258881; PeCIN1, KT258882; PeCIN2, KT258883; PeCIN3, KT258884; PeCIN4, KT258885; PeCIN5, KT258886; PeCIN6, KT258887; PeCIN7, KT258888; PeCIN8, KT258889; PeCIN9, KT258890; PeCYC1, KT258891; PeCYC2, KT258892; PeCYC3, KT258893.
Plant material
Phalaenopsis plants were grown in a greenhouse at National Cheng Kung University (NCKU) under natural light (photosynthetic photon-flux density 90 μmol m–2 s–1) and controlled temperature (23–27 °C).
RNA preparation
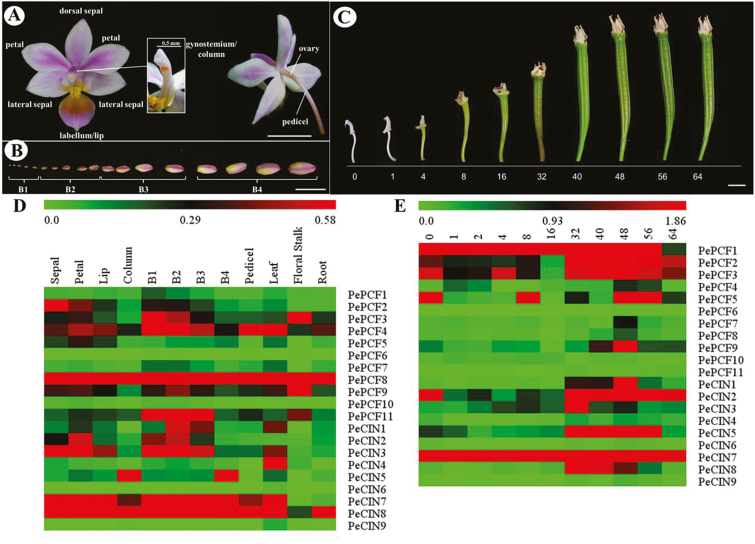
The flower of Phalaenopsis equestris has two main petals with three sepals behind them. The median petal is highly modified into an enlarged petal, called a lip or labellum. The male and female reproductive parts are fused in a structure, the gynostemium or column, located at the center of the flower (see Fig. 3A). Definition of the developing flower bud stage (stage 1: 0–1mm; stage 2: 1–2mm; stage 3: 2–5mm; stage 4: 5–10mm; Fig. 3B) is based on the description by Tsai et al. (2004). Morphological changes in the ovary and ovule development at various days after pollination (DAP) is based on the description by Chen et al. (2012). For RNA extraction, samples were collected, immersed in liquid nitrogen, and stored at –80 °C as described (Tsai et al., 2004).
Fig. 3.
Heat map representation for the expression profiles of PeTCP genes. (A) Structure of a P. equestris flower. (B) Floral bud development stages from B1 to B4. (C) Morphological changes in the ovary at various days after pollination. Expression patterns of (D) 11 PePCF genes and nine PeCIN genes in various organs and developmental stages of floral bud; (E) 11 PePCF genes and nine PeCIN genes in various developmental stages of ovule. PeActin4 was used as an internal control. The expression levels are represented by the color bar. B1: stage 1 flower bud (0~1mm); B2: stage 2 flower bud (1~2mm); B3: stage 3 flower bud (2~5mm); B4: stage 4 flower bud (5~10mm); Co: column; DAP, days after pollination; F: floral stalk; L: leaf; Li: lip; P: pedicel; Pe: petals; R: root; Se: sepals. Scale bar: 1cm.
Sequence retrieval for TCP proteins
The conserved TCP DNA-binding domain based on the hidden Markov model (HMM) (PF03634) was obtained from the Pfam protein family database (http://pfam.sanger.ac.uk). To identify the TCP TF coding genes of P. equestris, the HHM profile of the TCP domain was used as a query for an HMMER search (http://hmmer.janelia.org/) of the P. equestris genome sequence (Cai et al., 2015) (E-value=0.00001). Furthermore, to verify the reliability of the results, all candidate TCP sequences were analysed to confirm the presence of the conserved TCP domain by using InterproScan (Quevillon et al., 2005). The sequences of TCP family members in the genome of Arabidopsis and Oryza sativa were retrieved from the PlantTFDB TF database (http://planttfdb.cbi.pku.edu.cn/, v3.0).
Sequence alignment and phylogenetic analysis
Multiple sequence alignment of the amino acid sequences of TCP proteins in P. equestris, Arabidopsis and rice genomes involved use of Clustal W (Thompson et al., 1994) with default settings. Subsequently, MEGA 4.0 (Tamura et al., 2007) was used to construct an unrooted phylogenetic tree based on alignments with the neighbor-joining method and the parameters JTTmodel, pairwise gap deletion and 1000 bootstraps.
Real-time quantitative RT-PCR
RNA samples were treated with RNase-free DNase (New England Biolabs, Hitchin, UK) to remove remnant DNA, then underwent synthesis of first-strand cDNA by the use of the SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Gene-specific primers for PeTCP genes were designed with the use of Primer Express (Applied Biosystems, Foster City, CA, USA) and are listed in Supplementary Table S1 at JXB online. PeActin4 (5′-TTGTGAGCAACTGGGATG-3′ and 5′-GCCACGCGAAGTTCATTG-3′) and 18S rRNA (5′-TTAGGCCACGGAAGTTTGAG-3′ and 5′-ACACTTCACCG GACCATTCAA-3′) (Lin et al., 2014) were used as internal control. Each real-time RT-PCR contained 5ng of cDNA, 20mM primers and 12.5ml of SYBR GREEN PCR Master Mix (Applied Biosystems), and water was added to 25ml. Real-time PCR involved use of the ABI 7 500 Real-Time PCR Instrument (Applied Biosystems). For each real-time RT-PCR, each sample was analysed in triplicate. Data were analysed by the use of the Sequencing Detection System v1.2.3 (Applied Biosystems). The software MultiExperiment Viewer was used to construct heatmap representations for expression patterns.
Whole-mount in situ hybridization
Phalaenopsis equestris inflorescences were fixed in 4% (v/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde for 24h at 4 °C. They were dehydrated through a graded ethanol series. Digoxigenin-labeled sense and antisense RNA probes containing a partial C-terminal region and 3′-UTR were synthesized following the manufacturer’s instructions (Roche Applied Science). Hybridization and immunological detection of the signals with alkaline phosphatase were as described by Traas (2008).
Subcellular localization
Template-specific primers were designed by addition of attB1 adapter primer (5′-GGGGACAAGTTTGTACAA AAA AGCAGGCTCCACC-3′) to the 5′ end of the first 18–25 nt of each open reading frame (ORF), and attB2 adapter primer (5′-GGGGACCA CTTTGTACAAGAAAGCTGGGTT-3′) to the 3′ end of the first 18–25 nt of each ORF, which generated the full-length attB1 and attB2 sites flanking the ORFs (S1). Gateway-compatible amplified ORFs were recombined into the pDONR 221 vector (Invitrogen) by BP cloning. BP reactions were used directly for bacterial transformation. Entry clones were used for transformation of E. coli DH5α cells, and bacteria were plated on LB medium containing kanamycin. These entry clones were set up in LR reactions for recombining target genes into the destination vector. The 35S promoter-driven green fluorescent protein (GFP)-fusion construct for protoplast transfection was created by use of the high-copy plasmid p2GWF7, with C-terminal fusion (Karimi et al., 2007). Entry clones were used for generation of GFP-tagged clones. Then, the LR reactions were used for transformation of E. coli DH5α. Transformants were selected on plates containing ampicillin. The plasmids were transfected into Phalaenopsis protoplasts by PEG transformation. Signals were visualized by confocal laser microscopy (Carl Zeiss LSM780, Jena, Germany).
Bimolecular fluorescence complementation (BiFC) assay
Gateway-compatible vectors [pSAT5(A)-DEST-cEYFP-N1 and pSAT5-DEST-cEYFP-C1(B) (Lin et al., 2014)] were used to generate expression vectors by a Gateway cloning strategy. The N-terminus of yellow florescent protein (YFP) (YN) was cloned upstream of PePCF10 and PeCIN8 in the pE-SPYNE vector, and the C-terminus of yellow florescent protein (YC), was fused upstream of PePCF10 and PeCIN8 in the pE-SPYCE vector. The signals were visualized by confocal laser microscopy (Carl Zeiss LSM780).
Yeast two-hybrid analysis and Quantification of β-galactosidase activity
For the yeast two-hybrid, the full-length coding sequences of PePCF10 and PeCIN8 were cloned into either pGBKT7 bait vector (fused with Gal4 DNA-binding domain; BD) or pGADT7 prey vector (fused with GAL4 activation domain (AD), Clontech, http://clontech.com). The constructs were verified by sequencing and co-transformed into yeast strain Y187. To test for interactions, the bait and prey constructs were co-expressed in yeast. The yeast cells were plated on selection medium containing SD/–Trp–Leu and then incubated in a growth chamber at 30 °C for 2–5 d (Li et al., 2011). All primers used for cloning are listed in Supplementary Table S1. β-Galactosidase activity from cotransformed yeast was measured using the o-nitrophenyl β-D-galactopyranoside assay, where o-nitrophenyl produced from cleavage of o-nitrophenyl β-D-galactopyranoside by β-galactosidase was detected by spectrophotometry (Li et al., 2011, Li & Zachgo, 2013). For systematically analysing interaction behavior of PePCF10, PePCF10 was cloned into pDEST_GBKT7 bait vector, which was retrieved from Arabidopsis Biological Resource Center (ABRC) from the Gateway entry clone. The Y2H Gold yeast strain (Takara Bio Inc.) was employed for the assay. After selecting the yeast harboring bait plasmid, prey plasmids constructed on pDEST_GAD424 vector (Mitsuda et al., 2010) were additionally transformed and spotted onto positive control medium, which lacks leucine and tryptophan, and test medium, which lacks histidine, leucine, and tryptophan. Yeast growth was observed daily for several days.
