Highlight
Time-series RNA-seq data collected from etiolated maize and rice leaf tissues sampled during the greening process were used to identify potential cis-regulatory motifs that might be recruited into C4 genes from non-photosynthetic genes.
Key words: C4 photosynthesis, cell specificity, cis element, evolution, etiolation, systems biology.
Abstract
Identification of potential cis-regulatory motifs controlling the development of C4 photosynthesis is a major focus of current research. In this study, we used time-series RNA-seq data collected from etiolated maize and rice leaf tissues sampled during a de-etiolation process to systematically characterize the expression patterns of C4-related genes and to further identify potential cis elements in five different genomic regions (i.e. promoter, 5′UTR, 3′UTR, intron, and coding sequence) of C4 orthologous genes. The results demonstrate that although most of the C4 genes show similar expression patterns, a number of them, including chloroplast dicarboxylate transporter 1, aspartate aminotransferase, and triose phosphate transporter, show shifted expression patterns compared with their C3 counterparts. A number of conserved short DNA motifs between maize C4 genes and their rice orthologous genes were identified not only in the promoter, 5′UTR, 3′UTR, and coding sequences, but also in the introns of core C4 genes. We also identified cis-regulatory motifs that exist in maize C4 genes and also in genes showing similar expression patterns as maize C4 genes but that do not exist in rice C3 orthologs, suggesting a possible recruitment of pre-existing cis-elements from genes unrelated to C4 photosynthesis into C4 photosynthesis genes during C4 evolution.
Introduction
Many of the world’s most productive crop species, such as maize, sorghum, and miscanthus, use C4 photosynthesis (Brown, 1999). C4 photosynthesis has independently evolved from C3 photosynthesis in more than 66 lineages (Sage et al., 2011). Compared with C3 photosynthesis, C4 photosynthesis has higher water-, nitrogen-, and light-use efficiencies (Zhu et al., 2008; Sage and Zhu, 2011). This higher photosynthetic efficiency is achieved by concentrating CO2 at the site of RuBisCO, thereby minimizing the rate of photorespiration (Leegood, 2002). These modifications probably required the evolution of new regulatory mechanisms, in the form of either cis- or trans-regulatory factors or elements (Sheen, 1999; Hibberd and Covshoff, 2010; Kajala et al., 2012; Griffiths et al., 2013; Aubry, et al., 2014). Elucidation of these regulatory mechanisms underlying cell-specific expression of C4-related genes is a major focus of current C4 photosynthesis research.
Cell-specific expression of C4-related proteins and enzymes is governed by multiple layers of regulation (see reviews by Hibberd and Covshoff, 2010, and Williams et al., 2012). A number of cis-regulatory motifs controlling C4-specific expression have been identified (Hibberd and Covshoff, 2010). Both 5′UTR and 3′UTR regions can potentially be involved in mediating the cell-specific accumulation of C4-related genes (Marshall et al., 1997; Ali and Taylor 2001; Lai et al. 2002; Patel et al. 2004, 2006; Kajala et al., 2012; Williams et al., 2016). The bundle sheath-specific expression of both NAD-ME1 and NAD-ME2 genes is controlled by a segment of the coding sequence in NAD-ME and NADP-ME subtype species (Brown et al., 2011). Therefore, the regulatory motifs related to cell specificity of C4 genes might reside in all segments of the gene, i.e. promoter, coding sequence, 5′UTR, 3′UTR, and intron. Furthermore, many of these cis-regulatory elements have been reported to be recruited from pre-existing elements, such as the special coding segment in NAD-ME (Brown et al., 2011) and the regulatory elements in the UTR regions of CA and PPDK (Kajala et al., 2012).
Cis-regulatory elements controlling the spatially specific expression patterns of C4 genes have mainly been discovered using experimental approaches on the single-gene level, e.g. through deletion analysis (see reviews by Sheen, 1999; Hibberd and Covshoff, 2010). Recent progress in sequencing technology and computational approaches now offers an alternative method to identify candidate cis-regulatory motifs involved in the regulation of genes. Several methods have been developed for cis-element identification. These methods can be categorized into three major classes: (1) methods based on position weight matrix (PWM), e.g. TRAP (Roider et al., 2007), MATCH (Kel et al., 2003), and SIGNAL SCAN (Prestridge, 1996); (2) phylogenetic footprinting methods, e.g. FootPrinter (Blanchette, 2003), Phyloscan (Palumbo and Newberg, 2010), and PHYME (Sinha et al., 2004); and (3) standard motif-finding algorithms, e.g. Gibbs sampling (Jia and Li, 2012), MEME (Bailey et al., 2006), AlignACE (Roth, 1998), YMF (Sinha, 2003), and Weeder (Pavesi et al., 2006). It is a common practice to combine different approaches to increase the reliability and decrease the false discovery rates of the prediction.
In this study, we aimed to establish a basic routine to identify potential motifs that were recruited during C4 evolution. Specifically, we used time-series RNA-seq data from etiolated Zea mays (C4) and Oryza sativa (C3) leaf tissues sampled during a de-etiolation process. With this data, we first studied the responses of C4 genes in de-etiolated leaves during the greening process; then we used computational approaches to predict potential cis-regulatory elements in different segments of major C4-related genes; finally, we examined the likelihood of the recruitment of pre-existing cis-regulatory elements into C4 metabolic genes during C4 evolution, and we provide a list of potential recruited cis-elements that might serve for further experimental validation.
Material and methods
Plant material, RNA isolation, and mRNA sequencing
Zea mays ecotype B73 and Oryza sativa japonica seeds were sown and cultured in soil in darkness at 28/22 °C on a 16/8h cycle and at 60% humidity for 1 week. The 7-d-old etiolated seedlings were then exposed to continuous light (approx. 200 umol m–2 s–1 at the surface of the sampled leaves) and illuminated for 24h. Seeds for control experiments were sown and cultured in soil for 1 week with a 16-h (07:00–23:00h) light/8-h night cycle. Leaf sections of about 2cm length were taken from the end third of the leaf (i.e. near the tip), from the third leaf on the plant. Samples from etiolated plants were harvested before the start of illumination (termed 0h, at 09:00h) and then at six other time points into the light period, namely 0.5h, 1h, 3h, 6h, 12h, and 24h. Control samples were harvested at 09:00h. We used six pooled segments for each sample. These samples were immediately frozen in liquid nitrogen and stored at –80 °C until use. Total RNA was extracted employing the TRIzol® protocol and purified with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The RNA integrity was evaluated by agarose gel electrophoresis and the concentration was checked using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Quality-controlled RNA samples were prepared for sequencing on an Illumina HiSeq 2000 using the Illumina TruSeqTM RNA sample preparation v2 guide (Catalog # RS-122–2001). Library preparation and sequencing were conducted by the Beijing Genomics Institute (Shenzhen, China). Rawreads of Illumina 2000 sequencing data were submitted to the GenBank Short Read Archive (SRA) database (accession number SRX766219).
