Abstract
Background
PTEN is the most commonly deleted tumor suppressor gene in primary prostate cancer (PCa) and its loss is associated with poor clinical outcomes and ERG gene rearrangement.
Objective
We tested whether PTEN loss is associated with shorter recurrence-free survival (RFS) in surgically treated PCa patients with known ERG status.
Design, setting, and participants
A genetically validated, automated PTEN immunohistochemistry (IHC) protocol was used for 1275 primary prostate tumors from the Canary Foundation retrospective PCa tissue microarray cohort to assess homogeneous (in all tumor tissue sampled) or heterogeneous (in a subset of tumor tissue sampled) PTEN loss. ERG status as determined by a genetically validated IHC assay was available for a subset of 938 tumors.
Outcome measurements and statistical analysis
Associations between PTEN and ERG status were assessed using Fisher’s exact test. Kaplan-Meier and multivariate weighted Cox proportional models for RFS were constructed.
Results and limitations
When compared to intact PTEN, homogeneous (hazard ratio [HR] 1.66, p = 0.001) but not heterogeneous (HR 1.24, p = 0.14) PTEN loss was significantly associated with shorter RFS in multivariate models. Among ERG-positive tumors, homogeneous (HR 3.07, p < 0.0001) but not heterogeneous (HR 1.46, p = 0.10) PTEN loss was significantly associated with shorter RFS. Among ERG-negative tumors, PTEN did not reach significance for inclusion in the final multivariate models. The interaction term for PTEN and ERG status with respect to RFS did not reach statistical significance (p = 0.11) for the current sample size.
Conclusions
These data suggest that PTEN is a useful prognostic biomarker and that there is no statistically significant interaction between PTEN and ERG status for RFS.
Patient summary
We found that loss of the PTEN tumor suppressor gene in prostate tumors as assessed by tissue staining is correlated with shorter time to prostate cancer recurrence after radical prostatectomy.
Keywords: Biomarker, ERG, Immunohistochemistry, PTEN, Prostatic carcinoma, Radical prostatectomy
1. Introduction
PTEN is the most commonly deleted tumor suppressor gene in prostate cancer (PCa) [1–4] and its loss is associated with poor pathologic and clinical outcomes [5–20]. Since the PTEN gene is almost always lost by deletion in PCa, fluorescence in situ hybridization (FISH) has traditionally been used to detect PTEN loss and examine its association with outcomes [13,14,16,17,21,22]. However, we and others have demonstrated that PTEN loss is commonly subclonal and heterogeneous in primary prostate tumors [23–25], making its detection by FISH or techniques that require nucleic acid extraction technically challenging in some cases. Furthermore, there is emerging evidence that in addition to genetic deletion, PTEN protein levels may be regulated by microRNAs and epigenetic modifications [10,15,18]. To address these issues, we previously optimized and validated an immunohistochemistry (IHC) assay to detect PTEN protein loss [18]. We recently transferred this assay to an automated immunostaining platform that may be run in any Clinical Laboratory Improvement Amendments–certified pathology laboratory.
The ERG gene is rearranged in approximately half of all prostate tumors [26,27]. Most studies of surgically treated patients have shown that ERG gene rearrangements that lead to increased expression of ERG protein are not associated with poor outcomes on their own [27]. However, the presence or absence of ERG rearrangements may modify the association of other risk factors with PCa outcomes [28]. Notably, PTEN deletion is more common in ERG-rearranged prostate tumors [4,15–17,21,23,25,29–32], and PTEN loss almost certainly occurs subsequent to ERG rearrangement in most cases [23–25]. This fact led several groups to hypothesize that there may be a synergistic effect of ERG expression and PTEN loss on PCa progression [29,30,33]. However, results from human studies have been mixed. While early FISH-based studies suggested that ERG-rearranged PTEN-deleted tumors may have a higher risk of biochemical recurrence compared to PTEN-deleted tumors lacking ERG rearrangement [21], the largest FISH-based study did not replicate this finding [17]. In this study we used highly validated, clinical-grade assays to assess the association of PTEN and ERG protein status with recurrence-free survival (RFS) in a large multi-institutional cohort of surgically treated PCa patients. We show that PTEN protein loss is most strongly associated with shorter RFS if the loss is homogeneous in all tumor cores sampled, and that the interaction between PTEN and ERG with respect to RFS did not reach statistical significance.
