Abstract
In order to understand the importance of functional proteins in mosquito behavior, following blood meal, a baseline proteomic dataset is essential for providing insights into the physiology of blood feeding. Therefore, in this study as first step, in solution and 1-D electrophoresis digestion approach combined with tandem mass spectrometry (nano LC-MS/MS) and computational bioinformatics for data mining was used to prepare a baseline proteomic catalogue of salivary gland proteins of sugar fed An. culicifacies mosquitoes. A total of 106 proteins were identified and analyzed by SEQUEST algorithm against mosquito protein database from Uniprot/NCBI. Importantly, D7r1, D7r2, D7r4, salivary apyrase, anti-platelet protein, calreticulin, antigen 5 family proteins were identified and grouped on the basis of biological and functional roles. Secondly, differential protein expression and annotations between salivary glands of sugar fed vs blood fed mosquitoes was analyzed using 2-Delectrophoresis combined with MALDI-TOF mass spectrometry. The alterations in the differential expression of total 38 proteins was observed out of which 29 proteins like beclin-1, phosphorylating proteins, heme oxygenase 1, ferritin, apoptotic proteins, coagulation and immunity like, serine proteases, serpins, c-type lectin and protein in regulation of blood feeding behavior were found to be up regulated while 9 proteins related to blood feeding, juvenile hormone epoxide hydrolase ii, odorant binding proteins and energy metabolic enzymes were found to be down regulated. To our knowledge, this study provides a first time baseline proteomic dataset and functional annotations of An. culicifacies salivary gland proteins that may be involved during the blood feeding. Identification of differential salivary proteins between sugar fed and blood fed mosquitoes and their plausible role may provide insights into the physiological processes associated with feeding behavior and sporozoite transmission during the process of blood feeding.
Introduction
Malaria, a vector borne parasitic disease is caused by protozoa in the genus Plasmodium and affects 198 million cases and leads to an estimated 584,000 deaths worldwide in 2013 [1]. Among different anopheline vectors, Anopheles culicifacies is the most abundant rural vector of malaria in South East Asian region including India, where it contributes for about 70% of malarial cases [2–3]. For malaria transmission, various Plasmodium species are injected into the human host typically via the bites of female Anopheles mosquito. In order to transmit malaria, at least two bites are required by the mosquito, one for acquiring the parasite infection and other for the transmission of malaria parasites to a new human host [4].
The salivary glands of female Anopheles mosquitoes are important because infective form of malaria parasite must invade mosquito salivary glands, before they are transmitted to the human host. The salivary glands are also the site for the maturation of sporozoites, where a set of sporozoite-vector interactions takes place and these interactions determines the competence of parasite and success of malaria transmission [5]. Salivary glands of blood sucking insects have evolved to complement their blood feeding behaviour and produces large array of biochemically active molecules, which deactivates host’s hemostatic response triggered by biting and helps in food ingestion and digestion [6]. Salivary proteins of haematophagous arthropods have immunomodulatory, anti-inflammatory and anti-coagulant properties for successful blood feeding. In addition, saliva proteins are antigenic and immunogenic which boosts the infectivity of parasite [7–9]. For the intake of blood meal, female mosquitoes inject the concoction of these salivary molecules into the vertebrate host and this complex saliva mixture act as a transmission fluid for the parasite [10].
Earlier transcriptomic studies have contributed a lot in understanding the salivary gland tissue complex in various mosquito species [11–12] however today, rapid and more sensitive technological advances in proteomics approaches enable to identify large catalogues of annotated functional proteins and their molecular and biological characterization. Proteomics has its advantage over genomic studies as it fills the void between genome sequence and the fate of its expression at cellular level. In recent years by utilising the high throughput technique of mass spectrometry many proteomic maps of important mosquito species salivary gland have been generated which provided new insights into the functional role of various salivary proteins in An. stephensi [13], An. campestris-like [14], An. barbirostris species A2 [15], Aedes aegypti [16], An. funestus[17],An. gambiae [10, 18], Culex pipiens quinquefasciatus [19]. Studies on the differentially expressed salivary gland proteins of susceptible and insecticide resistant mosquitoes of An. stephensi have also been carried out earlier in our lab using 2D electrophoresis and mass spectrometry [20]. An. culicifacies, a rural vector of malaria in South East Asian region including India as a complex of five isomorphic sibling species A, B, C, D, and E [21]. Since few transcriptomic data are available for An. culicifacies mosquitoes and unknown genome sequences till date, therefore, a baseline information and detailed description of proteomics data of An. culicifacies salivary glands will serve as a platform for all the advanced future studies during development and transmission of malaria parasite.
In the present study, using an in solution and 1-DE in gel trypsin digestion approach followed by high throughput LC/MS/MS analysis and data mining, a first comprehensive proteomic catalogue of An. culicifacies salivary gland functional proteins with their functions and biological process is provided. We also provide data on annotated proteins of An. culicifacies salivary glandsusing 2-DE coupled with MALDI-TOF TOF mass spectrometry using blood fed mosquitoes. Our results revealed significant compositional differences in salivary gland proteins expressed after blood meal as compared with sugar fed control mosquitoes. This baseline proteomic catalogue of sugar fed An. culicifacies salivary glands may be further explored to elucidate a role of novel expressed proteins in parasite blood feeding and malaria transmission. This information will serve as a platform for all the advanced future studies related to blood feeding behaviors, parasite maturation in An. culicifacies mosquitoes to achieve the goal of developing malaria blocking strategies and other aspects of malaria transmission.
Methods
Chemicals
All chemicals were of analytical grade and were purchased from Sigma, St. Louis, MO, USA. Proteomic grade trypsin and protease inhibitors set was purchased from Roche Diagnostics, Germany. All kits and protein markers were purchased from Banglore Genei, Bangalore, India and G-biosciences, St. Louis, MO, USA. All the solutions were prepared using Milli-Q purified water, Milliore Corp., Bedford, MA, USA.
Mosquitoes
An. culicifacies mosquitoes were used in this study. This strain has been successfully reared through consecutive generations in our insectaries at National Institute of Malaria Research, New Delhi, INDIA. Female mosquitoes aged 2–3 days were maintained in stable conditions of temperature 27°C ± 2°C and 70%± 10% relative humidity and a photoperiod of 12:12 (light/dark) hours. These mosquitoes were divided into two groups i.e. sugar fed (SF) and blood fed (BF) mosquitoes. SF mosquitoes were fed on 10% sucrose diet only whereas the BF mosquitoes were fed on blood meal by feeding on rabbits. The protocol for mosquitoes feeding on rabbits was approved by the animal ethics committee of National Institute of Malaria Research, India.
Salivary glands extract (SGE) preparation
Salivary glands were dissected from 100 adult cold anesthetized An. culicifacies female mosquitoes (SF and BF each) under stereomicroscope (4X magnification) using fine needles and pooled in Phosphate Buffered Saline (PBS; 10mM Na2SO4, 145mM NaCl (pH 7.2) containing protease inhibitors (Complete, Roche Diagnostics, Germany). 100 pairs of pooled salivary glandswere homogenised by ultrasonication (3 pulses of 20 sec each) on ice. Salivary gland homogenate suspension was then centrifuged for 10 min at 5000 rpm at 4°C. Cell debris pellet was discarded and supernatant containing salivary gland protein extract (SGE) was stored at -80°C until further investigations. Protein concentration in the SGEs was quantified by Lowry’s method (GeNeiTM Protein Estimation Kit) using BSA as a standard [22].
Sample preparation for LC-MS/MS
In solution trypsin digestion
SGEs were reduced, alkylated and digested with trypsin for LC-MS/MS analysis. Briefly, 50 μg of SGE was denatured using 4M urea. After that disulphide bonds of the proteins were reduced by incubating at 560 C for 1 hour with dithiothreitol (DTT, 10mM). After reduction, proteins were alkylated with iodoacetamide (IAA, 25mM) in dark for 30 min at 250 C. Subsequently, ammonium bicarbonate (NH4HCO3, 100mM pH 8.1) was added to the protein solution to reduce the urea concentration to 0.5M. Finally, the protein lysates were digested in to peptides by incubating with trypsin: substrate ratio of 1:50 at 37°C overnight. This trypsin digest obtained was dried in a speed vac till complete dryness. Samples were cleaned and desalted using C18 packed ziptip for further analysis by nano LC-MS/MS.
One-dimensional gel electrophoresis (1-DE)
1-DE experiments were performed for fractionation of SGE samples of SF mosquitoes. Briefly, 27 μg of SGE sample was mixed with sample buffer (0.625M TrisHCl, 10% SDS, glycerol, and distilled water) containing β-mercaptoethanol (10% vol/vol) and boiled for 5 min at 95°C. Denatured protein sample were then loaded on to SDS PAGE mini gel consisting of 3% stacking and 12% resolving gel of 1-mm thickness along with the protein molecular weight marker (Genei protein range marker, Bangalore Genei) and subjected to electrophoresis (Bio-Rad electrophoresis apparatus, USA). After completion of the run, the gel was stained with FOCUS-FAST silverTM stain (G-Biosciences). Various bands were observed on the gel after silver staining. Each band was cut and collected in separate eppendorf tube in 50 μl of stop solution (2% acetic acid) and further stored at -20°C for trypsin digestion.