Expression profiles and co-expressed genes
The 90-bp pair-end sequencing reads were processed by the FASTX-toolkit pipeline version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/) to remove the adapters. Low-quality reads were then discarded to ensure that more than 70% of the bases in the retained reads possessed a Phred score greater than 30 (indicating a 1‰ sequencing error rate). Read quality was then examined by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were mapped to the B73 maize genome (http://www.phytozome.net/) with Bowtie 2 version 2.1.0 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) (Langmead and Salzberg, 2012) allowing at most three mismatches per read. RPKM values (reads per kilobase of transcript per million mapped reads) were then calculated for each gene. For each gene, the spline function in R (www.r-project.org) was applied to smooth the seven time points.
Regressed curves were then normalized by standard deviation before k-means clustering was conducted using R.
Identification of orthologous gene pairs and C4 isoforms
Genome-wide maize and rice orthologous gene pairs were identified by a combination of tools including BBH-LS (Zhang and Leong, 2012), orthoMCL (Chen et al., 2006), Inparanoid (Ostlund et al., 2010), MSOAR2 (Shi et al., 2010), and the Ensembl database (Hubbard et al., 2002). The results were organized based on the following principles. First, the results from BBH-LS, which is the most stringent method (Zhang and Leong, 2012), were used as the basis. Second, if a particular ortholog pair was not reported by BBH-LS but was by other methods, we retained the results of the other methods. Third, if the results from different approaches were in conflict with each other, genes with higher expression values were retained on the assumption that functional genes generally have higher expression. Fourth, only one-to-one orthologous gene pairs were retained. The list of identified orthologous gene pairs is given in Supplementary Table S1 at JXB online.
We used the 15 genes related to the C4 pathway for detailed analysis. To identify the particular isoform that was recruited into the C4 pathway, we first checked whether information on the cell-specificity of its expression was available. If not, the highest-expressed gene in maize among its paralogs was regarded as the C4 isoform. Multiple methods including Euclidean distance between expression curves, rank correlation, and mutual information (http://cran.r-project.org/web/packages/infotheo/index.html) were used to measure the level of similarities and differences in the expression patterns between maize genes and their corresponding rice orthologous genes.
Motif prediction and negative control
To obtain lists of genes that showed similar expression patterns with target C4 genes, we applied a k-means clustering algorithm to maize and rice genes that showed an average RPKM value greater than 1 (Supplementary Figs S1 and S2). Figure of merit (FOM) values were plotted to choose k (Supplementary Fig. S3). In this study, we applied k-means clustering with two k-values, k=80 and k=30 (hereafter termed as the k80 and k30 approaches, respectively) to decrease the false positive rate. Euclidean distances between 3rd-polynomial regressed expression curves of target genes and the rest of the genes falling into the same cluster were calculated, sorted, and the Z-score transformed. Genes with a Z-score value less than –1.644853 (5% tail) were retained as co-expressed genes for motif prediction. Thus, we were able to classify genes with similar expression patterns into different clusters. For both approaches, five genomic segments of the retained genes – i.e. the 3 kb upstream sequence of the transcription start site (TSS), 5′UTR, 3′UTR, coding sequence (CDS), and intron were extracted with the co-ordinates provided by the phytozome genome annotation (http://www.phytozome.net/). Introns and CDS segments were artificially concatenated by insertion of 10 Ns. Three different methods were combined to predict conserved regulatory motifs: TRAP (Roider et al., 2007) with the aid of TRANSFAC database release 2010.2 (Matys, 2003), Weeder2 (Pavesi et al., 2006; Zambelli et al, 2014), and MEME version 4.8.1 (Bailey et al., 2006). The enrichment P-value of identified motifs within input sequences compared with the genome background was set to 0.05 for the TRAP, MEME, and Weeder2 outputs. Predicted enriched motifs were mapped to genomic sequences to verify their existences using cisGenome version 2.0 (http://www.biostat.jhsph.edu/~hji/cisgenome) (Ji et al., 2006). During this process, at most two mismatches to the consensus sequence and zero mismatch to the degenerate consensus sequence were allowed (Ji et al., 2006). Alignments between motifs were done using STAMP version 1.1 (Mahony and Benos, 2007) with the maximum P-value set to 0.01. Negative controls were used to confirm the reliability of this combined approachby using three different sets of 50 randomly selected genes as the input list to predict motifs. No conserved DNA motifs were identified across the different methods (see Supplementary Table S2).
Results
Overview of the effects of illumination on the transcriptome of etiolated maize and rice leaves
In total, about 81.6 and 137.4 million reads were sequenced for maize and rice, respectively. Read counts and number of expressed genes for each time point are listed in Supplementary Table S3. The gross differences of expression patterns between maize and rice leaves during de-etiolation were first assessed by quantifying the transcript abundance in different pathways, as defined by MapMan bincodes (http://mapman.gabipd.org/). This was done by calculating the average RPKM value of all genes involved with a pathway. Comparing maize to rice, we found that the majority of pathways showed similar changes in expression patterns upon illumination. For example, pathways annotated as ‘cell activity’, ‘DNA activity’, ‘hormone metabolism’, and ‘secondary metabolism’ (see Supplementary Table S4) were slightly influenced by light. However, in many other pathways the responses to light exposure differed between maize and rice. For example, genes classified into the ‘cell activity’ pathway did not show much response to illumination in maize, whereas these genes showed slightly decreased expression in rice (Supplementary Table S4). In contrast, genes annotated as ‘C1 metabolism’ showed lower expression in rice in the dark and showed higher expression under illumination, while the expression of these genes was not influenced by illumination in maize (Supplementary Table S4). Genes annotated to be related to the ‘redox’ pathway showed similar expression levels between etiolated and control samples for maize, but their expression was suppressed in the etiolated samples compared with control samples for rice (Supplementary Table S4).
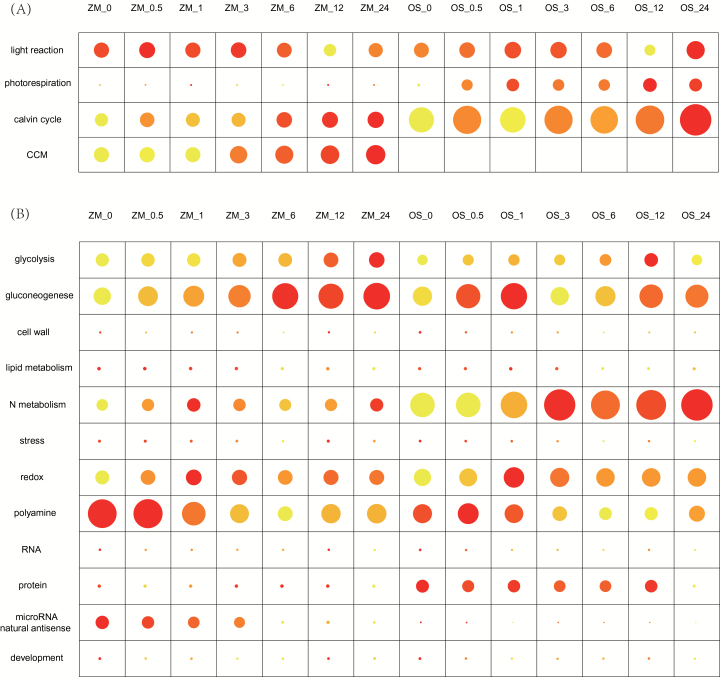
Photosynthesis-related pathways were generally up-regulated during greening (Fig. 1A). Genes involved in the light reactions were activated almost immediately in both species, but the expression level peaked much earlier in maize than in rice (Fig. 1A). Photorespiration-related genes showed higher expression in rice than in maize, which is consistent with a higher photorespiratory flux in C3 plants than in C4 plants. The genes involved in the Calvin–Benson cycle also showed higher expression under illumination in both species, with C3 (rice) showing higher expression as compared to C4 (maize). Maize showed much higher expression values for genes contributing to the C4 CO2-concentrating mechanism (CCM) while rice did not show detectable expression of CCM-associated genes. In maize, the expression of genes involved in both the Calvin–Benson cycle and CCM gradually increased and peaked after leaves were illuminated for 24h.
