Abstract
NR2E3 encodes the photoreceptor-specific nuclear hormone receptor that acts as a repressor of cone-specific gene expression in rod photoreceptors, and as an activator of several rod-specific genes. Recessive variants located in the ligand-binding domain (LBD) of NR2E3 cause enhanced short wavelength sensitive- (S-) cone syndrome (ESCS), a retinal degeneration characterized by an excess of Scones and non-functional rods. We analyzed the dimerization properties of NR2E3 and the effect of disease-causing LBD missense variants by bioluminescence resonance energy transfer (BRET2) protein interaction assays. Homodimerization was not affected in presence of p.A256V, p.R039G, p.R311Q and p.R334G variants, but abolished in presence of p.L263P, p.L336P, p.L353V, p.R385P and p.M407K variants. Homology modeling predicted structural changes induced by NR2E3 LBD variants. NR2E3 LBD variants did not affect interaction with CRX, but with NRL and rev-erbα/NR1D1. CRX and NRL heterodimerized more efficiently together, than did either with NR2E3. NR2E3 did not heterodimerize with TLX/NR2E1 and RXRα/NR2C1. The identification of a new compound heterozygous patient with detectable rod function, who expressed solely the p.A256V variant protein, suggests a correlation between LBD variants able to form functional NR2E3 dimers and atypical mild forms of ESCS with residual rod function.
Keywords: NR2E3, retinal degeneration, genotype-phenotype correlation, photoreceptor development
INTRODUCTION
The photoreceptor-specific nuclear receptor NR2E3 (nuclear receptor, class 2, subfamily E, member 3; MIM# 604485) is a member of the nuclear hormone receptor superfamily of ligand-modulated transcription factors (Kobayashi, et al., 1999). Nuclear receptors (NRs) share an evolutionarily conserved modular protein structure comprising a N-terminal A/B domain, a highly conserved DNA-binding domain (DBD) consisting of two Cys4 zinc fingers, a flexible hinge domain and a C-terminal ligand-binding domain (LBD) (Mangelsdorf, et al., 1995). The general structure of a NR LBD consists of a three-layer-antiparallel α-helical sandwich, where the incomplete central layer forms a hydrophobic pocket where lipophilic ligands dock to (Watson, et al., 2012). No NR2E3 ligands have been identified so far (Qin, et al., 2012), but structural modeling based on the NR2E3 LBD crystal structure demonstrated the potential of small ligands to bind to its hydrophobic pocket (Tan, et al., 2013). Generally, ligand-bound NR LBDs associate with transcriptional coactivators, whereas unliganded NRs bind to corepressors (Glass and Rosenfeld, 2000). However, the partial NR2E3 LBD crystal structure revealed an auto-repressed conformation, where the most C-terminal α-helix 12, also called activator function 2 (AF-2), occupied the canonical cofactor binding site (Tan, et al., 2013). Consistently, NR2E3 was initially reported as a transcriptional repressor (Chen, et al., 1999; Chen, et al., 2005). By repressing the generation of cone precursors during murine retinal development, NR2E3 is essential for proper rod and cone photoreceptor development (Cheng, et al., 2011; Haider, et al., 2006). Absence of NR2E3 in the mouse retina causes an excess of short wavelength-sensitive (S-) cones generated from post-mitotic photoreceptor precursor cells instead of the early-born rod precursor population (Cheng, et al., 2011). The excess of S-cones observed in the absence of NR2E3 disrupts the retinal architecture by formation of rosettes and whorls in the retina (Akhmedov, et al., 2000; Haider, et al., 2001; Jacobson, et al., 2004). In adult rod photoreceptor cells, NR2E3 is necessary to repress cone-specific transcription, but also to activate rhodopsin expression (Chen, et al., 2005; Cheng, et al., 2004; Peng, et al., 2005). Therefore, in the absence of NR2E3, rods become non-functional hybrid photoreceptors, expressing both rod- and cone-specific genes (Corbo and Cepko, 2005).
A high variability in clinical phenotypes of NR2E3-linked retinal degenerations has been reported (Audo, et al., 2012; Pachydaki, et al., 2009; Schorderet and Escher, 2009; Yzer, et al., 2013). Recessive variants in NR2E3 cause enhanced S-cone sensitivity syndrome (ESCS; MIM# 268100), also called Goldmann-Favre syndrome (GFS) (Haider, et al., 2000; Marmor, et al., 1990), although a number of patients had been initially diagnosed with autosomal recessive retinitis pigmentosa (ARRP) (Gerber, et al., 2000) or clumped pigmentary retinal degeneration (CPRD) (Sharon, et al., 2003). A c.166G>A (p.G56R) variant located in the DBD of NR2E3 caused autosomal dominant retinitis pigmentosa (ADRP; MIM# 611131) (Coppieters, et al., 2007). Patients with recessive NR2E3 variants often refer to the ophthalmologist for mixed cystoid/schisis-like maculopathy and may exhibit a vast spectrum of signs on funduscopy, including mid-peripheral white/yellow dots and flecks, colocalizing with the rosettes, nummular pigment deposits and helicoidal fibrosis (Bandah, et al., 2009; Jurklies, et al., 2001; Khan, et al., 2010; Marmor, 2006; Wang, et al., 2009; Yzer, et al., 2013). The increased number of S-cones results in a pathognomonic increase in the response to blue light, as assessed by spectral electroretinography (ERG), and in a markedly delayed 30-Hz flicker ERG (Audo, et al., 2008; Marmor, et al., 1990). The presence of non-functional hybrid rods leads to night-blindness and rod-specific ERG is undetectable in young infants already (Audo, et al., 2008; Cassiman, et al., 2013). However, two compound heterozygous patients affected with an atypical mild ESCS with residual rod function, harboring either [c.119-2A>C];[c.767C>T] ([c.119-2A>C];[p.A256V]) (Lam, et al., 2007) or [c.311G>A];[c.1000C>G] ([p.R104Q];[p.R334G]) variants (Hayashi, et al., 2005) have been reported. The high clinical variability of NR2E3-linked retinal degenerations suggested different potential disease mechanisms, including absence of NR2E3 protein (Haider, et al., 2000), absence of DNA-binding (Kanda and Swaroop, 2009; Roduit, et al., 2009) or impaired corepressor binding (Escher, et al., 2009; Takezawa, et al., 2007).
Class 2 nuclear receptors are thought to act as homodimers, and NR2E3 homodimers have been observed in non-denaturing protein gel migration conditions (Roduit, et al., 2009), by bioluminescence resonance energy transfer (BRET2) assays (Roduit, et al., 2009), in DNA-binding assays (Escher, et al., 2009; Kanda and Swaroop, 2009), and, more recently, in the dimeric LBD crystal structure (Tan, et al., 2013). Since disruption of dimers has been shown to impair the repressor function of NR2E3 (Kanda and Swaroop, 2009; Tan, et al., 2013), we resorted to BRET2 assays in order to analyze in detail homodimerization of NR2E3 and the effect of LBD missense variants identified in patients on protein dimerization. The effects of these variants on heterodimerization with the photoreceptor-expressed transcription factors cone-rod homeobox (CRX; MIM# 602225), neural retina leucine zipper (NRL; MIM# 162080) and NR1D1, called rev-erbα (MIM# 602408), were also assessed. Finally, the identification of a new ESCS patient with residual rod function suggests genotype/phenotype correlations for atypical mild forms of ESCS.
MATERIALS AND METHODS
Patient and Clinical Assessment
This study followed the tenets of the Declaration of Helsinki (1983 Revision) and was approved by the Ethics Committee of the University of Lausanne. Informed consent was obtained from the patient for the use of his DNA for research purposes. Complete clinical examination included refraction, best corrected visual acuity (BCVA), slit lamp assessment and dilated funduscopic examination. Visual fields (VF) were determined by Goldmann perimetry on an Octopus® 900 system (Haag-Streit AG, Bern, Switzerland). Ganzfeld ERG was performed with a RETIport 32 system (Roland Consult, Wiesbaden, Germany) and incorporated recommendations of the International Society for Clinical Electrophysiology of Vision (ISCEV) (Marmor, et al., 2009). Fundus autofluorescence was performed on a confocal scanning laser ophthalmoscope (cSLO) (Heidelberg Retina Angiograph 2, Heidelberg Engineering, Heidelberg, Germany). Lipofuscin autofluorescence was excited with an Argon laser light at 488 nm and a band-pass filter with a cut-off at 500 nm included in the system was inserted in front of the detector. Spectral domain optical coherence tomography (OCT) images were obtained using the Cirrus HD-OCT (Carl Zeiss Meditec AG, Jena, Germany). Molecular genetic analyses were approved by the Swiss Federal Department of Health (authorization #035.0003-48) and performed exactly as previously published (Escher, et al., 2012).