Two-dimensional gel electrophoresis (2-DE)
For 2-DE analysis, SGEs of both SF and BF mosquitoes were desalted and cleaned using the ReadyPrep 2D Cleanup Kit (Bio-Rad). 100μg of each sample was resuspended in 300μl of rehydration buffer (8M urea, 2M thiourea, 2% CHAPS, 50 mM DTT, 0.2% Bio-Lyte 3/10 ampholyte and a trace of bromophenol blue). Samples of both SF and BF mosquitoes were then immobilized on 17 cm IPG strips of pH 3–10 using a Protean IEF Cell (Bio-Rad) and proceeded for isoelectric focusing (IEF). Briefly, each sample was pipetted as a line along the edge of a rehydration tray channel. IPG strips were then gently placed on to the sample in rehydration tray (gel side down) and layered with 2–3 ml of mineral oil. The samples were rehydrated on IPG strips at 20°C overnight. After that IEF was run in three steps: 1) 250V for 20 min in linear mode; 2) 10000V for 2.5 hrs in linear mode; 3) 10000V for 5–7 hrs and 40000 V-hrs in rapid mode. After IEF run, the strips were equilibrated first in equilibration buffer I (6M urea, 0.375M Tris-HCl, pH 8.8, 2% SDS, 20% glycerol and 2% DTT) for 10 min and then in equilibration buffer II (6M urea, 0.375M Tris-HCl, pH 8.8, 2% SDS, 2 0% glycerol and 2.5%, IAA for 10 min. The second dimension separation was carried out on 10% SDS-PAGE on Mini Protean cell (Bio-Rad). The 2-DE gels were silver stained with FOCUS-FAST silverTM stain according to manufacturer’s instructions (G-Biosciences) after the run to visualize the spots and scanned using HP Scanjet G4010.
2-DE gel analysis
ImageMaster 2D Platinum 7.0 software (GE Healthcare Life Sciences) was used for 2-DE gel comparative analysis. Gels of SF and BF samples were analyzed and all the differentially expressed spots in blood fed samples were excised and collected in separate eppendorf tubes in 50 μl of stop solution (2% acetic acid) and stored at -20°C for trypsin digestion and MALDI-TOF analysis.
In gel trypsin digestion
Silver stained gel pieces of all the bands and selected 2-DE spots were destained until the gel became translucent. Gel pieces were dehydrated using acetonitrile (ACN) and completely dried in a speed vac. These dried gel pieces were incubated with 10mM DTT in 100mM NH4HCO3 for 1 hour at 560 C for reduction of proteins. For subsequent alkylation, DTT was removed and 55mM IAA in 100mM NH4HCO3 was added and the solution was kept for 45 min at ambient temperature in dark with occasional vortexing. Afterwards, the gel pieces were washed with 100mM NH4HCO3 (50μl) for 10 min and dehydrated with ACN two times and gel pieces were completely dried. Digestion buffer (50mM NH4HCO3 and 5mM CaCl2) was then added to gel pieces along with trypsin (12.5ng/μl) in an ice cold bath for 45 min. The gel pieces were extracted thrice with extraction buffer (5% formic acid in 50% acetonitrile, 20 min for each change) at room temperature. All the samples of SDS-PAGE gel bands were vacuum dried and reconstituted in 2% acetonitrile with 0.1% formic acid and all the 2D spots samples were suspended in Tris Acetate (TA) buffer for further LC/MS/MS and MALDI-TOF analysis.
Mass spectrometry and protein identification
Nano LC-MS/MS
Mass spectrometric analysis of in-solution and 1-DE in-gel protein digests (bands) of SF samples was performed on Thermo Scientific™ LTQ XL™ ion trap mass spectrometer. Each sample (15μl) was injected onto a New Objective PicoFrit C18 nanospray column of 360 um OD x 75μm ID x 15μm tip opening dimensions using a Thermo Scientific Surveyor Auto sampler operated in the no waste injection mode. The flow rate was 300nl/min and a linear acetonitrile gradient was used to separate the tryptic peptides based on their hydrophobicity. For in-solution digests, 2 to 35% linear acetonitrile gradient (98–65% water) with 0.1% formic acid was used over 210 min and for in-gel digests 2 to 32% acetonitrile gradient (98–68% water) with 0.1% formic acid was used over 85 min followed by high and low organic washes for another 5 min via a nanospray source. All spectra were obtained in a positive ion mode with the spray voltage set to 1.8kV and the ion transfer capillary set at 180°C. A data-dependent top 5 method was used for peptide sequencing where a full MS scan from m/z 350–1700 was followed by MS/MS scans of the five most abundant ions. Each ion was subjected to CID (Collision Induced Dissociation) for fragmentation and peptide identification. MS/MS threshold was 500 counts. Raw m/z data files derived from LC-MS/MS were analyzed using Proteome Discoverer 1.4 (Thermo Scientific). SEQUEST algorithm was used against the most recent species-specific fasta database for mosquito from NCBInr/UniProt (http://www.uniprot.org). The input parameters used for the search were dynamic modification: oxidation of methionine, static modification: carbamidomethylation of cysteine, enzyme: trypsin, missed cleavages: 2, precursor mass tolerance: ± 5000 ppm, fragment mass tolerance: ± 2 Da and the peptide level filter were used for high confidence peptides only. Results were further interpreted manually and proteins were selected on the basis of score, sequence coverage and number of peptides.
MALDI TOF/TOF MS
Trypsin digested 2 DE spots samples were first desalted and concentrated on C18 Zip Tips (Millipore, USA). Desalted peptides samples were mixed with α-cyano-4-hydroxy cinnamic acid matrix in 1:1 ratio and the 2μl of this mixture was spotted on to the MALDI plate. The plate was analyzed on MALDI TOF/TOF Bruker Daltonics UltraFlex III instrument operated in positive-ion reflector mode of 500–3000m/z detection range. Further analysis was done with Flex AnalysisTM software (Bruker-Daltonics) and calibrated internally for autoproteolysis of peptides with trypsin to obtain the peptide mass fingerprint. Peaklist data files obtained were analyzed using peptide mass fingerprinting (MatrixScience) search against most recent mosquito database from UniProt for identification of the proteins. Parameters used for search were: fixed modification (carbamidomethyl), variable modification (Methionine oxidation), enzyme (trypsin), peptide tolerance: 100-500ppm, Missed Cleavages: 1 or 2.
Bioinformatics and Data analysis
Bioinformatics analysis were carried out using Blast P and SMART algorithm (http://smart.embl-heidelberg.de/) in order to find out functions and conserved domains. Signal peptides were also depicted using Signal P 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Gene ontology (GO) search engines (http://www.geneontology.org/) were used for further identification of biological process and molecular function of all identified annotated putative functional proteins. The cellular component was predicted using CELLO (http://cello.life.nctu.edu.tw/)[23] and GO software.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [24] via the PRIDE partner repository with the dataset identifier PXD003450.
Results
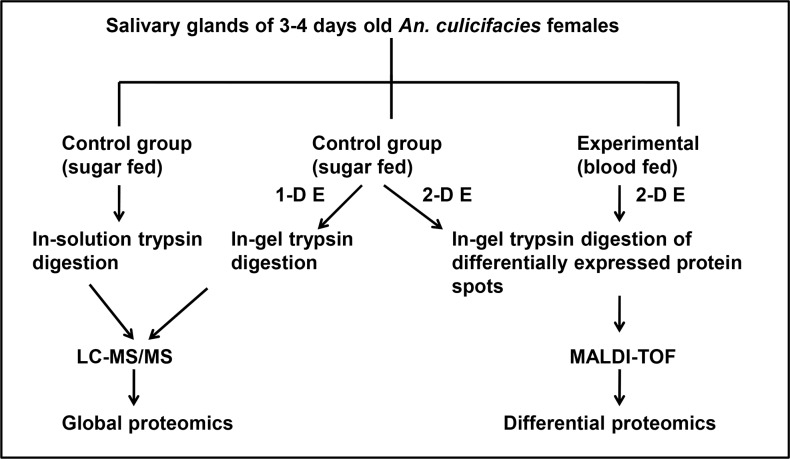
In order to understand the importance and functional role of novel expressed or annotated mosquito salivary gland proteins following blood meal, we have carried out our analyses on two aspects (Fig 1). Firstly, since no proteomic data is available for An. culicifacies, we aimed to prepare the baseline proteomic repertoire of salivary gland proteins of female An. culicifacies mosquitoes. In this study, a proteomic approach, 1-DE coupled with nano LC-MS/MS, was used for identification of salivary gland proteins of An. culicifacies. Secondly, differential proteomic analysis using 2-DE coupled with MALDI-TOF mass spectrometry was carried outto implicate the functional roles of novel expressed and annotated proteins following blood meal.
Fig 1. Experimental design.
A schematic depiction of global and differential proteomic experiments of salivary gland of sugar fed (SF) and blood fed (BF) An. culicifacies mosquitoes.
Characterization of salivary gland proteome of female An. culicifacies: in- solution and in-gel approach
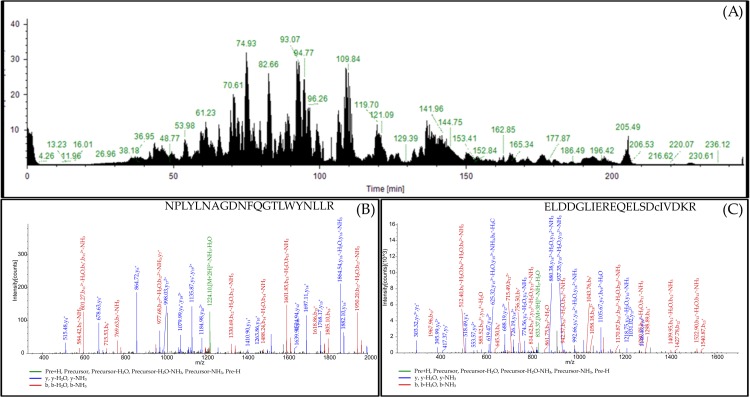
The aim of this study was to identify and characterize total salivary gland proteins of An. culicifacies to provide a baseline proteomic database as an initial step towards the cataloging of proteins and peptides. Analysis of salivary gland proteome of An. culicifacies using in-solution approaches leads to the identification of 81 major proteins. A total ion chromatogram (TIC) of the run obtained of in–solution digested sample is shown (Fig 2A). A representative MS/MS spectrum of two anti-hemostatic salivary gland specific proteins i.e. salivary apyrase protein peptide NPLYLNAGDNFQGTLWYNLLR with peak at m/z 1242.46 and antiplatelet protein peptide ELDDGLIEREQELSDcIVDKR with peak at m/z845.46 are depicted in Fig 2B and 2C.