Construction of transformed fusions and Arabidopsis transformation
cDNA fragments containing the coding regions of PePCF10 and PeCIN8 were cloned into the pBI121 vector (primers are in Supplementary Table S1). The constructs were then introduced into Agrobacterium tumefaciens (strain GV3101). To construct the transgene for the chimeric repressor, the coding sequences of PePCF10 and PeCIN8, except for the stop codon (primers are in Supplementary Table S1), were cloned into the SmaI site of p35SSRDXG in-frame to the region that encodes the SRDX repression domain (LDLDLELRLGFA) from SUPERMAN (Hiratsu et al., 2002, 2003). p35SSRDXG contains a CaMV 35S promoter followed by an Ω translation enhancer sequence, the SRDX repression domain sequence, Nos terminator and the attL1 and attL2 Gateway recombination sites (Invitrogen) outside the regions of the 35S promoter and the Nos terminator in the pUC119 vector. The transgene cassette was transferred into the destination vector pBCKK by the Gateway LR clonase reaction (Invitrogen). Each gene was used to transform Arabidopsis thaliana Col-0 by the floral dip method, as described previously (Clough and Bent, 1998). To select transformed Arabidopsis, seeds (T0) were screened on media supplemented with 50 µg ml–1 kanamycin (Sigma-Aldrich). After a 2-week selection, the kanamycin-resistant seedlings (T1) were transferred to soil and grown under the conditions described previously. Kanamycin segregation in the T1 generation was analysed by use of chi-square test. The homozygous, kanamycin-resistant T2 generation was used to confirm the integration fragment by PCR for each construct. Transformed lines with segregation ratio 3:1 were collected for analysis.
Scanning electron microscopy
Leaves and flowers were frozen by liquid nitrogen and observed by scanning electron microscopy (Hitachi TM3000, Tokyo, Japan). The data were analysed by using ImageJ (http://rsb.info.nih.gov/ij/).
Results
Identification and sequence analyses of TCP genes from P. equestris
To identify TCP TF coding genes, the HMM profile of the TCP domain (PF03634) was blast searched in the P. equestris genome (Cai et al., 2015). Full-length sequences of 23 TCP family genes were identified from the genome. All candidate TCP genes were confirmed to encode the conserved TCP domain with the use of InterproScan (Quevillon et al., 2005). The mean length of the 23 TCP TFs was 307 amino acids (range 220–425 amino acids). Other characteristics, including isoelectric point, molecular mass, and location at Phalaenopsis scaffolds are shown in Table 1.
Table 1.
TCP gene family in Phalaenopsis equestris
| Gene Name | Length (a.a.) | Molecular mass (Da) | pI | Scaffold location |
|---|---|---|---|---|
| PePCF1 | 253 | 26 846.3 | 5.71 | Scaffold000123:1543921~1544682 |
| PePCF2 | 289 | 30 740.3 | 7.96 | Scaffold000865:254675~255534 |
| PePCF3 | 333 | 34 736.9 | 9.51 | Scaffold000927:176~1177 |
| PePCF4 | 301 | 31 308.5 | 9.09 | Scaffold000053:108817~109722 |
| PePCF5 | 303 | 32 070.5 | 6.26 | Scaffold001019:11294~12205 |
| PePCF6 | 251 | 27 085.3 | 6.7 | Scaffold000382:232040~232795 |
| PePCF7 | 257 | 27 559.3 | 6.75 | Scaffold000552:2647726~2648558 |
| PePCF8 | 230 | 24 376.1 | 9.24 | Scaffold000002:70407263~70407955 |
| PePCF9 | 274 | 27 428.5 | 9.74 | Scaffold000002:66457383~66457997 |
| PePCF10 | 370 | 38 951.1 | 7.93 | Scaffold000058: 217817~218940 |
| PePCF11 | 274 | 29 655.9 | 8.75 | Scaffold000349:250533~251358 |
| PeCIN1 | 392 | 42 192 | 6.65 | Scaffold000552:3228304~3229491 |
| PeCIN2 | 372 | 40 543.5 | 9.06 | Scaffold000272:914770~915888 |
| PeCIN3 | 363 | 40 087.8 | 8.79 | Scaffold000113:35591~36682 |
| PeCIN4 | 277 | 30 765.9 | 7.79 | Scaffold000404:25049~25882 |
| PeCIN5 | 292 | 32 589.4 | 6.38 | Scaffold000873:1549183~1550061 |
| PeCIN6 | 425 | 44 992.4 | 9.89 | Scaffold000428:5478841~5480211 |
| PeCIN7 | 324 | 35 144.8 | 6.7 | Scaffold000198:252605~253579 |
| PeCIN8 | 220 | 24 505.2 | 6.21 | Scaffold000180:1361454~1362116 |
| PeCIN9 | 310 | 34 721.9 | 7.84 | Scaffold000408:191611~192543 |
| PeCYC1 | 288 | 32 774.3 | 8.94 | Scaffold000878:1078313~1079179 |
| PeCYC2 | 334 | 37 454.1 | 9.07 | Scaffold000428:1243571~1244575 |
| PeCYC3 | 326 | 36 663.6 | 9.06 | Scaffold000428:1253199~1254179 |
Sequence comparison and phylogenetic analysis
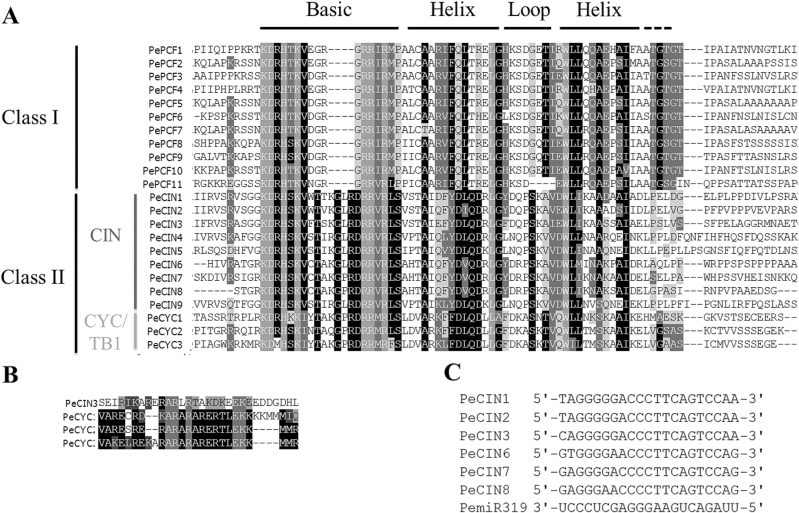
On multiple sequence alignment of the 23 PeTCP proteins, the sequences were found to encode a putative TCP-domain protein that contains a basic helix–loop–helix-type motif at the N-terminus (Supplementary Fig. S1). Orchid TCP proteins could be divided into two subfamilies, as for all species so far (Fig. 1A). The PCF or class I subfamily members showed an extended homology C-terminus from the TCP domain, and the class II subfamily had, as reported earlier, an extended four amino acids in the basic region. In addition, both subfamilies showed internally conserved but distinct loop region sequences (Cubas et al., 1999). According to this division, 11 genes belonged to the class I subfamily and 12 to the class II subfamily in the Phalaenopsis orchid genome. The result is also supported by the phylogenetic analysis (Fig. 2). In addition, the class II subfamily could be further divided into two subclades, class IIa or CYC/TB1 and class IIb or CIN (Fig. 2). The CYC/TB1 subclade contains the three orchid genes PeCYC1, PeCYC2, and PeCYC3 and CIN includes nine members, PeCIN1– PeCIN9 (Figs 1A and 2).
Fig. 1.
Multiple alignment of TCP protein sequences for Phalaenopsis equestris. (A) Alignment of the TCP domain and adjoining sequence for the predicted orchid TCP proteins. Overall conserved amino acids are in black. (B) Alignment of the R-domain of class II subfamily members. (C) Alignment of putative areas for miR319.
Fig. 2.
Phylogenetic relationships of TCP transcription factors from P. equestris, Arabidopsis and rice. The unrooted phylogenetic tree was created with MEGA 4.0 by the neighbor-joining method and the bootstrap test involved 1000 iterations. Blue, red and green lines indicate the PCF, CIN and CYC/TB1clades, respectively.
Previous reports showed that Arabidopsis class II CYC/TB1 genes AtTCP1, AtTCP12, and AtTCP18 as well as CIN genes AtTCP2 and AtTCP24 contain the R-domain at the C-terminus of the TCP domain (Yao et al., 2007). In Phalaenopsis, all three genes PeCYC1, PeCYC2, and PeCYC3 in CYC/TB1 and one gene, PeCIN3, in the CIN subclade encode proteins with the R-domain (Fig. 1B). In addition, in Arabidopsis, five of the CIN members are post-transcriptionally regulated by miRNA319 (AtTCP2, 3, 4, 10, and 24) (Palatnik et al., 2003, 2007; Ori et al., 2007). Six orchid TCP genes in the CIN subclade were identified to have a putative binding site for Phalaenopsis miR319 (Fig. 1C). This finding suggests the regulation of leaf development by a redundant set of miRNA-regulated homologous TCP genes in orchid.