2. Patients and methods
2.1. Subject selection and tissue microarray (TMA) design
The Canary Foundation retrospective PCa TMA resource has been described in detail elsewhere [34]. In brief, radical prostatectomy (RP) tumor tissue from 1275 patients from six academic centers was selected for the TMA using a quota sampling plan. Recurrent cases of Gleason score 3 + 3 and 3 + 4 and nonrecurrent cases with Gleason score 4 + 4 were oversampled in this cohort. While this strategy diminishes the prognostic significance of Gleason score, it improves power to discover biomarkers that provide prognostic information independent of Gleason score. Each tumor was sampled in triplicate using 1-mm cores. The study was approved by the institutional review board at each participating institution and was covered by a materials transfer agreement between institutions.
The TMA included samples from men with (1) recurrent PCa; (2) nonrecurrent PCa; and (3) unknown outcome because of inadequate follow-up time (ie, censoring). Recurrent PCa was defined as (1) a single serum prostate-specific antigen (PSA) level >0.2 ng/ml more than 8 wk after RP; and/or (2) receipt of salvage or secondary therapy after RP; and/or (3) clinical or radiologic evidence of metastatic disease after RP. Nonrecurrent PCa was defined as disease with none of the indicators of recurrence for at least 5 yr after RP. Patients with no evidence of recurrent PCa but less than 5 yr of follow-up after RP (ie, censored) were also included in the TMA. The median follow-up for patients alive was 7 yr (range 1 d–21 yr).
2.2. IHC assays
PTEN IHC was performed on the Ventana platform (Ventana Discovery Ultra, Ventana Medical Systems, Tucson, AZ, USA) using a rabbit anti-human PTEN antibody (Clone D4.3 XP; Cell Signaling Technologies, Danvers, MA, USA). We previously validated a manual version of this assay using the same primary antibody [18].
PTEN protein status was visually scored by a trained pathologist (T.L.L.) blinded to clinical data. A second reviewer (C.L.M.) independently scored all of the cases for evaluation of interobserver variability in scoring. A tissue core was considered to have PTEN protein loss if the intensity of cytoplasmic and nuclear staining was markedly lower or entirely negative across >10% of tumor cells compared to surrounding benign tissue and/or stroma, which provide internal positive controls [18]. If PTEN was lost in >10% and <100% of the tumor cells sampled in a given core, the core was annotated as showing heterogeneous PTEN loss. Alternatively, if the core showed PTEN loss in 100% of sampled tumor tissue, the core was annotated as showing homogeneous PTEN loss. Cores were scored as having ambiguous PTEN IHC results when the intensity of the tumor cell staining was light or absent in the absence of evaluable internal benign tissue or stromal staining.
For statistical analysis, each tumor was scored for the presence or absence of PTEN loss by summarizing scores for the cores sampled. A tumor was designated as having heterogeneous PTEN loss if at least one tumor core showed heterogeneous PTEN loss (intracore heterogeneity), or alternatively, if at least one core showed heterogeneous or homogeneous PTEN loss and at least one core showed intact PTEN in tumor cells (intercore heterogeneity). A tumor was scored as showing homogeneous PTEN loss if all tumor cores sampled showed homogeneous PTEN loss. Finally, a tumor was scored as having intact PTEN if all sample tumor cores showed intact PTEN.
ERG IHC was performed using a commercial rabbit monoclonal antibody to ERG (clone EPR3864; 1:100; Epitomics, Burlingame, CA, USA) as previously described [35]. One set of TMAs from a single institution (Eastern Virginia Medical School) was excluded because of technically insufficient staining. ERG staining was manually scored for each individual core as follows: 0 = no staining; 1 = faint nuclear staining visualized at high-power magnification; and 2 = strong nuclear reactivity easily seen at low-power magnification. In the current study, a tumor was considered ERG-positive in any tissue core showing strong nuclear reactivity for ERG. It has been shown that these dichotomous ERG scoring criteria correlate to fusion status [35,36].
2.3. Statistical analysis
Fisher’s exact test was used to assess association between PTEN IHC and ERG status. Kaplan-Meier estimates of RFS were plotted by biomarker. An RFS event is defined as any recurrence (clinical, biochemical, or salvage therapy), metastasis, or PCa death after surgery. A Cox proportional hazards model was used to correlate multiple factors and biomarkers with RFS. All tests were two-sided and p < 0.05 or less was considered statistically significant. Statistical analysis was carried out using SAS version 9 (SAS Institute, Cary, NC, USA). Graphs were generated using Spotfire S+ version 8 (TIBCO, Palo Alto, CA, USA).