Plasmids
QuikChange®II site-directed mutagenesis (Stratagene; Cedar Creek, TX) was performed on pcDNA3.1/HisC-hNR2E3 (Peng, et al., 2005) with primers listed in Supp. Table S1 to obtain mammalian expression vectors for NR2E3 LBD variant proteins. Variants are numbered in accordance to the GenBank entry NM_014249.2, where +1 corresponds to the A of the ATG translation initiation codon, i.e. nucleotide 191. For intronic sequences, human chromosome 15 sequence NCBI 36:15 was used. Amino acid changes are numbered in accordance to the SwissProt entry Q9Y5X4. All these NR2E3 variants are reported in the public LOVD NR2E3 database (www.lovd.nl/eye). A KpnI/XbaI fragment was excised off the pcDNA3.1/HisC-hNR2E3 variant plasmids and subcloned into pGFP2-C3 BRET2 vectors, linearized with KpnI and XbaI (PerkinElmer-BioSignal, Montreal, QC, Canada). The resulting plasmids encoded a fusion protein with the humanized green fluorescent protein 2 (hGFP2) in N-terminus and the different NR2E3 variant proteins in the C-terminus. QuikChange®II site-directed mutagenesis was performed on RlucN3-hNR2E3-wild-type BRET2 vector (Roduit, et al., 2009) with appropriate primers to obtain RlucN3-hNR2E3 expression vectors encoding a fusion protein with the NR2E3 variant protein in N-terminus, and the humanized Renilla luciferase (hRluc) in the C-terminus (Supp. Table S1). Coding sequences were amplified from pCMX-hTLX, pMT-NRL (Peng, et al., 2005) and pcDNA3.1/HisC-mNr1d1 (Mollema, et al., 2011) and subcloned into BglII and KpnI sites of linearized RLucN2 to obtain RLucN2-hNR2E1, RLucN2-hNRL and RLucN2-mNr1d1 expression vectors. To obtain GFPC3-hNR2E1, GFPC3-hRXRα, GFPC3-hCRX and GFPC3-hNRL, amplification was performed with the same forward primers, but reverse primers containing a stop codon, and subcloned into BglII and KpnI sites of linearized GFP2-C3. GFPC3-mNr1d1 vector was obtained by subcloning into XhoI and KpnI sites. Additional vectors used for BRET2 assays and transactivation studies were RlucN3-hCRX, GFPC3-hCRX, pcDNA3.1/HisC-hCRX, BR225-Luc and Mop250-Luc (Peng, et al., 2005; Roduit, et al., 2009). All constructs were verified by Sanger sequencing on an ABI 3130xl Genetic Analyzer using the Big Dye Terminator Labeling Kit (Applied Biosystems, Carlsbad, CA).
BRET2 Assays
BRET2 assays were elaborated on the protocol previously established in the lab (Roduit, et al., 2009). Human embryonic kidney (HEK) 293T cells were grown at 37° C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (PAA Laboratories GmbH, Pasching, Austria), supplemented with 10% FCS, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen AG, Basel, Switzerland). Cells were plated in 60-mm cell culture plates (TPP, Trasadingen, Switzerland) and transfected with 3–6 μg of plasmids at 30% confluence with the Calcium Phosphate method (ProFection®, Promega, Madison, WI). After 48 h (85–95 % confluence), cells were washed with 1x phosphate-buffered saline (PBS: 154 mM NaCl, 1 mM KH2PO4, 3 mM Na2HPO4 heptahydrate), incubated for 45 sec with 500 μl of trypsin/EDTA, transferred in 15-ml centrifuge tubes (Sarstedt AG, Sevelen, Switzerland), centrifuged for 2 min at 1000 rpm in an Eppendorf 5804 centrifuge, washed twice with 10 ml of Dulbecco’s Phosphate-Buffered Saline (D-PBS, GIBCO, Invitrogen AG, Basel, Switzerland), resuspended in 400 μl of D-PBS and stored on ice. As a transfection control, 45 μl of cell suspension was mixed with 55 μl of D-PBS in black 96-well microplates (Optiplate, PerkinElmer Life Sciences/Packard Biosciences) and emitted GFP2 fluorescence levels measured with a filter set excitation at 485 nm/ emission at 515 nm on an Envision® microplate reader (PerkinElmer Life Sciences/Biosignal, Montreal, QC). For luminescence assays, 45 μl of cell suspension was distributed in white 96-well microplates (Optiplate, PerkinElmer Life Sciences/Packard Biosciences). The luciferase substrate coelenterazine h (DeepBlueC, PerkinElmer/Biosignal) was added at a final concentration of 5 μM (volume 5 μl). Filters were 410 nm for luciferase emission and 515 nm for GFP2 emission. BRET2 ratio was defined as (emission at 515 nm)/(emission at 410 nm), higher ratios indicating increased dimerization activity.
To assess GFPC3-NR2E3 expression, HEK293T cells were cultured in 100-mm cell plates and transfected with 10 μg of GFPC3-NR2E3 expression plasmids. After 48 h, cells were washed in 1x PBS and resuspended in 100 μl of sample buffer containing 20 mM Hepes, 0.5 % Tween, phosphatase inhibitors cocktail 1 and 2 (Sigma, St-Louis, MO) and Complete® protease inhibitors (Roche Applied Science, Rotkreuz, Switzerland). Cells were lysed on ice by three 10-sec pulses at 40 % of power on a Sonicator 3000 (Misonix, Farmingdale, NY) and centrifuged for 5 min at 15000 rpm in a refrigerated Eppendorf benchtop centrifuge at 4° C. Supernatants were transferred and protein concentration determined with the Micro BCA Protein Assay Kit (Thermo Scientific) to proceed with Western blotting (see below).
Cell fractionation assay
HEK293T cells were cultured on 60 mm plates and transfected with 1.5 μg of pcDNA3.1/HisC-hNR2E3 expression vectors in a total of 3 μg of DNA. After 48h, cells were imaged by fluorescence microscopy (Axiovert 200, Carl Zeiss AG, Feldbach, Switzerland). Cells were then trypsinized, centrifuged at 1000 rpm at 25°C and pellets rinsed with 1x PBS, before proceeding with the preparation of cytoplasmic and nuclear extracts (NE-PER nuclear and cytoplasmic extraction reagents, Thermo Fisher Scientific, Lausanne, Switzerland). After Western blotting, images were quantified with ImageJ software (Schneider, et al., 2012) and NR2E3 expression normalized to α-tubulin expression, adjusted for cytoplasmic contamination of the nuclear fraction.
Western Blot Analysis
For Western blot analysis, 30 μg of proteins were transferred to PVDF membranes (Immobilon-P, Merck Millipore, Billerica, MA). For non-denaturing gel electrophoresis cell pellets were directly dissolved in an equal volume of 2x sample buffer (120 mM Tris-HCl pH 6.8, 20% glycerol, 0.1% bromophenol blue), and sodium dodecyl sulfate (SDS) removed from all gel solutions and electrophoresis buffers. Membranes were blocked for 1 h in TBS-Tween containing 5% nonfat dry milk. Rabbit polyclonal anti–NR2E3 antibody (#OAAB10504, Aviva Biosystems, San Diego, CA) was diluted 1:800, rabbit polyclonal anti-GFP 1:5’000 (#G1544, Sigma), mouse monoclonal anti-Xpress 1:500 (Invitrogen), rabbit polyclonal anti-alpha-tubulin (H300) 1:5000 (Santa Cruz Biotechnology, Santa Cruz, CA) and mouse monoclonal anti-α-tubulin 1:40’000 (#T5168, Sigma). The secondary ECL donkey anti-rabbit IgG horseradish peroxidase (HRP) and sheep anti-mouse IgG HRP-conjugated antibodies were diluted 1:25’000 (Amersham Biosciences, Otelfingen, Switzerland) and used to detect protein expression by chemiluminescence using LumiGlo (Amersham Biosciences) in a Fujifilm LAS-4000 mini imaging system (Bucher Biotec, Basel, Switzerland). Molecular weight markers were purchased at Fermentas (PageRuler™Plus).