To determine the phylogenetic relationships of PeTCP and other TCP genes, we reconstructed the phylogeny for the known TCP family using the conceptual amino acid sequences of the respective genes as input data. Phylogenetic analysis of the putative protein with TCP proteins from Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and Phalaenopsis (P. equestris) distinguished two types of TCP proteins based on differences within their TCP domains: class I and class II (Figs 1A and 2). In addition, PeCIN5 and PeCIN9 as well as PeCYC1, PeCYC2, and PeCYC3 may be duplicated genes in orchid (Fig. 2). Pairwise alignment analysis showed that PeCIN5 and PeCIN9 obtain the lowest E-value, 2e–51, compared with those of PeCIN5 or PeCIN9 to other PeCIN proteins. Indeed, PeCYC2 and PeCYC3 are located in the same assembled scaffold (Table 1).
Expression profiles of TCP genes in P. equestris
We determined the spatial and temporal expression profiles of TCP genes in P. equestris of different floral organs (sepal, petal, labellum/lip, and gynostemium/column) (Fig. 3A), various developmental stages of floral buds (B1, 2, 3, and 4) (Fig. 3B; Tsai et al., 2004), pedicel, leaf, floral stalk and root, and different developmental stages of ovule after pollination (Fig. 3C; Chen et al., 2012) by real-time RT-PCR analysis (Fig. 4 and Supplementary Figs S1–S8). The expression patterns of 11 PePCF-like and nine PeCIN-like genes are shown in Fig. 3. The expression in different tissues varied widely among orchid TCP genes and among different organs for individual TCP genes. The results suggested the functional divergence of orchid genes during plant developmental processes.
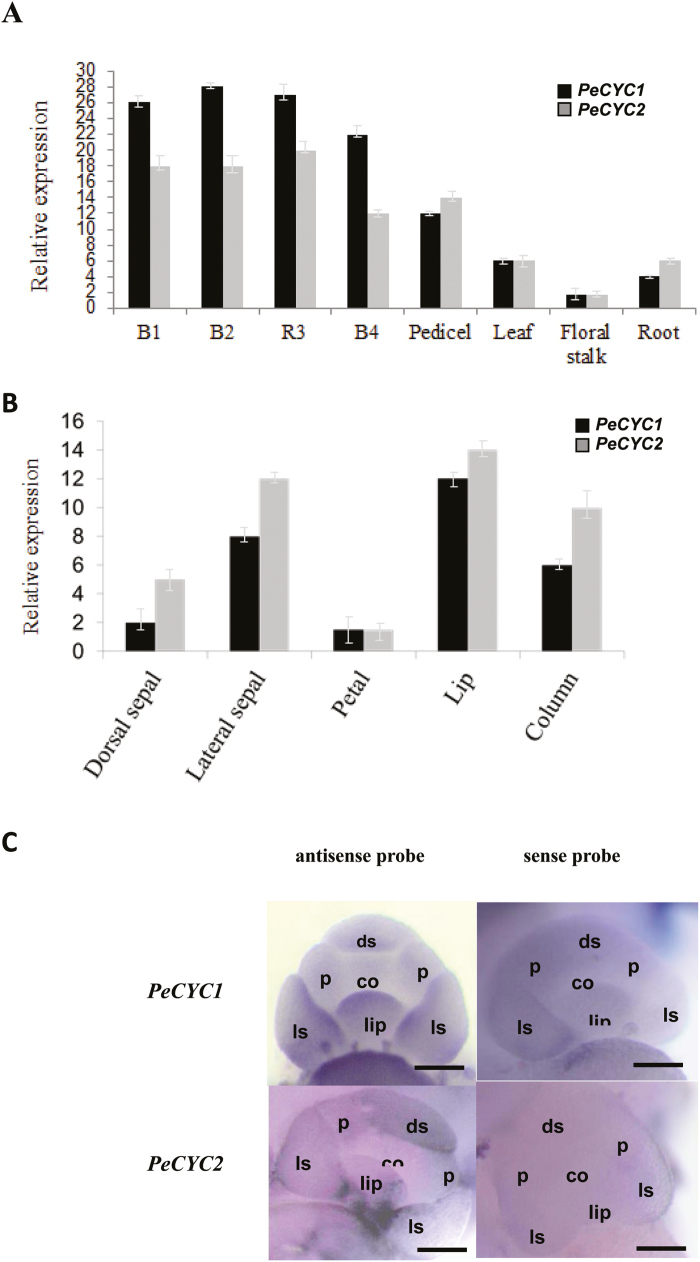
Fig. 4.
Spatial and temporal expression patterns of PeCYCs genes. (A) Real-time quantitative RT-PCR (qRT-PCR) analysis of the mRNA expression of PeCYC1 and PeCYC2 in various tissues and developmental stages of floral bud. (B) qRT-PCR analysis of the mRNA expression of PeCYC1 and PeCYC2 in different floral organs. (C) Whole mount in situ hybridization of PeCYC1 and PeCYC2 in early floral development stage. Upper left: antisense probe of PeCYC1; upper right: sense probe of PeCYC1; lower left: antisense probe of PeCYC2; lower right: sense probe of PeCYC2. co: column primordium; ds: dorsal sepal primordium; ls: lateral sepal primordium; p: petal primordium. Scale bar: 100 μm.
Most of the transcripts of PePCF-like genes could be detected in vegetative and reproductive tissues. However, PePCF6 was not expressed in any tissues, and the expression of PePCF11 was not detected in developing ovules (Fig. 3D, E and Supplementary Fig. S9). Many PeCIN-like genes showed wide expression in floral organs, developing floral bud, pedicel, leaf, floral stalk, and root (Fig. 3D and Supplementary Fig. S9A). Four genes (PeCIN2, 5, 7, and 8) expressed in developing ovule, especially in the late developmental stage of ovule (40–64 days after pollination [DAP]) (Fig. 3E and Supplementary Fig. S9B). However, the expression of PeCIN6 was not detected in any tissue (Fig. 3D, E and Supplementary Fig. S9), but PeCIN9 specifically expressed in leaf (Fig. 3D and Supplementary Fig. S9A).
As compared with the four PeCIN-like genes, nine PePCF genes were expressed in developing ovule (Fig. 3E and Supplementary Fig. S9B). This may imply that PePCF-like genes play more important roles in ovule differentiation and growth. Of note, PePCF10 predominantly expressed in ovules at early developmental stages (0–16 DAP) and PeCIN8 showed high expression at late developmental stages in ovule (16–64 DAP) (Fig. 3E and Supplementary Fig. S9B). Transcripts of PePCF6 and PeCIN6 were not detected in any organs, so they may be primarily expressed in other organs not tested or under special conditions (Fig. 3D, E and Supplementary Fig. S9).
PeCYC genes were predominantly expressed in developing floral buds
Expression profiles of three PeCYCs (PeCYC1, PeCYC2, and PeCYC3) were also analysed by real-time RT-PCR. Results showed that both PeCYC1 and PeCYC2 were predominantly expressed in developing floral buds (Fig. 4A). However, the expression of PeCYC3 was not detected. Expression analysis of PeCYC1 and PeCYC2 was further conducted in dissected floral organs (including the dorsal/lateral sepal, petal, lip, and column). Both PeCYC1 and PeCYC2 were strongly expressed in the lateral sepal, lip, and column (Fig. 4B). The detailed spatial expression patterns of PeCYC1 and PeCYC2 at the early stage of floral development were further investigated by whole mount in situ hybridization by antisense RNA probes. Results showed transcripts of PeCYC1 were detected at the primordia of lateral sepal and lip (Fig. 4C, upper left panel). However, transcripts of PeCYC2 were concentrated at the dorsal parts of the lateral sepal and lip primordia (Fig. 4C, lower left panel). Expression of PeCYCs at the floral dorsal region might correlate with orchid floral zygomorphic development.
Nuclear localization of class I PePCF10 and class II PeCIN8 proteins
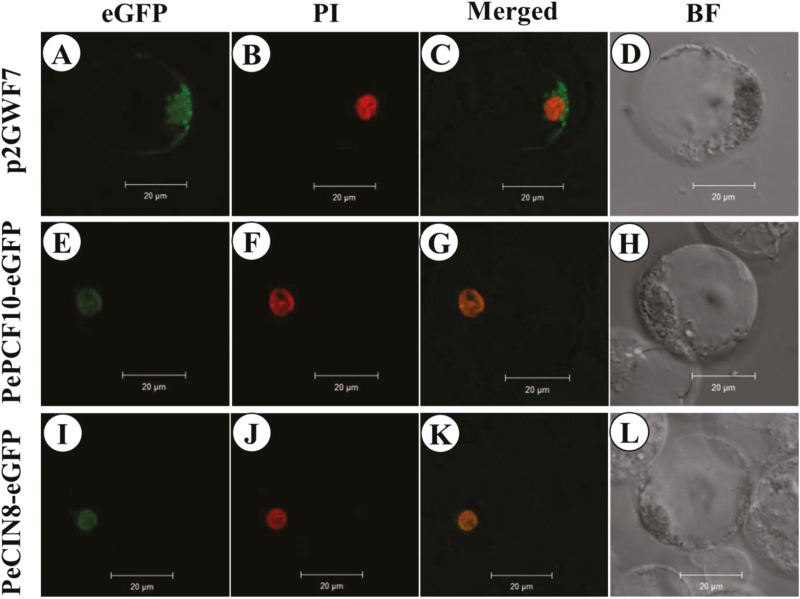
Orchid ovule development is precisely initiated by pollination. Because of the successive expression of PePCF10 and PeCIN8 during ovule development, we further characterized these two genes. To examine the subcellular localization of the two proteins, PePCF10–GFP and PeCIN8–GFP fusion proteins were constructed, respectively. Both PePCF10–GFP and PeCIN8–GFP fusion proteins were detected predominantly in nuclei derived from Phalaenopsis petal protoplasts (Fig. 5). The results are consistent with sequence-based prediction of the subcellular localization and putative transcription factor activity of PePCF10 and PeCIN8.