3. Results
Of the 1275 patients with tissue sampled for the TMAs, 1095 (86%) had evaluable PTEN status by IHC and 180 (14%) had missing data. Among the latter, 30/180 (17%) had ambiguous immunostaining results and 150/180 (83%) lacked tumor tissue in the TMA cores sampled. Of the tumors with evaluable staining, 258/1095 (24%) showed any PTEN protein loss, comprising 150 (14%) with heterogeneous PTEN loss (in some but not all tumor tissue sampled) and 108 (10%) with homogeneous PTEN loss (in all tumor tissue sampled; Fig. 1). The remaining 837/1095 (76%) cases had intact PTEN protein according to IHC for all tumor tissue sampled. Of the 150 cases with heterogeneous PTEN loss, 46 (31%) had only intercore heterogeneity (some cores with total loss and some with intact PTEN), nine (6%) had only intracore heterogeneity, and 95 (63%) had both intracore and intercore heterogeneity. A second reviewer scored all TMAs for evaluation of interobserver variability in PTEN IHC scoring. There was very low interobserver variability between the two independent reviewers, with 96.4% agreement over 2783 cores (κ = 0.905; 95% CI=0.887–0.923). Data for ERG in the cohort overall are reported elsewhere (Brooks JD et al, PLOS ONE, in press). ERG immunostaining results were available for 938 of the 1095 cases with interpretable PTEN IHC results (86%). Of these 938 cases, 401 (43%) were ERG-positive and the remainder were ERG-negative. PTEN loss (homogeneous or heterogeneous) was relatively enriched among the ERG-positive tumors, with 132/401 (33%) of ERG-positive tumors showing any PTEN loss compared to 99/537 (18%) of ERG-negative tumors (p < 0.0001; Table 1). ERG-negative tumors with any PTEN loss were slightly more likely to have homogeneous PTEN loss (48/99, 48%) than ERG-positive tumors with any PTEN loss (49/132, 37%), although this did not reach statistical significance (p = 0.11, Fisher’s exact test).
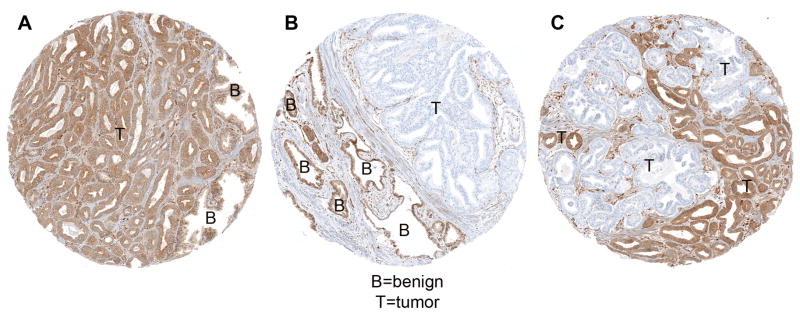
Fig. 1.
PTEN immunostaining examples from Canary the cohort (magnification 200×). (A) Intact PTEN. Tumor tissue has similar immunostaining intensity to surrounding benign tissue. (B) Homogeneous PTEN protein loss. All tumor tissue shows PTEN protein loss, with intact PTEN staining in surrounding benign tissue. (C) Heterogeneous PTEN protein loss. A subset of tumor tissue shows PTEN protein loss, while other intermingled tumor tissue shows intact immunostaining. T = tumor; B = benign.
Table 1.
