Abstract
A typical eukaryotic gene is comprised of alternating stretches of regions, exons and introns, retained in and spliced out a mature mRNA, respectively. Although the length of introns may vary substantially among organisms, a large fraction of genes contains short introns in many species. Notably, some Ciliates (Paramecium and Nyctotherus) possess only ultra-short introns, around 25 bp long. In Paramecium, ultra-short introns with length divisible by three (3n) are under strong evolutionary pressure and have a high frequency of in-frame stop codons, which, in the case of intron retention, cause premature termination of mRNA translation and consequent degradation of the mis-spliced mRNA by the nonsense-mediated decay mechanism. Here, we analyzed introns in five genera of Ciliates, Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia. Introns can be classified into two length classes in Tetrahymena and Ichthyophthirius (with means 48 bp, 69 bp, and 55 bp, 64 bp, respectively), but, surprisingly, comprise three distinct length classes in Oxytricha and Stylonychia (with means 33–35 bp, 47–51 bp, and 78–80 bp). In most ranges of the intron lengths, 3n introns are underrepresented and have a high frequency of in-frame stop codons in all studied species. Introns of Paramecium, Tetrahymena, and Ichthyophthirius are preferentially located at the 5' and 3' ends of genes, whereas introns of Oxytricha and Stylonychia are strongly skewed towards the 5' end. Analysis of evolutionary conservation shows that, in each studied genome, a significant fraction of intron positions is conserved between the orthologs, but intron lengths are not correlated between the species. In summary, our study provides a detailed characterization of introns in several genera of Ciliates and highlights some of their distinctive properties, which, together, indicate that splicing spellchecking is a universal and evolutionarily conserved process in the biogenesis of short introns in various representatives of Ciliates.
Introduction
Ciliates represent a unique and extremely heterogeneous group of protozoans containing many thousands of genetically diverse species [1]. Named after a hair-like organelle called cilia, they have a number of distinguishable features which make them important model organisms to study various biological phenomena such as RNA splicing, DNA rearrangements, and telomeres [2]. Unlike other eukaryotes, each ciliate possesses two types of nuclei, germline micronucleus (MIC) and somatic macronucleus (MAC) which derives from MIC during extensive developmental rearrangements of the genomic DNA. The diploid micronucleus serves as the germline source of DNA during reproduction and is transcriptionally silent, whereas the macronucleus expresses genes to support vegetative cell growth and cell proliferation [1]. Each nucleus contains distinct genomes, which are replicated, amplified, and eliminated at different stages of the ciliate life cycle [1–4].
The micronuclear DNA consists of extremely long molecules typical of eukaryotic chromosomes, containing genes interrupted by short non-coding elements, called internal eliminated sequences (IESs), and large stretches of the spacer DNA [2]. In contrast, the macronuclear DNA exists as short molecules ranging in size from a few hundred to several thousand base pairs (bp) with high gene density and without IESs and spacer DNA [2–4].
To date, macronuclear genomes of several ciliates have been completely sequenced and annotated (Class Oligohymenophorea, genera Tetrahymena, Ichthyophthirius, Paramecium; class Spirotrichea, genera Oxytricha, Stylonychia, Urostyla, Paraurostyla, Laurentiella) [5–10]. The MAC genome of Euplotes crassus (class Armophorea, genus Nyctotherus) had also been sequenced, but its annotation is not complete [11]. The numbers of genes as well as the general principles of the genome organization have been shown to be dramatically different between the organisms. The MAC genome of Tetrahymena thermophila, 104 MB in length, is comprised of 225 chromosomes and encodes almost 27000 genes [6]. During its differentiation it becomes 10–20% smaller than that of MIC, due to removal of the repetitive DNA and about 6000 IESs. The MAC genome of a closely related species, Ichthyophthirius multifiliis, is approximately three-fold smaller and has three-fold fewer genes (around 8000), which are significantly shorter than those of Tetrahymena [7]. These and other reductions have been suggested to occur due to adaptation of Ichthyophthirius to the parasitic lifestyle. On the contrary, the MAC genome of Paramecium tetraurelia has undergone numerous whole-genome duplications resulting in about 40000 genes with dozens of copies [5]. During MAC formation, around 45000 of IESs and other repetitive DNA are removed from MIC, which corresponds to ~25% of the MAC genome [5, 12]. Representatives of Spirotrichea, Oxytricha trifallax and Stylonychia lemnae, have a unique MAC genome organization comprised of thousands of so-called nanochromosomes, each typically encoding a single gene, and differently amplified up to ~2000 copies on average after extensive removal of ~96% of the MIC genome content [8]. Another distinctive feature of spirotricheans is a non-sequential or even reverse order of gene segments in the macronuclear and micronuclear DNA [2, 8]. The correct order and orientation of gene segments in the macronucleus are established during the process called genome unscrambling, which is guided by RNA [13].
The MIC-to-MAC genome reduction in Ciliates is also associated with fewer and smaller introns. In general, genomes with low intron densities also tend to have smaller introns and tend to be reduced in other ways as well, as seen for the yeast genome and more extremely compacted genomes such as those of microsporidia [14, 15]. The average number of introns per gene is low in Ciliates and varies from 1.6–2.3 in Paramecium and Oxytricha [5, 8] to 4.8 in Tetrahymena [7]. The alveolate ancestor was predicted to be relatively intron-rich with intron density comparable to that of human genes, indicating predominant introns loss during the evolution of alveolates [16]. For spirotricheans, one explanation of the observed introns scarcity could be transformation of introns into IESs leading to their permanent excision from the micronuclear genome during the MIC-to-MAC transition [17, 18]. However, the details of this mechanism in spirotricheans as well as its presence in other lineages remain unknown.
In Paramecium, the length of all introns lies in an extremely narrow range, 25–45 bp [5]. These are among the smallest introns known, likely reflecting a functional limit for spliceosomal recognition [19]. The intron size limitations for short introns recognition and exclusion from pre-mRNA are likely to be compensated by splicing spellchecking mechanisms such as nonsense-mediated decay (NMD). Indeed, in Paramecium, 3n introns (with length divisible by three) are under strong evolutionary pressure and have a high frequency of in-frame stop codons [20]. In the case of introns retention, the latter would guarantee termination of the translation of mis-spliced mRNAs and, hence eliminate production of potentially misfolded proteins.