Fig. 5.
Localization patterns of PePCF10–GFP and PeCIN8–GFP fusion proteins in P. aphrodite petal protoplasts. Pictures show fluorescence and bright-field confocal microscopy images. Images were merged with use of Axio Vision Rel.4.8 software. (A) Empty vector green fluorescence in the cytoplasm and nucleus of a flower cell. (B) Cell in (A) stained with propidium iodide (PI; red) to show the nucleus. (C) Merged image of (A) and (B) to show green fluorescence both in the cytoplasm and the nucleus of a flower cell. (D) Cell in (A), (B) and (C) visualized by bright-field confocal microscopy. (E) PePCF10–GFP green fluorescence in the nucleus of a flower cell. (F) Cell in (E) stained with PI (red) to show the nucleus. (G) Merged image of (E) and (F) to show the nuclear localization in a flower cell. (H) Cell in (E), (F) and (G) visualized by bright-field confocal microscopy. (I) PeCIN8–GFP green fluorescence in the nucleus of a flower cell. (J) Cell in (I) stained with PI (red) to show the nucleus. (K) Merged image of (I) and (J) to show the nuclear localization in a flower cell. (L) Cell in (I), (J) and (K) visualized by bright-field confocal microscopy. Scale bars: 20 μm.
Interaction behaviors of class I PePCF10 and class II PeCIN8 proteins
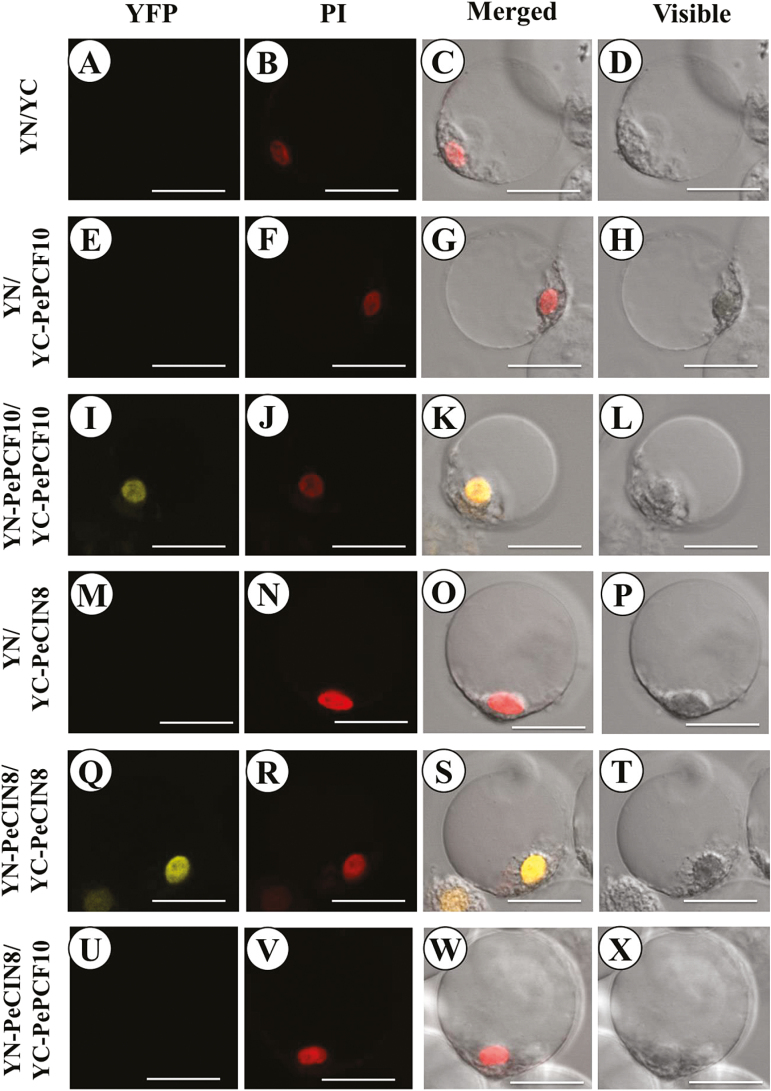
Several studies have provided evidence that TCP TFs can bind DNA as homo- or heterodimers. So far TCPs have been found to form heterodimers only between specific members of the same class (Kosugi and Ohashi, 2002; Viola et al., 2011; Danisman et al., 2012; Yang et al., 2012; Aguilar-Martinez and Sinha, 2013). To investigate the protein–protein interactions between PePCF10 and PeCIN8 proteins, we used bimolecular fluorescence complementation (BiFC) assay. The constructs were co-transfected and transiently expressed in the petal protoplasts of Phalaenopsis. Interaction fluorescence signals were observed in YN–PePCF10 and YC–PePCF10 (Fig. 6I, J, K, L) as well as YN–PeCIN8 and YC–PeCIN8 (Fig. 6Q, R, S, T), which suggests that both PePCF10 and PeCIN8 can form homodimers. In addition, the signals were localized in the nucleus, as indicated by the nuclear dye propidium iodide (PI). However, interaction was not observed with the combination of YN–PeCIN8 and YC–PCF10 (Fig. 6U, V, W, X). Therefore, PePCF10 and PeCIN8 may not form heterodimers. No fluorescence was detected with the empty vector control (Fig. 6A, B, C, D) and no yellow fluorescence was detected when only the C-terminal half of the YFP was co-bombarded with PePCF10 or PeCIN8 constructs (Fig. 6E, F, G, H, M, N, O, P). Yeast two-hybrid experiments were also performed to analyse the behavior of PePCF10 and PeCIN8. The results were similar to those of the BiFC assay, except that PePCF10 homodimer was not observed (Supplementary Fig. S10). It is possible that interaction between PePCF10 proteins was blocked by the steric hindrance of fused PePCF10–GAL4 protein. We also tested interactions between PePCF10 and 23 Arabidopsis TCPs systematically by yeast two-hybrid assay. Results showed that PePCF10 is considered to interact with many TCP proteins which belong to the PCF, CYC/TB1, or CIN clade (Supplementary Fig. S11), suggesting that PePCF10 may have broad specificity in terms of its interacting partners. Arabidopsis TCP14, which is closest to PePCF10, appears to interact with PePCF10 strongly, supporting the above-described BiFC data in which PePCF10 forms a homodimer. However, Arabidopsis TCP3, TCP4, and TCP10, which are close to PeCIN8, also showed interaction with PePCF10 (Supplementary Fig. S11). These data suggest that the specificity of protein–protein interaction of TCP could be somewhat different between Arabidopsis and orchid.
Fig. 6.
Bimolecular fluorescence complementation (BiFC) visualization of TCP dimerization in transiently transfected P. aphrodite petal protoplasts. (A–D) Co-expression of non-fused YN with non-fused YC was unable to reconstitute a fluorescent YFP chromophore. (E–H) Co-expression of non-fused YN with YC-PePCF10 was also unable to reconstitute a fluorescent YFP chromophore. (I–L) PeTCP10 interacts with PeTCP10 protein in nucleus of flower cell. (M–P) Co-expression of non-fused YN with YC-PeCIN8 was also unable to reconstitute a fluorescent YFP chromophore. (Q–T) PeCIN8 interacts with PeCIN8 protein in nucleus of flower cell. (U–X) Co-expression of YN-PeCIN8 with YC-PePCF10 was unable to reconstitute a fluorescent YFP chromophore. The fluorescence indicates interaction between the indicated partner proteins. The images were obtained from the yellow fluorescent protein channel or bright field or are a merged image of the yellow fluorescence, and cells stained with PI represented in red. Scale bars: 20 μm.
