Abstract
The most common causes of human infection from the arboviruses that are endemic in Australia are the arthritogenic alphaviruses: Ross River virus (RRV) and Barmah Forest virus (BFV). The most serious infections are caused by the neurotropic flaviviruses, Murray Valley encephalitis virus (MVEV) and the Kunjin subtype of West Nile virus. The greatest individual risk of arbovirus infection occurs in tropical/subtropical northern Australia because of the warm, wet summer conditions from December to June, where conventional arbovirus surveillance is difficult due to a combination of low population density, large distances between population centers, poor roads, and seasonal flooding. Furthermore, virus detection requires samples to be sent to Perth up to 2,000 km away for definitive analysis, causing delays of days to weeks before test results are available and public health interventions can be started. We deployed a portable molecular biology laboratory for remote field detection of endemic arboviruses in northern Queensland, then in tropical Western Australia and detected BFV, MVEV, and RRV RNA by polymerase chain reaction (PCR) assays of extracts from mosquitoes trapped in Queensland. We then used a field-portable compact real-time thermocycler for the samples collected in the Kimberley region of Western Australia. Real-time field PCR assays enabled concurrent endemic arbovirus distribution mapping in outback Queensland and Western Australia. Our deployable laboratory method provides a concept of operations for future remote area arbovirus surveillance.
Introduction
A range of arboviruses is present in northern Australia.1–3 The epidemiology of human disease they cause varies with their respective mosquito vectors, principal animal reservoir hosts, patterns of human settlement, and weather. The commonest causes of human disease are the arthritogenic alphaviruses, Ross River virus (RRV) and Barmah Forest virus (BFV), whereas the most serious diseases are caused by the flavivirus infections: Murray Valley encephalitis virus (MVEV), the Kunjin subtype of West Nile virus (KUNV), and dengue viruses (DENV).2,3 Surveillance for these arboviruses in northern Western Australia is carried out by either regular or opportunistic mosquito trapping, complemented by sentinel chicken flocks for the flaviviruses.4,5 The geographic coverage of these methods has been limited by the cost and limited access to surveillance sites. Mosquito samples and sentinel chicken serum samples are sometimes sent several thousand kilometres to Perth for definitive analysis.
A field survey approach using polymerase chain reaction (PCR)–based assays has been used for other infectious diseases in our region. We deployed a portable molecular laboratory to a mine site aid post to identify environmental melioidosis risk,6 and subsequently used a field deployable PCR assay for Burkholderia pseudomallei for environmental threat assessment in central Sri Lanka7 and peninsular Malaysia.8 We also developed and deployed a PCR assay for detection of influenza A/H1N1 during the global influenza pandemic in 2009 in the far north of Western Australia and shortly afterward in central Queensland.9 Successful use of PCR assays in austere, remote locations led us to ask whether a similar approach could be applied to surveillance of Australian arboviruses. In this study, we examined a series of portable PCR assay options for arbovirus surveillance operations in the Australian outback.
Materials and Methods
Reference center analysis.
Specificity of real-time PCR assays (primer and probe details shown in Table 1) was evaluated using 48 Western Australian mosquito homogenates known to contain MVEV, RRV, BFV, and KUNV. RNA was purified from each mosquito sample using QIAamp Viral RNA Mini Kits (Qiagen, Victoria, Australia) and tested with each primer/probe combination in a series of four separate singleplex assays. PCR mastermixes were based on SuperScript® III Platinum® One-Step qRT-PCR Kits amended to 0.3 units per reaction of Superscript III Reverse Transcriptase (ThermoFisher Scientific, Victoria, Australia), 0.5 units per reaction i-StarTaq™ DNA Polymerase (Intron Biotechnology, Gyeonggi, Korea), 10 unit per reaction RNAseOUT (ThermoFisher Scientific), and dl-Dithiothreitol to 0.9 mM (Sigma-Aldrich, New South Wales, Australia). Primers and probes were used at final concentration of 0.3 μM and 0.2 μM, respectively. Reaction volumes of 20 μL included 8 μL of RNA extract, non-template control, or positive control. Amplification was performed in Rotorgene6000 or Rotor-Gene Q thermal cyclers (Qiagen). Amplification profiles were as follows: reverse transcription at 50°C for 30 minutes and DNA polymerase activation at 95°C for 5 minutes followed by 50 cycles of denaturation at 94°C for 12 seconds, annealing at 55°C for 15 seconds, and extension at 72°C for 20 seconds. Florescence acquisition was performed at the end of extension, and analysis was performed with a fluorescence threshold of 0.05. Assay relative limits of detection were determined using titrations of known positive material (Tables 2 and 3).