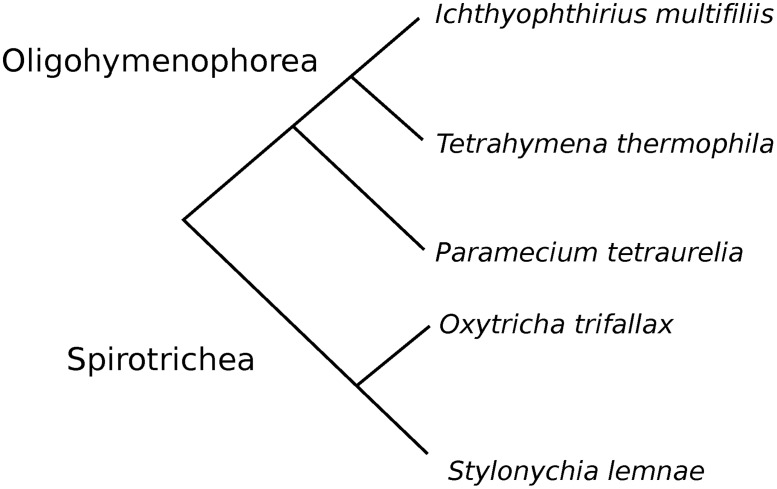
In many species, a substantial fraction of genes contains short introns [21]. From this perspective, Ciliates provide a useful model to explore the mechanisms of splicing and the evolution of short introns, as either all or most of their introns are very short [5–10]. Here, we considered introns in the genomes of several representatives of Ciliates, Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia (Fig 1). In each organism, we studied intron lengths, abundance, and distributions of 3n and 3n±1 introns, frequencies of in-frame stop-codons in introns, their splice sites, and evolutionary conservation of the introns positions and lengths.
Fig 1. Phylogenetic relationships between the studied species.
Results
Characterization of short introns in Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia
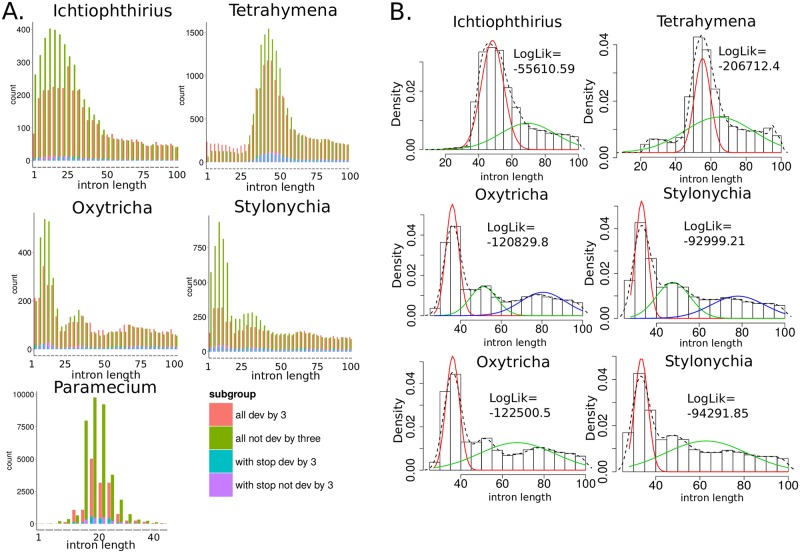
Fig 2 features the distributions of intron lengths in genes of Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia. Intron lengths of three closest species, Paramecium, Tetrahymena, and Ichthyophthirius, have unimodal distributions with modes 25 bp, 56 bp, and 48 bp, respectively. In the interval of 1–100 bp, the intron length distributions of Ichthyophthirius and Tetrahymena can be decomposed into two weighted normal distributions (Fig 3), which likely correspond to two different intron length classes with means 48 bp, 69 bp, and 55 bp, 64 bp, respectively. Interestingly, in the same length interval, the distributions of Oxytricha and Stylonychia intron lengths have three modes [8] and can be represented as a mixture of three normal distributions with means 33–35 bp, 47–51 bp, and 78–80 bp (Table 1). Decomposition of these distributions into a sum of two weighed normal distributions results in a worse approximation with a significantly lower log-likelihood (differences in the Akaike information criteria values between the 3-component and the 2-component mixture models equal −3190 and −2580 for Oxytricha and Stylonychia, respectively), indicating that these organisms possess three distinct classes of introns.
Fig 2.
A. Intron length distributions of Paramecium [5], Tetrahymena [6], Ichthyophthirius, Oxytricha [8], and Stylonychia. Introns containing “n” or “N” nucleotides were excluded. Pink and green colors show the numbers of 3n±1 and 3n introns, respectively. Blue and purple colors show the numbers of 3n±1 and 3n introns with in-frame stop codons, respectively. B. Decomposition of the intron length distributions of Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia in the length interval 1–100 bp into a sum of 2 or 3 weighted normal distributions. Histograms show the observed distributions without decomposition, whereas red, blue, and green curves denote the decomposed weighted normal distributions.
Fig 3. Densities of introns along the genes.
Genes containing at least five introns were selected and divided into five equal intervals; then numbers of introns in each interval were calculated. Different bar colors designate groups of introns (see the inserted legend). The same trend was observed for genes with ≥ 3 introns (S4 Fig).