Transactivation assays
HEK293T cells were plated in 12-well plates (TPP, Trasadingen, Switzerland) and each well was transfected with 30 ng of each of the expression vectors pcDNA3.1/HisC-hNR2E3 (wild-type and variants), pcDNA3.1/HisC-hCRX and pMT-hNRL, with 500 ng of the luciferase reporter constructs for rhodopsin (BR225-Luc) or M-opsin (Mop250-Luc) promoters (Peng, et al., 2005), and with 33 ng of pCMVβ (Clontech, Mountain View, CA) as internal standard. To keep the total of transfected DNA quantity constant, appropriate quantities of an empty pcDNA3.1/HisC vector were added in all experiments. Plasmids were prepared on NucleoBond®PC500 columns (Macherey-Nagel, Düren, Germany). Enzymatic activities were assessed with Luciferase Assay System (Promega) and standard β-Gal assay.
Protein Modeling
Protein modeling was based on the published X-ray diffraction data of a NR2E3 LBD dimer (PDB_1LOX) (Tan, et al., 2013) and a NR2A1 homodimer bound to DNA (PDB_1IQR) (Chandra, et al., 2013), available on Protein Data Bank (http://www.rcsb.org). Swiss PDB viewer version 4.1 in project mode was used to model and to visualize the NR2E3 LBD dimer on the DNA-bound NR2A1 homodimer template (Arnold, et al., 2006; Guex and Peitsch, 1997; Schwede, et al., 2003). The alignment of human NR2E3 (SwissProt_Q9Y5X4, 410 aa) and NR2A1 (SwissProt_B6ZGT3, 465 aa) amino acid sequences with Clustal W (www.ebi.ac.uk/Tools/msa/clustalw) and Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo) were used to manually curate the alignment in Swiss PDB viewer.
Statistical Analysis
All results were expressed as means ± SEM of the indicated number of experiments. Statistical analyses (one-way ANOVA with Dunnett’s multiple comparison test, two-way ANOVA) were performed with Prism 4.0.2 (GraphPad Software Inc., La Jolla, CA).
RESULTS
Structure of the NR2E3 LBD
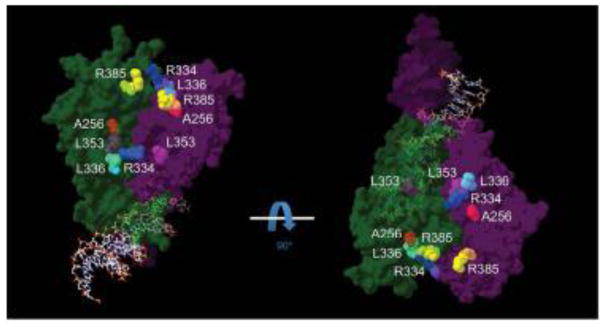
The recent crystallization of a large part of the NR2E3 LBD revealed a dimer arrangement (Tan, et al., 2013), with the dimer interface being formed by helices 10/11 of each monomer (Supp. Figure S1). Because NR2E3 and NR2A1, also called hepatocyte nuclear factor 4α (HNF4α; MIM# 600281), share a high sequence homology, with the exception of helices 1 and 2 that are not present in the LBD of NR2E3 (Supp. Figure S2), we superposed the crystal structure of the NR2E3 LBD dimer (Tan, et al., 2013) onto the crystal structure of the NR2A1 homodimer bound to DNA (Chandra, et al., 2013) to situate NR2E3 LBD missense variants in the context of an overall homodimer structure (Figure 1). A disease-causing variant had been reported at residue R385 that is located within the dimer interface formed by helices 10/11 (Supp. Table S2), but the protein model also indicated that ESCS-linked residues A256 (helix 4), R334 (loop between helix 8 and 9), L336 (ibidem) and L353 (helix 9) were located close to the dimer interface (Figure 1). Of note, ESCS-linked missense variants in the NR2E3 LBD affect residues almost strictly conserved among vertebrates, and even in sea urchin, suggesting a conserved function (Supp. Figure S3).
Figure 1.
General view of a NR2E3 homodimer bound to DNA. The crystal structure of the HNF4α/NR2A1 (isoform a) homodimer bound to DNA (PDB_4IQR) (Chandra, et al., 2013) served as a template to superpose the structure of the NR2E3 LBD dimer (PDB_4LOG) in an auto-repressed conformation (Tan, et al., 2013). One monomer is shown in dark green, the other in dark purple, both contacting the DNA double helix. ESCS-linked residues located close to the dimer interface are indicated in colors: A256 in red, R334 in blue, L336 in cyan, L353 in magenta and R385 in yellow. To facilitate visualization, two different views turned by 90°C are shown. For clarity, residues R309 and R311 that are not predicted to be in close proximity of the dimer interface are not indicated (see Supp. Figure S1).
Essential role of the NR2E3 LBD in dimerization
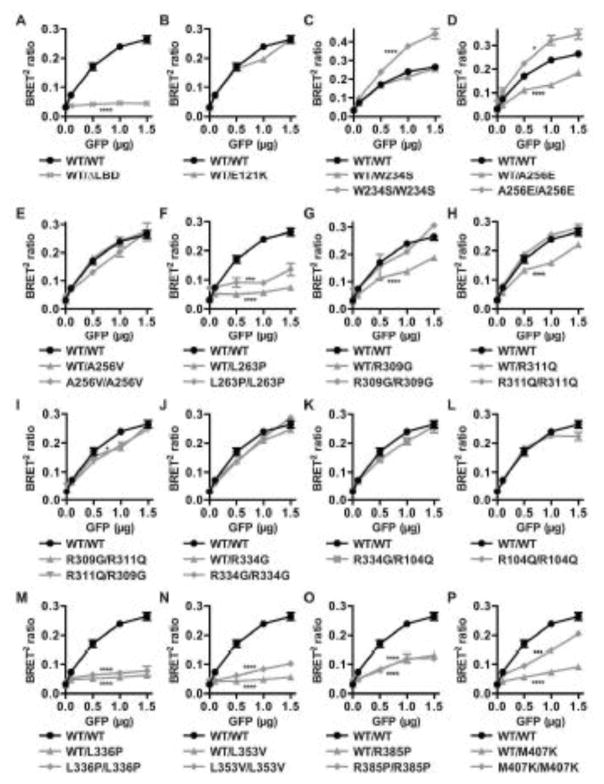
Consistent with the dimer arrangement of NR2E3 observed in the LBD crystal structure (Tan, et al., 2013), NR2E3 formed dimers in non-denaturing gel electrophoresis conditions (Roduit, et al., 2009). We tested the impact of some of the ESCS-linked NR2E3 LBD variants (Figure 1) in native conditions. Dimer formation appeared to be abolished in presence of the p.L263P variant, decreased in presence of the p.L353V and p.R385P variants and increased in presence of the p.W234S and p.A256E variants (Supp. Figure S4). To assess the dimerization potential of NR2E3 and the impact of some of the ESCS-linked NR2E3 LBD variants in further detail, we decided to elaborate on the BRET2 assays established in the lab by measuring protein interactions at several concentrations and not a single one as previously performed (Roduit, et al., 2009).
When expressing a constant amount of the RLuc-NR2E3 wild-type protein, and increasing amounts of the GFP-NR2E3 wild-type protein, an increase in the BRET2 ratio was observed, indicating an increased protein interaction between the RLuc-NR2E3 and GFP-NR2E3 fusion proteins (Figure 2A). We expressed a GFP-NR2E3-p.W234X variant protein, lacking the major part of the LBD (ΔLBD; Figure 2A). When co-expressing RLuc-NR2E3 wild-type protein, no dose-dependent increase in BRET2 ratio was observed, indicating that no protein interaction had occurred. Similar to what has been observed in other NRs, the LBD appears therefore to be essential for NR2E3 homodimerization and all disease-linked frameshift/truncation variants affecting the LBD are therefore expected to disrupt dimerization (Supp. Table S2). Of note, the BRET2 ratio profile of the NR2E3-p.E121K (c.361G>A) variant protein was not significantly different from that of the NR2E3 wild-type one (Figure 2B). This variant had initially been reported as a disease-causing variant (Haider, et al., 2000), but was later found in 15% of a Chinese control population, indicating that it is not pathogenic (Yang, et al., 2010).