Fig 2. Characterization of salivary gland proteome of An. culicifacies using in-solution approach.
(A) Total Ion chromatogram (LC/MS/MS) of in solution trypsin digest salivary gland proteins (B) Product ion MS/MS spectrum with peak at m/z1242.46 corresponds to the peptide sequence NPLYLNAGDNFQGTLWYNLLR which matched to known protein salivary apyrase of An. stephensi. (C) MS/MS spectrum of a peak at m/z845.46 corresponds to peptide ELDDGLIEREQELSDcIVDKR matched with antiplatelet protein of An. gambiae.
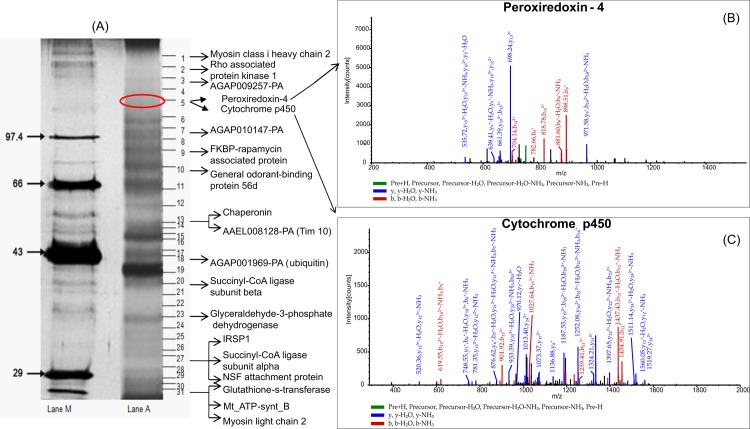
1-DE analysis of An. culicifacies salivary gland protein extract by in-gel approach showed 31 major well resolved bands on the gel after silver staining (Fig 3A). To identify the proteins in each band, each individual band was excised, digested with trypsin and subjected to nano LC/MS/MS analysis and subsequent analyses using SEQUEST algorithm leads to the identification of 36 proteins. Of the 36 proteins, 11 proteins were found to be common as identified by in solution approach. Important proteins identified by SEQUEST algorithm corresponding to each band are shown in Fig 3A. Representative MS/MS spectrum of proteins identified under band 5 with peak m/z i.e. AGAP002197-PA (cytP450) and Peroxiredoxin-4 were shown respectively (Fig 3B and 3C).
Fig 3. Characterization of salivary gland proteome of An. culicifacies using 1-DE gel based approach.
(A) 1-DE gel profile of salivary gland proteins after silver staining. Lane A: Salivary glands extract. Lane M: marker (size 14 to 100 kD) (B) MS/MS spectrum of peptide DNPEYFQKVMNSPHLLSKmDQYNFFR with peak at m/z1089.99 corresponds to protein AGAP002197-PA (cytP450) which is orthologous protein of An. gambiae. (C) MS/MS spectrum of peptide KEGGLGKINIPLVSDITHSIAK with peak at m/z765.33 corresponds to protein orthologous to a Peroxiredoxin-4 of Culex tarsalis.
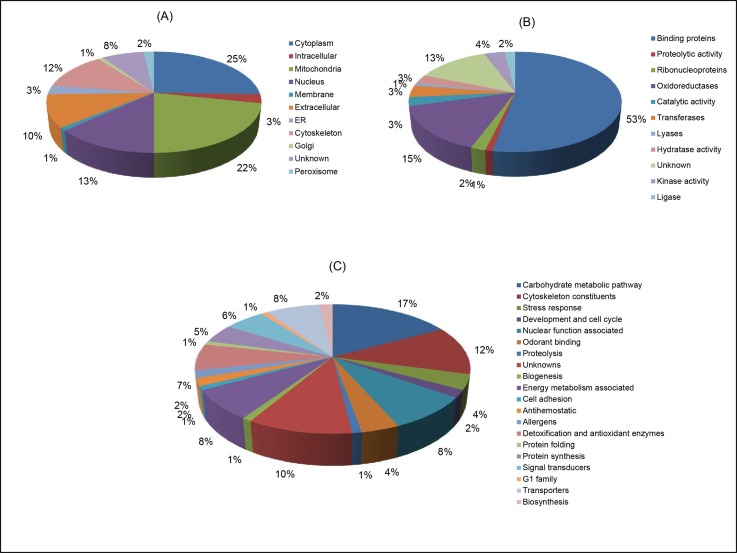
Hence, a total 106 putative functional proteins were identified using both in solution and in gel digestion approach in An. culicifacies salivary glands. Importantly, salivary gland specific proteins like D7r1, D7r2, D7r4, salivary apyrase, anti-platelet protein, antigen 5 family proteins, G1 family long form salivary protein 3, salivary maltase and other proteins like calreticulin, prohibitin, catalase, peroxiredoxin-4, cytochrome P450, thioredoxin, Glutathione S transferase were identified. Molecular functions, biological processes and sub cellular localization of proteins were further studied using various bioinformatics algorithms and gene ontology assignments. Most of the proteins were found to be present at cytoplasm (25%) followed by mitochondria (22%), nucleus (13%), extracellular (10%), cytoskeleton (12%), unknown (8%), etc. (Fig 4A). On the basis of molecular function almost 53% proteins were scrutinized to be the binding proteins followed by oxidoreductases (15%), unknowns, transferases, hydratases, kinases, ligases, catalytic enzymes, ribonucleoproteins and proteolytic enzymes (Fig 4B). Functional putative proteins were classified into 20 groups according to the biological role and among them mostly proteins were assigned to the carbohydrate metabolic pathway (17%) followed by cytoskeleton constituents (12%), unknowns (10%) etc. (Fig 4C). All the identified 106 proteins were also subjected to domain based functional analysis and are summarized below in Table 1.
Fig 4. Functional annotations of An. culicifacies salivary gland proteome using Gene Ontology tools.
(A) Sub cellular location (B) Molecular function (C) Biological process
Table 1. Salivary gland protein profile of sugar fed An. culicifacies using in-solution and in-gel approach.
| Accession | Protein | In- solution | In-gel | Organism | % Sequence coverage | Peptide matches | Comment / Band No. (in-gel) |
|---|---|---|---|---|---|---|---|
| Cytoskeletal: 13 | |||||||
| T1DPM1 | Putative actin indirect flight muscle | ✓ | An. aquasalis | 6.22 | 17 | Actin | |
| Q7PJV2 | AGAP010147-PA | ✓ | ✓ | An. gambiae | 40.68 | 82 | Myosin head / Band 14 |
| Q7PSI4 | AGAP010929-PA | ✓ | An. gambiae | 52.80 | 15 | Tubulin | |
| F5HME9 | AGAP001797-PE | ✓ | ✓ | An. gambiae | 46.32 | 13 | Tropomyosin/ Band 13 |
| F5HMN8 | AGAP001497-PB | ✓ | An. gambiae | 19.73 | 13 | Calponin homology domain | |
| B0WAN1 | Paramyosin, long form | ✓ | Cu. quinquefasciatus | 16.78 | 12 | Myosin tail | |
| Q7PNL5 | AGAP005459-PA | ✓ | An. gambiae | 30.15 | 3 | Chitin | |
| B0WVV0 | Putative uncharacterized protein | ✓ | Cu. quinquefasciatus | 4.47 | 2 | Spc97_Spc98 domain | |
| T1E7U1 | Putative troponin t skeletal muscle | ✓ | ✓ | An. aquasalis | 14.44 | 5 | Troponin/ Band 18 |
| B3RH54 | Lava lamp protein | ✓ | Cu. quinquefasciatus | 2 | 3 | Microtubule associated | |
| Q7Q7K5 | AGAP011515-PA | ✓ | An. gambiae | 24.43 | 6 | Actin/ Band 18 | |
| E9P4K4 | Myosin light chain 2 | ✓ | Cu. pipiens | 26.67 | 5 | EF-Hand domain/ Band 31 | |
| Q7Q978 | AGAP004877-PA | ✓ | An. gambiae | 14.17 | 3 | Myosin | |
| Energy metabolism related: 08 | |||||||
| Q17FL3 | ATP synthase subunit beta | ✓ | ✓ | Aedes aegypti | 65.48 | 21 | ATP binding/ Band 18 |
| Q7PHI8 | ATP synthase subunit alpha | ✓ | ✓ | An. gambiae | 27.22 | 11 | ATP binding / Band 17 |
| Q9XYC8 | AAEL005798-PA | ✓ | Aedes aegypti | 29.44 | 8 | V ATPase | |
| A7UTS9 | AGAP005627-PC | ✓ | An. gambiae | 23.10 | 6 | ATP guanido P Transferase | |
| Q7PWZ7 | AGAP001138-PA | ✓ | ✓ | An. gambiae | 12.71 | 2 | Mt. ATP synthase/ Band 31 |
| Q7PNG6 | AGAP007841-PA | ✓ | An. gambiae | 28.40 | 3 | ATP synthase epsilon | |