Functional characterization of the two PeTCP genes in transgenic Arabidopsis
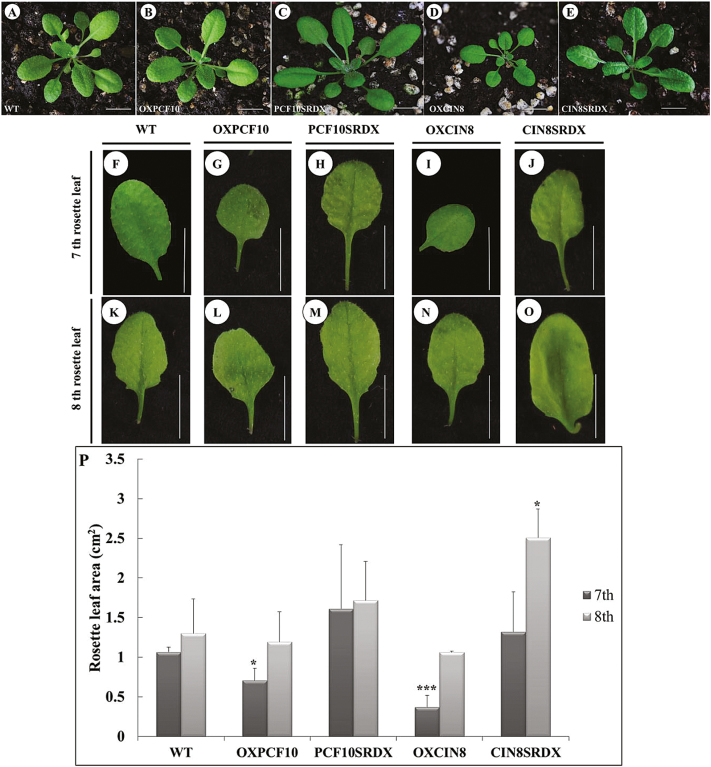
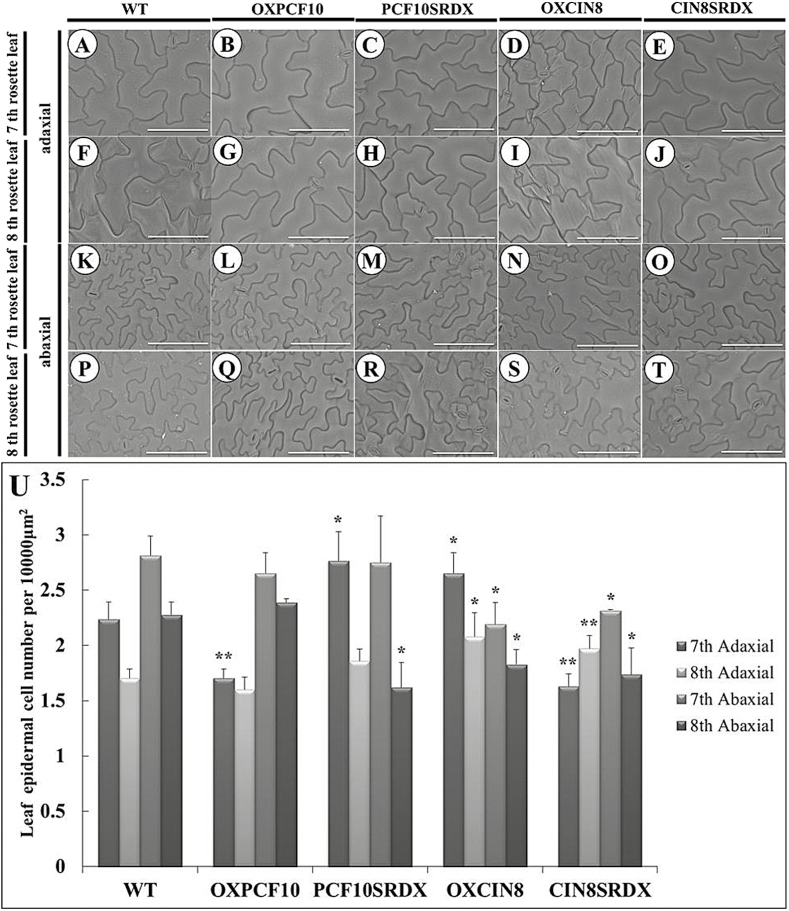
We constructed transgenic Arabidopsis plants expressing PePCF10 (OXPCF10) under the control of the CaMV 35S promoter via Agrobacterium-mediated transformation. Twenty-five of the 100 independently transformed OXPCF10 transgenic T1 lines showed kanamycin resistance and a similar phenotype. Among 25 T2 lines, 12 showed a 3:1 segregating kanamycin resistance phenotype. The rosette size of OXPCF10 plants was normal (Fig. 7B), and we chose the seventh and eighth rosette leaf for observation. OXPCF10 plants showed a round-leaf phenotype (Fig. 7G, L). The leaf area of the seventh rosette leaf was smaller for OXPCF10 than wild-type (WT) plants (Fig. 7P). A previous study indicated that Arabidopsis leaf size regulation is mainly controlled by cell number and size (Tsukaya, 2006). To assess the contributions of cell number and size to the reduced leaf size of the seventh rosette leaf in OXPCF10 plants, we compared the seventh and eighth rosette leaves of WT and transgenic plants by scanning electron microscopy. For seventh rosette leaves, the adaxial epidermal cell size was larger for OXPCF10 than WT plants, and the adaxial epidermal cell number was significantly reduced (Fig. 8A, B, U). For the eighth rosette leaves, the adaxial epidermal cell size and cell number did not differ between WT and transgenic plants (Fig. 8F, G, U); the abaxial epidermal cell size and cell number of seventh rosette leaves (Fig. 8K, L, U) and eighth rosette leaves did not differ (Fig. 8P, Q, U). The altered leaf size and round leaves of OXPCF10 plants were related to decreased adaxial epidermal cell number and increased cell size.
Fig. 7.
Rosette leaves of wild-type (WT) and transgenic Arabidopsis plants. (A–E) rosette of WT (A), OXPCF10 (B), PCF10SRDX (C), OXCIN8 (D) and CIN8SRDX (E). (F–J) Seventh rosette leaf from WT (F), OXPCF10 (G), PCF10SRDX (H), OXCIN8 (I) and CIN8SRDX (J). (K–O) Eighth rosette leaf from WT (K), OXPCF10 (L), PCF10SRDX (M), OXCIN8 (N) and CIN8SRDX (O). (P) Seventh and eighth rosette leaf area of WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX. Asterisks indicate statistically significant differences (*P<0.05, ***P<0.001 compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats. Scale bars: 1cm.
Fig. 8.
Scanning electron micrographs of leaf epidermal cells of WT and transgenic Arabidopsis plants. (A–E) Adaxial epidermis in the seventh rosette leaf from WT (A), OXPCF10 (B), PCF10SRDX (C), OXCIN8 (D) and CIN8SRDX (E). (F–J) Adaxial epidermis in the eighth rosette leaf from WT (F), OXPCF10 (G), PCF10SRDX (H), OXCIN8 (I) and CIN8SRDX (J). (K–O) Abaxial epidermis in the seventh rosette leaf from WT (K), OXPCF10 (L), PCF10SRDX (M), OXCIN8 (N) and CIN8SRDX (O). (P–T) Abaxial epidermis in the eighth rosette leaf from WT (P), OXPCF10 (Q), PCF10SRDX (R), OXCIN8 (S) and CIN8SRDX (T). (U) Seventh and eighth rosette leaf epidermal cell number for WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX. Asterisks indicate statistically significant differences (*P<0.05, **P<0.01, compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats (n=200 each). Scale bars: 100 μm.
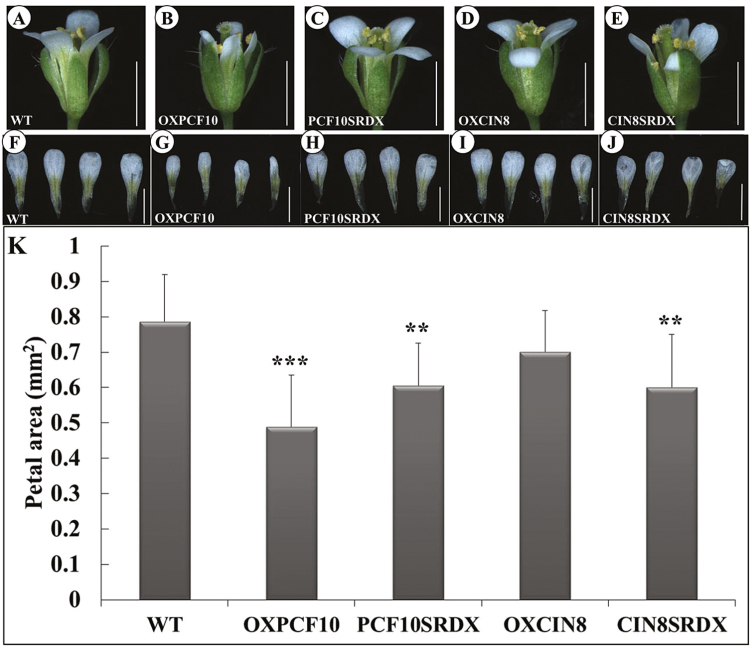
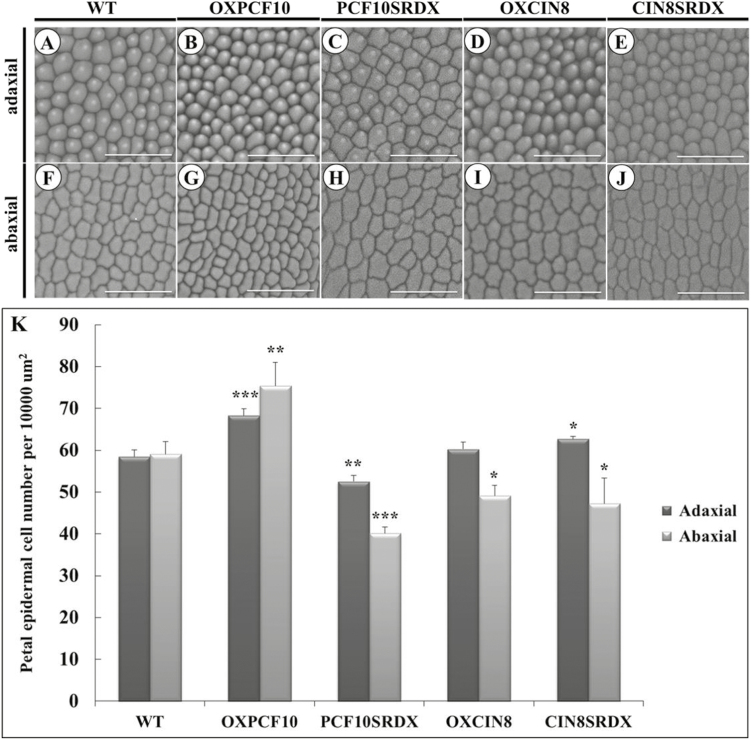
The flowers were smaller and less expanded for OXPCF10 than WT plants (Fig. 9A, B). Flower petals were smaller (Fig. 9F, G, K) and the adaxial petal epidermis contained cells with reduced size (Fig. 10A, B) but increased number (Fig. 10K). A similar phenotype was observed for abaxial epidermal cells (Fig. 10F, G, K). The altered petal size of OXPCF10 plants was related to the reduced epidermal cell size and increased epidermal cell number.