Fig 1.
Pathway-level gene expression of maize and rice during the de-etiolation process. The dot size represents the gene expression level across the whole genome. The color code indicates the relative gene expression level within a given pathway, from low (yellow) to high (red). (A) Photosynthetic pathways, and (B) non-photosynthesis related pathways. CCM, CO2 concentration mechanism.
Many pathways other than photosynthesis were also influenced by illumination. For example, ‘micro RNA and natural antisense’ genes showed higher expression in the dark and then decreased upon illumination (Fig. 1B). Similarly, pathways annotated as ‘development’, ‘stress’, ‘RNA activity’, ‘protein activity’, ‘polyamine metabolism’, and ‘tetrapyrrole synthesis’ also showed higher expression in the dark as compared to illumination in both species (Fig. 1B).
Transcript abundance of C4 genes during greening
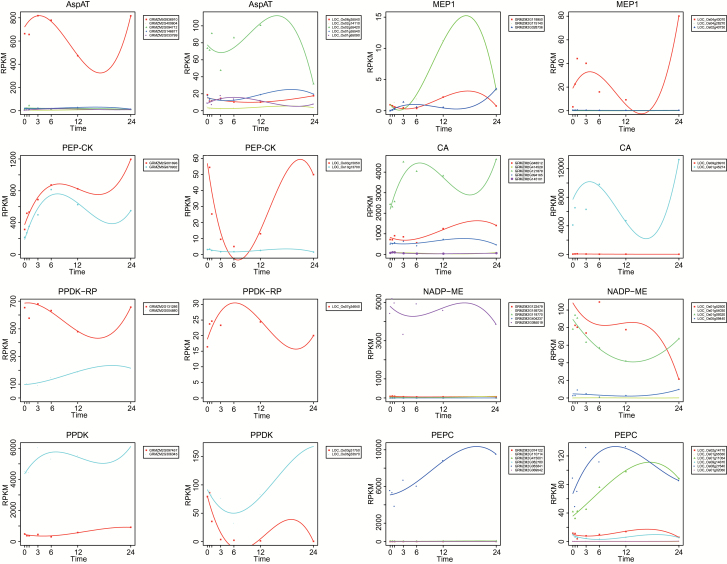
For the majority of genes involved in the C4 pathway in maize and their orthologous genes in rice, only one member of each gene family showed a high level of expression in the mature leaf section during the greening process (Fig. 2). The only exception in maize is PEP-CK, i.e. both GRMZM2G001696 and GRMZM5G870932 were detected with high expression, and similar expression patterns were observed (Fig. 2). While we cannot rule out that technical issues related to read mapping could be the cause of this gene expression pattern, it is also possible that either both genes might be involved in the C4 pathway in maize or one is playing an important role in housekeeping. In rice, both NADP-ME and PEPC gene families hold two members with high expression in the leaf during greening (Fig. 2). In addition, these two members in each pair showed different expression patterns. For both NADP-ME and PEPC, one of the two highly expressed members, i.e., LOC_Os01g52500 for NADP-ME and LOC_Os01g11054 for PEPC, showed a similar expression pattern to its maize counterpart, while the other member, i.e., LOC_Os01g09320 and LOC_Os08g27840, did not.
Fig 2.
Expression curves of C4 gene families. The x-axis represents different time points and the y-axis represents the RPKM value. Expression curves were 3rd-order polynomial regressed, whilst points indicate actual RPKM values.
Taking into consideration that C4 evolved from the ancestral C3 state by recruiting pre-existing components (Sheen, 1999; Hibberd and Covshoff, 2010), we further examined whether these highly expressed genes in the C4 gene families were orthologous between maize and rice. We applied a combination of methods including BBH-LS (Zhang and Leong, 2012), orthoMCL (Chen et al., 2006), Inparanoid (Ostlund et al., 2010), MSOAR2 (Shi et al., 2010), and the Ensembl database (Hubbard et al., 2002). When comparing the expression patterns of C4 orthologous genes of 15 C4 photosynthesis-related genes (Pick et al., 2011) between maize and rice, we categorized the different expression patterns into three different types, namely similar, distinct, and shifted (Fig. 3).
Fig 3.
Expression patterns of C4 orthologous gene pairs between maize (red) and rice (blue). RPKM values were normalized after 3rd-order polynomial regression.
Orthologous genes for PEPC, PPDK, PPT, Mep3, and AMK showed similar expression patterns between maize and rice if they were normalized to the same scale of expression level (Table 1). However, for all these five genes the maize orthologous genes showed higher expression than their rice counterparts before normalization, in particular for PPT and Mep3. Although the expression levels of maize PEPC and PPDK were elevated by approximately two-fold after 24h illumination, we detected relatively high expression of PEPC and PPDK in maize seedlings under dark conditions, which is in contrast to some previous studies (Sheen and Bogorad, 1987; Langdale et al., 1988).
Table 1.