Figure 2.
Homodimerization of NR2E3 wild-type and variant proteins. HEK293T cells were transiently transfected with 1 μg of expression plasmids for RLuc-NR2E3 wild-type (WT) or variant fusion proteins and with increasing amounts (0.1–1.5 μg) of expression plasmids for GFP-NR2E3 wild-type (WT) or variant fusion proteins (GFP μg). BRET2 ratios for wild-type homodimers are represented by black circles in all panels (A–P). The localization of the different variants is described in the main text. All experiments were performed four times in duplicates and SEM indicated. Two-way ANOVA (interaction) was used to determine variant BRET2 ratio profiles to be significantly different from the wild-type profile. *: p<0.05; ***: p<0.001; ****: p<0.0001.
Differential homodimerization properties of NR2E3 LBD variant proteins
We then tested dimerization of all NR2E3 LBD variant proteins with NR2E3 wild-type protein, as well as the homo- and hetero-dimerization potential of NR2E3 variant proteins described in patients (Figure 2). The c.701G>A (p.W234S) variant located in helix 3 did not affect dimerization with NR2E3 wild-type protein, but the p.W234S/p.W234S dimer was even more efficient in dimer formation than the wild-type counterpart (Figure 2C). The acidic c.767C>A/p.A256E substitution, located in the C-terminus of helix 4, reduced the BRET2 ratio of the wild-type/variant heterodimer, but increased that of the variant homodimer (Figure 2D). In contrast, the hydrophobic p.A256V substitution located at the same residue did not affect dimerization (Figure 2E). The c.788T>C (p.L263P) variant located in helix 5 abolished dimerization (Figure 2F). BRET2 ratios for this latter variant could also be altered because of a significant cytoplasmic localization, in contrast of a nuclear localization comparable to the NR2E3 wild-type protein for all other analyzed variants (Supp. Figure S5). For both c.925C>G (p.R309G) and c.932G>A (p.R311Q) variants located in the C-terminus of helix 7, dimer formation was similar to the wild-type protein, but dimerization between the variant and the wild-type protein was affected (Figure 2G, H). Because a compound heterozygous [p.R309G];[p.R311Q] patient has been reported (Wright, et al., 2004), we also tested dimerization of p.R309G/p.R311Q variant proteins: a slight decrease at the limit of statistical significance was detected only in the RLuc-NR2E3-p.R311Q/GFP-NR2E3-p.R309G configuration, but not in the RLuc-NR2E3-p.R309G/GFP-NR2E3-p.R311Q one (Figure 2I). Two variants located in the loop between helices 8 and 9 had opposite effects: p.R334G did not impair dimerization (Figure 2J), whereas c.1007T>C (p.L336P) completely abolished it (Figure 2M). Because the p.R334G variant had been identified in a compound heterozygous patient together with a p.R104Q variant (Hayashi, et al., 2005), we tested this heterodimer and observed no significant difference in comparison with the wild-type homodimer (Figure 2K). Also, the p.R104Q variant alone did not affect dimerization (Figure 2L). The c.1057C>G (p.L353V) variant located in helix 9 abolished dimerization (Figure 2N), as well as the c.1154G>C (p.R385P) variant, located in helix 10/11 (Figure 2O). Decreased BRET2 ratios for the p.R385P variant protein could also be attributed to decreased protein expression, as this was the only variant fusion protein consistently detected at lower expression levels by Western blotting (Supp. Figure S6). Finally, dimer formation was abolished between the c.1220T>A (p.M407K) variant, located in helix 12, and NR2E3 wild-type protein, and dimerization was also severely impaired for p.M407K homodimers (Figure 2P).
Heterodimerization of NR2E3 with retinal transcription factors
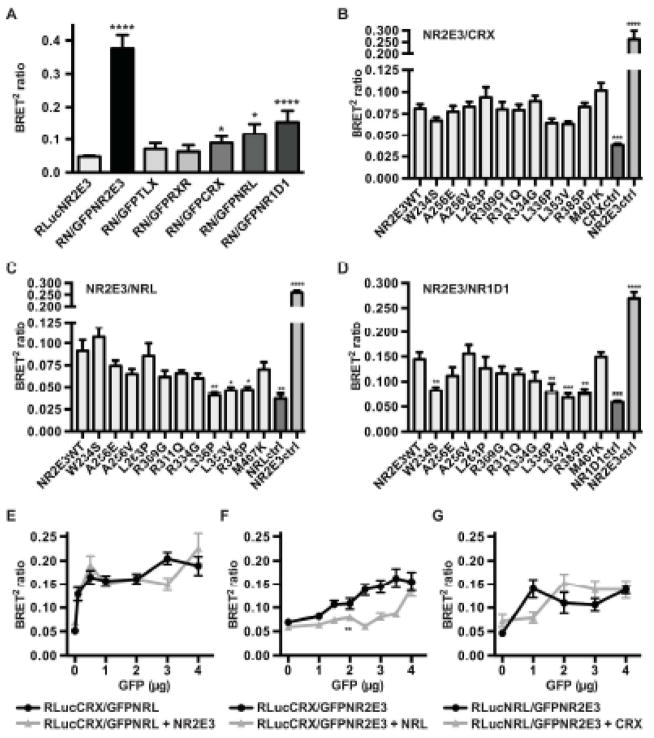
NR2E3 readily forms homodimers as indicated by a high BRET2 ratio (Figure 2). We used BRET2 assays to test the potential of NR2E3 to dimerize with other retina-expressed nuclear receptors, NR2E1, called tailless (TLX; MIM# 603849) and NR2C1, called retinoid X receptor (RXRα; MIM# 180245), and also with the photoreceptor-expressed transcription factors CRX, NRL (neural retina leucine zipper) and NR1D1, that had been previously shown to interact with NR2E3 (Cheng, et al., 2004; Kanda and Swaroop, 2009) (Figure 3A). NR2E3 did not form dimers with NR2E1 that had been reported to act as a monomer (Yu, et al., 1994), nor with NR2C1, the obligate heterodimerization partner of class I nuclear receptors (Mangelsdorf and Evans, 1995). Interactions with CRX and NRL were low and at the limit of statistical significance, but increased BRET2 ratios were observed in presence of NR1D1 (Figure 3A). To test whether NR2E3 LBD variants affected interaction with CRX, NRL and NR1D1 we also resorted to BRET2 assays. Consistent with CRX interacting with the DBD of NR2E3 (Peng, et al., 2005), NR2E3 LBD variants did not affect heterodimerization (Figure 3B). Interaction with NRL was affected by NR2E3 LBD variants p.L336P, p.L353V and p.R385P (Figure 3C), and heterodimerization with NR1D1 by p.W234S, p.L336P, p.L353V and p.R385P (Figure 3D).
Figure 3.
Heterodimerization of NR2E3 with retinal transcription factors. A) HEK293T cells were transfected with 3 μg of RLuc-NR2E3 wild-type (RLucNR2E3 or RN) and with 3 μg of GFP-NR2E3 wild-type (GFPNR2E3), GFP-TLX/NR2E1 (GFPTLX), GFP-RXRα/NR2C1 (GFPRXR), GFP-CRX (GFPCRX), GFP-NRL (GFPNRL) and GFP- rev-erbα/NR1D1 (GFP-NR1D1) expression plasmids. Experiments were performed four times in duplicates and SEM indicated. For statistical analysis, one-way ANOVA followed by Dunnett’s multiple comparison test comparing the mean of each column with the mean of the control RLucNR2E3 column was performed. B–D) Differential interaction of NR2E3 variant proteins with CRX, NRL and NR1D1. HEK293T cells were transiently transfected with 3 μg of RLuc-CRX (A), RLuc-NRL (B) and RLuc-rev-erbα/NR1D1 (RLuc-NR1D1) (C) expression plasmids, and, 3 μg of GFPC3-NR2E3 wild-type (NR2E3WT) or NR2E3 variant expression plasmids. RLuc expression vectors alone (respectively CRXctrl, NRLctrl and NR1D1ctrl) were used as negative controls, and Rluc-NR2E3WT/GFP-NR2E3WT transfected as positive controls (NR2E3ctrl). Experiments were performed three times in duplicates and SEM indicated. For statistical analysis, one-way ANOVA followed by Dunnett’s multiple comparison test, comparing the mean of each column with the mean of the control NR2E3 wild-type (NR2E3WT) column was performed. E–G) Characterization of CRX/NRL/NR2E3 interactions. HEK293T cells were transiently transfected with 2 μg of RLuc-CRX (E, F) and RLuc-NRL (G) expression plasmids, and increasing amounts of GFPC3-NRL (E), GFPC3-NR2E3 (F, G), in presence or not of 2 μg of pcDNA3.1/HisC-hNR2E3 (E), pMT-NRL (F), and pcDNA3.1/HisC-hCRX (G) expression plasmids. Experiments were performed three times in duplicates and SEM indicated. For statistical analysis, two-way ANOVA was performed. *: p<0.05; **<0.01; ***: p<0.001; ****: p<0.0001.