Summary of PTEN immunohistochemistry results stratified by ERG status
| ERG status, n (%)
|
||
|---|---|---|
| Negative | Positive | |
| Intact PTEN | 438 (81.6) | 269 (67.1) |
| Heterogeneous PTEN loss | 51 (9.5) | 83 (20.7) |
| Homogeneous PTEN loss | 48 (8.9) | 49 (12.2) |
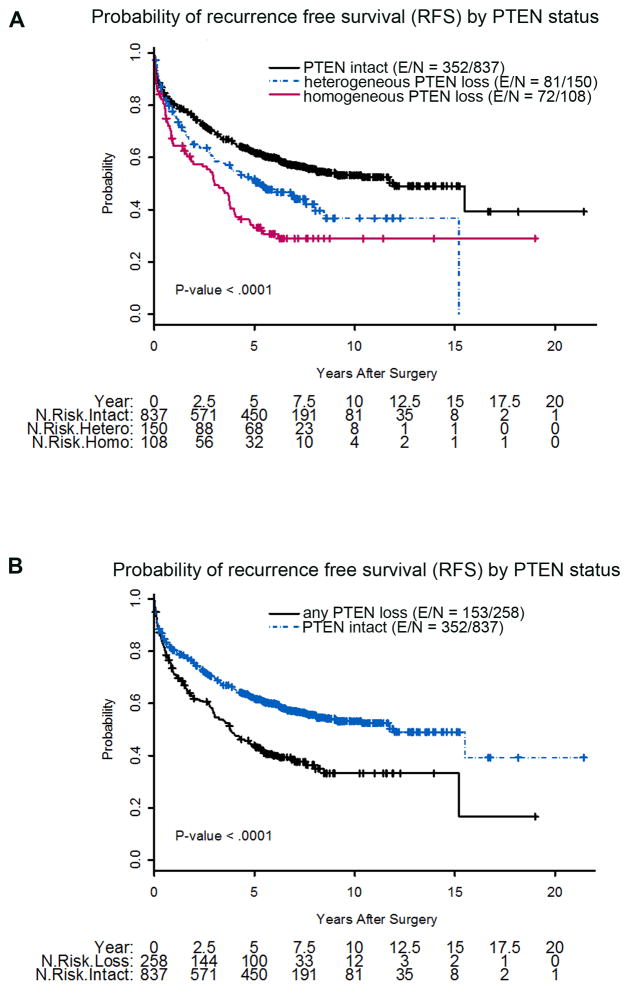
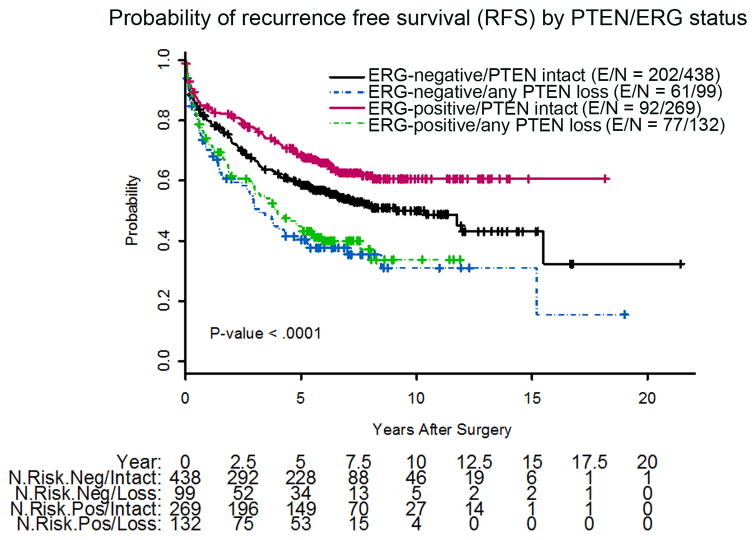
PTEN IHC status was associated with a number of clinicopathologic factors, including Gleason score and pathologic stage. Because information on pelvic lymph node status was missing for nearly 40% of the cohort, we were not able to assess correlation of PTEN loss with this parameter. Homogeneous PTEN loss was seen in only 4% of tumors with Gleason score ≤6, compared to 18% of tumors with Gleason score 8–10 (p < 0.0001; Table 2). PTEN loss was also associated with extraprostatic extension (p < 0.0001) and seminal vesicle invasion (p = 0.0009), and was thus associated with overall pathologic stage (p < 0.0001). However, PTEN loss was not associated with preoperative PSA, patient age, or surgical margin status (Tables 2 and 3). In univariate models, homogeneous PTEN loss was significantly associated with shorter RFS compared to intact PTEN (hazard ratio [HR] 2.04; p < 0.0001) and heterogeneous PTEN loss (HR 1.43; p = 0.03; Table 4, Fig. 2A). When grouped together, any PTEN loss (heterogeneous or homogeneous) was significantly associated with shorter RFS compared to intact PTEN (HR 1.66, p < 0.0001; Table 4, Fig. 2B). When stratified by ERG status, any PTEN loss (heterogeneous or homogeneous) was significantly associated with shorter RFS for both ERG-positive (HR 2.06, p < 0.0001) and ERG-negative tumors (HR 1.62, p = 0.001; Table 4, Fig. 3).
Table 2.