| Q7Q3N8 | ATP synthase subunit gamma | ✓ | ✓ | An. gambiae | 14.14 | 3 | ATP synthase/ Band 27 |
| Q7Q5G0 | AGAP006456-PA | ✓ | An. gambiae | 15.65 | 3 | NADH dehydrogenase (DH)/ Band 29 | |
| Carbohydrate metabolism: 18 | |||||||
| Q7Q1U8 | Glyceraldehyde-3-phosphate DH | ✓ | ✓ | An. gambiae | 31.63 | 6 | Phosphorylation/ Band 23 |
| Q7Q3F6 | AGAP007852-PA | ✓ | An. gambiae | 24.30 | 13 | Aconitase | |
| Q7PSR9 | AGAP011159-PA | ✓ | An. gambiae | 50.98 | 5 | Cytochrome C | |
| Q7Q3L6 | Phosphorylase | ✓ | An. gambiae | 8.31 | 4 | Phosphorylation | |
| Q7PYE7 | AGAP001903-PA | ✓ | ✓ | An. gambiae | 50.74 | 9 | L_DH/ Band 27 |
| Q7Q3D8 | AGAP007827-PA | ✓ | An. gambiae | 22.40 | 4 | Enolase | |
| T1DQ50 | Putative isocitrate DH | ✓ | An. aquasalis | 15.36 | 3 | Oxidoreductase | |
| T1E9K5 | Putative 2-oxoglutarate DH e1 | ✓ | An. aquasalis | 7.14 | 5 | Oxidoreductase | |
| F5HKV6 | Fructose-bisphosphate aldolase | ✓ | An. gambiae | 13.77 | 3 | Aldolase | |
| Q7PPE7 | Pyruvate kinase | ✓ | An. gambiae | 16.99 | 4 | Phosphorylation | |
| Q7PYD5 | AGAP001884-PA | ✓ | An. gambiae | 14.91 | 4 | Fumarase C | |
| B0VZW3 | Glycerol-3-phosphate DH | ✓ | Cu. quinquefasciatus | 18.36 | 4 | Dehydrogenase | |
| Q06DJ2 | Salivary maltase | ✓ | An. funestus | 19.90 | 3 | Glycoside hydrolase family 13 | |
| Q7PV48 | Citrate synthase | ✓ | An. gambiae | 18.45 | 6 | Transferase | |
| Q7PQM3 | 6-phosphogluconate DH | ✓ | An. gambiae | 14.73 | 5 | Oxidoreductase | |
| A7URV6 | AGAP006936-PB | ✓ | An. gambiae | 30.64 | 4 | Cytochrome C1 | |
| B0XGN1 | Succinyl-CoA ligase subunit alpha | ✓ | Cu. quinquefasciatus | 10.53 | 2 | Ligase/ Band 27 | |
| Q7PMT2 | Succinyl-CoA ligase subunit beta | ✓ | An. gambiae | 9.64 | 2 | Ligase/ Band 20 | |
| Transport: 08 | |||||||
| Q7PPA5-2 | Isoform A of Calcium-transporting ATPase | ✓ | An. gambiae | 32.87 | 22 | Cation Atpase | |
| Q27238 | ADP, ATP carrier protein 1 | ✓ | An. gambiae | 39.20 | 10 | Mt carrier protein | |
| Q7PMF3 | AGAP010025-PA | ✓ | An. gambiae | 7.45 | 2 | Rab GDP dissociation inhibitor | |
| F5HK77 | AGAP002354-PB | ✓ | An. gambiae | 5.67 | 2 | Membrane trafficking | |
| T1EB45 | Putative endocytosis/signaling protein ehd1 | ✓ | An. aquasalis | 9.11 | 4 | Dynamin domain | |
| Q7PW34 | AGAP009105-PA | ✓ | An. gambiae | 4.77 | 2 | HEAT repeats | |
| Q1HRD0 | AAEL008128-PA | ✓ | Aedes aegypti | 21.35 | 1 | Tim10 domain Band 13 | |
| B0X105 | Soluble NSF attachment protein | ✓ | Cu. quinquefasciatus | 8.53 | 2 | NSF attachment protein/ Band 27 | |
| Biosynthesis: 02 | |||||||
| Q7QA89 | AGAP004366-PA | ✓ | An. gambiae | 15.88 | 6 | Aldehyde DH | |
| Q7Q3R0 | AGAP007990-PA | ✓ | An. gambiae | 6.51 | 2 | UDP-glucuronosyl transferase | |
| Protein synthesis: 05 | |||||||
| T1DN37 | Elongation factor 1alpha | ✓ | ✓ | An. aquasalis | 30.48 | 5 | Band 18 |
| P33514 | 40S ribosomal protein | ✓ | An. gambiae | 27.60 | 2 | Ribosomal_S7e | |
| Q7PNJ7 | AGAP000883-PA | ✓ | An. gambiae | 10.54 | 3 | Elongation Factor | |
| T1DJH7 | Putative elongation factor 2 | ✓ | An. aquasalis | 8.54 | 5 | GTP binding | |
| T1DFZ6 | 60s acidic ribosomal protein | ✓ | An. aquasalis | 15.96 | 2 | Ribosomal L10/ Band 25 | |
| Nuclear function Associated: 09 | |||||||
| Q7Q2C5 | Histone H2B | ✓ | An. gambiae | 37.36 | 3 | Nucleosome assembly | |
| B6DE21 | Histone H3 | ✓ | An. darling | 30.15 | 2 | Nucleosome assembly | |
| Q7QAJ4 | AGAP003671-PA | ✓ | An. gambiae | 9.28 | 2 | Homeobox domain | |
| F5HM53 | AGAP004028-PB | ✓ | An. gambiae | 2.28 | 2 | ResIII domain | |
| B8RJF1 | Histone H4 | ✓ | Culex tarsalis | 50.52 | 5 | Nucleosome assembly | |
| Q7QE14 | AGAP010700-PA | ✓ | An. gambiae | 4.68 | 3 | HAND domain | |
| Q7QAK7 | DNA-directed RNA polymerase | ✓ | An. gambiae | 3.83 | 3 | Beta subunit | |
| B0X8G8 | RNA-binding motif protein | ✓ | Cu. quinquefasciatus | 13.87 | 2 | Band 29 | |
| B0XH66 | Guanine nucleotide binding | ✓ | Cu. quinquefasciatus | 10.80 | 4 | NCD*/ Band 31 | |
| Development and cell cycle related related: 02 | |||||||
| Q7PMG2 | AGAP009642-PA | ✓ | An. gambiae | 7.74 | 2 | Prohibitin | |
| B0WN45 | Putative uncharacterized protein | ✓ | Cu. quinquefasciatus | 10.65 | 2 | Borealin domain | |
| Detoxification and antioxidant enzymes: 07 | |||||||
| Q8MUR9 | Glutathione S-transferase S1-2 | ✓ | An. gambiae | 48.21 | 5 | Transferase/ Band 31 | |
| Q5TX96 | AGAP002197-PA | ✓ | An. gambiae | 7.07 | 2 | Cyt P450/ Band 5 | |
| B0XGK0 | Glutathione S-transferase, theta | ✓ | Cu. quinquefasciatus | 10.96 | 2 | Transferase/ Band 31 | |
| T1E7E5 | Dihydrolipoyl dehydrogenase | ✓ | An. aquasalis | 15.22 | 3 | oxidoreductase | |
| B0XCN4 | L (2) long form | ✓ | Cu. quinquefasciatus | 8.96 | 5 | Thioredoxin/ Signal P: 1–29 | |
| Q6RBZ5 | Catalase | ✓ | An. gambiae | 2 | Peroxidase | ||
| B8RJA9 | Peroxiredoxin-4 | ✓ | Cu. tarsalis | 16.30% | 2 | Peroxidase/ Band 5 | |
| Stress response: 04 | |||||||
| Q7PQK5 | AGAP004192-PA | ✓ | An. gambiae | 13.81 | 6 | HSP 70/ Signal P: 1–20 | |
| Q7PT10 | Heat shock protein 83 | ✓ | An. gambiae | 12.36 | 6 | HSP 90 | |
| B0X2F8 | FKBP-rapamycin associated protein | ✓ | Cu. quinquefasciatus | 18.95 | 1 | Kinase/ Band 9 | |
| B0WWW | Chaperonin | ✓ | Cu. quinquefasciatus | 11.73 | 1 | HSP 60/ Band 13 | |
| Signal Transduction: 06 | |||||||
| B0WM74 | Putative uncharacterized protein | ✓ | Cu. quinquefasciatus | 3.99 | 2 | Pleckstrin homology domain | |
| A0NBC2 | AGAP007643-PA | ✓ | An. gambiae | 17.34 | 3 | 14-3-3 family | |
| Q16ZM1 | AAEL008141-PA | ✓ | Aedes aegypti | 3.38 | 3 | PAS domain | |