Functional analysis of NR2E3/CRX/NRL interactions
To regulate rod- and cone-specific gene expression, NR2E3 has been shown to functionally interact with CRX and NRL (Cheng, et al., 2004; Peng, et al., 2005). This prompted us to test pairwise by BRET2 assays heterodimer formation in the presence or not of the third partner. As suggested by rapidly increasing BRET2 ratios at low concentrations, CRX and NRL readily formed heterodimers that were not affected by the presence of NR2E3 (Figure 3E). In contrast, CRX and NR2E3 formed heterodimers only at higher concentrations, when NRL was co-expressed (Figure 3F). Finally, NRL/NR2E3 heterodimer formation was not significantly delayed in the presence of CRX (Figure 3G).
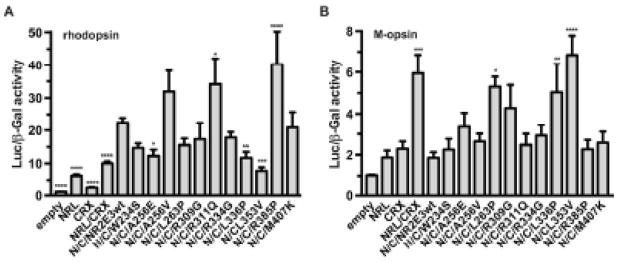
The effects of NR2E3 LBD variants on the function of the CRX/NRL/NR2E3 were then assessed by HEK293T cell-based luciferase reporter assays (Figure 4). Whereas the NR2E3 wild-type protein potentiated NRL/CRX mediated transactivation of the rhodopsin promoter (Cheng, et al., 2004), this activator function was significantly impaired in presence of p.A256E, p.L336P and p.L353V variants and potentiated in presence of p.R311Q and p.R385P variants (Figure 4A). Conversely, NR2E3 wild-type protein repressed NRL/CRX-mediated transactivation of the cone-specific M-opsin promoter (Peng, et al., 2005), and this repressor function was significantly impaired in presence of p.L263P, p.L336P and p.L353V variants (Figure 4B).
Figure 4.
Differential transcriptional activity of NR2E3 LBD variant proteins. HEK293T cells were transiently transfected with rhodopsin (A) or M-opsin (B) luciferase reporter constructs. NRL (N) and CRX (C) were cotransfected when indicated. Luciferase activity was normalized to β-galactosidase activity. Four to six independent experiments were performed in duplicates and SEM indicated. For statistical analysis, one-way ANOVA followed by Dunnett’s multiple comparison test, comparing the mean of each column with the mean of the NRL/CRX/NR2E3 wild-type (N/C/NR2E3wt) column was performed. *: p<0.05; **<0.01; ***: p<0.001; ****: p<0.0001.
Clinical description of an atypical compound heterozygous NR2E3 patient
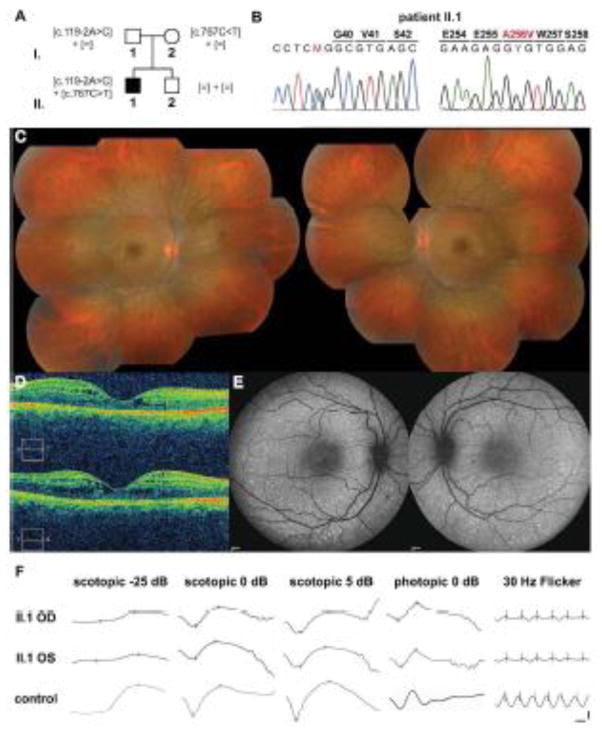
An 18-year-old patient of Italian origin was referred for blurred vision without any other ocular symptoms (Figure 5). Best corrected visual acuity (BCVA) was 0.8 in the right eye (OD) and 1.0 in the left eye (OS). Visual fields (VF) presented a slight supero-temporal dissociation of isopters V4e and I4e (Supp. Figure S7). Funduscopy showed pin-point fleck-like lesions along the vascular arcades (Figure 5C) and optical coherence tomography (OCT) confirmed an early foveal schisis-like maculopathy (Figure 5D). On fundus autofluorescence, small hyperautofluorescent dots were observed along the vascular arcades (Figure 5E). ISCEV full-field ERGs demonstrated a 3-fold decrease in the rod-specific responses (scotopic -25 dB) in OD that was still within the normal limit, and a 4-fold decrease in OS, with physiological implicit times (Figure 5F, Table 1). In scotopic ERGs at 0 dB and 5 dB stimulation, where mixed rod and cone responses can be elicited, reduced amplitudes in a and b waves and markedly delayed a wave implicit times were observed, suggesting involvement of the S-cone system. Delayed large and prolonged b-wave in the transient photopic cone ERG (photopic 0 dB), and 30 Hz flicker ERG being both of reduced amplitudes and delayed implicit times, were also compatible with ESCS/GFS. Subsequent molecular analyses identified the proband as a compound heterozygous patient harboring the frequent splice-site variant c.119-2A>C located in intron 1 of NR2E3 and a c.767C>T (p.A256V) variant, located in exon 6 of NR2E3, recessively inherited respectively from his father and his mother (Figures 5A, B). Taken together, these data identified the proband as an atypical, mildly affected ESCS/GFS patient, with residual rod function.
Figure 5.
Compound heterozygous NR2E3 [c.119-2A>C];[p.A256V] patient. A) Pedigree of the Italian family with recessive inheritance of the [c.119-2A>C] variant from the unaffected father and the [c.767C>T] (p.A256V) variant from the unaffected mother. None of the variants was detected in the unaffected brother II.2. B) Electropherograms of the heterozygous substitutions c.119-2A>C in exon 2 and c.767C>T (p.A256V) in exon 6 present in patient II.1. Sequences were aligned with the reference genomic NR2E3 sequence NW_001838218.2. C) Composite fundus photograph of the 18-year-old proband II.1. Small white/yellowish dots and flecks along the vascular arcades and around the macula are observed (OD: left panel; OS: right panel). D) Optical coherence tomography (OCT) of the right (upper panel) and left eyes (lower panel) of the proband. Foveal schisis is visible in both eyes. E) Fundus autofluorescence examination revealed hyperautofluorescent pin-points along the vascular arcades in both eyes (OD: left panel; OS: right panel). F) ISCEV scotopic and photopic full-field ERGs revealed residual rod-specific responses (scotopic -25 dB), decreased scotopic maximal responses (scotopic 5 dB), delayed transient photopic responses with normal amplitudes (photopic 0 dB). The photopic 30-Hz flicker was delayed and of decreased amplitude. Horizontal bar: 20 msec; vertical bar: 50 μV.
Table 1.