Table 1.
Real-time PCR arbovirus assay design
| Virus | Primer identifier | Primer function | Primer sequence | Product size |
|---|---|---|---|---|
| BFV* | BFV-27 | Forward | 5′-CCTCAGGGCCACTCACCATA-3′ | 85 bp |
| BFV-111 | Reverse | 5′-TTCTGCTAGTGTCTTGCACATACTACA-3′ | ||
| BFV-49 | Probe | 5′-6FAM-ACAACAACGACTATAATAGCTACCACA-BHQ1-3′ | ||
| KUNV† | KUN-53 | Forward | 5′-CATTGGTTGAGGATACAGTATTG-3′ | 132 bp |
| KUN-184 | Reverse | 5′-CCTGACTTCCTCACTAAAATTTT-3′ | ||
| KUN-105 | Probe | 5′-VICAAGCTAAACTTCTACACATAATAACA-MGBNFQ-3′ | ||
| MVEV‡ | MVE-503 | Forward | 5′-AGCCGAAGCGGTCATAGGT-3′ | 72 bp |
| MVE-574 | Reverse | 5′-CATGTGCGGACTGCAAATTT-3′ | ||
| MVE-527 | Probe | 5′-6FAM-TTTCAATGCTTTCAATGTCA-MGBNFQ-3′ | ||
| RRV§ | RRV-9 | Forward | 5′-CGATGACGTGGGTACAGAGGAT-3′ | 79 bp |
| RRV-87 | Reverse | 5′- GTTACCAAGACCAGCACAACCA-3′ | ||
| RRV-33 | Probe | 5′-6FAM-TAGAGGGCCAGCCCACCTAACCCACTG-BHQ1-3′ |
BFV = Barmah Forest virus; bp = base pair; KUNV = Kunjin virus; MVEV = Murray Valley encephalitis virus; PCR = polymerase chain reaction; RRV = Ross River virus.
BFV target was the E1 region -3′ UTR junction of the structural polyprotein gene (accession U73745; 10,967–11,051 nt).
KUNV target was the NS5 region -3′ UTR junction of the non-structural polyprotein gene (accession D00246.1; 10,352–10,483 nt).
MVEV target was the NS5 region of the nonstructural polyprotein gene (accession AF161266.1; 9,563–9,634 nt).
RRV target was the E1 region of the structural polyprotein gene (accession GQ433359; 11,234–11,312 nt).
Table 2.
Sensitivity of real-time PCR assays compared with the reference method
| Analyte | Real-time PCR assay limits of detection* | ||||
|---|---|---|---|---|---|
| Reference method | Test 1 | Test 2 | Test 3 | Average | |
| BFV | 10e2 | 10e1 | 10e2 | 10e4 | 10e2 |
| MVEV | 10e6 | 10e4 | 10e6 | 10e5 | 10e5 |
| RRV | 10e1 | 10e0 | 10e0 | 10e0 | 10e0 |
| KUNV | 10e4 | 10e3 | 10e5 | 10e4 | 10e4 |
BFV = Barmah Forest virus; KUNV = Kunjin virus; MVEV = Murray Valley encephalitis virus; PCR = polymerase chain reaction; RRV = Ross River virus.
Using positive reference laboratory control material as the comparison and three replicates each in a 10-fold dilution series, lowest dilution of serial 10-fold dilution produce a positive PCR result. Singleplex real-time arbovirus PCR assay targets BFV, MVEV, RRV, and KUNV.
Table 3.
Real-time PCR assay performance of Australian arbovirus PCR field assays under reference laboratory conditions
| Test performance* | ||||
|---|---|---|---|---|
| BFV | KUN | MVEV | RRV | |
| Sensitivity | 1.00 | 1.00 | 0.62 | 0.50 |
| Specificity | 1.00 | 0.95 | 1.00 | 1.00 |
| PPV | 1.00 | 0.82 | 1.00 | 1.00 |
| NPV | 1.00 | 1.00 | 0.93 | 0.91 |
BFV = Barmah Forest virus; KUNV = Kunjin virus; MVEV = Murray Valley encephalitis virus; PCR = polymerase chain reaction; PPV = positive predictive value; RRV = Ross River virus; NPV = negative predictive value.