Table 1. Parameters of the weighted normal decomposition components of the introns length distributions of Ichthyophthirius, Tetrahymena, Oxytricha, and Stylonychia in the introns length interval 1–100 bp.
| Parameter | First component | Second component | Third component | Organism |
|---|---|---|---|---|
| Mean | 48.3 | 69.5 | – | Ichthyophthirius |
| SD | 6.7 | 16.5 | – | |
| Weight | 0.6 | 0.4 | – | |
| Log-likelihood | −55611 | |||
| Mean | 55.2 | 64.0 | – | Tetrahymena |
| SD | 4.6 | 19.5 | – | |
| Weight | 0.4 | 0.6 | – | |
| Log-likelihood | −206712 | |||
| Mean | 36.0 | 51.2 | 80.5 | Oxytricha |
| SD | 3.2 | 6.6 | 11.0 | |
| Weight | 0.4 | 0.2 | 0.3 | |
| Log-likelihood | −120830 | |||
| Mean | 33.1 | 47.8 | 78.1 | Stylonychia |
| SD | 2.9 | 7.8 | 12.5 | |
| Weight | 0.4 | 0.3 | 0.3 | |
| Log-likelihood | −92999 | |||
From the intron length distributions in Fig 2 it is clear that 3n introns are distributed differently in all studied species. We observed that 3n introns are significantly underrepresented in 33–63 bp introns of Ichthyophthirius, 45–62 bp introns of Tetrahymena, 27–69 bp introns of Stylonychia, and 27–38 bp introns of Oxytricha (as described in Methods, “Calculation of 3n introns underrepresentation”). Therefore, 3n introns depletion was intrinsic for short introns of all studied species, whereas no significant under-representation of 3n introns was identified for longer introns. Interestingly, in Tetrahymena, 3n introns in the range 22–44 bp were significantly more frequent, and, relative to 3n±1 introns, depleted in in-frame stop codons (Table 2).
Table 2. Depletion of 3n introns in Ichthyophthirius, Tetrahymena, Oxytricha, and Stylonychia.
| Introns length interval, bp | Organism | Over- or underrepresentation of 3n introns in the length interval | P-value |
|---|---|---|---|
| 33–63 | Ichthyophthirius | 1.4-fold underrepresented | < 2.2x10-16 |
| 45–62 | Tetrahymena | 1.3-fold underrepresented | < 2.2x10-16 |
| 22–44 | Tetrahymena | 1.5-fold overrepresented | < 2.2x10-16 |
| 27–38 | Oxytricha | 1.3-fold underrepresented | < 2.2x10-16 |
| 27–69 | Stylonychia | 1.7-fold underrepresented | < 2.2x10-16 |
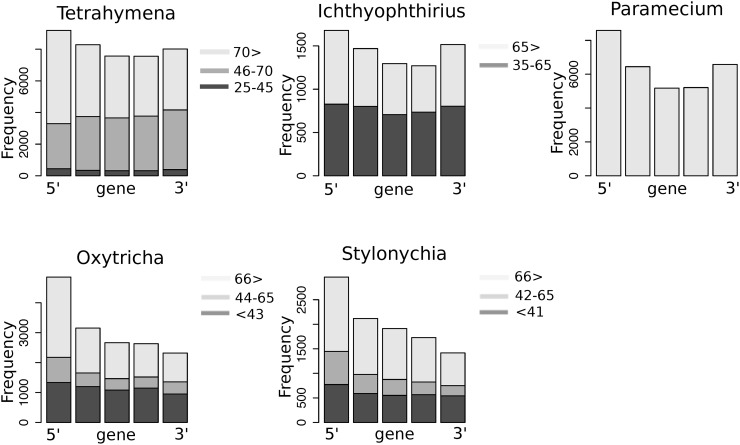
In order to analyze densities of introns along the genes with respect to the introns length, we divided introns into several discrete groups by their length. For Tetrahymena and Ichthyophthirius, the groups were defined based on the inflection points of the intron length densities and comprised of 25–45 bp (ultra-short), 46–70 bp (short), and >70 bp (long) groups for Tetrahymena, and 35–65 bp (short) and >65 bp (long) groups for Ichthyophthirius. For Oxytricha and Stylonychia, the groups were <43 bp (ultra-short), 44–65 bp (short), and >66 bp (long), as defined based on the local minima of the fitted introns length distributions.
Intron positions in Oxytricha and Stylonychia were strongly skewed towards the 5' end of a gene, especially in the groups of short and long introns (Fig 3). Interestingly, none of the intron groups of Tetrahymena and Ichthyophthirius are uniformly distributed along the genes, but peaked at their 5' and 3' ends to a different extent. The pattern is most distinct in Paramecium and less so in Tetrahymena, although long introns of Tetrahymena tend to be more non-evenly distributed than short introns (Fig 3). Tetrahymena is most intron-rich among the studied organisms, and its introns density across the genes most closely resembles the uniform distribution, whereas Paramecium has almost twofold fewer introns per gene.
Splice sites in short introns
The initial stage of splicing, precise introns recognition, depends on a complex organization of regulatory motifs, which interact with different parts of the spliceosomal complex and are located both in introns and exons [22]. However, although short introns prevail in the studied genomes, it is not clear how these extremely small introns are recognized and processed by the spliceosomal machinery [15]. Therefore, we first explored the nucleotide composition at the 5' and 3' splice sites.
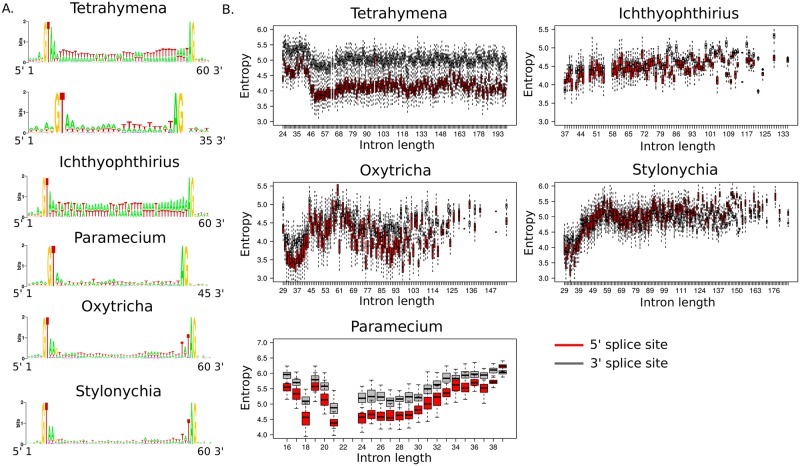
Like all eukaryotes, Ciliates have conserved dinucleotides at the intron termini [5–10], hence satisfying the GT—AG, or Breathnach-Chambon, rule [23]. However, introns of Oxytricha and Stylonychia have longer splice motifs with five conserved nucleotides at both ends (5'–GTAAG…TATAG–3') [8, 9, 24] (Fig 4A). Exon regions adjacent to the exon-intron boundaries are not conserved in any of the studied species.