Electroretinographic findings in the patient II.I
| OD II.I | OS II.I | control (mean) | limit | |
|---|---|---|---|---|
| scotopic ERG -25 dB | ||||
| b-wave (μV) | 90.3 | 59.8 | 244.8±79.7 | 85.4 |
| implicit time (msec) | 87.8 | 93.3 | 86.9±7.5 | 101.9 |
| scotopic ERG 0 dB | ||||
| a-wave (μV) | 221.0 | 184.0 | 233.0±62.0 | 109.0 |
| implicit time (msec) | 22.3 | 25.8 | 18.4±2.6 | 23.6 |
| b-wave (μV) | 446.0 | 492.0 | 411.7±101.6 | 208.5 |
| implicit time (msec) | 57.8 | 54.9 | 50.3±3.2 | 56.7 |
| scotopic ERG 5 dB | ||||
| a-wave (μV) | 267.0 | 270.0 | 282.8±72.3 | 138.2 |
| implicit time (msec) | 24.1 | 24.7 | 15.5±0.7 | 16.9 |
| b-wave (μV) | 492.0 | 437.0 | 444.4±106.2 | 232.0 |
| implicit time (msec) | 54.9 | 58.7 | 52.2±4.4 | 61.0 |
| photopic ERG 0 dB | ||||
| a-wave (μV) | 44.6 | 67.7 | 41.2±12.8 | 15.6 |
| implicit time (msec) | 17.0 | 17.9 | 14.9±1.3 | 17.5 |
| b-wave (μV) | 146.0 | 147.0 | 144.3±41.8 | 60.7 |
| implicit time (msec) | 38.2 | 38.2 | 31.9±1.1 | 34.1 |
| Flicker 30 Hz | ||||
| amplitude (μV) | 65.3 | 62.0 | 119.5±36.1 | 47.3 |
| implicit time (msec) | 34.1 | 30.5 | 28.2±1.1 | 30.4 |
International Society for Clinical Electrophysiology of Vision (ISCEV) standard photopic and scotopic ERG responses (Marmor, et al., 2009). Normal limit (limit) corresponds to: mean – 2 x SD. Off limit results of the patient are in italic.
DISCUSSION
BRET2 analysis identified NR2E3 LBD variants that i) disrupted NR2E3 homodimerization (p.L263P, p.L336P, p.L353V, p.R385P), ii) disrupted NR2E3 heterodimerization (p.W234S, p.L336P, p.L353V, p.R385P), iii) disrupted NR2E3 homo- and heterodimerization (p.L336P, p.L353V, p.R385P), iii) increased homodimerization (p.W234S, p.A256E), and, iv) did not affect dimerization (p.A256V, p.R309G, p.R311Q, p.R334G) (Supp. Table S3). BRET2 ratios could be altered by misfolding of variant proteins, for instance by introducing Proline residues that cause a kink in the peptidic chain. Therefore, in addition to testing for correct protein expression and cellular localization, we also performed functional assays on a rod- and a cone-specific promoter. Of note, the differential dimerization potential of NR2E3 did not necessarily correlate with transcriptional activity of NR2E3 on the rhodopsin and the M-opsin promoters in a heterologous system where CRX and NRL were co-expressed (Supp. Table S3). In presence of the p.L336P and p.L353V variants no homodimer formation and no interaction with NRL was observed, and, consistently, transactivation of the rhodopsin promoter and transrepression of the M-opsin promoter was impaired. No homodimers were observed in presence of the p.L263P variant, but, there, only transrepression was affected. Furthermore, for the dimerization-incompetent p.R385P and p.M407K variants, transactivation and transrepression were within normal levels, if not increased. Complex interactions in NR2E3-associated transcription factors are further illustrated by the p.A256E variant, where an increased homodimer formation was observed, but rhodopsin transactivation was affected. Our observation that residues L336, L353 and R385 critical for NR2E3 homodimerization are also involved in the interaction with NRL and NR1D1, suggest a potential competition between NR2E3 homodimer formation and heterodimerization with other photoreceptor-specific transcription factors. In support of this hypothesis, the p.W234S variant indeed exhibited increased homodimerization but impaired heterodimerization with NR1D1. Finally, given the low BRET2 ratios observed in presence of CRX and NR2E3, as compared to CRX and NRL, further experimental data on promoter co-occupancy might be needed to assess the physiological relevance of CRX/NR2E3 interactions.
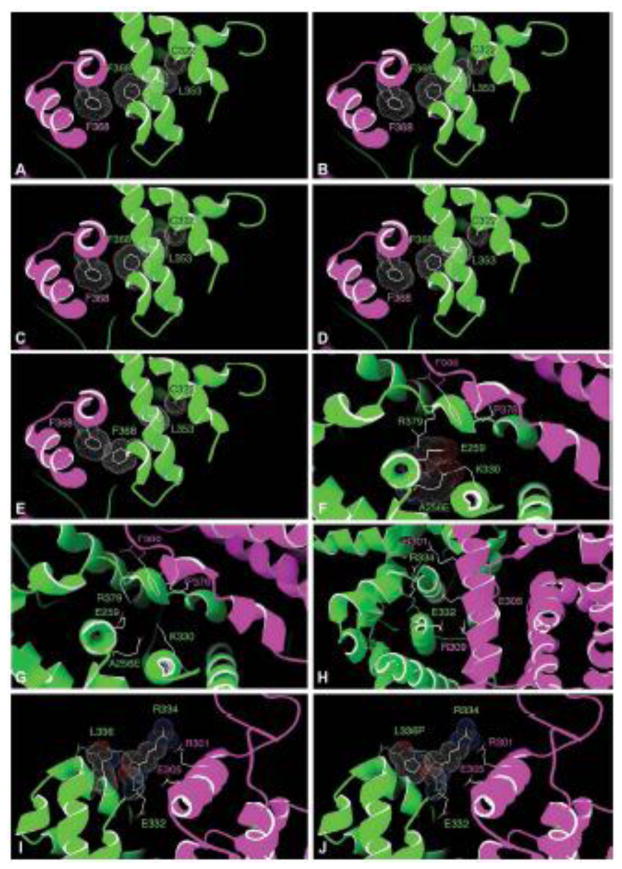
The homology model provides some additional insight for absence of dimer formation. R385 is located at the kink connecting helix 10 and helix 11 and was predicted to be in close proximity of the LBD of its NR2E3 partner (Figure 1). Consistently, the p.R385P variant severely reduced the dimerization of both the homo- and the heterodimer. L353 is located in helix 9 and its sidechain can adopt the conformation of a rotamer, frequently observed in α-helices (e.g. 30%). The packing with C322 and F368, located in helices 8 and 10/11 respectively, was optimal (Figure 6A). In contrast, the p.L353V variant could not fit without inducing a rearrangement of the structure. In particular, the two most frequent Valine rotamers observed 90% (Figure 6B) and 7% of the time (Figure 6C), were predicted to severely clash with the Cα of C322, and in the third possible conformation to clash into F368 (Figure 6D). Consistent with our functional data, this variant was predicted in all cases to disrupt the dimerization interface provided by the evolutionarily conserved F368 and G369 residues of both partners (Figure 6E).
Figure 6.
Structural contexts of disease-causing NR2E3 LBD variants based on the model shown in Figure 1 (see discussion). Detailed views of the contexts of the A256 residue (A–B), the R334 residue (C), the R334 and L336 residues (D–E) and the L353 residue (F–J). One NR2E3 LBD monomer is shown in purple, the other in green. For clarity, only the two last helices of the green monomer are shown in E–H.
Interestingly, the p.A256V and p.R334G variants were comparable to the NR2E3 wild-type protein in all functional assays (Supp. Table S3). Alanine 256 is located at the hinge between helices 4 and 5 (Figure 6F). Consistent with the functional data, there is just enough space to tolerate the p.A256V variant without perturbing the structure and the dimerization interface. However, the Glutamate of the above-mentioned p.A256E variant, which bears a longer negatively charged side chain, could not be accommodated at this same position within the structure. All rotamers (Lovell, et al., 2000) were predicted to clash with either the backbone or the sidechains of other residues (e.g. L252, L253, L260, V327, L328). Only two of the 15 possible rotamers (Lovell, et al., 2000) could accommodate this negative charge buried within a highly hydrophobic environment by pointing it outward to the immediate vicinity of the negative and positive charges geared by E259 and K330, respectively (Figure 6F). However, when pointing outward, p.A256E is clashing into E259 and K330, which will have to move slightly as a result. Since both residues are located in close proximity of the dimer interface, in a region where sidechain rearrangement can be accommodated, the increased dimerization observed in presence of the p.A256E homodimer might be due to further stabilization of the homodimer interface, for example through a stacking of R379 with F380 of the other monomer (Figure 6G).