Association of PTEN immunohistochemistry status with clinicopathologic factors
| PTEN status, n (%)
|
p valuea | ||||
|---|---|---|---|---|---|
| Missing | Intact | Heterogeneous loss | Homogeneous loss | ||
| Margins | |||||
| Missing | 23 (12.9) | 125 (69.8) | 20 (11.2) | 11 (6.2) | |
| Positive | 53 (13.8) | 253 (65.7) | 44 (11.4) | 35 (9.1) | 0.9 |
| Negative | 104 (14.6) | 459 (64.6) | 86 (12.1) | 62 (8.7) | |
| Pathologic stage | |||||
| Missing | 22 (10.6) | 142 (68.6) | 25 (12.1) | 18 (8.7) | |
| III/IV | 48 (13.8) | 206 (59.2) | 43 (12.4) | 51 (14.7) | <0.0001 |
| I/II | 110 (15.3) | 489 (67.9) | 82 (11.4) | 39 (5.4) | |
| Seminal vesicle invasion | |||||
| Missing | 0 (0.0) | 15 (88.2) | 0 (0.0) | 2 (11.8) | |
| No | 169 (14.4) | 780 (66.3) | 137 (11.6) | 91 (7.7) | 0.0009 |
| Yes | 11 (13.6) | 42 (51.9) | 13 (16.1) | 15 (18.5) | |
| Extraprostatic extension | |||||
| Missing | 3 (17.7) | 12 (70.6) | 0 (0.0) | 2 (11.8) | |
| No | 124 (14.1) | 601 (68.5) | 101 (11.5) | 51 (5.8) | <0.0001 |
| Yes | 53 (13.9) | 224 (58.8) | 49 (12.9) | 55 (14.4) | |
| Gleason score | |||||
| Missing | 3 (30.0) | 5 (50.0) | 2 (20.0) | 0 (0.0) | |
| ≤6 | 98 (17.9) | 392 (71.4) | 35 (6.4) | 24 (4.4) | <0.0001 |
| 3 + 4 | 51 (11.1) | 285 (62.2) | 79 (17.3) | 43 (9.4) | |
| 4 + 3 | 13 (9.1) | 96 (67.1) | 14 (9.8) | 20 (14.0) | |
| 8–10 | 15 (13.0) | 59 (51.3) | 20 (17.4) | 21 (18.3) | |
χ2 test.
Table 3.
Association of PTEN immunohistochemistry status with age and preoperative PSA
| n | Mean | SD | Minimum | Median | Maximum | p valuea | |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| Missing PTEN | 174 | 62.2 | 7.1 | 42 | 63 | 80 | |
| Intact PTEN | 761 | 61.1 | 7.2 | 35 | 62 | 77 | 0.34 |
| Heterogeneous PTEN loss | 137 | 62.3 | 6.7 | 44 | 63 | 76 | |
| Homogeneous PTEN loss | 97 | 61.3 | 6.9 | 45 | 61 | 75 | |
| Preoperative PSA | |||||||
| Missing PTEN | 158 | 8.7 | 7.1 | 0.98 | 6.62 | 52.6 | |
| Intact PTEN | 752 | 8.5 | 8.1 | 0.02 | 6.32 | 124 | 0.6 |
| Heterogeneous PTEN loss | 139 | 8.5 | 7.9 | 0.71 | 6.4 | 64.85 | |
| Homogeneous PTEN loss | 98 | 9.9 | 11.2 | 1 | 6.85 | 78.3 | |
PSA = prostate-specific antigen; SD = standard deviation.
Kruskal-Wallis test.
Table 4.
Summary of univariate Cox models correlating PTEN immunohistochemistry status with recurrence-free survival (RFS)
| Comparison | HR (95% CI) | p value | Events | Censored | Total |
|---|---|---|---|---|---|
| PTEN status | |||||
| Homogeneous loss vs intact | 2.04 (1.59–2.63) | <0.0001 | 500 | 590 | 1095 |
| Homogeneous vs heterogeneous loss | 1.43 (1.04–1.96) | 0.03 | |||
| Any loss vs intact | 1.66 (1.37–2.01) | <0.0001 | 505 | 590 | 1095 |
| ERG-positive PTEN status | |||||
| Any loss vs intact | 2.06 (1.52–2.90) | <0.0001 | 169 | 232 | 401 |
| ERG-negative PTEN status | |||||
| Any loss vs intact | 1.62 (1.22–2.16) | 0.001 | 263 | 274 | 537 |
HR = hazard ratio; CI = confidence interval.
Fig. 2.
Kaplan-Meier probability of recurrence-free survival stratified by (A) homogeneous PTEN loss, heterogeneous PTEN loss, and intact PTEN; and (B) any PTEN loss (homogeneous or heterogeneous) and intact PTEN. N = total number of patients; E = events; hetero = heterogeneous; homo = homogeneous.
Fig. 3.
Kaplan-Meier probability of recurrence-free survival stratified by PTEN and ERG status. N = total number of patients; E = events; ERG− = ERG-negative; ERG+ = ERG-positive.