Fig 4.
A. Conserved nucleotides at 5' and 3' ends of introns of Tetrahymena, Ichthyophthirius, Paramecium, Oxytricha, and Stylonychia [5–10]. For each intron, a 5 bp long exon part was also considered. B. Strength of the 5' and 3' splice site motif as a function of the intron length. The strength of the splice site motif was calculated as the entropy of its frequency matrix, see the main text. Red and black boxplots designate the entropy measure of 5' and 3' splice sites, respectively. In order to account for uneven sample size, for each intron length up to 200 bp, we randomly sampled 50 introns 50 times and estimated their average splice sites entropy. Intron length intervals with less than 50 introns were excluded.
We analyzed the relation between the strength of the 3' and 5' splice site motifs and the intron length. For this purpose, we considered 5 bp-long 5' and 3' boundaries of each intron, constructed frequency matrices, and calculated the entropy of the obtained matrices by the formula:
where pi,j is the frequency of a nucleotide i at a motif position j.
In both Oxytricha and Stylonychia, the shortest introns (30–45 bp) tend to have strongest splice sites among other introns of these organisms. In Tetrahymena, the strongest 3' and 5' splice sites are found in introns with the length in the range 45–70 bp. These observations are in agreement with the intron-definition mechanism being predominantly used for splicing of short introns [25]. A reverse pattern has been observed for 24–45 bp long introns of Tetrahymena, that tend to have weaker splice sites. Similar to other introns of Tetrahymena, they possess the same conserved nucleotides at their 5' and 3' ends, (GT…AG), but are associated with gaps in protein alignments with orthologs from other studied species, indicating that these introns could be gene annotation artifacts (S2 Fig).
Unlike other species, Ichthyophthirius demonstrates no dependency between the strength of the splice sites and the intron length (Fig 4B).
Frequencies of in-frame stop codons in ultra-short introns
We next analyzed frequencies of in-frame stop codons in 3n introns in comparison with 3n±1 introns across the studied species. The background frequencies of in-frame stop codons were generated by Monte-Carlo simulations as described in Methods (“Estimation of in-frame stop-codon frequencies in introns”).
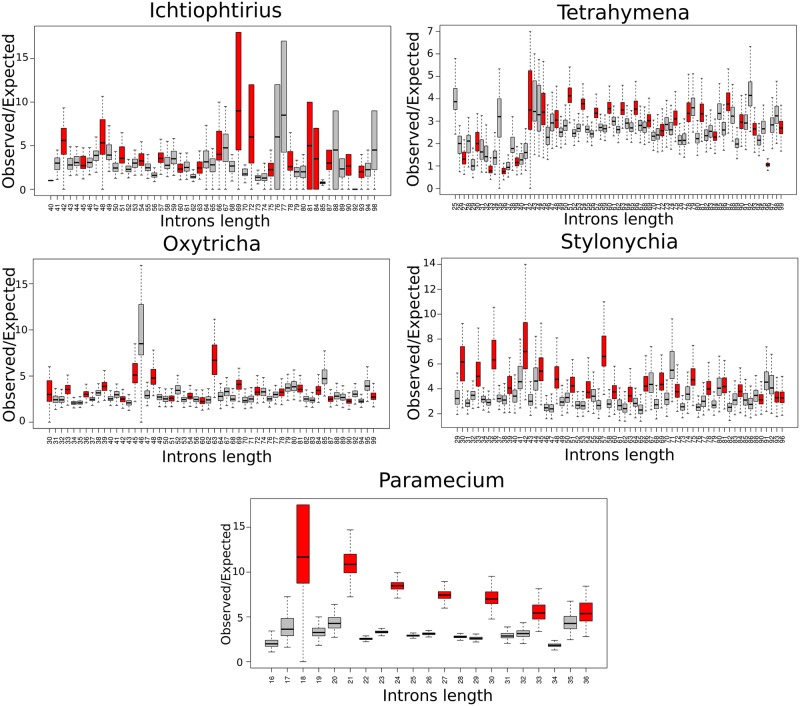
Our results indicate that in-frame stop-codons in short 3n introns are enriched in all studied ciliates. The ratio of the observed and expected numbers of 3n introns with in-frame stop codons was the highest in all introns of Paramecium, 50–70 bp introns of Tetrahymena, 30–40 bp introns of Oxytricha, and 30–70 bp introns of Stylonychia (Fig 5), overlapping with the regions of significant 3n introns underrepresentation in these species. Again, in contrast to other studied ciliates, in Ichthyophthirius 40–60 bp 3n introns were only slightly enriched in in-frame stop codons, compared to 3n±1 introns. The observed underrepresentation of 3n introns and their enrichment in in-frame stop-codons indicate that splicing of the short introns is an error-prone mechanism in Ciliates, leading to the inclusion of introns into mRNAs that should be degraded by the nonsense-mediated decay.
Fig 5. Enrichment of in-frame stop codons in introns of Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia.
Red and gray boxplots designate O/E values for 3n and 3n±1 introns, respectively. Boxplots were obtained by combining O/E values from 1000 replicates. P-value of stop-codons enrichment was less than 10-4 (by Monte-Carlo 1000-fold simulations) for all introns of Paramecium, Oxytricha, Stylonychia, 41–100 bp long introns of Tetrahymena, and almost all introns of Ichthyophthirius.