Fig. 9.
Flowers of WT and transgenic Arabidopsis plants. (A–E) Side views of WT (A), OXPCF10 (B), PCF10SRDX (C), OXCIN8 (D) and CIN8SRDX (E). (F–J) Petals of WT (F), OXPCF10 (G), PCF10SRDX (H), OXCIN8 (I) and CIN8SRDX (J). (K) Petal area of WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX. Asterisks indicate statistically significant differences (**P<0.01, ***P<0.001, compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats (n=10 each). Scale bars: 1mm.
Fig. 10.
Scanning electron micrographs of epidermal cells of WT and transgenic Arabidopsis petals. (A–E) Adaxial epidermal cells of WT (A), OXPCF10 (B), PCF10SRDX (C), OXCIN8 (D) and CIN8SRDX (E). (F–J) Abaxial epidermal cells of WT (F), OXPCF10 (G), PCF10SRDX (H), OXCIN8 (I) and CIN8SRDX (J). (K) Epidermal cell number of WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX. Asterisks indicate statistically significant differences (*P<0.05, **P<0.01, ***P<0.001, compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats (n=200 each). Scale bars: 50 μm.
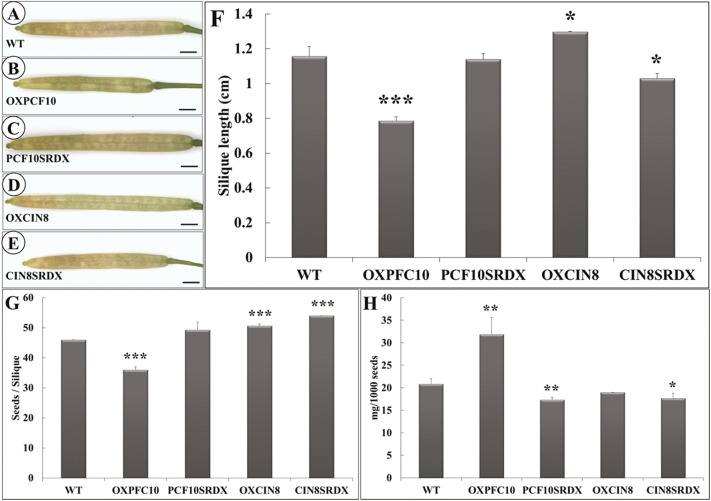
Siliques were shorter for OXPCF10 than WT plants (Fig. 11A, B, F). The total seed number per silique of OXPCF10 plants was significantly reduced (Fig. 11G). Big reduction of seeds observed in the overexpressed plants might relate to the abnormal gametogenesis (Takeda et al., 2006; Sarvepalli and Nath, 2011; Viola et al., 2011). The seed size was larger for OXPCF10 than WT plants (data not shown) and the seed weight was significantly increased (Fig. 11H). The altered silique length and seed size and weight of OXPCF10 plants suggested that PePCF10 may be involved in reproductive development.
Fig. 11.
Siliques and seeds of WT and transgenic Arabidopsis plants. (A–E) Siliques of WT (A), OXPCF10 (B), PCF10SRDX (C), OXCIN8 (D) and CIN8SRDX (E). (F) Silique length of WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX. Asterisks indicate statistically significant differences (*P<0.05, ***P<0.001, compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats (n=20 each). (G) Seeds per silique of WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX. Asterisks indicate statistically significant differences (***P<0.001, compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats (n=20 each). (H) Weight (mg) of 1000 seeds of WT, OXPCF10, PCF10SRDX, OXCIN8 and CIN8SRDX plants. Asterisks indicate statistically significant differences (*P<0.05, **P<0.01 compared with WT by Student’s t-test). Errors bar represent the SD of three biological repeats. Scale bars: 1mm.
In addition to overexpression of PePCF10 in Arabidopsis, we converted PePCF10 to a chimeric repressor by fusing it with the SRDX repression domain (PCF10SRDX). Twenty-six of the 100 independently transformed PCF10SRDX transgenic T1 lines showed kanamycin resistance and a similar phenotype. Among 26 T2 lines, 13 showed a 3:1 segregating kanamycin resistance phenotype. The rosette size of PCF10SRDX plants was reduced and leaves showed a wrinkled surface and downward curl (Fig. 7C). The leaf area of the seventh and eighth rosette leaf did not differ between WT and transgenic plants, but transgenic lines showed slightly enlarged leaves (Fig. 7F, H, K, M, P). Similar phenotypes were reported in mutant and transgenic lines, with altered proliferation of leaf blade cells (Serrano-Cartagena et al., 2000; Bemis and Torii, 2007; Kieffer et al., 2011). For seventh rosette leaves, the adaxial epidermis contained cells with reduced size but increased number in PCF10SRDX plants (Fig. 8A, C, U). For eighth rosette leaves, the adaxial epidermal cell size and cell number did not differ between WT and transgenic plants (Fig. 8F, H, U) and the abaxial epidermal cell size and cell number of seventh rosette leaves did not differ (Fig. 8K, M, U). The abaxial epidermal cell size of the eighth rosette leaves was larger for transgenic than WT plants and the cell number was less (Fig. 8P, R, U). Although the leaf area was not altered in PCF10SRDX plants (Fig. 7P), the adaxial epidermal cell size was smaller and cell number greater in the seventh rosette leaf and the abaxial epidermal cell size was larger and cell number lower in the eighth rosette leaf. PePCF10 may regulate cell proliferation and differentiation and thus generate a wrinkled surface and downwardly curled rosette leaves.
The morphology of the flowers did not differ between PCF10SRDX and WT plants (Fig. 9A, C). However, petals were smaller for PCF10SRDX than WT flowers (Fig. 9H, K). The adaxial epidermal cell size of petals did not differ (Fig. 10A, C), but the adaxial epidermal cell number was lower (Fig. 10K). In addition, the abaxial epidermal cell size of petals was larger for PCF10SRDX than WT plants (Fig. 10F, H) and the abaxial epidermal cell number was less (Fig. 10K). The silique length, total seed number per silique, and seed size did not differ between the WT and PCF10SRDX plants (Fig. 11A, C, F, G), but the seed weight of PCF10SRDX plants was significantly decreased (Fig. 11H). These results suggest a function for PePCF10 in cell proliferation in vegetative and reproductive tissues.
The same strategy was adopted to functionally characterize PeCIN8. Twenty-four of the 100 independently transformed OXCIN8 transgenic T1 lines showed kanamycin resistance and a similar phenotype. Among 24 T2 lines, 11 showed a 3:1 segregating kanamycin resistance phenotype. The rosette size of OXCIN8 plants was reduced (Fig. 7D). The rosette leaves showed a round-leaf phenotype (Fig. 7I, N). The leaf area of the seventh rosette leaf was smaller compared with the WT plants. The leaf area of the eighth rosette leaf did not differ when compared with WT (Fig. 7P). Similar phenotypic effects were observed on transgenic lines with altered proliferation of leaf blade cells (Schommer et al., 2008; Sarvepalli and Nath, 2011). The adaxial epidermal cell size of the seventh and eighth rosette leaf was smaller for OXCIN8 than WT plants (Fig. 8A, D, F, I) and the adaxial epidermal cell number was greater (Fig. 8U). The abaxial epidermal cells of the seventh and eighth rosette leaf of OXCIN8 plants showed the opposite pattern. The abaxial epidermal cell size of the seventh and eighth rosette leaf was larger for OXCIN8 than WT plants (Fig. 8K, N, P, S) and the abaxial epidermal cell number was less (Fig. 8U). The altered leaf size and round leaves of OXCIN8 plants were related to changed cell number and cell size in both the adaxial and abaxial sides of the leaf.
The adaxial epidermal cell size and cell number did not differ between OXCIN8 and WT petals (Fig. 10A, D, K). However, the abaxial epidermal cell size was larger for OXCIN8 than WT (Fig. 10F, I) and the abaxial epidermal cell number was less (Fig. 10K). Although the petal area was not modified in OXCIN8 plants (Fig. 9K), the abaxial epidermal cell size was larger and cell number lower than those of WT petals. The siliques were longer for OXCIN8 than WT plants (Fig. 11A, D, F). The total seed number per silique was increased in OXCIN8 plants (Fig. 11G). However, the seed size did not differ between OXCIN8 and WT seeds and seed weight did not differ (Fig. 11H).