One sample was found to be inhibitory for the PCR assay.
Cold chain logistics.
All laboratory equipment, consumables, and reagents were shipped from Perth, Western Australia, to north Queensland by scheduled flight, then transported to a training area by road in less than 24 hours from start to finish. Refrigeration was sustained en route by (wet) ice brick packing in sealed polystyrene containers. Frozen reagents were packed in dried ice on arrival on location. Chilled items were stored in portable refrigerators (Waeco, Queensland, Australia). No cold chain was available for the return journey. During the second series of trapping activities in Western Australia, the reagent cold chain was maintained with prefrozen ice bricks during travel to location as above, and sustained in a clinical laboratory reagent freezer connected to a mains power supply in Kununurra, Western Australia. Dried ice was shipped in by road only for the mosquito traps and was not available for reagent storage.
Mosquito trapping and identification.
The first round of trapping was conducted in May 2010 at Townsville Field Training Area, 50 km west of Townsville, by environmental health officers supervised by an experienced entomologist using CO2-baited encephalitis virus surveillance traps. Each trap had a small light and contained pelleted dry ice. Traps were placed between 4 pm and 5 pm in each evening, and retrieved between 7 am and 8 am in the following morning. Four traps were placed per night in locations within the training area, other than on the last night where three traps were set. Trapped mosquitoes were immobilized in a dry ice container. Mosquito identification was conducted to species level using a field microscope with fiber optic light source, and the keys of Lee and others.10 The second round of trapping was conducted in the vicinity of Kununurra during a 9-day period in 2012 using CO2-baited traps of similar design to those used in Queensland, using up to six traps per night. The mosquitoes in this series were pooled into sterile containers on emptying the traps each morning, and 25 insect aliquots were dispensed for homogenization and subsequent arbovirus detection.
In-field PCR assays.
During the field laboratory deployment to north Queensland, mosquitoes were sorted into pools of up to 10 insects of the same species and ground in 1.0 mL viral transport medium using a macerator with a sterile, disposable tip. The coarse particles were pelleted by short centrifugation, and 50 μL supernatant was purified by magnetic bead extraction (MagMax-24; Applied Biosystems, Carlsbad, CA). We ran the previously developed arbovirus PCR assays in the Queensland field laboratory, (Table 1) on a standard laboratory real-time thermocycler (StepOne; Applied Biosystems). The same extraction methods and PCR assays were used for the subsequent field laboratory deployment in Western Australia, on a smaller, portable real-time thermocycler (MiniOpticon; Bio-Rad, New South Wales, Australia). Molecular grade ultrapure water was used as a non-template control at every eighth well, and positive control template was added to an adjacent well as a positive reaction control. On the first trapping run, mosquito numbers were unexpectedly low, with a predicted low virus yield. We used a test triage in which we carried out limited testing on the first run to avoid reagent wastage and preserve reagent supplies. We therefore tested only for BFV in the first series. Reagents for the other three viruses were used only after a second round of trapping that produced a higher yield of insects in some traps. The remaining BFV reagents were used to test insects from three of these trap locations.
Assay results were analyzed in field on a laptop computer running the thermocycler software. Positive results were plotted on the detailed training area map by the corresponding mosquito trap grid reference. The trap locations in Western Australia were recorded using a global positioning system receiver (eTrex; Garmin Ltd.).
Results
Real-time PCR assays using the deployable equipment and reagents under fixed laboratory conditions detected all four reference arboviruses in a panel of 48 mosquito homogenates (Table 2) at a sensitivity within a 10-fold dilution of the reference laboratory method.
Mosquito traps set in the first 2 days of Queensland trapping in an area of wooded upland slopes contained low numbers of mosquitoes and were tested for BFV only to conserve reagents, as described above. These tests detected BFV in nine of 18 traps (Table 4) and none of the negative control wells. In the later series of trapped mosquito pools, more insects were obtained from sites sampled for a second time, thus increasing the quantity of material for arbovirus RNA extraction. KUNV, MVEV, and RRV were detected in two, eight, and six of 15 traps, respectively. Because of depletion of BFV master mix during the first test series, only three additional trap contents were available for testing thereafter. All three extracts from all three traps were BFV positive. All negative controls returned negative results for all viruses in all test runs.