Identification of conserved introns
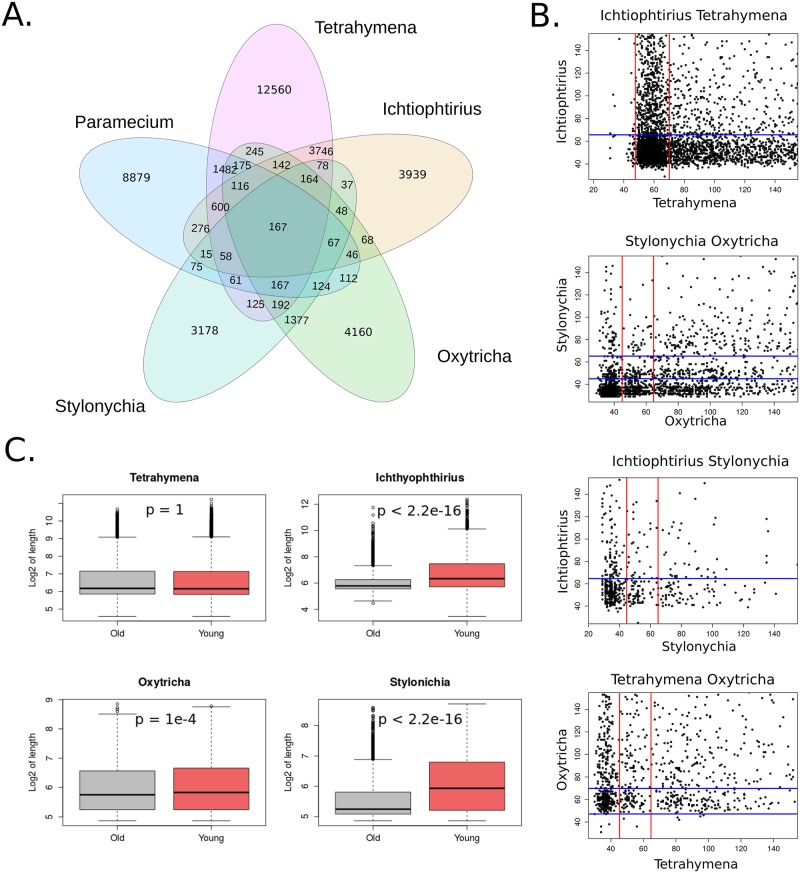
Conserved introns were identified as described in Methods (“Identification of conserved introns”). As expected, given the branching order in the phylogenetic tree (Fig 1), the largest number of common introns was found in two pairs of the most closely related species, Tetrahymena/Ichthyophthirius (5022 introns), and Oxytricha/Stylonychia (2266 introns) (Fig 6). Of note, however, is that the Stylonychia genome annotation was made using the Oxytricha annotation as a model, and thus the conservation results might be slightly biased for these species. Paramecium had most of the common introns with Tetrahymena and almost two-fold fewer with Ichthyophthirius and Oxytricha. In total, 167 intron positions were conserved between all studied species. In general, 28%, 37%, 59%, 43%, and 46% of intron positions were conserved in at least one of the remaining species for Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia, respectively.
Fig 6.
A. The numbers of introns conserved between the orthologs of Paramecium, Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia. B. Scatter-plots of orthologous introns length of Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia. The intron length range up to 150 bp is shown. Red and blue lines separate classes of the intron lengths. C. L distributions of the conserved and young introns in orthologous genes for Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia (in the log2 scale). Conserved introns are significantly shorter than young introns in Ichthyophthirius, Oxytricha, and Stylonychia (by the Wilcoxon rank sum test, p-value < 2.2×10-16, < 10-4, and < 2.2×10-16, respectively).
The numbers of introns in orthologous genes were moderately correlated between Oxytricha and Stylonychia (0.52, Spearman's rank correlation coefficient, SCC), and between Tetrahymena and Ichthyophthirius (0.66, SCC); a weak correlation was observed between the numbers of introns in orthologs of Paramecium with either Tetrahymena or Ichthyophthirius (0.38 and 0.31 SCC, respectively).
Contrary to our expectations, we found that the length of conserved introns did not correlate between both closest and distant organisms (Fig 6). The intron length classes were also independent of each other between the species (S3 Fig). Comparing length distributions of the conserved (old) and not conserved (young) introns in orthologs of the studied species, we found that old introns tend to be significantly shorter than young introns in Ichthyophthirius, Oxytricha, and Stylonychia, but not in Tetrahymena (Fig 6).
Discussion and Conclusions
The first distinctive feature of ciliate introns is the multimodality of their length distributions and the presence of intron length classes that are different between oligohymenophoreans and spirotricheans. These features of the ciliate introns were initially identified when annotating their genome sequences [5, 6, 8], but not yet explored in detail.
The bimodality of intron length distributions has been observed in many eukaryotic species. Two classes of introns, with a high peak of short (termed 'minimal length') introns and a much flatter peak of longer introns, ranging up to thousands of base pairs are present in humans [26, 27]. The functional link between intron length classes and gene expression suggests that introns tend to be smaller in highly expressed genes to provide for better transcriptional efficiency [27, 28]. The effect of intron length is even more important when only one copy of the gene is expressed, e.g. for imprinted genes [29] or genes expressed in a haploid pollen [30].
The bimodality of intron lengths is also associated with two major modes of recognition and pairing between adjacent splice sites, so-called “intron definition” and “exon definition” models [25, 31]. In vertebrate genomes, recognition of short introns primarily relies on intronic motifs such as splice sites, which are paired across the introns during excision, whereas the excision of longer introns is additionally supported by cis-acting elements yielding splice-site pairing across the exons [32]. In lower eukaryotes, where the genome architecture is characterized by small introns and long exons, the intron definition mechanism is probably predominant [32]. In addition to the splice motifs, intron length is critical for splicing efficiency in the organisms with the shortest introns, such as nucleomorphs. While the majority of introns of these species are 18–20 bp long and are efficiently removed, 21–27 bp introns show a substantial drop in their splicing efficiency [15]. It has been suggested that due to intron size and, consequently, information content reduction, intron size itself becomes a recognition signal, restricting the spliceosome ability to recognize introns over a certain size range [15].