The modeling of the p.R334G variant also correlated with the functional data that showed a dimerization potential comparable to the wild-type one. R334 is located in the loop connecting helices 8 and 9 and its sidechain is in contact with R301 and R385 (Figure 6H). Although arginine side chains had been reported to be seen in close proximity (Neves, et al., 2012), their positive charge was balanced by negatively charged counterions in about 90% of the cases, a role that the not too distant E305 might play here. By introducing the variant p.R334G, the loop connecting helices 8 and 9 was predicted to become very flexible because of the two successive Glycines (G334 and G335). This and the absence of the long R334 sidechain might favor a rearrangement of R301 and E305, which could potentially reach the backbone of the loop, thus stabilizing the dimer. It is also possible that in absence of R334, the negative charge of E255 might favor a long range attraction of R301 of the dimerization partner. The homology model provides also a mechanism by which the p.L336P variant also located in the loop connecting helix 8 and helix 9, may abolish dimerization. Whereas L336 fits very well in the loop environment through packing with V342 (Figure 6I), this packing was lost in presence of a Proline (p.L336P) (Figure 6J). Furthermore, the Proline ring was predicted to clash with the backbone of T333 and R334. Thus, to accommodate the p.L336P variant, the loop would have to move toward the dimerization partner, thus pushing R334 discussed earlier even closer to the dimerization partner, likely to the point of disruption. The two only patients that have been reported so far to be affected with an atypically mild form of ESCS with detectable rod responses were compound heterozygotes [c.119-2A>C];[p.A256V] (Lam, et al., 2007) or [p.R104Q];[p.R334G] (Hayashi, et al., 2005). The first patient is expected to express only the p.A256V variant protein, because the c.119-2A>C splice-site mutation results in missplicing of exon 2 (Bernal, et al., 2008), yielding potentially in nonsense-mediated RNA decay. The second patient could potentially express p.R104Q and p.R334G homodimers or p.R104Q/p.R334G heterodimers, that all showed BRET2 ratios similar to the wild-type protein. The present description of an additional compound heterozygote [c.119-2A>C];[p.A256V] atypical patient with residual rod function, and of a different geographic origin from the previous ones, suggests that NR2E3 variant proteins efficiently forming dimers are able to mediate partial physiological activity by repressing cone-specific genes in rod photoreceptors and by activating rhodopsin expression. Whether this partial activity restored some function in all rods or only in a portion of the otherwise non-functional rods remains elusive. The foveal schisis-like maculopathy observed in the patient provides circumstantial evidence that the NR2E3 activity might not have been sufficient to block the generation of some excess of S-cones, instead of the early rod progenitors (Cheng, et al., 2011), given the excess of cones has been recognized to disrupt the retinal architecture (Chen and Nathans, 2007). Of note, even if the p.R104Q/p.R334G heterodimer were DNA-binding defective (Escher, et al., 2009), activation of rhodopsin could still be effective, as no NR2E3 binding sites have been identified on the proximal rhodopsin promoter (Cheng, et al., 2004).
In addition to the p.A256V and p.R334G variants, the p.R309G and p.R311Q variants that are not located at the dimer interface, were also comparable to the NR2E3 wild-type protein in our functional assays. However, these variants cause typical ESCS with night blindness, excluding dimerization potential as a disease mechanism (Schorderet and Escher, 2009). Additionally, for these two variants, dimerization was impaired in presence of NR2E3 wild-type and variant proteins, but all reported heterozygous carriers are healthy (Schorderet and Escher, 2009). Impaired binding to the corepressors Ret-CoR (Takezawa, et al., 2007) or atrophin-1 (Escher, et al., 2009) had been suggested as a disease mechanism for the p.R311Q variant, and additional BRET2 analysis could contribute to substantiate this hypothesis.
Supplementary Material
Acknowledgments
Funding: This work was supported by Swiss National Science Foundation grants 31003A-122269 (P.E. and D.F.S.) and 31003A_138492 (P.E.) as well as the Gottfried-und-Julia-Bangerter-Rhyner-Stiftung (P.E.).
The excellent help of Mrs Tatiana Favez in expression vector construction and of Mrs Nathalie Voirol in cell culture experiments is greatly acknowledged, as well as technical and administrative help of Dr. François-Xavier Borruat, Dr. Fabienne Marcelli, Angélique Schmidt, Barrett Leahy, Yann Leuba, Sue Houghton, Luca Marchionno and Josiane Keller. Plasmids pcDNA3.1/HisC-hNR2E3, pcDNA3.1/HisC-hCRX, pMT-NRL, BR225-Luc and Mop250-Luc were kindly provided by Dr. Shiming Chen, pSG5-hRXRα by Dr. Pierre Chambon and pCMX-hTLX by Dr. Chih-Cheng Tsai. We thank Dr. Marie-Claire Gaillard for critical reading of the manuscript. The authors declare no conflict of interest.
References
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, et al. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci USA. 2000;97(10):5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Audo I, Bujakowska KM, Leveillard T, Mohand-Said S, Lancelot ME, Germain A, Antonio A, Michiels C, Saraiva JP, Letexier M, Sahel JA, Bhattacharya SS, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis. 2012;7(1):8. doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audo I, Michaelides M, Robson AG, Hawlina M, Vaclavik V, Sandbach JM, Neveu MM, Hogg CR, Hunt DM, Moore AT, Bird AC, Webster AR, et al. Phenotypic variation in enhanced S-cone syndrome. Invest Ophthalmol Vis Sci. 2008;49(5):2082–2093. doi: 10.1167/iovs.05-1629. [DOI] [PubMed] [Google Scholar]
- Bandah D, Merin S, Ashhab M, Banin E, Sharon D. The spectrum of retinal diseases caused by NR2E3 mutations in Israeli and Palestinian patients. Arch Ophthalmol. 2009;127(3):297–302. doi: 10.1001/archophthalmol.2008.615. [DOI] [PubMed] [Google Scholar]
- Bernal S, Solans T, Gamundi MJ, Hernan I, de Jorge L, Carballo M, Navarro R, Tizzano E, Ayuso C, Baiget M. Analysis of the involvement of the NR2E3 gene in autosomal recessive retinal dystrophies. Clin Genet. 2008;73(4):360–366. doi: 10.1111/j.1399-0004.2008.00963.x. [DOI] [PubMed] [Google Scholar]
- Cassiman C, Spileers W, De Baere E, de Ravel T, Casteels I. Peculiar fundus abnormalities and pathognomonic electrophysiological findings in a 14-month-old boy with NR2E3 mutations. Ophthalmic Genet. 2013;34(1–2):105–108. doi: 10.3109/13816810.2012.726395. [DOI] [PubMed] [Google Scholar]
- Chandra V, Huang P, Potluri N, Wu D, Kim Y, Rastinejad F. Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature. 2013;495(7441):394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Figueroa DJ, Marmorstein AD, Zhang Q, Petrukhin K, Caskey CT, Austin CP. Retina-specific nuclear receptor: A potential regulator of cellular retinaldehyde-binding protein expressed in retinal pigment epithelium and Muller glial cells. Proc Natl Acad Sci USA. 1999;96(26):15149–15154. doi: 10.1073/pnas.96.26.15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nathans J. Genetic ablation of cone photoreceptors eliminates retinal folds in the retinal degeneration 7 (rd7) mouse. Invest Ophthalmol Vis Sci. 2007;48(6):2799–2805. doi: 10.1167/iovs.06-0922. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25(1):118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khan NW, Roger JE, Swaroop A. Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3, originate from early-born photoreceptor precursors. Hum Mol Genet. 2011;20(21):4102–4115. doi: 10.1093/hmg/ddr334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13(15):1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Coppieters F, Leroy BP, Beysen D, Hellemans J, De Bosscher K, Haegeman G, Robberecht K, Wuyts W, Coucke PJ, De Baere E. Recurrent mutation in the first zinc finger of the orphan nuclear receptor NR2E3 causes autosomal dominant retinitis pigmentosa. Am J Hum Genet. 2007;81:147–157. doi: 10.1086/518426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1(2):e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher P, Gouras P, Roduit R, Tiab L, Bolay S, Delarive T, Chen S, Tsai C-C, Hayashi M, Zernant J, Merriam JE, Mermod N, et al. Mutations in NR2E3 can cause dominant or recessive retinal degenerations in the same family. Hum Mutat. 2009;30:342–351. doi: 10.1002/humu.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher P, Tran HV, Vaclavik V, Borruat FX, Schorderet DF, Munier FL. Double concentric autofluorescence rings in NR2E3-p. G56R-linked autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2012;53(8):4754–4764. doi: 10.1167/iovs.11-8693. [DOI] [PubMed] [Google Scholar]