Table 4.
Arbovirus detection in mosquitoes trapped in May 2010 in Far North Queensland uplands using field deployable PCR methods
| Laboratory no. | Date of collection | Date of testing | Description | Trap no. | Mosquito species in pool | No. of insects tested | Real-time PCR | |||
|---|---|---|---|---|---|---|---|---|---|---|
| BFV† | KUNV† | MVEV† | RRV† | |||||||
| 1 | May 14 | May 17 | Base camp vicinity | 3 | Mixed culicines | 5 | BFV 39* | NA‡ | NA‡ | NA‡ |
| 2 | May 15 | May 17 | Airstrip | 5 | Culex annulirostris | 12 | Neg | NA‡ | NA‡ | NA‡ |
| 3 | May 15 | May 17 | Camp area | 6 | Mixed | 10 | BFV | NA‡ | NA‡ | NA‡ |
| 4 | May 15 | May 17 | Camp area | 6 | Cx. annulirostris | 5 | BFV | NA‡ | NA‡ | NA‡ |
| 5 | May 15 | May 17 | Creek junction | 7 | Culex quinquefasciatus | 12 | BFV | NA‡ | NA‡ | NA‡ |
| 6 | May 15 | May 17 | Creek junction | 7 | Mixed culicines including Aedes | 7 | BFV 36 | NA‡ | NA‡ | NA‡ |
| 7 | May 15 | May 17 | Creek junction | 7 | Cx. annulirostris | 20 | BFV 38 | NA‡ | NA‡ | NA‡ |
| 8 | May 16 | May 17 | Homestead | 9 | Mixed mosquitoes | 18 | BFV 39 | NA‡ | NA‡ | NA‡ |
| 9 | May 16 | May 17 | Homestead | 9 | Cx. annulirostris | 20 | BFV 39 | NA‡ | NA‡ | NA‡ |
| 10 | May 15 | May 17 | Range control buildings | 8 | Mixed culicines | 6 | Neg | NA‡ | NA‡ | NA‡ |
| 11 | May 16 | May 17 | Creek | 11 | Mixed culicines | 6 | Neg | NA‡ | NA‡ | NA‡ |
| 12 | May 16 | May 17 | Creek | 11 | Cx. annulirostris | 12 | Neg | NA‡ | NA‡ | NA‡ |
| 13 | May 16 | May 17 | Creek | 12 | Cx. annulirostris | 10 | BFV | NA‡ | NA‡ | NA‡ |
| 14 | May 16 | May 17 | Creek | 12 | Mixed mosquito spp. | 9 | Neg | NA‡ | NA‡ | NA‡ |
| 15 | May 14 | May 17 | Creek base camp vicinity | 2 | Cx. annulirostris | 10 | Neg | NA‡ | NA‡ | NA‡ |
| 16 | May 14 | May 17 | Case camp vicinity | 2 | Mixed Aedes spp. | 6 | BFV 38 | NA‡ | NA‡ | NA‡ |
| 17 | May 16 | May 17 | – | 10 | Cx. annulirostris | 12 | Neg | NA‡ | NA‡ | NA‡ |
| 18 | May 17 | May 17 | Non-template control | – | NTC | – | Neg | NA‡ | NA‡ | NA‡ |
| 19 | May 17 | May 17 | Control tower | 13 | Cx. annulirostris | 5 | NA§ | Neg | Neg | Neg |
| 20 | May 17 | May 17 | Hut | 15 | Cx. annulirostris | 12 | NA§ | Neg | Neg | Neg |
| 21 | May 17 | May 17 | Airfield | 5 | Anopheles annulipes | 14 | NA§ | Neg | Neg | Neg |
| 22 | May 14 | May 17 | Base camp vicinity | 2 | An. annulipes | 9 | NA§ | Neg | Neg | RRV |
| 23 | May 14 | May 17 | Woodland | 4 | An. annulipes | 2 | NA§ | Neg | Neg | Neg |
| 24 | May 16 | May 17 | Camp site | 10 | Cx. annulirostris | 25 | NA§ | Neg | Neg | Neg |
| 25 | May 17 | May 17 | Hut | 15 | Aedes linneatipennis | 13 | NA§ | Neg | Neg | Neg |
| 26 | May 17 | May 17 | Control tower | 13 | Aedes tremulus | 17 | NA§ | Neg | Neg | Neg |