| Q17HK0 | AAEL002654-PA | ✓ | Aedes aegypti | 4.33 | 2 | Growth factor receptor/ Signal P: 1–24 | |
| Q16J24 | AAEL013466-PA | ✓ | Aedes aegypti | 2.82 | 2 | Ankyrin repeats | |
| Q7PUN2 | AGAP001969-PA | ✓ | An. gambiae | 32.36 | 2 | Ubiquitin domain/ Band 18 | |
| Anti-hemostatic proteins:02 | |||||||
| Q8I6Q2 | Salivary apyrase | ✓ | An. stephensi | 7.48 | 4 | 5'-Nucleotidase/ Signal P: 1–22 | |
| B3VDI9 | Anti-platelet protein | ✓ | An. gambiae | 8.97 | 4 | Collagen binding | |
| Chemosensory/Odorant binding: 04 | |||||||
| Q06DJ4 | Short form D7r4 | ✓ | An. funestus | 17.58 | 3 | Signal P: 1–22 | |
| O97414 | D7r1 protein | ✓ | An. gambiae | 6.31 | 2 | Signal P: 1–22 | |
| Q95V98 | Short form D7r2 salivary protein | ✓ | An. arabiensis | 27.98 | 3 | GOBP | |
| B0X0G3 | General odorant-binding protein 56d | ✓ | Cu. quinquefasciatus | 9.16 | 2 | GOBP/ Band 10 | |
| SG1 family: 01 | |||||||
| Q06DI5 | G1 family long form salivary protein 3 | ✓ | An. funestus | 13.01 | 4 | NCD* | |
| Allergens: 02 | |||||||
| Q8I6R0 | Salivary antigen-5 related protein | ✓ | An. stephensi | 11.58 | 2 | CAP domain/ Signal P: 1–21 | |
| L7RJB8 | IRSP1 | ✓ | An. gambiae | 13.48 | 1 | CAP domain/ Band 27 | |
| Protein folding: 01 | |||||||
| J7EQD2 | Calreticulin | ✓ | An. stephensi | 35.47 | 8 | Multiple functions/ Signal P: 1–16 | |
| Cell adhesion: 01 | |||||||
| Q17HV9 | AAEL002565-PA | ✓ | Aedes aegypti | 2% | 5 | fibronectin 3 | |
| Proteolysis: 01 | |||||||
| Q5TP08 | AGAP009917-PA | ✓ | An. gambiae | 2.74 | 5 | Rhs repeat | |
| Biogenesis: 01 | |||||||
| Q17I93 | AAEL002433-PA | ✓ | Aedes aegypti | 13.92 | 2 | Peroxisomal biogenesis factor 11/ Band 27 | |
| Unknown role: 11 | |||||||
| Q7PUV3 | AGAP001622-PA | ✓ | ✓ | An. gambiae | 27.83 | 5 | EF- hand domain/ Band 31 |
| Q5TR40 | AGAP006179-PC | ✓ | An. gambiae | 43.79 | 7 | EF-hand domain | |
| Q7Q515 | AGAP006686-PA | ✓ | An. gambiae | 6.16 | 9 | EF-hand domain | |
| F5HJ34 | AGAP001023-PA | ✓ | An. gambiae | 40.65 | 4 | NCD* | |
| Q174U1 | AAEL006790-PA | ✓ | Aedes aegypti | 3.90 | 3 | NCD* | |
| A7URJ0 | AGAP007249-PB | ✓ | An. gambiae | 19.85 | 2 | NCD* | |
| Q16FT3 | AAEL014638-PA | ✓ | Aedes aegypti | 16.24 | 2 | NCD*/ Band 27 | |
| B0X0C8 | Putative uncharacterized protein | ✓ | Cu. quinquefasciatus | 13.51 | 2 | NCD*/ Band 31 | |
| Q7PS70 | AGAP003775-PA | ✓ | An. gambiae | 10.82 | 2 | NCD*/ Band 27 | |
| T1EB87 | Uncharacterized protein | ✓ | An. aquasalis | 10.46 | 2 | NCD*/ Band 13 | |
| Q17N70 | AAEL000785-PA | ✓ | Aedes aegypti | 4.69 | 1 | DUF1168/ Band 2 | |
*NCD- No conserved domain
Comparison of salivary gland proteins between Sugar Fed (SF) and Blood Fed (BF) An. culicifacies mosquitoes by 2-DE
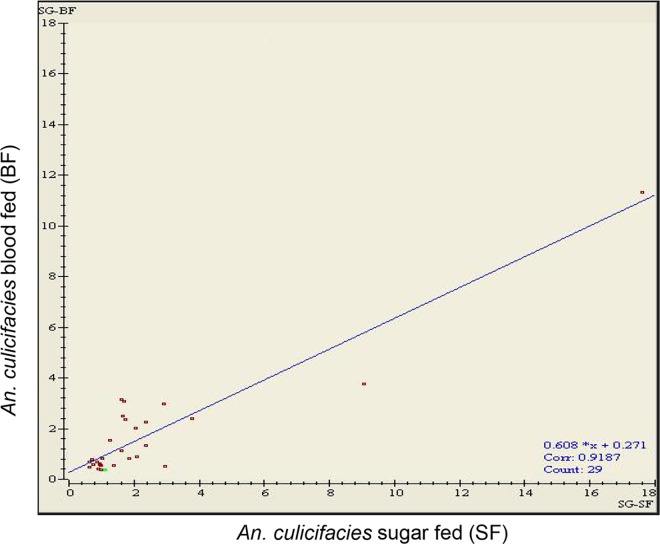
In order to identify the plausible implications of blood meal on the expression and annotations of salivary gland proteins, a comparative study between SF and BF mosquitoes using 2-DE coupled with MALDI-TOF TOF mass spectrometry was conducted in An. culicifacies. A total of 45 spots in SF and 71 spots in BF between a pI range of 4–10 and MW 6–80 kDa were detected using ImageMaster 2D Platinum software (Fig 5A and 5B). Among these total spots, 29 spots were found to be differential expressed between SF and BF mosquitoes. These differential spots in SF and BF salivary gland profile were compared on the basis of pixel volume of each spot. Pixel volume of spots of both SF and BF An. culicifacies are calculated on the basis of spot area and intensity and shown as a scatter plot with a correlation coefficient of > 0.91 between SF and BF (Fig 6).
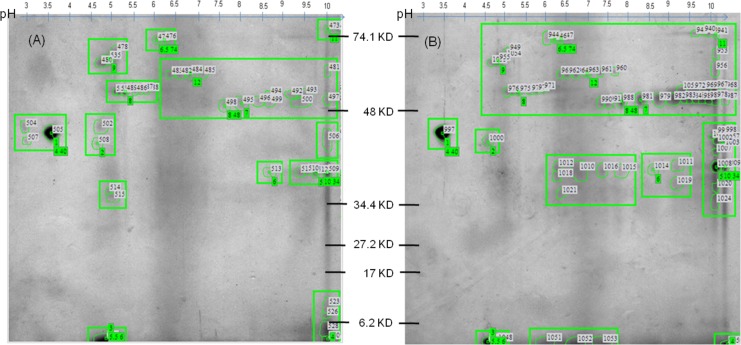
Fig 5. 2-DE gel picture of An. culicifacies salivary gland proteins.
(A) Gel picture of sugar fed species show total spot id number (473–535,black colored) (B) Gel picture of blood fed species show total spot id number (940–1058,black colored)
Fig 6. Scatter plot showing differential spots between SF and BF of An. culicifacies.
Blue line shows linear regression. X axis: volumes of protein spots in SF species. Y axis: volumes of protein spots in BF species. Correlation coefficient was calculated and indicated at the bottom.
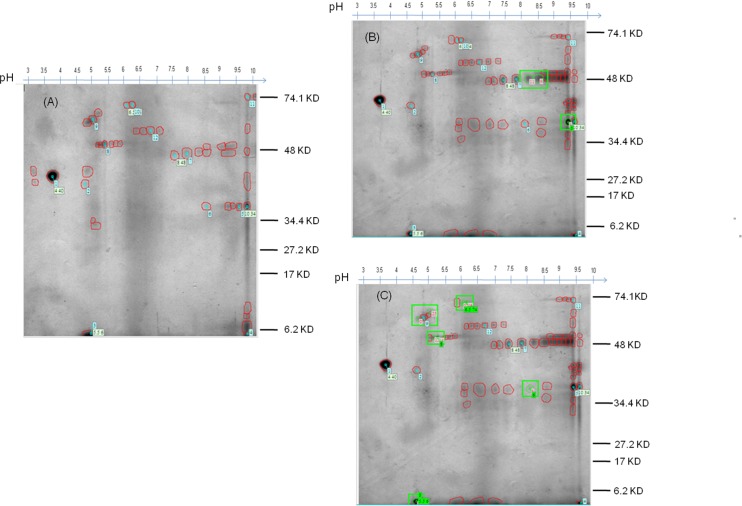
Among 29 annotated spots, only 11 spots were further processed for MALDI as remaining spots did not match the criteria for MALDI-TOF TOF mass spectrometry. Of the 11 differentially expressed spots 3 spots were found to be over expressed (match ID 5, 9, 11) and 8 spots were found to be under expressed (match ID 0, 2, 3, 12, 14, 25, 26, 27) in BF as compared to SF (Fig 7A–7C). Analysis of raw data files of all the 11 annotated spots using peptide mass fingerprinting (PMF) leads to identification of total 29 up regulated proteins and 9 down regulated proteins and their description with fold change are given in Table 2 and Table 3 respectively. All these proteins were further analysed using Gene ontology and other bioinformatics algorithms for the identification of molecular function.
Fig 7. 2-DE gel picture of annotated salivary gland proteins of BF An. culicifacies.
(A) 2-DE salivary gland protein profile of sugar fed (SF) mosquitoes. (B) 2-DE salivary gland protein profile of over expressed spots (5, 9 11) in BF mosquitoes (green squared and numbered in red). (C) 2-DE salivary gland protein profile of under expressed spots (0, 2, 3, 12, 14, 25, 26, 27) in BF mosquitoes (green squared and numbered in red).
Table 2. Annotated up regulated salivary proteins in An. culicifacies upon blood feeding.