Interestingly, in both Tetrahymena and Ichthyophthirius, bimodal intron length distributions are observed, suggesting that, similar to higher eukaryotes, they may possess two different mechanisms of the intron recognition and splicing, whereas, Paramecium has likely reduced or completely lost the exon definition mechanism. In general, the variability of the maxima of the intron length distributions and length classes across the studied oligohymenophoreans suggest that the evolutionary constrains imposed on intron length vary between the species possibly leading to the existence of several alternative mechanisms of introns recognition in these species. To our knowledge, spirotricheans are the only organisms with trimodal intron length distributions. Although the extent of this phenomenon in other species is not known, this could be evolutionarily associated with genome rearrangements and extensive IES removal during the MIC-to-MAC genome transition.
In most intron classes of all studied organisms, the intron length distributions had a deficit in introns whose length is divisible by three (3n introns). Here, we show that 3n introns are significantly underrepresented in short introns of Tetrahymena and Ichthyophthirius, ultra-short introns of Oxytricha, and both ultra-short and short introns of Stylonychia. The results indicate that 3n introns are under the strongest evolutionarily pressure in Paramecium [20], Tetrahymena, and Stylonychia, whereas in Oxytricha and Ichthyophthirius they are significantly underrepresented only among short introns. Then we analyzed frequencies of in-frame stop codons in introns of the studied species and found that 3n introns were enriched in stop codons in those intron length classes, where 3n introns were significantly underrepresented. Similar to the previous result, the pattern was most pronounced in three organisms, Paramecium, Tetrahymena, and Stylonychia. Further analysis of splice motifs at the introns termini revealed that short introns of Tetrahymena and ultra-short introns of Stylonychia and Oxytricha tended to have stronger splice sites than long introns of the respective species, suggesting a selection towards improved site-specific recognition and effective splicing of short introns.
Different properties have been observed for ultra-short introns of Tetrahymena. Unlike short and ultra-short introns of Tetrahymena and Oxytricha/Stylonychia, respectively, they are significantly overrepresented among 3n introns, and ultra-short 3n introns are depleted in in-frame stop-codons. These ultra-short introns also tend to have weaker splice sites. Hence, in the case of their retention, these introns would cause frequent read-through during the translation of a mis-spliced mRNA. However, we found that only a few ultra-short introns of Tetrahymena are conserved, and that ultra-short introns are associated with insertions in alignments with orthologous proteins of other studied species. The latter indicates that these introns are probably annotation artifacts, which would also explain their low conservation and weak splice sites.
There are several distinctive features between the studied oligohymenophoreans (Paramecium, Tetrahymena, Ichthyophthirius) and spirotricheans (Oxytricha and Stylonychia). They possess different numbers of intron classes (two and three, respectively). Oligohymenophoreans have two conserved nucleotides at their intron termini (5'–GT…AG–3'), whereas spirotricheans have three additional conserved nucleotides at the introns boundaries (5'–GTAAG…TATAG–3') and have stronger splice sites. Introns of oligohymenophoreans have two peaks of the introns densities, at 5' and 3' ends of genes, whereas introns of Oxytricha and Stylonychia tend to occur at the 5' end of a gene.
Introns of protein-coding genes are also unevenly distributed towards the 5′ ends of genes in Saccharomyces cerevisiae [33] and other intron-poor genomes, but are evenly distributed within coding sequences of genes in intron-rich genomes [34]. The explanation of this trend suggests the preferential loss of introns from the 3' regions of genes, possibly due to 5'-truncated transcripts subject to reverse transcription and causing intron loss via homologous recombination [33, 35, 36]. The model assumes formation of a stem-loop between DNA and cDNA, with intron loss dependent on the length of a 3' terminus folding back during homologous recombination and yielding introns losses from either the 3' end or the middle of a gene [37]. While spirotricheans demonstrate a strong 5' bias of the introns density, which is also observed in other intron-poor organisms, oligohymenophoreans show a different trend, suggesting an alternative or complementary mechanism of introns loss in oligohymenophoreans presumably from middle regions of the genes. The observed differences in the characteristics of short introns between oligohymenophoreans and spirotricheans may derive from peculiarities of their MAC genome structure and the mechanisms of its formation, which, in spirotricheans, involve extensive genome re-organization and scrambling, at some part mediated by RNA [13].
We found that a substantial fraction of introns is conserved in orthologs, ranging from 28% in Paramecium and 59% in Ichthyophthirius, reflecting the phylogenetic distance between the studied species. However, despite that, introns lengths and the introns length classes are not correlated between either distant or close species. The latter indicates that, unlike intron position in a gene, intron length is subject to fast changes in Ciliates.
In summary, our study provides a detailed characterization of the introns in two classes of Ciliates and highlights some of their distinctive properties, which, together, indicate that splicing spellchecking is a universal and highly evolutionary important process in the biogenesis of short introns in various representatives of Ciliates.
Methods
Data sources
Genome, proteome, and annotation data of Paramecium tetraurelia was obtained from [5, 38] (“Ptetraurelia_genes_v1.fasta”,“Ptetraurelia_peptides_v1.fasta”, and “parameciumDB.gff3” files); Tetrahymena thermophila, from [6, 39] (“T_thermophila_oct2008_gene.fasta”, “T_thermophila_oct2008_proteins.fasta”, and “T_thermophila_dec2011.gff3” files); Oxytricha trifallax, from [8, 40] (“Oxytricha_trifallax_022112_assembly.fasta”, “Oxytricha_trifallax_022112_aa.fasta”, and “Oxytricha_trifallax_022112.gff3” files); Ichthyophthirius multifiliis, from [7, 41] (“img1_0407.assembly.fsa”, “img1_0407.aa.fsa”, and “img1_0407.gff3” files); Stylonychia lemnae, from [9, 42] (“stylo_assembly.fa”,”stylo_protein.fa”, and”stylo.gff3” files).
Calculation of 3n introns underrepresentation
For the intron length distributions of Tetrahymena, Ichthyophthirius, Oxytricha, and Stylonychia, we determined intervals of the intron length, where 3n introns are underrepresented, as follows: starting from the minimum intron length of the organism up to 100 bp, with a 3b-long sliding window, introns of the corresponding range of lengths were selected. Next, the proportion of 3n introns within each window was calculated. Underrepresentation of 3n introns in a window was calculated as the ratio between the observed proportion of 3n introns and the expected one, which is 0.33, calculated as described above. Next, we combined 3 bp long intervals, where 3n were underrepresented, into longer continuous intervals. Similarly, we calculated the proportion of 3n introns in these longer intervals and calculated significance with the two-sample proportion test between the observed proportion of 3n introns in the longer intervals, and the expected one, equal to 0.33.