| 27 | May 18 | May 18 | Buildings | 16 | Cx. annulirostris | 4 | NA§ | Neg | MVEV 37 | RRV 34 |
| 28 | May 18 | May 18 | Buildings | 16 | An. annulipes | 7 | NA§ | Neg | MVEV 26 | Neg |
| 29 | May 18 | May 18 | Camp site | 17 | Cx. annulirostris | 11 | NA§ | Neg | MVEV 33 | Neg |
| 30 | May 18 | May 18 | Creek | 18 | Cx. annulirostris | 3 | BFV 16 | Neg | MVEV 36 | RRV 37 |
| 31 | May 18 | May 18 | Creek | 18 | Culex bitaeniorhynchus | 10 | BFV 17 | Neg | MVEV 33 | RRV 34 |
| 32 | May 18 | May 18 | Creek | 18 | Coquillettidia xanthogaster | 20 | BFV 17 | Neg | MVEV 34 | RRV 36 |
| 33 | May 18 | May 18 | Camp site | 19 | Cx. annulirostris | 9 | NA§ | Neg | MVEV 30 | RRV 32 |
BFV = Barmah Forest virus; KUNV = Kunjin virus; MVEV = Murray Valley Encephalitis virus; NA = not applicable; Neg = negative; PCR = polymerase chain reaction; RRV = Ross River Virus.
Positive qPCR assay results expressed as cycling threshold (Ct) values when < 40.0, and negative when Ct ≥ 40.0.
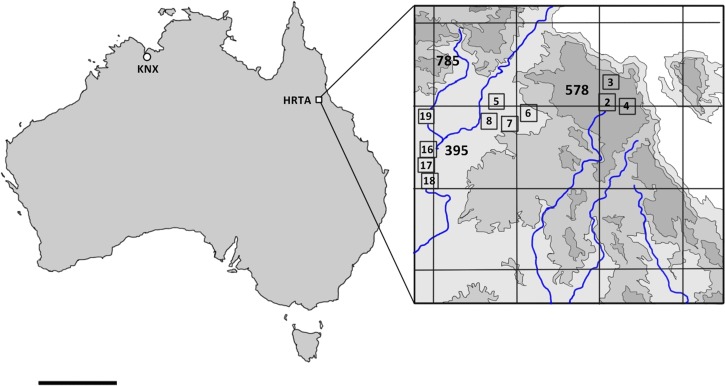
Traps per arbovirus were BFV = 16, KUNV = 10, MVEV = 10, and RRV = 10. Trap numbers correspond to numbered boxes in Figure 1.
Limited analysis of tested mosquito extracts for only BFV due to expected low sensitivity of testing small numbers of trapped insects (below preferred minimum aliquot of 25 insects).
Exhaustion of BFV PCR master mix prevented analysis of all second series of mosquito pools.
MVEV and RRV were detected only in insect pools obtained within 1 km of the main creek line running through the area (Figure 1 , Table 4) while BFV, by contrast, was mainly detected in insects from upland wooded areas away from the main creek line. Interestingly, there was an overlap in the mosquito vector species in which different endemic arboviruses were detected. The dominant species, Culex annulirostris, was shown to carry BFV, MVEV, and RRV at multiple locations along the creek line. More than one type of arbovirus (e.g., RRV and MVEV) was also detected in pools of other species collected from these locations (Table 4).
Figure 1.
Location of arbovirus surveillance mosquito trapping in Australia (left) and detail of survey area in vicinity of Harvey Range (right) based on the inset diagram from the Australian Defense Force Topographic Survey, 1:40,000 High Range Training Area (Special) (HRTA). The area is located in the vicinity of Dotswood, Queensland; −19.624423, 146.289802. Grid lines demarcate 10 × 10-km squares. Blue lines represent creeks. Numbers outside boxes in bold indicate height in meters above mean sea level. Numbers in small boxes indicate mosquito trap locations and correspond to trap numbers in Table 4. Four traps were located outside the topographic series map. KNX = trapping in vicinity of Kununurra, Western Australia; scale bar = 1,000 km.