Multivariate models were constructed for a subset of 808 patients with complete clinicopathologic information available. There was no difference between patients included in these models and those excluded because of incomplete information for any clinicopathologic variable measured (Supplementary Tables 1 and 2). In multivariate models, homogeneous PTEN loss was associated with shorter RFS compared to intact PTEN (HR 1.66, p = 0.001; Table 5). Heterogeneous PTEN loss showed a nonsignificant trend towards shorter RFS compared to intact PTEN (HR 1.24, p = 0.14; Table 5). To assess the additive value of PTEN for RFS prediction when combined with clinicopathologic factors, area under the receiver operating characteristic curve (AUC) plots were constructed (Supplementary Fig. 1). Inclusion of clinicopathologic factors yielded AUC of 0.72, while addition of two and three PTEN IHC status categories increased AUC to 0.73 and 0.74, respectively.
Table 5.
Multivariate Cox proportional hazards models for recurrence-free survival in association with homogeneous/heterogeneous PTEN loss by ERG status
| Model and factor | Comparison | HR (95% CI) | p value |
|---|---|---|---|
| All patients | |||
| (n = 808, 371 events) | |||
| log(PSA) | 1-unit increase | 1.44 (1.22–1.70) | <0.0001 |
| PTEN status | Homogeneous loss vs intact | 1.66 (1.22–2.24) | 0.001 |
| Heterogeneous loss vs intact | 1.24 (0.93–1.65) | 0.14 | |
| Margins | Positive vs negative | 1.73 (1.39–2.16) | <0.0001 |
| Seminal vesicle invasion | Yes vs no | 1.93 (1.38–2.71) | 0.0001 |
| Extraprostatic extension | Yes vs no | 1.30 (1.03–1.63) | 0.03 |
| Gleason score | 3 + 4 vs 6 | 1.15 (0.89–1.48) | 0.29 |
| 4 + 3 vs 6 | 1.87 (1.38–2.54) | <0.0001 | |
| 8–10 vs 6 | 1.50 (1.06–2.11) | 0.02 | |
| ERG-positive | |||
| (n = 284, 120 events) | |||
| log(PSA) | 1-unit increase | 1.56 (1.17–2.07) | 0.002 |
| PTEN status | Homogeneous loss vs intact | 3.07 (1.94–4.84) | <0.0001 |
| Heterogeneous loss vs intact | 1.46 (0.93–2.30) | 0.10 | |
| Margins | Positive vs negative | 1.88 (1.30–2.72) | 0.0008 |
| Seminal vesicle invasion | Yes vs no | 3.55 (1.85–6.79) | 0.0001 |
| ERG-negative | |||
| (n = 454, 220 events) | |||
| log(PSA) | 1-unit increase | 1.51 (1.22–1.86) | 0.0001 |
| Gleason score | 3 + 4 vs 6 | 1.24 (0.89–1.73) | 0.20 |
| 4 + 3 vs 6 | 2.01 (1.37–2.96) | 0.0004 | |
| 8–10 vs 6 | 1.78 (1.16–2.72) | 0.008 | |
| Margins | Positive vs negative | 1.82 (1.38–2.41) | <0.0001 |
| Extraprostatic extension | Positive vs negative | 1.45 (1.09–1.92) | 0.01 |
HR = hazard ratio; CI = confidence interval; PSA = prostate-specific antigen.
Multivariate models were also constructed for ERG-positive and ERG-negative tumors separately. Among ERG-positive tumors, homogeneous (HR 3.07, p < 0.0001) but not heterogeneous PTEN loss (HR 1.46, p = 0.10) was significantly associated with shorter RFS compared with intact PTEN. Among ERG-negative tumors, PTEN loss did not reach significance for inclusion in the final model (p = 0.08), although the effect of PTEN loss was in the same direction as seen for the ERG-positive group. The interaction term between PTEN and ERG status did not reach statistical significance in a multivariate Cox model for RFS (p = 0.11), although post hoc bootstrapping simulations indicated that at least 1000 patients are required to detect an interaction with 80% power. In multivariate models in which homogeneous and heterogeneous PTEN loss were grouped together, any PTEN loss was associated with shorter RFS (HR 1.40, p = 0.004; Table 6). When ERG-positive and ERG-negative tumors were considered separately in multivariate models, the association between any PTEN loss and shorter RFS was significant for ERG-positive tumors (HR 1.98, p = 0.0003) and nonsignificant for ERG-negative tumors, so was not included in the final model. When PTEN status was modeled as intact or loss, the interaction term between PTEN and ERG status was not statistically significant for RFS (p = 0.25).