Estimation of in-frame stop-codon frequencies in introns
In order to estimate the enrichment, the background (expected) frequencies were generated as follows: for each length of introns, starting from the minimal intron length of the organism, through 100 bp, a random set of the same size, with the nucleotide frequencies as in the corresponding introns, of the length N–4 was generated 1000 times, where N is the intron length and 4 corresponds to two absolutely conserved dinucleotides at the intron termini. Next, the ratio between the numbers of the observed and randomly generated introns with in-frame stop-codons (TGA) was calculated. P-values were calculated as the number of times from 1000 replications when the expected ratio was larger or equal to the observed one.
Construction of COGs (Clusters of Orthologous Groups)
First, two rounds of psi-BLAST protein search were performed, with and without low-complexity filtering and composition-based statistics (“psiblast -comp_based_stats F -seg no” and “psiblast -comp_based_stats T -seg yes”), between putative proteins of Paramecium, Tetrahymena, and Ichthyophthirius. Next, COGs (Clusters of Orthologous Groups) were constructed by the COGtriangles software (Update 20.04.12 [43]) and assignment of Oxytricha and Stylonychia putative proteins to these COGs was performed by COGnitor [44, 45] after running two passes of psi-BLAST with and without low-complexity filtering. Further, in-paralogs were filtered out, and one-to-one best hits between the organisms were selected using the best psi-blast (for Paramecium, Tetrahymena, and Ichthyophthirius) and Cognitor (for Oxytricha and Stylonychia) scores.
Identification of conserved introns
Conserved introns were identified by the following algorithm: (1) protein sequences of orthologous genes in the same COG cluster were aligned with the “muscle” R package [46]; (2) for each intron position, 5 bp boundary regions at each side were selected; (3) for the alignment in the selected region, two parameters were estimated: a, the total number of gaps in the aligned proteins, and b, the distance between two introns positions in the alignment. Intron insertion was considered to be conserved if a ≤ 3 and b ≤ 2. The algorithm is schematically outlined in S1 Fig.
Statistical analysis and software
Locations of introns and their sequences were obtained from the annotation and genome sequence data with custom scripts written in R. Sequence logos were constructed using the WebLogo on-line tool [47]. Decomposition of introns length distributions into a sum of normal distributions was performed with the standard Expectation Maximization algorithm for normal mixtures implemented in “mixtools” R package [48]. All statistical analyses were performed in the R programming language.
Supporting Information
(1) Protein sequences of orthologous genes in the same COG cluster were aligned with “muscle” R package [40]; (2) for each intron position, 5 bp at each side were selected; (3) for alignment in a selected region, two parameters were estimated: number of gaps in both aligned proteins (a) and distance between two intron positions in an alignment (b). An intron position was considered to be conserved if a≤3 and b≤2.
(TIFF)
For comparison, two intron groups are shown: ultra-short (20–45 bp) and short (45–70 bp). In total, 254 ultra-short and 5199 short introns were considered. Orthologs were taken from COGs, which were constructed as described in Methods (“Construction of COGs”).
(TIFF)
The percent of the total number of conserved introns in a pair of species is indicated in each cell.
(TIFF)
Genes containing at least three introns were selected and divided into three equal intervals; then numbers of introns in each interval were calculated.
(TIFF)
The introns coordinates are given in numbers of base pairs and numbers of aminoacids for genes and proteins, respectively.
(ZIP)
Tables include COG IDs, protein IDs, and corresponding intron IDs. Conserved introns were identified as described in Methods (“Identification of conserved introns”).
(ZIP)
(TXT)
Acknowledgments
This study was supported by the Russian Science Foundation (grant 14-24-00155). It was initiated at the Winter School “Modern Biology—Future Biotech—2013”.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Russian Science Foundation (grant 14-24-00155).
References
- 1.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994; 58(2): 233–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prescott DM. Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat Rev Genet. 2000; 1(3): 191–8. [DOI] [PubMed] [Google Scholar]
- 3.Prescott DM. Cutting, splicing, reordering, and elimination of DNA sequences in hypotrichous ciliates. Bioessays. 1992; 14(5): 317–24. [DOI] [PubMed] [Google Scholar]
- 4.Prescott DM. Evolution of DNA organization in hypotrichous ciliates. Ann N Y Acad Sci. 1999; 870: 301–13. [DOI] [PubMed] [Google Scholar]
- 5.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006; 444(7116): 171–8. [DOI] [PubMed] [Google Scholar]
- 6.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006; 4(9): e286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyne RS, Hannick L, Shanmugam D, Hostetler JB, Brami D, Joardar VS, et al. Comparative genomics of the pathogenic ciliate Ichthyophthirius multifiliis, its free-living relatives and a host species provide insights into adoption of a parasitic lifestyle and prospects for disease control. Genome Biol. 2012;12(10): R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, et al. The Oxytricha trifallax Macronuclear Genome: A Complex Eukaryotic Genome with 16,000 Tiny Chromosomes. PLoS Biol. 2013; 11(1): e1001473 10.1371/journal.pbio.1001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aeschlimann SH, Jönsson F, Postberg J, Stover NA, Petera RL, Lipps HJ, et al. The draft assembly of the radically organized Stylonychia lemnae macronuclear genome. Genome Biol Evol. 2014; 6(7): 1707–1723. 10.1093/gbe/evu139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Jung S, Beh LY, Eddy SR, Landweber LF. Combinatorial DNA Rearrangement Facilitates the Origin of New Genes in Ciliates. Genome Biol Evol. 2015; 7(10): 2859–70. 10.1093/gbe/evv172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinogradov DV, Tsoĭ OV, Zaika AV, Lobanov AV, Turanov AA, Gladyshev VN, et al. Draft macronuclear genome of a ciliate Euplotes crassus. Mol Biol (Mosk). 2012; 46(2): 361–6 [Article in Russian]. [PubMed] [Google Scholar]