Euclidean distances between maize and rice orthologous gene pairs. For columns headed 1–7, the 1st column indicates the pattern observed in Fig. 3, where ‘s’ stands for similar and ‘d’ stands for different; the 2nd column is the Euclid distance between two clusters that maize and rice genes fall into; the 3rd column is the rank correlation coefficient between maize and rice RPKM vectors ordered across time points; the 4th column is the rank correlation coefficient between maize and rice RPKM vectors ordered across genes; the 5th column is the mutual information value calculated by the R package ‘infotheo’ when setting the bin number to be 3; the 6th column is the maximum mutual information value; the 7th column is the random mutual information value by taking the average of 100 permutations of RPKM values across time points
| Gene ID | Maize ID | Rice ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|---|
| PEPC | GRMZM2G083841 | LOC_Os01g11054 | s | 2.21 | 0.82 | 0.21 | 0.73 | 1.00 | 0.39 |
| PPDK | GRMZM2G097457 | LOC_Os05g33570 | s | 0.00 | 0.54 | 0.77 | 0.26 | 1.00 | 0.38 |
| NADP-ME | GRMZM2G085019 | LOC_Os01g09320 | d | 12.25 | 0.46 | 0.31 | 0.26 | 1.00 | 0.38 |
| PEP-CK | GRMZM2G001696 | LOC_Os03g15050 | d | 8.26 | -0.61 | -0.74 | 0.46 | 1.00 | 0.38 |
| TPT | GRMZM2G070605 | LOC_Os01g13770 | d | 10.47 | -0.11 | -0.12 | 0.26 | 1.00 | 0.39 |
| CA | GRMZM2G121878 | LOC_Os01g45274 | s | 3.76 | 0.29 | 0.03 | 0.26 | 1.00 | 0.39 |
| PPT | GRMZM2G174107 | LOC_Os08g25624 | s | 2.14 | 0.68 | 0.86 | 0.46 | 1.00 | 0.36 |
| PP | GRMZM2G090718 | LOC_Os02g52940 | d | 10.26 | 0.18 | 0.28 | 0.46 | 1.00 | 0.40 |
| AlaAT | GRMZM2G028379 | LOC_Os03g48080 | d | 8.14 | 0.64 | 0.31 | 0.46 | 1.00 | 0.37 |
| DiT1 | GRMZM2G383088 | LOC_Os12g33080 | d | 13.74 | -0.79 | -0.81 | 1.00 | 1.00 | 0.39 |
| Mep3 | GRMZM2G138258 | LOC_Os01g72710 | s | 4.15 | 0.50 | 0.59 | 0.26 | 1.00 | 0.40 |
| AMK | GRMZM2G178192 | LOC_Os08g01770 | s | 3.54 | 0.82 | 0.75 | 0.73 | 1.00 | 0.39 |
| PPDK-RP | GRMZM2G131286 | LOC_Os07g34640 | s | 7.46 | -0.50 | -0.17 | 0.73 | 1.00 | 0.38 |
| MEP1 | GRMZM2G175140 | LOC_Os04g43070 | d | 13.13 | 0.25 | 0.23 | 0.26 | 1.00 | 0.44 |
| AspAT | GRMZM5G836910 | LOC_Os02g55420 | d | 12.91 | -0.82 | -0.42 | 0.73 | 1.00 | 0.39 |
The majority of the 15 gene pairs, namely NADP-ME, PEP-CK, TPT, CA, PP, AlaAT, PPDK-RP, MEP1, and AspAT showed different expression patterns between maize and rice (Fig. 3), with most of them showing lower expression in the dark in both species (Fig. 3). CA, AlaAT, and MEP1 were the only three genes out of the 15 genes for which the expression levels were lower in maize as compared to rice (Fig. 3). DiT1, a dicarboxylate transporter that translocates 2-Oxoglutarate (2-OG) or malate across the plastid envelope membrane, showed similar expression patterns but with a ‘phase shift’ between maize and rice (Fig. 3), as reflected by the high mutual information value between the maize and rice transcriptomics data (Table 1).
Systematic identification of potential regulatory motifs in various genomic regions of C4 genes
For each of the 15 C4 genes, two gene lists, i.e. derived from either the k80 or k30 approaches, were used as input for three different methods (TRAP, MEME, and Weeder2) for motif predictions. The predicted short DNA elements for both maize and rice are listed in Supplementary Tables S5 and S6, respectively.
Motifs predicted by the k80 and k30 approaches covered the majority (179 out of 180 for k80 and 180 out of 180 for k30) of motifs identified from photosynthesis-enriched gene clusters by Wang et al. (2014) (see Supplementary Table S7 and S8), suggesting the validities of the data, routine, and algorithm used in this study. Consistency of the predicted motifs using these two gene lists, i.e. either from the k80 or k30 results of k-means clustering, were checked by motif mapping using STAMP (Table 2, Supplementary Table S9). Overall, about 60% of motifs (for the promoter region, around 85% of the motifs) predicted by the k80 approach overlapped with those motifs predicted by the k30 approach (Table 2, Supplementary Table S9), which represents about 36% of the total number of motifs predicted by the k30 approach (Supplementary Tables S5 and S6). To make the predictions more reliable, we based our further analysis on the overlapped motifs predicted by both the k80 and k30 approaches.
Table 2.
Mapping motifs predicted by the k80 and k30 approaches. Total predicted motifs is the total number of motifs predicted by the gene list obtained by k-mean clustering using the k80 approach. Mapped motifs is the number that could be mapped to motifs predicted by the k30 approach by STAMP with a P-value cut-off set at 0.01
| Genomic section | Total predicted motifs | Mapped motifs | Mapped rate |
|---|---|---|---|
| Maize promoter | 747 | 645 | 86.3% |
| Rice promoter | 545 | 461 | 84.6% |
| Maize 5UTR | 311 | 175 | 56.3% |
| Rice 5UTR | 208 | 101 | 48.6% |
| Maize 3UTR | 385 | 217 | 56.4% |
| Rice 3UTR | 311 | 217 | 69.8% |
| Maize CDS | 531 | 382 | 71.9% |
| Rice CDS | 440 | 333 | 75.7% |
| Maize intron | 538 | 436 | 81.0% |
| Rice intron | 556 | 184 | 33.1% |
Motifs identified for maize C4 genes were aligned against motifs identified for rice orthologous genes (see Supplementary Table S10). Analysis of the identified motifs showed that except for maize AMK 5′UTR, maize PP 3′UTR, maize MEP1 introns, rice MEP1 introns, and DiT1 5′UTR, all other segments of C4-related genes contain conserved motifs between maize and rice (Tables 3 and 4, Supplementary Table S10, and Fig. 4). Interestingly, although no cis-regulatory motifs have been validated experimentally, a large number of conserved motifs were identified in intron regions (Supplementary Table S10), further indicating that introns might be involved with active regulation in agreement with previous findings (Chang and Sun, 2002; Le Hir et al., 2003). The motifs predicted for C4 genes conserved between maize and rice are shown in Table 3. Diagrams showing the relative number of motifs in different genomic segments are shown in Fig 4. The identified motifs conserved between C3 and C4 orthologs might be related to photosynthesis or to the morphogenesis during the de-etiolation process in general.
Table 3.
Likelihood of identifying cis elements in genomic regions of C4 orthologous genes. ‘√’ indicates that conserved motifs were identified between the different methods; ‘X’ indicates that no conserved motifs were identified
The numbered columns are as follows: 1, PEPC; 2, PPDK; 3, NADP-ME; 4, PEP-CK; 5, TPT; 6, CA; 7, PPT; 8, PP; 9, AlaAT; 10, DiT1; 11, Mep3; 12, AMK; 13, PPCK-RP; 14, MEP1; 15, AspAT.
| Section | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maize | Promoter | √ | √ | √ | √ | √ | √ | √ | X | √ | √ | √ | √ | √ | √ | √ |
| 5′UTR | √ | X | √ | √ | √ | √ | X | √ | √ | √ | X | X | X | √ | √ | |
| 3′UTR | √ | √ | √ | X | √ | √ | X | √ | X | √ | √ | √ | √ | √ | √ | |
| Intron | √ | X | √ | X | √ | √ | √ | √ | √ | √ | X | X | √ | √ | √ | |
| CDS | √ | √ | √ | X | √ | √ | √ | X | √ | √ | √ | √ | √ | √ | √ | |
| Rice | Promoter | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | X | √ | √ | √ |
| 5′UTR | √ | √ | √ | X | √ | X | √ | √ | √ | X | √ | X | √ | X | √ | |
| 3′UTR | √ | √ | √ | √ | √ | √ | √ | X | √ | √ | √ | √ | √ | √ | X | |
| Intron | √ | √ | X | X | √ | √ | √ | X | √ | X | √ | X | √ | X | √ | |
| CDS | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | X | √ | √ | √ |
Table 4.