Table 6.
Multivariate Cox proportional models for recurrence-free survival in association with any PTEN loss by ERG status
| Model and factor | Comparison | HR (95% CI) | p value |
|---|---|---|---|
| All patients | |||
| (n = 808, 371 events) | |||
| log(PSA) | 1-unit increase | 1.44 (1.22–1.70) | <0.0001 |
| PTEN status | Any loss vs. intact | 1.40 (1.12–1.77) | 0.004 |
| Margins | Positive vs negative | 1.73 (1.39–2.16) | <0.0001 |
| Extraprostatic extension | Yes vs no | 1.32 (1.06–1.66) | 0.02 |
| Seminal vesical invasion | Yes vs no | 1.95 (1.39–2.74) | 0.0001 |
| Gleason score | 3 + 4 vs ≤6 | 1.14 (0.89–1.47) | 0.31 |
| 4 + 3 vs ≤6 | 1.88 (1.38–2.55) | <.0001 | |
| 8–10 vs ≤6 | 1.47 (1.04–2.07) | 0.03 | |
| ERG-positive | |||
| (n = 284, 120 events) | |||
| log(PSA) | 1-unit increase | 1.60 (1.19–2.14) | 0.002 |
| PTEN status | Any loss vs intact | 1.98 (1.37–2.87) | 0.0003 |
| Margins | Positive vs negative | 1.89 (1.30–2.73) | 0.0008 |
| Seminal vesical invasion | Yes vs no | 2.92 (1.54–5.54) | 0.001 |
| ERG-negative | |||
| (n = 454, 220 events) | |||
| log(PSA) | 1-unit increase | 1.51 (1.22–1.86) | 0.0001 |
| Margins | Positive vs negative | 1.82 (1.38–2.41) | <0.0001 |
| Extraprostatic extension | Yes vs no | 1.45 (1.09–1.92) | 0.01 |
| Gleason score | 3 + 4 vs ≤6 | 1.24 (0.89–1.73) | 0.20 |
| 4 + 3 vs ≤6 | 2.01 (1.37–2.86) | 0.0004 | |
| 8–10 vs ≤6 | 1.78 (1.16–2.72) | 0.008 | |
HR = hazard ratio; CI = confidence interval; PSA = prostate-specific antigen.
4. Discussion
There is a growing need for biomarkers that help to distinguish indolent from aggressive prostate tumors and add to current clinicopathologic risk stratification measures. We recently developed and validated an IHC assay to assess PTEN protein loss in PCa [18]. The original assay involved manual staining of slides, but we have now adapted this assay for automated performance on a Ventana autostainer system and demonstrated equivalence to the manual assay. In a subset of 551 tumors for which IHC (by the automated assay) and FISH data were available [22], we found that intact PTEN immunostaining was 91% specific for the absence of PTEN gene deletion by FISH, and 98% and 62% sensitive for detection of homozygous and hemizygous gene deletion, respectively, by FISH [37].
Using manual IHC, our group previously demonstrated that PTEN protein loss is associated with higher risk of biochemical recurrence in a nested case-control cohort of surgically treated patients [20]. Similar to the current findings with the automated protocol, PTEN loss correlated with higher Gleason grade and stage, and homogeneous PTEN loss was independently associated with biochemical recurrence in multivariate models with HR of approximately 2. Of note, heterogeneous PTEN loss (in some but not all tumor tissue sampled) was a weaker prognostic indicator compared to homogeneous loss, as seen in the current study. Some cases of heterogeneous loss will likely be missed when the IHC assay is applied to prostate biopsies because of sampling error (and may have been similarly missed in the current TMA sampling). However, since heterogeneous loss is more weakly associated with poor outcomes, these false negatives may be less clinically significant. It remains unclear why homogeneous PTEN loss is more tightly associated with shorter RFS. Homogeneous loss of PTEN protein may signify increased selection for (and expansion of) a single PTEN-null clone, a finding that has been associated with PCa progression in a recent single-cell analysis [38]. Equally plausible is the possibility that tumors with a higher mass of PTEN-null cells have a higher risk of local or disseminated spread for stochastic reasons. The current study adds insights to work identifying a putative interaction between PTEN loss and ERG rearrangements. Mouse models have suggested that PTEN loss and TMPRSS2:ERG gene rearrangement synergize to drive cell migration and invasion, perhaps explaining the tendency towards co-occurrence in human PCa [29,30]. Furthermore, in the mouse prostate, ERG expression may restore decreased androgen signaling due to reciprocal feedback between PI3K and androgen receptor in the context of PTEN loss [33]. At least four studies have examined the interaction of PTEN and ERG in association with PCa progression in clinical series. The first study to explore the interaction between ERG and PTEN used FISH to assess PTEN gene status in 125 patients [21] and found that PTEN loss was more strongly associated with biochemical recurrence after RP among ERG-positive compared to ERG-negative tumors. However, a larger study of 1895 patients [17] found no influence of ERG status on the association of PTEN deletion assessed by FISH with postoperative biochemical recurrence, and this result was replicated in an expanded cohort including more than 5000 patients [39]. In a study of 262 patients [40], loss of PTEN protein expression by IHC was more strongly associated with biochemical recurrence among ERG-positive compared to ERG-negative tumors. Similar findings have been reported for a cohort of patients treated with brachytherapy [41]. Only one study has examined the interaction of PTEN and ERG and their association with PCa-specific mortality in a cohort of 308 patients managed conservatively [16]. Interestingly, PTEN deletion detected by FISH was associated with higher risk of PCa mortality among ERG-negative but not ERG-positive tumors. However, in a subsequent study of 652 patients (including the original 308 patients), the authors failed to validate this interaction between PTEN deletion and ERG status with respect to PCa death [42].