In the subsequent northwestern Australian deployment of a lighter, field-portable real-time thermocycler and supporting equipment, the same assays delivered two RRV positive results (Cts = 29.2 and 29.7, respectively) from mosquito homogenate extracts within 24 hours of collection by the vector control team at two locations in irrigated agricultural areas close to the East Kimberley town of Kununurra. All samples were negative for BFV, KUNV, and MVEV. Template controls were consistently negative. Positive controls for all four viruses were all positive.
Discussion
A modular molecular biology laboratory was deployed into the field, successfully operating nucleic acid purification, detection, and confirmation methods on two successive occasions. The field laboratory was able to give entomology and environmental health staff same-day support for their health risk assessments. This capability allows a rapid assessment of the specific arbovirus threat and initiation of an early environmental health response. This study provided new information on arbovirus activity in two remote locations in northern Queensland and Western Australia.
The alphaviruses RRV and BFV cause the majority of notified human arbovirus infections in Australia, resulting in a substantial burden of acute and chronic illness.2 The flavivirus infections caused by MVEV and KUNV are less common but can cause severe encephalitis with poor outcomes.2,11–14 Arbovirus activity occurs in periods of heavy rainfall or flooding, and during warm periods provided when there are suitable amplifying animal hosts, usually macropods such as kangaroos and wallabies. These conditions were met and endemic arbovirus infections were abundant during the unusually wet climatic conditions 2011 and 2012 summer/autumn periods that resulted in extensive flooding across much of northern and central Australia and, in 2011, to southeastern Australia with high levels of arbovirus activity including the first outbreak of MVEV encephalitis in southeastern Australia since 1974.11,12
DENV activity is confined to northern Queensland where periodic epidemics occur following transient reintroduction of the virus from overseas,2 during which there is a virus–human cycle without an animal host.
The military training areas of northern Australia contain pristine wilderness with little human habitation and an abundant native fauna, and therefore represent islands of preserved biodiversity, protected under Australian Commonwealth legislation.15 A consequence of this protected natural environment is the maintenance of natural reservoirs for endemic arboviruses. Recognizing a theoretical risk to future visitors to the training area, we included mosquito trapping and Australian arbovirus detection in our environmental threat assessment.16 However, at that time there was no surveillance information for that area, and data on local distribution of endemic Australian arboviruses were limited to the reports of three confirmed human cases of MVE in the 1990s and two in the following 5 years.17
Demonstration of unanticipated levels of BFV, MVEV, and RRV in culicine mosquitoes from the training area exposed a previously unrecognized health threat and enabled development of a preventive plan.15 MVEV- and RRV-infected mosquitoes were recovered from a more restricted distribution than BFV-infected mosquitoes. The differences in distributions patterns appear to reflect the ecology of these infections, since all three arboviruses were found close to creek lines while BFV was also detected on the wooded upland slopes. There was no clear difference in tested vector populations that could explain these differences, and data are lacking on the behavior of locally prevalent native fauna such as macropods that may act as mammalian reservoirs. In future, use of PCR detection of viruses in expectorated mosquito saliva in conjunction with honey-baited Flinders Technical Associates cards that can be sent to a central laboratory for subsequent testing may confirm our preliminary findings and develop our understanding of how these endemic Australian arboviruses persist in wilderness locations.18 There may be a case for periodic, targeted deployable PCR-enhanced surveillance in Western Australia where sentinel chicken flocks and mosquito trapping expeditions are currently in use.4,5 Small localized infection cluster in remote areas could also be investigated using our approach. Deployment of a more compact version of the portable laboratory demonstrated the flexibility of PCR assay support for entomological field surveys in wilderness areas.
Although we were able to show that these tests can be successfully deployed to detect both alphaviruses and flaviviruses in mosquito populations, the logistic challenges of these remote locations prevented completion of a reference center posttest evaluation. Early depletion of critical reagents meant that we could not complete a second round of tests for all arboviruses of interest. Also, the lack of a reliable cold chain on the return journey prevented us from returning samples from the trapped mosquito pools to our reference laboratory for confirmatory testing and subsequent sequencing to determine the molecular epidemiology of the viruses we detected in the field and provide definitive evidence of borderline PCR assay results. We were therefore unable to confirm the sensitivity of deployed field PCR assays on residual mosquito homogenates and to recognize this as a priority for future field assay development. The high rate of BFV detection was notable, and the inability to perform confirmatory tests means that we cannot completely exclude cross-contamination. In particular, borderline PCR results with high Ct may represent false-positive results. Their significance will require further field studies, augmented by sequencing of the amplified products. However, all negative control samples remained negative, and as we found no evidence of cross-contamination with other viruses, this appears unlikely. Further work is needed to optimize arbovirus RNA transport during a long-distance return journey before we can use in-field PCR assays for routine arbovirus surveillance.