The most conserved motifs predicted for maize and rice. The motifs listed in this table satisfy the following criteria: (1) conserved between at least two prediction methods; (2) conserved between maize and rice orthologous genes; and (3) conserved across maize or rice genes. ‘–’ indicates that no motifs were identified under the specified conditions. Overlapping results between the k80 and k30 approaches are indicated in bold. Numbers in brackets indicate the numbers of copies of this particular motif in the corresponding maize and rice genomic segments, respectively. M, A or C; R, A or G; W, A or T; S, C or G; Y, C or T; K, G or T; V, not T; H, not G; D, not C; B, not A
| Section | PEPC | PPDK | PPT | Mep3 | AMK |
|---|---|---|---|---|---|
| Promoter | CGTTGC (1,7) | AAAANG (34,27) | CGCCGN (14,31) | CGTCRC (4,5) | AACTNTGK (3,6) |
| CNTAAA (6,24) | AAANGA (41,17) | CGGCAG (4,5) | TCGAGCAG (2,0) | AARGGW (30,42) | |
| GTAACG (1,2) | AACAAG (10,3) | CGNCGA (12,4) | TCGCGCAC (0,1) | CCAMTA (6,8) | |
| TTTKTTTT (3,16) | AGSAGG (9,12) | CYGCCG (8,21) | TTGACG (1,3) | CCANAT (24,18) | |
| CAAGTA (3,1) | GACGWA (2,8) | CCCATA (3,8) | |||
| CNTTTC (22,11) | TCCGTC (6,8) | GAAAGGCA (3,2) | |||
| GTACTT (2,3) | GAAAMR (33,62) | ||||
| KGCTAC (12,6) | GTGTAG (9,4) | ||||
| RGCTAT (3,3) | NTACCC (15,12) | ||||
| TCKTTT (20,9) | TAACAN (30,8) | ||||
| TGTACTTY (1,1) | TAACCA (6,12) | ||||
| TTYKTTTC (12,4) | |||||
| TYKTTTCT (8,4) | |||||
| 5′UTR | – | – | CTCGNC (2,6) | – | – |
| SNCCTC (8,27) | |||||
| TCGNCC (2,3) | |||||
| 3′UTR | – | GCCTGC (3,2) | – | – | AMCCAA (1,2) |
| Intron | ATATRTT (7,14) | ACGTTY (5,9) | AAYNTC (22,113) | – | – |
| CGTTNC (13,16) | TTGAAR (14,62) | ||||
| GANGTG (13,41) | |||||
| TTGYNC (27,115) | |||||
| CDS | CAAGNA (18,21) | AGCASC (10,16) | GNAAGA (14,12) | - | AATNTA (3,8) |
| GCTGNTG (6,12) | GTAACA (6,3) | CTCAAC (6,2) | |||
| YGAAGA (6,12) |
Fig 4.
Diagram showing numbers and species of conserved DNA motifs between maize and rice. Conserved DNA motifs identified with the k80 appraoch are marked with color as indicated in the keys, and the number of mapped sites are shown. Overlapping results between the k80 and k30 approaches are marked in bold in the keys.
Furthermore, we also identified those motifs that might be recruited into C4 metabolic genes from genes unrelated to C4 photosynthesis. These motifs were selected based on the following criteria: (1) this particular short DNA motif is enriched in both a C4 gene and a non-C4 gene that show similar expression patterns with the target C4 gene; and (2) this particular short DNA motif exists in a maize C4 gene but not in its rice C4 orthologous genes. The first criterion ensures that this chosen motif might be associated with the regulation of this particular expression pattern of a C4 gene. The second criterion ensures that this particular motif does not exist in the C3 orthologous gene. The potential recruited cis-regulatory motifs are listed in Supplementary Table S11 and their distribution in different genomic segments is shown in Fig. 5.
Fig 5.
The number of recruited motif sites in different segments of C4 genes. The total number of mapped sites for potential recruited motifs in maize identified using the k80 approach are given in the corresponding genomic segments, and the number of overlapping motifs is indicated in brackets.
We also found conservation between the predicted motifs in this study and the motifs identified by previous experimental approaches (Sheen, 1999, Gowik et al., 2004; Supplementary Table S12). In addition, to facilitate the identification of potential transcription factors that might bind to these candidate motifs, we have further listed the motifs with information for binding transcription factors available from the PLACE database (Higo et al., 1999) in Supplementary Table S13.
Discussion
This study reports the changes in the transcriptomics of etiolated leaves of maize and rice upon illumination and also systematically predicts potential cis-regulatory motifs in C4-related genes. We provide evidence for potential recruitment of cis-regulatory motifs from non-photosynthesis genes into C4 metabolic genes during evolution of C4 photosynthesis. In this section, we first discuss the rationale of using the de-etiolation process as a model system for studying C4 photosynthesis. Next, we discuss a number of differences in the transcriptome responses during the de-etiolation process between maize and rice. Finally, we discuss the evidence from this study supporting potential recruitment of pre-existing regulatory elements from non-photosynthetic genes into C4 photosynthesis.
The de-etiolation system as a model to study regulation of C4 photosynthesis
We chose the greening process of etiolated leaves for this study for the following reasons. First, light serves both as the energy source for photosynthesis and as an environmental signal during photosynthesis development (Deng and Quail, 1999), and many photosynthetic genes show altered expression levels during light induction (Shen et al., 2009) together with changes of many other genes, which makes it possible to conduct clustering analysis and to perform motif identification based on genes in the same cluster. Second, with multiple sampling times throughout the de-etiolation process, time-series expression patterns for each gene can be established and used for inferring regulator–target gene pairs. With maize being a C4 species and rice being a C3 species, comparison of the motifs over-enriched in the C4 target genes and the associated genes in the same cluster, but not in the C3 orthologous genes, provides an opportunity to identify motifs that were potentially recruited during C4 evolution. Using expression data based on the de-etiolation process is not a new idea. In fact, this system was one of the most widely used to identify cis-elements controlling individual enzymes (Schaffner and Sheen, 1991; Sheen, 1991; Kausch et al., 2001). Recently, this system has also been used to demonstrate that during C4 evolution the histone modification code was recurrently recruited into different lineages of C4 lineages (Heimann et al., 2013).