PeCIN8 fused with SRDX-overexpressed transgenic plants (CIN8SRDX) were also produced. Twenty-five of the 100 independently transformed CIN8SRDX transgenic T1 lines showed kanamycin resistance and a similar phenotype. Among 25 T2 lines, 12 showed a 3:1 segregating kanamycin resistance phenotype. The rosette size of CIN8SRDX plants was normal (Fig. 7E). However, the rosette leaves showed a wrinkled surface and a downward curl (Fig. 7E, J, O). The leaf area of the seventh rosette leaf seemed slightly larger for CIN8SRDX than WT plants; however, the eighth rosette leaf area was significantly larger (Fig. 7P). For seventh rosette leaves, the adaxial epidermal cell size was larger for CIN8SRDX than WT plants, and the adaxial epidermal cell number was smaller (Fig. 8A, E, U). For eighth rosette leaves, the adaxial epidermal cell size seemed smaller for CIN8SRDX than WT plants and the adaxial epidermal cell number was higher (Fig. 8F, J, U). For both seventh and eighth rosette leaves, the abaxial epidermal cell size was larger for CIN8SRDX than WT plants and the abaxial epidermal cell number was lower (Fig. 8K, O, P, T, U). These results suggest that the wrinkled surface and downwardly curled rosette leaves of CIN8SRDX plants were due to increased adaxial epidermal cell number and abaxial epidermal cell size.
The flowers of CIN8SRDX plants were less expanded (Fig. 9E) and significantly smaller than those of WT plants (Fig. 9F, J, K). The adaxial epidermal cell size seemed smaller for CIN8SRDX than WT petals (Fig. 10A, E) and the adaxial epidermal cell number was significantly increased (Fig. 10K). The abaxial epidermal cells were larger for CIN8SRDX than WT petals because of elongated cells (Fig. 10F, J), and the abaxial epidermal cell number was significantly decreased (Fig. 10K). The siliques were shorter for CIN8SRDX than WT plants (Fig. 11A, E, F) and the total seed number per silique was significantly increased (Fig. 11G). The seed weight was greater for CIN8SRDX than WT plants (Fig. 11H).
Discussion
In the present study, we identified 23 TCP genes from P. equestris. The number of TCP genes in P. equestris was similar to that in Arabidopsis (24) and rice (22) (Martín-Trillo and Cubas, 2010). As well, the gene number in each subfamily of TCP genes was similar among the three species (Fig. 2). Although the genome size of Phalaenopsis is larger than that of Arabidopsis and rice—approximately 9 and 2.8 times, respectively—the number of genes among the genomes is similar (Cai et al., 2015). This fact suggests that the TCP family did not expand along with genome expansion of P. equestris during orchid evolution (Cai et al., 2015). TCP genes in orchids, like those Arabidopsis, rice and other plants, may participate in regulating multiple aspects of plant development such as gametophyte development (Takeda et al., 2006; Sarvepalli and Nath, 2011), hormone signal transduction (Aguilar-Martinez et al., 2007; Guo et al., 2010; Yanai et al., 2011), mitochondria biogenesis (Hammani et al., 2011), regulation of circadian clock (Pruneda-Paz et al., 2009; Giraud et al., 2010), lateral branching (Takeda et al., 2003; Aguilar-Martinez et al., 2007), flower development (Koyama et al., 2011; Uberti-Manassero et al., 2012), seed germination (Tatematsu et al., 2008; Rueda-Romero et al., 2012), and leaf development (Kieffer et al., 2011). Actually, the expression of TCP genes was broadly detected in various vegetative, reproductive tissues and different developmental stages of Phalaenopsis orchid (Fig. 3).
In most orchid flowers, ovary and ovule development is precisely triggered by pollination (Tsai et al., 2008). Auxin is a positive regulator for the orchid ovule initiation (Tsai et al., 2005; Novak et al., 2014). We found PePCF10 predominantly expressed in ovules at early developmental stages (0–16 DAP) and PeCIN8 showed high expression at late developmental stages in ovules (16–64 DAP) (Fig. 3E and Supplementary Fig. S9B), with overlapping expression at 16 DAP. At about 16 DAP, when PePCF10 and PeCIN8 are co-expressed, placental protuberances develop from a single epidermal layer of the placenta (Chen et al., 2012). Archesporial cells are differentiated at the terminus of the nucellar filament. Later, periclinal divisions in epidermal cells surrounding the archesporial cells generate the inner integument (Arditti and Pridgeon, 1997). Thus, the function of the homodimer TF PePCF10 might be related to placenta growth and continued initiation of archesporial cells and inner integument. In addition, a homodimer of PeCIN8 might associate with the later developmental process of megagametogenesis. Further study of the relationship between PePCF10 and PeCIN8 will allow us to gain new insights into the regulation of the unique system of orchid ovule development.
Functional analysis by overexpressing PePCF10 or a chimeric repressor gene in Arabidopsis suggested that PePCF10 plays a role in regulating epidermal cell proliferation and differentiation. Previous study suggested that TCP proteins could be dual-function transcription factors able to positively or negatively regulate expression of target genes (Herve et al., 2009; Wang et al., 2013). Although the seventh and eighth rosette leaf areas of PCF10SRDX lines slightly differed from those of the WT, the epidermal cell number was increased or decreased in PCF10SRDX lines depending on the different epidermal cell axis. PePCF10 may act as a dual functional TF in regulating cell proliferation in the developing leaf, and the distinctive activities of PePCF10 may depend on its interaction with other proteins in the adaxial or abaxial side of the leaf. Our results are consistent with the function of TCP proteins being dependent on the organ, tissue, or cellular context (Herve et al., 2009).
The class II TCP gene CIN was reported to act as a repressor of cell proliferation in leaves. High levels of miR319 or low miR319-targeted TCP activity might cause excess cell proliferation, thus resulting in a crinkled leaf in Arabidopsis, snapdragon, and tomato or larger leaves in monocotyledonous plants (rice and creeping bentgrass) (Nath et al., 2003; Palatnik et al., 2003; Ori et al., 2007; Yang et al., 2013). In this study, we found that the expression of PeCIN8 was broadly distributed in orchid vegetative and reproductive tissues (Fig. 3). Overexpressed PeCIN8 or Arabidopsis with the chimeric repressor gene showed a varied leaf morphology associated with altered cell number and cell size of epidermal cells. In addition, the function of PeCIN8 was exhibited by the petal size. Furthermore, the epidermal cell number in the affected organs of transgenic plants was increased or decreased depending on different epidermal cell axis, so PeCIN8 may act as a dual-function TF in regulating cell proliferation in the developing leaf or petal.
Previous studies have reported that Arabidopsis TCP genes including TCP2, TCP3, TCP4, TCP10, and TCP24 possess miR319-binding sites. The cleavage activity of miR319 on these AtTCP transcripts has been demonstrated (Nag et al., 2009; Palatnik et al., 2003). A similar finding was later shown in tomato LANCEOLATE (LA, SlTCP2), the closest homolog of AtTCP4 (Ori et al., 2007). Although PeCIN8 was predicted to possess an miR319 targeting site, it seems that cleavage of PeCIN8 transcripts did not occur in the PeCIN8-overexpressing Arabidopsis. Koyama et al. (2007) reported that the ectopic expression of AtTCP3 under the control of CaMV 35S promoter (35S:AtTCP3) did not show visible abnormalities compared with Arabidopsis wild-type. However, overexpression of a mutant form of AtTCP3 without the miR319-binding sequence (35S:mAtTCP3) induced fusion of cotyledons, defects in the formation of shoots and elongation of hypocotyls in Arabidopsis. Similar results were found when miR319-resistant or -sensitive versions of AtTCP2 and AtTCP4 were expressed, suggesting the phenotype differences are likely due to the presence of miR319 (Palatnik et al., 2003, 2007). In this study, overexpression of wild-type form of PeCIN8 induces small rosette size and rounder leaves, indicating a higher level of PeCIN8 transcripts in the transgenic Arabidopsis plants. These data, which contradict previous results, might be due to no cleavage of the PeCIN8 transcripts occurring or a lower sensitivity of PeCIN8 transcripts to Arabidopsis miR319 regulation. It was reported that only one point mutation at the miR319-binding site of tomato LA alleles could confer partial resistance of LA transcripts to miR319 cleavage activity (Ori et al., 2007). This dominant mutation of LA in tomato resulted in small simple leaves instead of the large compound ones of wild type. In addition, the AtTCP4 hyper-activation lines that expressed 35S:AtTCP4:VP16 under Arabidopsis Col-0 background show much smaller leaf sizes and rounder leaves (Sarvepalli and Nath, 2011). This suggests that the regulation of TCP genes for plant development depends on the miR319 level or the ratio of TCP transcripts and miR319. Furthermore, we could not exclude the possibility that the post-transcriptional regulation of CIN-type TCP genes is different between Arabidopsis and P. equestris.
In conclusion, we identified 23 members of the orchid TCP TF gene family. These genes have broad expression profiles corresponding to multiple aspects of growth and development. Orchid PePCF10 and PeCIN8 showed successive expression patterns during ovule development. Additionally, both proteins could form homodimers and localize in the nucleus. Functional analysis suggested that both genes might regulate cell proliferation. These two genes may contribute to the unique functions of TCP TFs in orchids, controlling ovule initiation and development.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Figure S1. The expression patterns of PePCF genes in various organs and developmental stages of floral bud of Phalaenopsis equestris by real-time RT-PCR analysis. Quantification was normalized to Actin4.