- 12.Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury JM, Denby Wilkes C, et al. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012; 8(10): e1002984 10.1371/journal.pgen.1002984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowacki Mariusz, Vijayan Vikram, Zhou Yi, Schotanus Klaas, Doak Thomas G & Landweber Laura F. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2007; 451(7175): 153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001; 414(6862): 450–3. [DOI] [PubMed] [Google Scholar]
- 15.Slamovits CH, Keeling PJ. Evolution of ultrasmall spliceosomal introns in highly reduced nuclear genomes. Mol Biol Evol. 2009; 26(8): 1699–705. 10.1093/molbev/msp081 [DOI] [PubMed] [Google Scholar]
- 16.Csurös M, Rogozin IB, Koonin EV. Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol Biol Evol. 2008; 25(5): 903–11. 10.1093/molbev/msn039 [DOI] [PubMed] [Google Scholar]
- 17.Chang WJ, Bryson PD, Liang H, Shin MK, Landweber LF. The evolutionary origin of a complex scrambled gene. Proc Natl Acad Sci U S A. 2005; 102(42): 15149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang WJ, Addis VM, Li AJ, Axelsson E, Ardell DH, Landweber LF. Intron Evolution and Information processing in the DNA polymerase alpha gene in spirotrichous ciliates: a hypothesis for interconversion between DNA and RNA deletion. Biol Direct. 2007; 2:6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavalier-Smith T. The tiny enslaved genome of a rhizarian alga. Proc Natl Acad Sci U S A. 2006; 103: 9379–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, et al. Translational control of intron splicing in eukaryotes. Nature. 2008; 451(7176): 359–62. 10.1038/nature06495 [DOI] [PubMed] [Google Scholar]
- 21.Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001; 98(20): 11193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewey Colin N, Rogozin Igor B, and Koonin Eugene V. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. BMC Genomics. 2006; 7: 311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978;75(10): 4853–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott DM, Prescott JD, Prescott RM. Coding properties of macronuclear DNA molecules in Sterkiella nova (Oxytricha nova). Protist. 2002; 153(1): 71–7. [DOI] [PubMed] [Google Scholar]
- 25.Robberson BL, Cote GJ, Berget SM. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990; 10(1): 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Yang Z, Kibukawa M, Paddock M, Passey DA, Wong GK. Minimal introns are not ‘junk’. Genome Res. 2002; 12: 1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belshaw R and Bensasson D. The rise and falls of introns. Heredity. 2006; 96: 208–213. [DOI] [PubMed] [Google Scholar]
- 28.Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003; 13: 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurst LD, McVean G, Moore T. Imprinted genes have few and small introns. Nat Genet. 1996; 12: 234–237. [DOI] [PubMed] [Google Scholar]
- 30.Seoighe C, Gehring C, Hurst LD. Gametophytic selection in Arabidopsis thaliana supports the selective model of intron length reduction. PLoS Genet. 2005; 1: e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talerico M, Berget SM. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994; 14(5): 3434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013; 4(1): 49–60. 10.1002/wrna.1140 [DOI] [PubMed] [Google Scholar]
- 33.Fink GR. Pseudogenes in yeast? Cell. 1987; 49: 5–6. [DOI] [PubMed] [Google Scholar]
- 34.Lin K, Zhang DY. The excess of 5' introns in eukaryotic genomes. Nucleic Acids Res. 2005; 33(20): 6522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derr LK. 1998. The involvement of cellular recombination and repair genes in RNA-mediated recombination in Saccharomyces cerevisiae. Genetics 148: 937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mourier T, Jeffares DC. Eukaryotic intron loss. Science. 2003; 300(5624): 1393 [DOI] [PubMed] [Google Scholar]
- 37.Niu DK, Hou WR, Li SW. mRNA-mediated intron losses: evidence from extraordinarily large exons. Mol Biol Evol. 2005; 22(6): 1475–81. [DOI] [PubMed] [Google Scholar]
- 38.ParameciumDB. http://paramecium.cgm.cnrs-gif.fr/.
- 39.Tetrahymena Genome Database. http://ciliate.org/index.php/.
- 40.Oxytricha Genome Database. http://oxy.ciliate.org/.
- 41.Ichthyophthirius Genome Database. http://ich.ciliate.org/.
- 42.Stylonychia Genome Database. http://stylo.ciliate.org/.
- 43.COGsoft. ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/COGsoft/.
- 44.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997; 278(5338): 631–7. [DOI] [PubMed] [Google Scholar]
- 45.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, et al. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010;26(12): 1481–7. 10.1093/bioinformatics/btq229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Research. 2004;14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benaglia T, Chauveau D, Hunter DR, Young D. mixtools: An R Package for Analyzing Finite Mixture Models. Journal of Statistical Software. 2009;32(6): 1–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(1) Protein sequences of orthologous genes in the same COG cluster were aligned with “muscle” R package [40]; (2) for each intron position, 5 bp at each side were selected; (3) for alignment in a selected region, two parameters were estimated: number of gaps in both aligned proteins (a) and distance between two intron positions in an alignment (b). An intron position was considered to be conserved if a≤3 and b≤2.
(TIFF)
For comparison, two intron groups are shown: ultra-short (20–45 bp) and short (45–70 bp). In total, 254 ultra-short and 5199 short introns were considered. Orthologs were taken from COGs, which were constructed as described in Methods (“Construction of COGs”).
(TIFF)
The percent of the total number of conserved introns in a pair of species is indicated in each cell.
(TIFF)
Genes containing at least three introns were selected and divided into three equal intervals; then numbers of introns in each interval were calculated.
(TIFF)
The introns coordinates are given in numbers of base pairs and numbers of aminoacids for genes and proteins, respectively.
(ZIP)
Tables include COG IDs, protein IDs, and corresponding intron IDs. Conserved introns were identified as described in Methods (“Identification of conserved introns”).
(ZIP)
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.