| Match ID/ Fold increase | Accession | Protein | Organism, % Sequence coverage | Peptide matches | Function/ Signal peptide if any |
|---|---|---|---|---|---|
| 5 / 1.94 | A0A023EV62 | Putative ceramide kinase | Ae. albopictus, 15% | 13 | Phosphorylation |
| A0A023EU04 | Serine/threonine kinase | Ae. albopictus, 15% | 9 | Phosphorylation | |
| B0XFC4 | Phosphatidylinositol transfer protein SEC14 | Culex quinquefasciatus,14% | 6 | Signal transduction | |
| B0WIF0 | Serine protease inhibitor | Culex quinquefasciatus,15% | 9 | Proteolytic | |
| C4N137 | 30 kDa salivary antigen family | An. darling, 16% | 6 | Antigen / Signal P: 1–29 | |
| A0A023EDZ3 | Putative secreted protein | Ae. albopictus, 35% | 7 | Signal P: 1–26 | |
| 9/ 1.79 | A0A084W2T3 | Ribokinase | An. sinensis, 61% | 8 | Phosphorylation |
| W5JB49 | Calcium/calmodulin-dependent protein kinase 1 | An. darlingi 30% | 15 | Phosphorylation | |
| A0A023EV08 | Putative creatine kinase | Ae. albopictus, 28% | 12 | Phosphorylation | |
| B0X4Y5 | Polo kinase | Culex quinquefasciatus, 19% | 26 | Phosphorylation | |
| A0A023EIY7 | Tyrosine phosphatase iva1 | Ae. albopictus, 32% | 6 | Phosphatase | |
| A0A084W4J8 | MRAS2, putative | An. sinensis, 37% | 14 | Signal transduction | |
| Q7QGU9 | Oxysterol-binding protein | An. gambiae, 23% | 11 | Signal transduction | |
| Q0IEK7 | Lipoyltransferase 2, Mt | Ae. aegypti, 32% | 11 | Protein modification | |
| B0X918 | Ferritin subunit | Culex quinquefasciatus, 37% | 7 | Iron homeostasis | |
| D3KAG7 | CLIPB14 | An. gambiae, 32% | 9 | Proteolytic | |
| A0A084VN17 | AGAP004754-PA | An. sinensis, 63% | 10 | Caspase like domain | |
| W5J7X6 | Glucose dehydrogenase | An. darling, 24% | 9 | Oxidoreductase | |
| J9E8U1 | AAEL017136-PA | Ae. aegypti, 32% | 13 | Cyt P450 | |
| A0A084WNZ9 | Putative antennal carrier protein TOL-2 | An. sinensis, 73% | 4 | Chemosensory/ Signal P: 1–19 | |
| W5J836 | Takeout | An. darling, 17% | 3 | Chemosensory/ Signal P: 1–19 | |
| 11/ 1.5 | B8RJ80 | 5'-AMP-activated protein kinase, alpha-2 | Culex tarsalis, 30% | 5 | Phosphorylation |
| B0WVM0 | Beclin-1 | Culex quinquefasciatus, 12% | 6 | Autophagy | |
| A0A023EM51 | Putative heme oxygenase 1 | Ae. albopictus, 20% | 8 | Heme oxidation | |
| W5JD00 | 26S proteasome regulatory subunitS3 | An. darling, 15% | 9 | Proteolytic | |
| Q7QKL3 | AGAP003249-PA | An. gambiae, 11% | 8 | Serine protease/ Signal P: 1–30 | |
| A0A023EFV7 | Putative galactose-specific c-type lectin | Ae. albopictus, 34% | 4 | Immune/ Signal P: 1–22 | |
| B0WQX3 | Aldehyde dehydrogenase | Culex quinquefasciatus, 10% | 5 | Oxidation reduction | |
| Q8I8P8 | OBP39 | An. gambiae, 14% | 5 | Chemosensory |
Table 3. Annotated down regulated salivary proteins in An. culicifacies upon blood feeding.
| Match ID/ Fold decrease | Accession | Protein | Organism, % sequence coverage | Peptide matches | Function/ Signal peptide if any |
|---|---|---|---|---|---|
| 0/0.17 | B0WIV8 | Putative uncharacterized protein | Culex quinquefasciatus, 22% | 7 | Caspase recruitment domain |
| A0A084VT63 | Isocitrate dehydrogenase | An. sinensis, 29% | 12 | Oxidation reduction | |
| 3/0.38 | A0A023EUU0 | Putative juvenile hormone epoxide hydrolase ii | Ae. albopictus, 22% | 8 | Physiological change/ Signal P: 1–19 |
| Q6TRY1 | Putative salivary OBP 2 | Culex quinquefasciatus, 27% | 2 | Signal P: 1–20 | |
| A0A023EVV5 | Putative pftaire-interacting factor 1a | Ae. albopictus, 23% | 14 | Unknown | |
| 12/0.37 | B0W7K8 | Chemosensory protein 1 | Culex quinquefasciatus, 71% | 8 | OBP 10 |
| 14/0.42 | A0A023EFB5 | Putative 11 kDa salivary protein | Ae. albopictus, 60% | 4 | Magnesium transport/ Signal P: 1–19 |
| 27/0.34 | W5JJH0 | Sphingosine phosphate lyase | An. darling, 13% | 8 | Apoptosis regulation |
| O17491 | Iron regulatory protein | An. gambiae, 17% | 5 | Iron homeostasis |
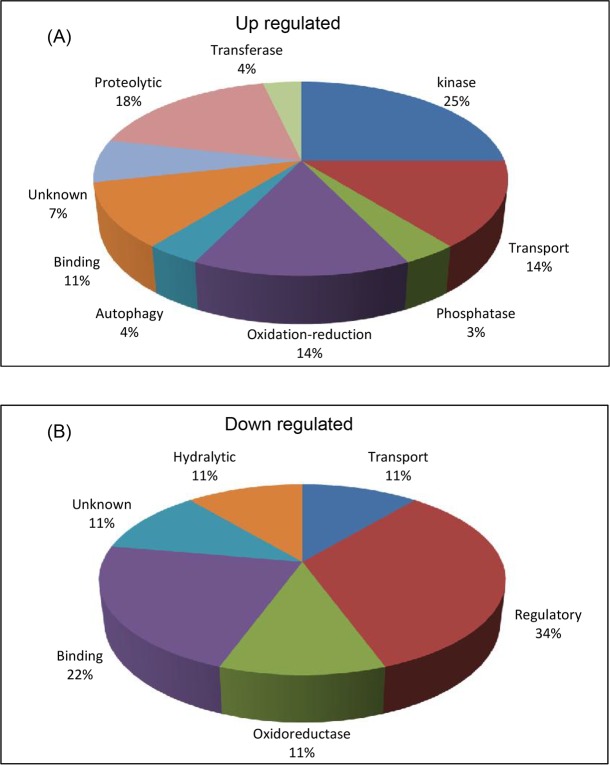
Among up regulated proteins, majority of the proteins fell into categories of kinase function (25%) followed by proteolytic function (18%). These kinase proteins, serpins, signal transduction proteins, CLIP B protein, putative secreted proteins and 30kDa salivary antigen protein were found to be expressed 2 fold higher after blood meal as compared to sugar fed mosquitoes. Further, oxidoreductive proteins like glucose dehydrogenase, aldehyde dehydrogenase and detoxifying protein like cytochrome 450 were found to be 1.5 fold higher expressed in blood fed salivary glands than sugar fed. The down regulated proteins were mostly found to be having physiological functions (like chemosensory protein, odorant bindng protein etc) and regulatory functions namely apoptosis regulation, iron regulatory etc. Details of the molecular functions of up regulated and down regulated proteins in BF mosquitoes are shown in a pie chart (Fig 8A and 8B).
Fig 8. Molecular functions of differentially regulated salivary gland proteins analyzed using gene ontology tool.
(A) GO function of identified up regulated proteins. (B) GO function of identified down regulated proteins.
Discussion
Salivary glands of mosquitoes are important organs and are the first target during the process of parasite invasion, maturation and subsequent transmission to the human host. Because of the direct contact of the salivary glands with the human host during biting and release of antiplatelet aggregation components and anti-inflammatory proteins to facilitate blood feeding, their importance in studying host, vector and parasite interactions is well known. Various studies have reported altered salivary proteins after blood meals using 1-DE and 2-DE in different mosquito species however few transcriptomic and no proteomic studies have been carried out in the salivary glands of An. culicifacies species and also genome sequence is unknown till date hence, in the current study, as a first step proteomic studies were carried out using 1-DE combined with LC-MS/MS and secondly, the differential expression and annotations of SF and BF compositions were compared using 2-DE coupled with MALDI-TOF mass spectrometry in salivary gland of An. culicifacies.
Our findings by both in-solution and in-gel approach identified a total of 106 putative salivary proteins of An. culicifacies and a baseline catalogue was prepared. Significant identified proteins in the catalogue included the members of D7 short form secreted proteins i.e. D7 r1, D7r2 and D7r4. These D7 family proteins are well described salivary gland proteins unique to dipterans and are distant relatives of the odorant binding protein superfamily [25–27] which actually bind host biogenic amines to antagonize vasoconstriction and platelet aggregating [25] and facilitate the blood feeding and potentially parasite ingestion from the vertebrate host. These proteins were also reported earlier in Aedes aegypti and An. stephensi using transcriptomic and proteomic approach [11, 13, 28]. Our results, together with other earlier studies in Aedes aegypti and An. stephensi indicate that this family of proteins are involved in hematophagy and potentially in parasite transmission during blood meal.
Salivary gland specific antigen 5 (Ag5) family proteins are extracellular proteins of ubiquitous family and are known to act as functional allergens in insects [29]. Another salivary gland specific family of enzymes reported in the insects and ticks are antihemostatic protein i.e. apyrases that act as vasodilator. During blood feeding nucleotidase activity of apyrase prevents ADP-induced platelet aggregation in the host and thereby helps in hematophagy [30]. Moreover the end product generated in the process i.e. AMP gets converted in to adenosine which acts as a powerful anti-inflammatory substance [31]. In addition, another enzyme salivary maltase was also identified which helps in the digestion of sugar meal [32]. Protein AGAP009642-PA with prohibitin domain was also identified in the profile. In insects like Drosophila and silkworm prohibitin is known to have role in normal development [33]. In a recent study, prohibitin was reported to act as a virus receptor during dengue transmission. It interacts with dengue E protein and is associated with dengue serotype 2 infection in Aedes aegypti and Aedes albopictus [34]. Various proteins involved in detoxification of xenobiotics and antioxidant were observed like glutathione S transferase (GST), theta and GST S1-2, peroxiredoxin 4, catalase, thioredoxine and dihydrolipoyl dehydrogenase. Results from the proteomic approach used in this study confirmed that An. culicifacies also possess anti-platelet aggregation (apyrase and anti-platelet protein) and anti-inflammatory (D7 and D7 related) proteins that may facilitate in blood feeding process. A detailed catalogue of these related proteins is presented in Table 1.