Transcriptomic responses of maize and rice during the de-etiolation process
Alhough the de-etiolation process has been reported extensively in the literature (Schaffner and Sheen, 1991; Sheen, 1991; Kausch et al., 2001), so far there has been no comparative transcriptomic data for maize and rice during the de-etiolation process. A number of distinguishing features in the transcriptomes between maize and rice were identified here. Firstly, as expected, we observed decreased expression of genes involved in photorespiration and increased expression of genes involved in the CO2-concentrating mechanism (CCM) in the C4 plant, consistent with earlier reports for other C4 species (Bräutigam et al., 2011, 2014; Gowik et al., 2011) (Fig. 1A). The expression of enzymes in the Calvin–Benson cycle showed a slightly lower level in maize compared to rice. The decreased expression of RuBisCO reflects a decreased demand for this enzyme under elevated CO2 levels in bundle sheath cells. In addition, we found that expression of genes involved in other Calvin–Benson cycle enzymes, such as glyceraldehyde-3-phophate dehydrogenase (GAPDH), RuBisCO activase (RCA), and fructose-bisphosphate aldolase, also showed decreased levels. We also observed decreased expression of enzymes involved in nitrogen metabolism and protein synthesis (Fig. 1B), which have also been observed in earlier comparative studies of C3 and C4 transcriptomics (Bräutigam et al. 2011, 2014; Gowik et al. 2011). This decrease might be related to the decreased content of RuBisCO, one of the most nitrogen-costly proteins in the leaf (Ellis, 1979; Dhingra et al., 2004), and hence a decreased demand for protein synthesis (Piques et al., 2009).
Nearly all genes involved in photosynthesis showed up-regulation during the de-etiolation process (Fig. 1A) (Bradbeer, 1969; Kobayashi et al., 1980; Shen et al., 2009). Interestingly, although genes encoding components of the photosynthetic light reaction in both rice and maize were up-regulated upon exposure to light, the enzymes involved in the light reactions showed faster responses to light in maize compared to rice, i.e. they reached their peak expression levels faster than in rice (Fig. 1A). In contrast, the response speeds of the Calvin–Benson cycle enzymes in maize were similar to C3 leaves (Fig. 1A). This might indicate that the expression patterns of genes in the light reaction may be an essential step before light-induced establishment of cell-specific accumulations of Calvin–Benson cycle enzymes, as suggested by an earlier study (Langdale et al., 1988).
Different genes involved in the C4 cycle showed distinct expression patterns between rice and maize. Five C4 photosynthesis-related enzymes, PEPC, PPDK, PPT, Mep3, and AMK, showed similar expression patterns between maize and rice (Fig. 3), suggesting potentially conserved regulatory mechanisms for these genes between the two species. DiT1, aspartate aminotransferase (AspAT), MEP1, triose phosphate transporter (TPT), and PEP carboxykinase (PEP-CK) showed shifted expression patterns between maize and rice. DiT1, AspAT, MEP1, and PEP-CK are key enzymes related to the operation of the C4 pathway in mature maize leaves (Pick et al., 2011). DiT1 has been reported as a crucial protein at the interface between carbon and nitrogen metabolism (Schneidereit et al., 2006). In addition, PEP-CK and AspAT also play an important role in the interaction of carbon and nitrogen metabolism (Walker et al., 1999). Considering that both maize and rice N metabolism and gluconeogenesis pathways showed a strong circadian rhythm (Fig. 1B), it is possible that some of the regulatory mechanisms of the current C4 pathway might have recruited pre-existing mechanisms from the circadian rhythms.
Cis-regulatory motifs related to C4 photosynthesis
As a common theme of evolution, C4 evolved from C3 photosynthesis by recruiting pre-existing elements. For example, all C4 metabolic enzymes exist in C3 plants and play important house-keeping roles in their C3 host (Aubry et al., 2011). The increased bundle sheath size and chloroplast number in bundle sheath cells in some plants growing in arid regions might represent an adaptation strategy to cope with drought stress (Griffiths et al., 2013). Recent evidence has suggested that even the regulatory elements, such as the 240-nt element in the coding sequence of NAD-ME and the regulatory elements in the UTR region of CA and PPDK, also pre-exist in C3 ancestor enzymes as well (Brown et al., 2011; Kajala et al., 2012). This raises an intriguing hypothesis that recruitment of pre-existing cis-motifs might have been a common mechanism for evolution of regulatory elements during C4 emergence. This study provides new evidence supporting this hypothesis.
We systematically identified potential cis-regulatory motifs that might be involved in regulating C4 photosynthesis genes. Given that earlier reports have shown that promoter regions (Gowik et al., 2004), coding sequence (Brown et al., 2011), 5′UTR (Marshall et al. 1997; Patel et al. 2004, 2006), 3′UTR (Ali and Taylor, 2001; Lai et al., 2002; Kajala et al., 2012), and intron regions can harbor cis-regulatory motifs controlling cell-specific expression, we examined all these genomic regions. Consistent with previous reports, potential candidate motifs were identified in all these different regions of genes (Tables 3 and 4, Figs 4 and 5). Nearly all the motifs identified previously through experimental approaches in maize (Sheen, 1999) were also identified in our predictions (see Supplementary Table S12). However, motifs identified earlier in dicots (e.g. Gowik et al., 2004; Williams et al., 2016) were not identified in this analysis, possibly due to different regulatory mechanisms controlling expression of C4 genes between monocots and dicots.
The distributions of the identified cis-motifs show some distinct features. First, a cis motif can reside in more than one segment of a gene. For example, the Dof1 binding motif AAAAGG was predicted to reside in both the promoter region and the intron region of PEPC (see Supplementary Table S12). Second, a cis-regulatory motif may regulate more than one gene. For example, the Dof1 binding motif AAAAGG was predicted to exist in 5′ flanking sequence of PEPC (as reported by Sheen, 1999), in the maize AlaAT promoter sequences (Supplementary Table S12), and in the intron regions of maize PEPC and NADP-ME (Supplementary Table S12), suggesting reuse of the same motif in regulating multiple C4 genes, a phenomenon shown earlier in C4 genes in dicots (Williams et al., 2016).
With the identified cis-regulatory motifs, we examined whether it is possible for a cis-regulatory motif to be recruited from a gene unrelated to C4 photosynthesis to a C4 gene. To do this, we identified motifs that exist in maize C4 genes and genes showing the same expression patterns but not in their rice orthologous genes. Considering that these motifs were identified based on sequence information for genes in the same cluster, these motifs might have been potentially recruited into C4 metabolic genes from those genes unrelated to them (Supplementary Tables S10 and S11, Fig. 5). These data suggest that genes sharing similar expression patterns with C4 metabolic genes might have been a rich source of cis-regulatory elements recruited into C4 genes during the evolution of C4 photosynthesis. Indeed, many genes in the bundle sheath cells of C3 plants showed highly specialized cell-specific expression and play particular metabolic roles, e.g. the bundle sheath cells of Arabidopsis thaliana show a strong preference for sulfur and glucosinolate metabolism (Aubry et al., 2014). Therefore, the required regulatory metabolism for establishing cell specificity, including both the cis-element and the trans-factors, is in place in C3 plants. It should be much easier to recruit them into genes encoding metabolic enzymes related to C4 photosynthesis, as compared to evolving de novo mechanisms for conferring cell specificity. Our recent anlaysis showed that transposons might have participated in such processes to recruit motifs in the promoter regions (Cao et al., 2016). Here, we show that the potentially recruited motifs reside in all regions of C4 genes (Fig. 5). In fact, many of our potentially recruited motifs overlap with bundle-sheath cell-specific motifs identified by Wang et al. (2014) (see Supplementary Tables S7 and S8). However, it is worth pointing out a caveat that the potentially recruited motifs identified here could perhaps represent differences between BEP clade and panicoid grasses (Brutnell et al., 2010), rather than differences between C3 and C4 photosynthesis. Detailed functional and evolutionary studies of these identified cis-motifs are now needed to clarify their significance to C4 gene expression, in particular to establish their cell-specific expression patterns.