In conclusion, we demonstrated the feasibility of deploying and operating in-field PCR assays for endemic Australian arboviruses to field laboratory locations in tropical Australia. MVEV, RRV, and BFV detected in culicine mosquitoes trapped in a remote wilderness location assisted location of natural reservoirs for endemic arboviruses, and identified priorities for future disease surveillance and control measures.
ACKNOWLEDGMENTS
We thank our colleagues at PathWest Laboratory medicine for help dispatching reagents and equipment from Perth to Townsville, and David Porter for his assistance with the equipment return. We also thank Jim Frames of Thermo Fisher Scientific for expert assistance with equipment and reagent logistics. We are grateful to Maj Brady McPherson, OIC Health Assessment Team for his support in the field throughout the laboratory deployment, and to Col Andrew Williams for command support for the exercise. We are grateful to the Joint Health Command of the Australian Defence Force (ADF) for permission to publish the results of this investigation.
Disclaimer: The opinions expressed in this paper are those of the authors and do not represent the official view of the ADF.
Footnotes
Authors' addresses: Timothy J. J. Inglis, Department of Microbiology, PathWest Laboratory Medicine WA, Nedlands, Australia, School of Pathology and Laboratory Medicine, University of Western Australia, Crawley, Australia, and 3rd Health Support Battalion, Adelaide, Australia, E-mail: tim.inglis@health.wa.gov.au. Richard S. Bradbury, School of Medical and Applied Sciences, Central Queensland University, Rockhampton, Australia, and 3rd Health Support Battalion, Adelaide, Australia, E-mail: r.bradbury@cqu.edu.au. Russell L. McInnes, Genomics, Agilent Technologies Australia, Mulgrave, Australia, E-mail: russell_mcinnes@agilent.com. Stephen P. Frances, Department of Vector Surveillance and Control, Australian Army Malaria Institute, Queensland, Australia, E-mail: steve.frances@defence.gov.au. Adam J. Merritt, Avram Levy, and David W. Smith, Department of Microbiology, PathWest Laboratory Medicine WA, Nedlands, Australia, and School of Pathology and Laboratory Medicine, The University of Western Australia, Crawley, Australia, E-mails: adam.merritt@health.wa.gov.au, avram.levy@health.wa.gov.au, and david.smith@health.wa.gov.au. Jay Nicholson, Environmental Health Hazards Unit, Western Australian Department of Health, Claremont, Australia, E-mail: jay.nicholson@health.wa.gov.au. Peter J. Neville, Vector Control, Western Australian Department of Health, Claremont, Australia, E-mail: peter.neville@health.wa.gov.au. Michael Lindsay, Environmental Health Hazards Unit, Western Australian Department of Health, Perth, Australia, E-mail: michael.lindsay@health.wa.gov.au.