In our efforts to identify differentially expressed and annotated proteins in response to blood feeding, we have compared salivary gland proteins of sugar fed (SF) An. culicifacies species with blood fed (BF) mosquitoes by 2-DE which was combined with MALDI TOF to enlighten the important roles of salivary proteins in hematophagy. Interestingly, among the 29 upregulated proteins large array of phosphorylating /kinase enzymes namely ceramide kinase, serine/threonine kinase, calcium/calmodulin-dependent protein kinase type 1 etc were identified in An. culicifacies that are mainly involved in the function of autophagy and its regulation. It is known that autophagy process in response to various cellular stresses like starvation, pathogen infection etc. get upregulated and hence protect organism against cellular distress [35]. Various authors have demonstrated that these identified kinase enzymes like ceramide kinase [36], serine/threonine kinase and calcium/calmodulin-dependent protein kinase type 1 [37] etc. generates several proapoptotic signals that induce autophagy [38]. Therefore, our observations indicate that these identified proteins in salivary glands of blood fed mosquitoes might play a role as a survival factor against unwanted microorganism or pathogen and external stress enhanced during blood feeding. The role of autophagy was also implicated in epithelium protection against the products of blood digestion in insect as mentioned earlier by Rost-Roszkowskan et al [39]. In addition, Beclin-1/autophagy-related protein 6 was also identified to be up regulated in our studies that might play a central role in autophagy as suggested by earlier studies in Drosophila by Sharavage et al [40]. The phosphorylation of beclin-1 promotes disassociation of beclin 2-beclin 1 complex which activates autophagy [35, 41]. In a recent observations, the transcriptomics studies on An. culicifacies salivary glands also reports the upregulation of autophagy related proteins in response to blood feeding [42].
Oxysterol binding protein (OSBP) is known to be involved in signal transduction pathways and cellular lipid metabolism [43]. A study in Aedes agypti reported the up regulation of OSBP transcription after a blood meal [44,45]. In Anopheles culicifacies, OSBP protein was over expressed in blood fed confirming that OSBP plays a role in blood feeding. Up regulation of heme/iron assimilating proteins i.e. heme oxygenase enzyme 1 (HO1) and ferritin suggested that blood meal is responsible for iron overload [46]. It is known that autophagy process in response to various cellular stresses like starvation, pathogen infection etc. get upregulated and hence protect organism against cellular distress [47–48]. Caspases are cysteine proteases involved in the initiation and execution of apoptosis [49]. Elevated C-type lectin protein with carbohydrate recognition domain functions as receptor in pathogen recognition and play important role in insect immune response [50]. Enzymes related to energy metabolism i.e. aldehyde dehydrogenase and glucose dehydrogenase was also over expressed post blood meal. It has been reported that aldehyde dehydrogenase enzyme play a critical role in regulation of juvenile hormone (JH) synthesis in blood fed mosquitoes [51]. In adult female mosquitoes, JH controls the reproductive maturation [52] and induces the vitellogenesis [53]. Other over expressed proteins like antennal carrier protein TOL-2 (JH binding domain), takeout protein (JH binding domain) and odorant-binding proteins (OBP39) have role in regulation of blood feeding behaviours [54–55]. Reduced blood feeding after knockdown of takeout RNA in vivo in An. gambiae had been reported [56].
Over expression of 30 kDa salivary antigen family protein in An. culicifacies also correlates with the earlier studies in Aedes aegypti [57] and may provide valuable information on immune responses. The function of these 30 kDa salivary allergens remains unknown so far however; aegyptin a member of this family has been reported to inhibit collagen-induced human platelet aggregation [58].
Further, our findings showed the under expression of Juvenile hormone epoxide hydrolase (JHEH) II post blood meal. JHEHs are family of enzymes involved in irreversible degradation of juvenile hormones and have organ specific regulation [59]. Decreased expression of kreb cycle enzymes i.e. isocitrate dehydrogenase and Aconitase/IRP also correlates to the earlier publication in mosquitoes An. gambiae [27] and Aedes [57] respectively. However the importance of these down regulated enzymes associated with ATP synthesis and proteins that utilize ATP and their biological meaning related to blood feeding yet to be determined. It is also reported that these enzymes of kreb cycle were decreased after 24 hrs of blood meal [27].Down regulation of OBP 10 and OBP 2 identified in An. culicifacies justified the earlier publications by Wasinpiyamongkol et al that reported same depletion of OBP transcripts in SG of Aedes aegypti [57]. OBP’s are actually important for sensing however it was reported that after taking blood meal further OBP are not required until next blood meal and as the mosquito transitions to oviposition behavior [60].
Almost all the differentially expressed proteins in blood fed mosquito salivary glands seem to have key role in successful blood feeding. Blood feeding in mosquitoes depends on a number of factors namely host platelet activation, aggregation and coagulation and immune systems [6]. However, more studies are needed to further investigate the functions of novel and annotated salivary proteins to examine their functional role in mosquito feeding and host probing behaviour. Such studies using RNAi gene silencing assays on salivary gland transcribed genes may provide explanations on the secretion mechanisms of the mosquitos’ salivary glands. Since no such information is available in An. culicifacies mosquitoes such studies combined with this proteomic dataset will help to identify and characterise role of specific proteins whether in blood feeding or not because proteomic annotations does not clearly conclude their functional role.
Conclusion
To our knowledge, this study presents the first proteomic baseline map and cataloging of the salivary glands of sugar fed female An. culicifacies with detailed putative functional annotation of all the identified proteins. The differences in the proteins suggest a plausible role in facilitating blood feeding but may also be implicated in the transmission of malaria parasites. Identification of novel expressed proteins and up and down regulated annotated proteins in blood fed mosquitoes using differential 2 DE method particularly several proteins related to autophagy like beclin-1, phosphorylating related proteins, heme oxygenase 1, ferritin, apoptotic, coagulation and immunity related proteins and proteins involved in regulation of blood feeding behavior and juvenile hormone may relate to the functions in the hematophagy. These correlations however, do not provide direct proof in enhancing of the blood feeding behavior. Understanding of these identified differentially regulated proteins after blood meal that may directly or indirectly be associated with development and egg laying capacity will provide insights into the physiological changes associated with feeding behavior, parasite transmission during blood feeding and may open the way for the development of novel malaria blocking strategies.
Acknowledgments
We thank our technical staff Mr. Bhanu Arya (Technical Officer), Mrs. Poonam Gupta (Technical Assistant) for their excellent technical help in conducting of experiments. Thanks are due to Mr. Alakh Dev Prasad for dissection of mosquitoes tissues. We will also like to thank Dr. S.P.Singh, Research Scientist and all the technical staff of NIMR Insectary for providing us mosquitoes. We also thank Dr. Yash Gupta for his useful insight in the paper. This paper bears the NIMR publication screening committee approval no. 006/2016.
Data Availability
All relevant data are within the paper.
Funding Statement
This work has been financially supported by the grant from Department of Science and Technology, Government of India (Ritu Rawal, SR/WOS-A/LS-590/2012 and Department of Biotechnology, New Delhi, Government of India (Dr. Arun Sharma, BT/PR8221/MED/12/617/2013).
References
- 1.World Health Organization “World Malaria Report 2014, Tech. Rep” WHO, Geneva, Switzerland, 2014.
- 2.Sharma AK, Tyagi V, Singh S, Veer V, Agrawal OP, Sulumaran D. Distribution of Anopheles culicifacies and detection of its sibling species E from Madhya Pradesh: Central India. J Arthropod-Borne Dis 2014, 8(2):186–196. [PMC free article] [PubMed] [Google Scholar]
- 3.Goswami G, Singh OP, Nanda N, Raghavendra K, Gakhar SK, Subbarao SK. Identification of all members of the Anopheles culicifacies complex using allele-specific polymerase chain reaction assays. Am J Trop Med Hyg. 2006; 75:454–460. [PubMed] [Google Scholar]
- 4.Paaijmans KP1, Cator LJ, Thomas MB. Temperature-dependent pre-blood meal period and temperature-driven asynchrony between parasite development and mosquito biting rate reduce malaria transmission intensity. PLoS One. 2013; 10.1371/journal.pone.0055777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez MH, Hernandez-Hernandez FC. Insect–malaria parasites interactions: the salivary gland. Insect Biochem and Mol Bio. 2004; 34:615–624. [DOI] [PubMed] [Google Scholar]
- 6.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003; 48:73–88. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro JM, Mans BJ, Arca B. An insight into the sialome of blood feeding Nematocera. Insect Biochem Mol Biol. 2010; 40(11):767–784. 10.1016/j.ibmb.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribeiro JM, Arca B. From sialomes to the sialoverse: An insight into salivary potion of blood-feeding insects. Adv in Insect Phys. 2009; 37:59–118. [Google Scholar]
- 9.Titus RG, Ribeiro JM: Salivary gland lysates from the sand fly Lutzomyia longipalpisenhance Leishmania infectivity. Science. 1988; 239(4845):1306–1308. [DOI] [PubMed] [Google Scholar]
- 10.Choumet V, Carmi-Leroy A, Laurent C, Lenormand P, Rousselle JC, Namane A et al. The salivary glands and saliva of Anopheles gambiae as an essential step in the Plasmodium life cycle: A global proteomic study. Proteomics. 2007; 7:3384–3394. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003; 33:717–732. [DOI] [PubMed] [Google Scholar]