False positive discovery rate and negative control for cis-element identification
Computational identification of cis-elements has the caveat of having a high false-positive rate. In this study, we took a number of measures to overcome this shortcoming of computational approaches, as follows.
(1) Combining multiple approaches for cis-element identification. In this study we applied three approaches, namely TRAP, MEME, and Weeder, to predict enriched short DNA motifs. Candidate motifs were retained only if they were conserved between at least two methods. (2) Conservation between species. Given that genes with similar expression patterns are likely to share common regulatory mechanisms, we have predicted enriched motifs for maize and rice orthologous genes, which were then used to detect the shared motifs between PEPC, PPDK, PPT, Mep3, and AMK. (3) Conservation between different genes. Given that almost all C4 genes showed higher expression in maize compared to rice, it is likely that they might share some conserved regulatory mechanisms to achieve this higher gene expression. Thus, we required identified short DNA motifs to exist in at least two C4 genes to increase their likelihood to be real functional cis-elements. (4) Appearance frequencies. Given that motifs with multiple occurrences are more likely to act as binding sites of transcription factors, we checked the appearance frequencies of the identified cis-regulatory motifs. (5) We also conducted a negative control and the results showed that although some motifs could be identified by randomly selected sequences, no results were retained after a conservation check across different methods (see Supplementary Table S2). (6) Finally, when we used the k-means clustering approach to identify the genes in the same cluster, we used two different cluster numbers. Only motifs identified by analysis with both clustering numbers were retained in this study in order to decrease the false-positive rate. Although these stringent measures will effectively decrease the false-positive rates for mis-identifying new motifs, they might also lead to the possible loss of potential cis-regulatory motifs. Therefore, the reported motifs in this study might represent only a conserved list of the potential motifs in these C4 genes.
Conclusions
Studying the molecular mechanisms controlling C4 genes is a major focus of current C4 photosynthesis research. This study systematically characterized the expression patterns of photosynthesis genes during the de-etilation process, and further identified cis-regulatory motifs potentially related to C4 photosynthesis genes. Although most of the C4 photosynthesis genes showed similar expression patterns between maize and rice, many C4 photosynthesis genes, in particular DiT1, aspartate aminotransferase, PEP-CK, and triose phosphate transporter, showed shifted expression patterns, suggesting a possible recruitment of pre-existing regulatory mechanisms controlling the circadian rhythm during C4 emergence. During the process of identifying cis-regulatory elements, we took several measures to decrease the potential false-positive rate by using a number of motif prediction methods and using more than one clustering number in the k-means clustering. Our analysis shows the widespread existence of cis-motifs in different segments of C4 genes. Finally, considering that many motifs reside in C4 genes and in genes showing similar expression patterns to C4 genes in maize while they do not reside in their C3 orthologs in rice, we suggest the possibility of recruitment of such motifs from genes other than photosynthesis genes into C4 photosynthesis genes.
Supplementary Data
Supplementary data are available at JXB online.
Figure S1. Eighty clusters of maize and rice expressed genes, with the expression curve of each member of a cluster plotted together with the average value of all genes falling into the same cluster.
Figure S2. Thirty clusters of maize and rice expressed genes, with the expression curve of each member of a cluster plotted together with the average value of all genes falling into the same cluster.
Figure S3. Figure of merits of randomly selected genes.
Table S1. One-to-one orthologous gene pairs used in this study identified by a combination of methods.
Table S2. Negative control for cross-methods prediction of DNA motifs.
Table S3. Statistics for RNA-seq samples.
Table S4. Average expression levels of MapMan pathways for maize and rice.
Table S5. Identified motifs for each genomic section of 15 C4 gene pairs using the gene list obtained by k80 of the k-means clustering approach.
Table S6. Identified motifs for each genomic section of 15 C4 gene pairs using the gene list obtained by k30 of the k-means clustering approach.
Table S7. Comparison of identified motifs between the k80 approach and the leaf gradient data in Wang et al. (2014).
Table S8. Comparison of identified motifs between the k30 approach and the leaf gradient data in Wang et al. (2014).
Table S9. Comparison of identified motifs between the k80 and k30 approaches.
Table S10. Comparison of identified motifs between maize and rice C4 orthologous genes for the overlapping results between the k80 and k30 approaches.
Table S11. Potentially recruited motifs and their number of matching sites in corresponding genomic segments.
Table S12. Comparison of identified overlapping motifs between the k80 and k30 approaches with the motifs summarized in Sheen (1999).
Table S13. Comparison of identified motifs with the PLACE database.
Acknowledgements
The authors thank anonymous reviewers for constructive comments that helped us to improve the earlier submissions of this manuscript. XGZ thank the Chinese Academy of Sciences and the Max Planck Society for support. Funding for authors’ research is from the Bill & Melinda Gates Foundation (Grant No. OPP1014417; 1129902), National Science Foundation of China (Grant No. 30970213, C020401), Ministry of Science and Technology of China (Grant No. 2011DFA31070 and 2014AA101601) and EU project 3to4 (EU 289582). APMW appreciates funding by the DFG (grants WE 2231/9-2; EXC 1028, IRTG 1525).