References
- 1.Mackenzie JS, Lindsay MD, Coelen RJ, Broom AK, Hall RA, Smith DW. Arboviruses causing human disease in the Australasian zoogeographic region. Arch Virol. 1994;136:447–467. doi: 10.1007/BF01321074. [DOI] [PubMed] [Google Scholar]
- 2.Smith DW, Speers DJ, Mackenzie JS. The viruses of Australia and the risk to tourists. Travel Med Infect Dis. 2011;9:113–125. doi: 10.1016/j.tmaid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Wright P, Fitzsimmons GJ, Johansen CA, Whelan PI. National Arbovirus and Malaria Advisory Committee Arboviral diseases and malaria in Australia, 2009–10: annual report of the National Arbovirus and Malaria Advisory Committee. Commun Dis Intell Q Rep. 2012;36:70–81. doi: 10.33321/cdi.2012.36.2. [DOI] [PubMed] [Google Scholar]
- 4.Spencer JD, Azoulas J, Broom AK, Buick TD, Currie B, Daniels PW, Doggett SL, Hapgood GD, Jarrett PJ, Lindsay MD, Lloyd G, Mackenzie JS, Merianos A, Moran RJ, Ritchie SA, Russell RC, Smith DW, Stenhouse FO, Whelan PI. Murray Valley encephalitis virus surveillance and control initiatives in Australia: a report on behalf of National Arbovirus Advisory Committee of the Communicable Diseases Network Australia. Commun Dis Intell Q Rep. 2001;25:33–47. doi: 10.33321/cdi.2001.25.7. [DOI] [PubMed] [Google Scholar]
- 5.Broom AK, Whelan PL. Sentinel chicken surveillance programme in Australia, July 2003 to June 2004. Commun Dis Intell Q Rep. 2005;29:65–70. doi: 10.33321/cdi.2005.29.3. [DOI] [PubMed] [Google Scholar]
- 6.Inglis TJ, Levy A, Merritt AJ, Hodge M, McDonald R, Woods DE. Melioidosis risk in a tropical industrial environment. Am J Trop Med Hyg. 2009;80:78–84. [PubMed] [Google Scholar]
- 7.Inglis TJ, Merritt A, Montgomery J, Jayasinghe I, Thevanesam V, McInnes R. Deployable laboratory response to emergence of melioidosis in central Sri Lanka. J Clin Microbiol. 2008;46:3479–3481. doi: 10.1128/JCM.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inglis TJJ. The lab without walls: a deployable approach to tropical infectious diseases. Am J Trop Med Hyg. 2013;88:614–618. doi: 10.4269/ajtmh.12-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inglis TJ, Merritt AJ, Levy A, Vietheer P, Bradbury R, Scholler A, Chidlow G, Smith DW. Deployable laboratory response to influenza pandemic; PCR assay field trials and comparison with reference methods. PLoS One. 2011;6:e25526. doi: 10.1371/journal.pone.0025526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee DJ, Hicks MM, Griffiths M, Russell RC, Debenham ML, Bryan JH, Marks EN. Entomology Monograph No. 2. vol. 1. Canberra, Australia: Australian Government Publishing Service; 1980. The Culicidae of the Australasian Region. [Google Scholar]
- 11.van den Hurk AF, Craig SB, Tulsiani SM, Jansen CC. Emerging tropical diseases in Australia. Part 4. Mosquito-borne diseases. Ann Trop Med Parasitol. 2010;104:623–640. doi: 10.1179/136485910X12851868779984. [DOI] [PubMed] [Google Scholar]
- 12.Selvey LA, Dailey L, Lindsay M, Armstrong P, Tobin S, Koehler AP, Markey PG, Smith DW. The changing epidemiology of Murray Valley encephalitis in Australia: the 2011 outbreak and a review of the literature. PLoS Negl Trop Dis. 2014;8:e2656. doi: 10.1371/journal.pntd.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speers DJ, Flexman J, Blyth CC, Rooban N, Raby E, Ramaseshan G, Benson S, Smith DW. Clinical and radiological predictors of outcome for Murray Valley encephalitis. Am J Trop Med Hyg. 2013;88:481–489. doi: 10.4269/ajtmh.12-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray TJ, Burrow JN, Markey PG, Whelan PI, Jackson J, Smith DW, Currie BJ. West Nile virus (Kunjin subtype) disease in the northern territory of Australia—a case of encephalitis and review of all reported cases. Am J Trop Med Hyg. 2011;85:952–956. doi: 10.4269/ajtmh.2011.11-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Department of the Environment, Australian Government . Federal Environment Protection and Biodiversity Conservation Act (EPBC Act) Canberra, Australia: Australian Government; 1999. [Google Scholar]
- 16.Brumpton BM, McPherson BA, Frances SP, Inglis TJJ, McCall BJ. Townsville field training area health assessment. ADF Health J. 2011;12:45–50. [Google Scholar]
- 17.Queensland Health . 2010. Murray Valley encephalitis.http://access.health.qld.gov.au/hid/InfectionsandParasites/ViralInfections/murrayValleyEncephalitis_fs.asp Available at. Accessed August 1, 2010. [Google Scholar]
- 18.Hall-Mendelin S, Ritchie SA, Johansen CA, Zborowski P, Cortis G, Dandridge S, Hall RA, van den Hurk AF. Exploiting mosquito sugar feeding to detect mosquito-borne pathogens. Proc Natl Acad Sci USA. 2010;107:11255–11259. doi: 10.1073/pnas.1002040107. [DOI] [PMC free article] [PubMed] [Google Scholar]