Figure S2. The expression patterns of PePCF genes in various developmental stages of ovule of P. equestris by real-time RT-PCR analysis. Quantification was normalized to Actin4.
Figure S3. The expression patterns of PeCIN genes in various organs and developmental stages of floral bud of P. equestris by real-time RT-PCR analysis with use of PeActin4 as internal control.
Figure S4. The expression patterns of PeCIN genes in various developmental stages of ovule of P. equestris by real-time RT-PCR analysis with use of PeActin4 as internal control.
Figure S5. The expression patterns of PePCF genes in various organs and developmental stages of floral bud of P. equestris by real-time RT-PCR analysis with use of Pa18S rRNA as internal control.
Figure S6. The expression patterns of PePCF genes in various developmental stages of ovule of P. equestris by real-time RT-PCR analysis with use of Pa18S rRNA as internal control.
Figure S7. The expression patterns of PeCIN genes in various organs and developmental stages of floral bud of P. equestris by real-time analysis with use of Pa18S rRNA as internal control.
Figure S8. The expression patterns of PeCIN genes in various developmental stages of ovule of P. equestris by real-time RT-PCR analysis with use of Pa18S rRNA as internal control.
Figure S9. Heat map representation for the expression profiles of PePCF and PeCIN in various organs and developmental stages of floral bud (A), and in various developmental stages of ovule (B).
Figure S10. Analysis of protein interactions between PePCF10 and PeCIN8 by quantitative β-galactosidase activity assays.
Figure S11. Result of yeast two-hybrid assay between PePCF10 and Arabidopsis TCPs (A). Matrix describing experimental design shown in (B) and (C).
Acknowledgements
We thank Dr Tzyy-Jen Chiou (Agricultural Biotechnology Research Center, Academia Sinica, Taipei, Taiwan) for vector construction of BiFC assay. We also thank Dr Tzong-Yueh Chen (Department of Biotechnology and Bioindustry Sciences, National Cheng Kung University, Tainan, Taiwan) for assisting real-time RT-PCR experiment. This work was supported by the Ministry of Science and Technology, Taiwan [grants NSC 99-2311-B-006-004-MY3 and MOST 103-2313-B-006-001-MY3].
Glossary
Abbreviations:
- BiFC
bimolecular fluorescence complementation
- CaMV
cauliflower mosaic virus
- CYC
CYCLOIDEA
- DAP
days after pollination
- GFP
green florescent protein
- HMM
hidden Markov model
- ORF
open reading frame
- OX
overexpressing
- PCF
proliferating cell factor
- TB1
TEOSINTE BRANCHED1
- TCP
TEOSINTE-BRANCHED/CYCLOIDEA/PCF
- TF
transcription factor
- PI
propidium iodide
- RT-PCR
reverse transcription–PCR
- WT
wild-type
- YC
C-terminal half of YFP
- YFP
yellow florescent protein
- YN
N-terminal half of YFP.
References
- Aguilar-Martinez JA, Poza-Carrion C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant Cell 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez JA, Sinha N. 2013. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Frontiers in Plant Science 4, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arditti J, Pridgeon AM. 1997. Orchid biology: Reviews and perspectives, VII. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Balsemão-Pires E, Andrade LR, Sachetto-Martins G. 2013. Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiology and Biochemistry 67, 120–125. [DOI] [PubMed] [Google Scholar]
- Bemis SM, Torii KU. 2007. Autonomy of cell proliferation and developmental programs during Arabidopsis aboveground organ morphogenesis. Developmental Biology 304, 367–381. [DOI] [PubMed] [Google Scholar]
- Cai J, Liu X, Vanneste K, et al. 2015. The genome sequence of the orchid Phalaenopsis equestris . Nature Genetics 47, 65–72. [DOI] [PubMed] [Google Scholar]
- Chen YY, Lee PF, Hsiao YY, Wu WL, Pan ZJ, Lee YI, Liu KW, Chen LJ, Liu ZJ, Tsai WC. 2012. C- and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant and Cell Physiology 53, 1053–1067. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. 1999. The TCP domain: a motif found in proteins regulating plant growth and development. The Plant Journal 18, 215–222. [DOI] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, et al. 2012. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiology 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler RL. 1993. Phylogeny and classification of the orchid family. Portland: Dioscorides Press. [Google Scholar]
- Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J. 2010. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana . The Plant Cell 22, 3921–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J. 2010. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana . The Plant Cell 22, 1161–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P. 2011. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. The Plant Cell 23, 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. 2009. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiology 149, 1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis . The Plant Journal 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. 2002. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Letters 514, 351–354. [DOI] [PubMed] [Google Scholar]
- Ingram GC, Waites R. 2006. Keeping it together: co-ordinating plant growth. Current Opinion in Plant Biology 9, 12–20. [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. 2007. Building blocks for plant gene assembly. Plant Physiology 145, 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. 2011. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis . The Plant Journal 68, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. 2002. DNA binding and dimerization specificity and potential targets for the TCP protein family. The Plant Journal 30, 337–348. [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. 2007. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis . The Plant Cell 19, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. 2010. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis . The Plant Cell 22, 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Ohme-Takagi M, Sato F. 2011. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana . Plant Signaling & Behavior 6, 697–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gutsche N, Zachgo S. 2011. The ROXY1 C-terminal L**LL motif is essential for the interaction with TGA transcription factors. Plant Physiology 157, 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S. 2013. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana . The Plant Journal 76, 901–913. [DOI] [PubMed] [Google Scholar]
- Lin HY, Chen JC, Wei MJ, Lien YC, Li HH, Ko SS, Liu ZH, Fang SC. 2014. Genome-wide annotation, expression profiling, and protein interaction studies of the core cell-cycle genes in Phalaenopsis aphrodite . Plant Molecular Biology 84, 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JA, Sun Y, Blair PB, Mukhtar MS. 2015. TCP three-way handshake: linking developmental processes with plant immunity. Trends in Plant Science 20, 238–245. [DOI] [PubMed] [Google Scholar]
- Manassero NG, Viola IL, Welchen E, Gonzalez DH. 2013. TCP transcription factors: architectures of plant forms. Biomelecular Concepts 4, 111–127. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P. 2010. TCP genes: a family snapshot ten years later. Trends in Plant Science 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, Fujita M, Shinozaki K, Matsui M, Ohme-Takagi M. 2010. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana . Plant and Cell Physiology 51, 2145–2151. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Trontin C. 2011. High time for a roll call: gene duplication and phylogenetic relationships of TCP-like genes in monocots. Annals of Botany 107, 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, King S, Jack T. 2009. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America 106, 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E. 2003. Genetic control of surface curvature. Science 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Novak SD, Luna LJ, Gamage RN. 2014. Role of auxin in orchid development. Plant Signaling & Behavior 9, e972277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, et al. 2007. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nature Genetics 39, 787–791. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, et al. 2007. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319 . Developmental Cell 13, 115–125. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. 2009. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Research 33, W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, Carbonero P, Oñate-Sánchez L. 2012. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. Journal of Experimental Botany 63, 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U. 2011. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. The Plant Journal 67, 595–607. [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. 2008. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena J, Candela H, Robles P, Ponce MR, Pérez-Pérez JM, Piqueras P, Micol JL. 2000. Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156, 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Amano K, Ohto M-a, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C. 2006. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Molecular Biology 61, 165–177. [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C. 2003. The OsTB1 gene negatively regulates lateral branching in rice. The Plant Journal 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E. 2008. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana . The Plant Journal 53, 42–52. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J. 2008. Whole-mount in situ hybridization of RNA probes to plant tissues. CSH Protocols, doi: 10.1101/pdb.prot4944. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Chuang MH, Kuoh CS, Chen WH, Chen HH. 2004. Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant and Cell Physiology 45, 831–844. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Hsiao YY, Pan ZJ, Kuoh CS, Chen WH, Chen HH. 2008. The role of ethylene in orchid ovule development. Plant Science 175, 98–105. [Google Scholar]
- Tsai WC, Lee PF, Chen HI, Hsiao YY, Wei WJ, Pan ZJ, Chuang MH, Kuoh CS, Chen WH, Chen HH. 2005. PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant and Cell Physiology 46, 1125–1139. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2006. Mechanism of leaf-shape determination. Annual Review of Plant Biology 57, 477–496. [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH. 2012. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. Journal of Experimental Botany 63, 809–823. [DOI] [PubMed] [Google Scholar]
- Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH. 2011. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochemical Journal 435, 143–155. [DOI] [PubMed] [Google Scholar]
- Wang MY, Zhao PM, Cheng HQ, et al. 2013. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiology 162, 1669–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Russ D, Ori N. 2011. Gibberellin partly mediates LANCEOLATE activity in tomato. The Plant Journal 68, 571–582. [DOI] [PubMed] [Google Scholar]
- Yang C, Li D, Mao D, Liu XUE, Ji C, Li X, Zhao X, Cheng Z, Chen C, Zhu L. 2013. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant, Cell & Environment 36, 2207–2218. [DOI] [PubMed] [Google Scholar]
- Yang X, Pang HB, Liu BL, Qiu ZJ, Gao Q, Wei L, Dong Y, Wang YZ. 2012. Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. The Plant Cell 24, 1834–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Ma H, Wang J, Zhang D. 2007. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa . Journal of Integrative Plant Biology 49, 885–897. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.