References
- Ali S, Taylor WC. 2001. Quantitative regulation of the Flaveria Me1 gene is controlled by the 3′-untranslated region and sequences near the amino terminus. Plant Molecular Biology 46, 251–261. [DOI] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. Journal of Experimental Botany 62, 3049–3059. [DOI] [PubMed] [Google Scholar]
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM. 2014. Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant Journal 78, 659–673. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research, 34, W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M. 2003. FootPrinter: a program designed for phylogenetic footprinting. Nucleic Acids Research 31, 3840–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer JW. 1969. The activities of the photosynthetic carbon cycle enzymes of greening bean leaves. New Phytologist 68, 233–245. [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Schliesky S, Külahoglu C, Osborne CP, Weber APM. 2014. Towards an integrative model of C4 photosynthesis subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. Journal of Experimental Botany 65, 3579–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HR. 1999. Agronomic implications of C4 photosynthesis. In: Sage RF, Monson RK, eds. C4plant biology. San Diago, CA: Academic Press. [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439. [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu XG, Kellogg E, Van Eck J. 2010. Setaria viridis: a model for C4 photosynthesis. Plant Cell 22, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Xu J, Zheng G, Zhu XG. 2016. Evidence for a role of transposon in the recruitment of cis-regulatory motifs during evolution of C4 photosynthesis. BMC Genomics 17, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Sun T. 2002. Characterization of cis-regulatory regions responsible for developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana . Plant Molecular Biology 49, 579–589. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Research 34, D363–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH. 1999. Signalling in light-controlled development. Seminars in Cell & Developmental Biology 10, 121–129. [DOI] [PubMed] [Google Scholar]
- Dhingra A, Portis AR, Daniell H. 2004. Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proceedings of the National Academy of Sciences, USA 101, 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. 1979. The most abundant protein in the world. Trends in Biochemical Science 4, 241–244. [Google Scholar]
- Gowik U, Bräutigam A, Weber KL, Weber APM, Westhoff P. 2011. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? The Plant Cell 23, 2087–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. 2004. cis-regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. The Plant Cell 16, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths H, Weller G, Toy LFM, Dennis RJ. 2013. You’re so vein: Bundle sheath physiology, phylogeny and evolution in C3 and C4 plants. Plant, Cell and Environment 36, 249–261. [DOI] [PubMed] [Google Scholar]
- Heimann L, Horst I, Perduns R, Dreesen B, Offermann S, Peterhansel C. 2013. A common histone modification code on C4 genes in maize and its conservation in Sorghum and Setaria italica . Plant Physiology 162, 456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, et al. 2002. The Ensembl genome database project. Nucleic Acids Research 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Vokes SA, Wong WH. 2006. A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors. Nucleic Acids Research 34, e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Li J. 2012. Finding transcription factor binding motifs for coregulated genes by combining sequence overrepresentation with cross-species conservation. Journal of Probability and Statistics 2012, 830575. [Google Scholar]
- Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, Hibberd JM. 2012. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. The Plant Journal 69, 47–56. [DOI] [PubMed] [Google Scholar]
- Kausch AP, Owen Jr TP, Zachwieja SJ, Flynn AR, Sheen J. 2001. Mesophyll-specific, light and metabolic regulation of the C4 PPCZm1 promoter in transgenic maize. Plant Molecular Biology, 45, 1–15. [DOI] [PubMed] [Google Scholar]
- Kel A, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis O, Wingender E. 2003. MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Research 31, 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Asami S, Akazawa T. 1980. Development of enzymes involved in photosynthetic carbon assimilation in greening seedlings of maize (Zea mays). Plant Physiology 65, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Wang L, Nelson TM. 2002. Distinct but conserved functions for two chloroplastic NADP-malic enzyme isoforms in C3 and C4 Flaveria species. Plant Physiology 128, 125–139. [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Zelitch I, Miller E, Nelson T. 1988. Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. The EMBO Journal 7, 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. 2003. How introns influence and enhance eukaryotic gene expression. Trends in Biochemical Sciences 28, 215–220. [DOI] [PubMed] [Google Scholar]
- Leegood RC. 2002. C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. Journal of Experimental Botany 53, 581–590. [DOI] [PubMed] [Google Scholar]
- Mahony S, Benos PV. 2007. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Research 35, W253–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Chitty JA, Surin B, Taylor WC. 1997. Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5′ and 3′ sequences. The Plant Cell 9, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Research 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund G, Schmitt T, Forslund K, et al. 2010. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Research 38, D196–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo MJ, Newberg LA. 2010. Phyloscan: locating transcription-regulating binding sites in mixed aligned and unaligned sequence data. Nucleic Acids Research 38, W268–W274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Corey AC, Yin L, Ali S, Taylor WC, Berry JO. 2004. Untranslated regions from C4 Amaranth AhRbc S1 mRNAs confer translational enhancement and preferential bundle sheath cell expression in transgenic C4 Flaveria bidentis . Plant Physiology 136, 3550–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Siegel AJ, Berry JO. 2006. Untranslated regions of FbRbcS1 mRNA mediate bundle sheath cell-specific gene expression in leaves of a C4 plant. The Journal of Biological Chemistry 281, 25485–25491. [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Zambelli F, Stefani M, Mauri G, Pesole G. 2006. MoD Tools: regulatory motif discovery in nucleotide sequences from co-regulated or homologous genes. Nucleic Acids Research 34, W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant Cell 23, 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M. 2009. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Molecular Systems Biology 5, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS. 1996. SIGNAL SCAN 4.0: additional databases and sequence formats. Computer Applications in the Biosciences (CABIOS), 12, 157–160. [DOI] [PubMed] [Google Scholar]
- Roider HG, Kanhere A, Manke T, Vingron M. 2007. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics, 23, 134–141. [DOI] [PubMed] [Google Scholar]
- Roth FP. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nature Biotechnology 16, 939–945. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin PA, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany, 62, 3155–3169. [DOI] [PubMed] [Google Scholar]
- Sage RF, Zhu XG. 2011. Exploiting the engine of C4 photosynthesis. Journal of Experimental Botany 62, 2989–3000. [DOI] [PubMed] [Google Scholar]
- Schaffner AR, Sheen J. 1991. Maize rbcS promoter activity depends on sequence elements not found in dicot. The Plant Cell 3, 997–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit J, Häusler RE, Fiene G, Kaiser WM, Weber APM. 2006. Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. The Plant Journal 45, 206–224. [DOI] [PubMed] [Google Scholar]
- Sheen J. 1991. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. The Plant Cell 3, 225–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. 1999. C4 gene expression. Annual Review of Plant Physiology and Plant Molecular Biology 50, 187–217. [DOI] [PubMed] [Google Scholar]
- Sheen JY, Bogorad L. 1987. Differential expression of C4 pathway genes in mesophyll and bundle sheath cells of greening maize leaves. The Journal of Biological Chemistry, 262, 11726–11730. [PubMed] [Google Scholar]
- Shen Z, Li P, Ni RJ, et al. 2009. Label-free quantitative proteomics analysis of etiolated maize seedling leaves during greening. Molecular & Cellular Proteomics 8, 2443–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Zhang L, Jiang T. 2010. MSOAR 2.0: incorporating tandem duplications into ortholog assignment based on genome rearrangement. BMC Bioinformatics 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. 2003. YMF: a program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Research 31, 3586–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Blanchette M, Tompa M. 2004. PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RP, Chen ZH, Tecsi LI, Faminani F, Lea PJ, Leegood RC. 1999. Phosphoenolpyruvate carboxykinase plays a role in interactions of carbon and nitrogen metabolism during grape seed development. Planta 210, 9–18. [DOI] [PubMed] [Google Scholar]
- Wang L, Czedik-Eysenberg A, Mertz RA, et al. 2014. Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nature Biotechnology 32, 1158–1165. [DOI] [PubMed] [Google Scholar]
- Williams BP, Aubry S, Hibberd JM. 2012. Molecular evolution of genes recruited into C4 photosynthesis. Trends in Plant Science 17, 213–220. [DOI] [PubMed] [Google Scholar]
- Williams BP, Burgess SJ, Reyna-Llorens I, Knerova J, Aubry S, Stanley S, Hibberd JM. 2016. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. The Plant Cell 28, 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. 2014. Using Weeder, Pscan, and PscanChIP for the discovery of enriched transcription factor binding site motifs in nucleotide sequences. Current Protocols in Bioinformatics 47, 2. [DOI] [PubMed] [Google Scholar]
- Zhang M, Leong HW. 2012. BBH-LS: an algorithm for computing positional homologs using sequence and gene context similarity. BMC Systems Biology 6(Suppl 1), S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR. 2008. What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Current Opinion in Biotechnology 19, 153–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.