- 12.Dixit R, Sharma A, Mourya DT, Kamaraju R, Patole MS, Shouche YS, et al. Salivary gland transcriptome analysis during Plasmodium infection in malaria vector Anopheles stephensi. Int J Infect Dis. 2009; 13(5):636–646. 10.1016/j.ijid.2008.07.027 [DOI] [PubMed] [Google Scholar]
- 13.Vijay S, Rawat M, Sharma A. Mass spectrometry based proteomic analysis of salivary glands of urban malaria vector Anopheles stephensi. BioMed Res Int. 2014. 12 10.1155/2014/686319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sor-suwan S, Jariyapan N, Roytrakul S. K, Paemanee A, Saeung A, Thongsahuan S, Phattanawiboon B, Bates P. A, Poovorawan Y, Choochote W. Salivary gland proteome of the human malaria vector, Anopheles campestris -like (Diptera: Culicidae). Parasitol Res. 2013; 112:1065–1075. 10.1007/s00436-012-3233-y [DOI] [PubMed] [Google Scholar]
- 15.Jariyapan N, Roytrakul S, Paemanee A, Junkum A, Saeung A, Thongsahuan S, Sor-suwan S, Phattanawiboon B. Proteomic analysis of salivary glands of female Anopheles barbirostrisspecies A2 (Diptera: Culicidae) by two-dimensional gel electrophoresis and mass spectrometry. Parasitol Res. 2012; 111:1239–1249. 10.1007/s00436-012-2958-y [DOI] [PubMed] [Google Scholar]
- 16.Almeras L, Fontaine A, Belghazi M, Bourdon S, Boucomont-Chapeaublanc E, Orlandi-Pradines E et al. Salivary gland protein repertoire from Aedes aegypti mosquitoes. Vector Borne Zoonotic Dis. 2010; 10(4):391–402. 10.1089/vbz.2009.0042 [DOI] [PubMed] [Google Scholar]
- 17.Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol 2007; 37:164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalume DE, Okulate M, Zhong J, Reddy R, Suresh S, Deshpande N et al. A proteomic analysis of salivary glands of female Anopheles gambiae mosquito. Proteomics. 2005; 5:3765–3777 [DOI] [PubMed] [Google Scholar]
- 19.Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004; 34:543–563. [DOI] [PubMed] [Google Scholar]
- 20.Vijay S, Rawal R, Kadian K, Raghavendra K, Sharma A. Annotated Differentially expressed salivary proteins of susceptible and insecticide resistant mosquitoes of Anopheles stephensi. PLoS One. 2015; 10.1371/journal.pone.0119666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adak T, Kaur S, Singh OP. Comparative susceptibility of different members of the Anopheles culicifacies complex to Plasmodium vivax. Trans R Soc Trop Med Hyg.1999; 93:573–77. [DOI] [PubMed] [Google Scholar]
- 22.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951; 193:265–275. [PubMed] [Google Scholar]
- 23.Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins: Structure, Function and Bioinformatics. 2006; 64:643–651. [DOI] [PubMed] [Google Scholar]
- 24.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, et al. 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotech. 2014; 30(3):223–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Boil Chem. 2006; 281(4):1935–42. [DOI] [PubMed] [Google Scholar]
- 26.Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009; 10:57 10.1186/1471-2164-10-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, Radtke A, Choi YJ, Mendes AM, Valenzuela JG, Dimopoulos G. Transcriptomic and functional analysis of the Anopheles gambiae salivary gland in relation to blood feeding. http://www.ncbi.nlm.nih.gov/pubmed/20946652BMC Genomics. 2010; 11:566 10.1186/1471-2164-11-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James AA, Blackmer K, Marinotti O, Ghosn CR, Racioppi JV. Isolation and characterization of the gene expressing the major salivary gland protein of the female mosquito, Aedes aegypti. Mol Biochem Parasitol. 1991; 44:245–253. [DOI] [PubMed] [Google Scholar]
- 29.Caljon G, Broos K, De Goeyse I, De Ridder K, Sternberg JM, Coosemans M. Identification of a functional Antigen 5-related allergen in the saliva of a blood feeding insect, the tsetse fly. Insect Biochem Mol Biol. 2009; 39(5–6):332–341. [DOI] [PubMed] [Google Scholar]
- 30.Champagne DE, Smartt CT, Ribeiro JM, James AA.The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5'-nucleotidase family. Proc Natl Acad Sci. 1995; 92(3):694–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez de Leon AA, Tabachnick WJ. Apyrase activity and adenosine diphosphate induced platelet aggregation inhibition by the salivary gland proteins of Culicoides variipennis, the North American vector of bluetongue viruses. Vet Parasitol. 1996; 61(3–4):327–38. [DOI] [PubMed] [Google Scholar]
- 32.James AA, Blackmer K, Racioppi JV. A salivary gland specific, maltase-like gene of the vector mosquito, Aedes aegypti. Gene. 1989; 75: 73–83. [DOI] [PubMed] [Google Scholar]
- 33.Lv Z, Zhang X, Liu L, Chen J, Nie Z, Sheng Q et al. Characterization of a gene encoding prohibitin in silkworm, Bombyx mori. Gene. 2012; 502(2):118–124. 10.1016/j.gene.2012.03.035 [DOI] [PubMed] [Google Scholar]
- 34.Kuadkitkan A, Wikan N, Fongsaran C, Smith DR. Identification and characterization of prohibitin as a receptor protein mediating DENV-2 entry into insect cells. Virology. 2010; 406(1):149–161. 10.1016/j.virol.2010.07.015 [DOI] [PubMed] [Google Scholar]
- 35.He C and Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009; 43: 67–93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up regulation of Beclin 1. J Biol Chem. 2004; 279:18384–18391. [DOI] [PubMed] [Google Scholar]
- 37.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol.2002; 157:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B and Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005; 115(10):2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost-Roszkowska MM, Świątek P, Poprawa I, Rupik W, Swadźba E, Kszuk-Jendrysik M. Ultrastructural analysis of apoptosis and autophagy in the midgut epithelium of Piscicola geometra (Annelida, Hirudinida) after blood feeding. Protoplasma. 2015; 252(5):1387–1396. 10.1007/s00709-015-0774-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013; 140(6):1321–1329. 10.1242/dev.089490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009; 10:285–292. 10.1038/embor.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma P, Mishra AK, Thomas P, Das De T, Rohilla SL, Singh N et al. Unraveling dual feeding associated molecular complexity of salivary glands in the mosquito Anopheles culicifacies. Biology Open. 2015; 4:1002–1015. 10.1242/bio.012294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olkkonen V, Johansson M, Suchanek M, Yan D, Hynynen R, Ehnholm C et al. The OSBP-related proteins (ORPs): global sterol sensors for co-ordination of cellular lipid metabolism, membrane trafficking and signalling processes? Biochem Soc Trans. 2006; 34(3):389–391. [DOI] [PubMed] [Google Scholar]
- 44.Fu Q, Lynn-Miller A, Lan Q. Characterization of the oxysterol-binding protein gene family in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2011; 20(4):541–552. 10.1111/j.1365-2583.2011.01087.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham DQD and Winzerling Joy J. Insect Ferritins: typical or atypical? Biochim Biophys Acta.2010; 1800(8):824–833. 10.1016/j.bbagen.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou G, Kohlhepp p, Geiser D, Rasquillo MDC, Vazquez-Moreno L, Winzerling JJ. Fate of blood meal iron in mosquitos. J Insect Physiol. 2007; 53(11):1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gulley MM, Zhang X, Michel K. The roles of serpins in mosquito immunology and physiology. J Insect Physiol. 2013; 59(2):138–147. 10.1016/j.jinsphys.2012.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorman MJ and Paskewitz SM. Serine proteases as mediators of mosquito immune responses. Insect Biochem and Mol Bio. 2001; 31:257–262. [DOI] [PubMed] [Google Scholar]
- 49.Cooper DM, Granville DJ, Lowenberger C. The insect caspases. Apoptosis. 2009; 14(3):247–56. 10.1007/s10495-009-0322-1 [DOI] [PubMed] [Google Scholar]
- 50.Schnitger AKD, Yassine H, Kafatos FC, Osta MA. Two C-type lectins cooperate to defend Anopheles gambiae against gram-negative bacteria. JBC. 2009; 284:17616–17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivera-Perez C, Nouzova M, Clifton ME, Garcia EM, LeBlanc E, Noriega FG. Aldehyde dehydrogenase 3 converts farnesal into farnesoic acid in the corpora allata of mosquitoes. Insect Biochem Mol Biol. 2013; 43(8):675–682. 10.1016/j.ibmb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Busche JM, Zhang X. Identification of juvenile hormone target genes in the adult female mosquitoes. Insect Biochem and Mol Biol. 2010;40(1):23–29. [DOI] [PubMed] [Google Scholar]
- 53.Amsalem E, Malka O, Grozinger C, Hefetz A. Exploring the role of juvenile hormone and vitellogenin in reproduction and social behavior in bumble bees. BMC Evol Biol. 2014; 14:45 10.1186/1471-2148-14-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.So WV, Sarov-Blat L, Kotarski CK, McDonald MJ, Allada R, Rosbash M. Takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol Cell Biol. 2000; 20:6935–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rund SSC, Bonar NA, Champion MM, Ghazi JP, Houk CM, Leming MT. Daily rhythms in antennal protein and olfactory sensitivity in the malaria mosquito Anopheles gambiae. Scientific reports. 2013; 10.1038/srep02494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das S, Dimopoulos G: Molecular analysis of photic inhibition of blood feeding in Anopheles gambiae. BMC Physiol. 2008; 8:23 10.1186/1472-6793-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wasinpiyamongkol A, Patramool S, Luplertlop N, Surasombatpattana P, Doucoure S, Mouchet F et al. Blood-feeding and immunogenic Aedes aegypti saliva proteins. Proteomics. 2010; 10:1–11. [DOI] [PubMed] [Google Scholar]
- 58.Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JMC, Francischetti IMB. Aegyptin, a novel mosquito salivary gland protein specifically binds to collagen and prevents its interaction with glycoprotein VI, integrin α2β1 and von willebrand factor. J Biol Chem. 2007; 282(37):26928–26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seino A, Ogura T, Tsubota T, Shimomura M, Nakakura T, Tan A et al. Characterization of juvenile hormone epoxide hydrolase and related genes in the larval development of the silkworm bombyx mori. Biosci Biotechnol Biochem. 2010; 74(7):1421–1429. [DOI] [PubMed] [Google Scholar]
- 60.Rinker DC, Pitts RJ, Zhou X, Suh E, Rokas A, Zwiebel LJ. Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc Natl Acad Sci. 2013; 110:8260–8265. 10.1073/pnas.1302562110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.