- Gerber S, Rozet JM, Takezawa SI, dos Santos LC, Lopes L, Gribouval O, Penet C, Perrault I, Ducroq D, Souied E, Jeanpierre M, Romana S, et al. The photoreceptor cell-specific nuclear receptor gene (PNR) accounts for retinitis pigmentosa in the Crypto-Jews from Portugal (Marranos), survivors from the Spanish Inquisition. Hum Genet. 2000;107(3):276–284. doi: 10.1007/s004390000350. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14(2):121–141. [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Haider NB, Demarco P, Nystuen AM, Huang X, Smith RS, McCall MA, Naggert JK, Nishina PM. The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis Neurosci. 2006;23(6):917–929. doi: 10.1017/S095252380623027X. [DOI] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24(2):127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001;10(16):1619–1626. doi: 10.1093/hmg/10.16.1619. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Gekka T, Goto-Omoto S, Takeuchi T, Kubo A, Kitahara K. Novel NR2E3 mutations (R104Q, R334G) associated with a mild form of enhanced Scone syndrome demonstrate compound heterozygosity. Ophthalmology. 2005;112(12):2115. doi: 10.1016/j.ophtha.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Sumaroka A, Aleman TS, Cideciyan AV, Schwartz SB, Roman AJ, McInnes RR, Sheffield VC, Stone EM, Swaroop A, Wright AF. Nuclear receptor NR2E3 gene mutations distort human retinal laminar architecture and cause an unusual degeneration. Hum Mol Genet. 2004;13(17):1893–1902. doi: 10.1093/hmg/ddh198. [DOI] [PubMed] [Google Scholar]
- Jurklies B, Weismann M, Kellner U, Zrenner E, Bornfeld N. Clinical findings in autosomal recessive syndrome of blue cone hypersensitivity. Ophthalmologe. 2001;98(3):285–293. doi: 10.1007/s003470170164. [DOI] [PubMed] [Google Scholar]
- Kanda A, Swaroop A. A comprehensive analysis of sequence variants and putative disease-causing mutations in photoreceptor-specific nuclear receptor NR2E3. Mol Vis. 2009;15:2174–2184. [PMC free article] [PubMed] [Google Scholar]
- Khan AO, Aldahmesh MA, Al-Harthi E, Alkuraya FS. Helicoid subretinal fibrosis associated with a novel recessive NR2E3 mutation p. S44X Arch Ophthalmol. 2010;128(3):344–348. doi: 10.1001/archophthalmol.2010.15. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam BL, Goldberg JL, Hartley KL, Stone EM, Liu M. Atypical mild enhanced Scone syndrome with novel compound heterozygosity of the NR2E3 gene. Am J Ophthalmol. 2007;144(1):157–159. doi: 10.1016/j.ajo.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40(3):389–408. [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor MF. A teenager with nightblindness and cystic maculopathy: enhanced S cone syndrome (Goldmann-Favre syndrome) Doc Ophthalmol. 2006;113(3):213–215. doi: 10.1007/s10633-006-9031-z. [DOI] [PubMed] [Google Scholar]
- Marmor MF, Fulton AB, Holder GE, Miyake Y, Brigell M, Bach M Vision. ISfCEo. ISCEV Standard for full-field clinical electroretinography (2008 update) Doc Ophthalmol. 2009;118(1):69–77. doi: 10.1007/s10633-008-9155-4. [DOI] [PubMed] [Google Scholar]
- Marmor MF, Jacobson SG, Foerster MH, Kellner U, Weleber RG. Diagnostic clinical findings of a new syndrome with night blindness, maculopathy, and enhanced S cone sensitivity. Am J Ophthalmol. 1990;110(2):124–134. doi: 10.1016/s0002-9394(14)76980-6. [DOI] [PubMed] [Google Scholar]
- Mollema NJ, Yuan Y, Jelcick AS, Sachs AJ, von Alpen D, Schorderet DF, Escher P, Haider NB. Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS ONE. 2011;6(3):e17494. doi: 10.1371/journal.pone.0017494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves MA, Yeager M, Abagyan R. Unusual arginine formations in protein function and assembly: rings, strings, and stacks. J Phys Chem B. 2012;116(23):7006–7013. doi: 10.1021/jp3009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachydaki SI, Klaver CC, Barbazetto IA, Roy MS, Gouras P, Allikmets R, Yannuzzi LA. Phenotypic features of patients with NR2E3 mutations. Arch Ophthalmol. 2009;127(1):71–75. doi: 10.1001/archophthalmol.2008.534. [DOI] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14(6):747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Qin Q, Knapinska A, Dobri N, Madoux F, Chase P, Hodder P, Petrukhin K. In pursuit of synthetic modulators for the orphan retina-specific nuclear receptor NR2E3. J Ocul Pharmacol Ther. 2012;29(3):298–309. doi: 10.1089/jop.2012.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roduit R, Escher P, Schorderet DF. Mutations in the DNA-binding domain affect in vivo NR2E3 dimerization and interaction with CRX. PLoS One. 2009;4(10):e7379. doi: 10.1371/journal.pone.0007379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet DF, Escher P. NR2E3 mutations in enhanced S-cone sensitivity syndrome (ESCS), Goldmann-Favre syndrome (GFS), clumped pigmentary retinal degeneration (CPRD), and retinitis pigmentosa (RP) Hum Mutat. 2009;30(11):1475–1485. doi: 10.1002/humu.21096. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja T. Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol. 2003;121:1316–1323. doi: 10.1001/archopht.121.9.1316. [DOI] [PubMed] [Google Scholar]
- Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yanagi Y, Ito H, Takada I, Kishimoto M, Miyajima A, Takeyama K, Umesono K, et al. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 2007;26(3):764–774. doi: 10.1038/sj.emboj.7601548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tan MH, Zhou XE, Soon FF, Li X, Li J, Yong EL, Melcher K, Xu HE. The crystal structure of the orphan nuclear receptor NR2E3/PNR ligand binding domain reveals a dimeric auto-repressed conformation. PLoS One. 2013;8(9):e74359. doi: 10.1371/journal.pone.0074359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NK, Fine H, Chang S, Chou CL, Cella W, Tosi J, Lin CS, Nagasaki T, Tsang SH. Cellular origin of fundus autofluorescence in patients and mice with defective NR2E3 Gene. Br J Ophthalmol. 2009;93(9):1234–1240. doi: 10.1136/bjo.2008.153577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Schwabe JW. Nuclear hormone receptor co-repressors: structure and function. Mol Cell Endocrinol. 2012;348(2):440–449. doi: 10.1016/j.mce.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AF, Reddick AC, Schwartz SB, Ferguson JS, Aleman TS, Kellner U, Jurklies B, Schuster A, Zrenner E, Wissinger B, Lennon A, Shu X, et al. Mutation analysis of NR2E3 and NRL genes in enhanced S-cone syndrome. Hum Mutat. 2004;24(5):439–450. doi: 10.1002/humu.9285. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang X, Chen LJ, Chiang SWg, Tam PO, Lai TY, Chan CK, Wang N, Lam DS, Pang CP. NR2E3 but not NRL mutations are associated with retinitis pigmentosa in the Chinese population. Invest Ophthalmol Vis Sci. 2010;51(1) doi: 10.1167/iovs.09-4299. [DOI] [PubMed] [Google Scholar]
- Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370(6488):375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- Yzer S, Barbazetto I, Allikmets R, van Schooneveld MJ, Bergen A, Tsang SH, Jacobson SG, Yannuzzi LA. Expanded clinical spectrum of enhanced Scone syndrome. JAMA Ophthalmol. 2013;131(10):1324–1330. doi: 10.1001/jamaophthalmol.2013.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.