Taken together, our study and previous work suggest that PTEN loss is associated with biochemical recurrence in both ERG-positive and ERG-negative tumors. In our study, the interaction term for PTEN and ERG status with respect to RFS did not reach statistical significance in the Cox models. While post hoc power calculations indicate that we would have needed at least 1000 samples to achieve 80% power to detect a significant interaction, such analyses must be interpreted with caution. While some prior studies have found that PTEN loss is associated with poor prognosis only for ERG-positive tumors, this finding could be because of the relative enrichment of PTEN loss among ERG-positive tumors. Indeed, this may be why PTEN loss was not significant in the final multivariate models for ERG-negative tumors, but was significant for ERG-positive tumors. In previous studies, only between five and 19 ERG-negative tumors with PTEN loss were available for follow-up [21,40,41], so the studies may also have been underpowered for observation of an association with outcome in this subgroup. By contrast, the largest FISH-based study that found no effect of ERG status on the association of PTEN loss with progression examined 97 and 356 ERG-negative PTEN-loss tumors in the original and expanded series [17,39].
There are a number of important limitations to the current study. Because of the multi-institutional design, some data for the cohort are incomplete, including the lymph node status of patients (missing for >40% of cases) and racial and family history information. In addition, the outcome measured in the current study is RFS rather than PCa-specific mortality. Of patients experiencing biochemical recurrence (as was seen in the majority of the recurrent cases in the current study), only a minority will die from PCa, so this remains a surrogate outcome measure with well-described limitations. Finally, the degree to which PTEN adds to established clinicopathologic factors for prediction of prognosis in the RP setting remains unclear. Receiver operating characteristic analysis demonstrated that PTEN shifted the area under the curve (AUC) from 0.72 (with clinicopathologic factors alone) to 0.74 (combined three-category PTEN score and clinicopathologic factors). This effect size is similar to that observed for newly available genomic classifiers such as Decipher in the RP setting [43]. However, as seen in studies of genomic classifiers, even marginal shifts in AUC can have a significant impact on decision curve analysis. Perhaps more importantly, complete grading and pathologic staging information is not available in the setting of needle biopsies, and thus biomarkers such as PTEN are likely have more added value for prediction of prognosis.
5. Conclusions
Using a highly validated and automated IHC assay for a diverse and multi-institutional set of PCa tumors, we found that homogeneous rather than heterogeneous PTEN protein loss is most strongly associated with a higher risk of recurrence after RP, even after adjusting for other clinicopathologic parameters. In univariate analyses, PTEN loss was associated with poor outcomes among both ERG-positive and ERG-negative tumors and we did not find evidence for a statistically significant interaction between PTEN and ERG status in predicting RFS in multivariate models. If reproduced in additional cohorts, these data suggest that PTEN IHC may be a simple and relatively inexpensive test to aid in stratification of PCa risk.
Supplementary Material
Take Home Message.
We used highly validated, clinical-grade assays to assess the association of PTEN and ERG protein status with recurrence-free survival (RFS) in a large multi-institutional cohort of surgically treated prostate cancer patients. We show that PTEN protein loss is most strongly associated with shorter RFS if the loss is homogeneous in all tumor tissue sampled. In addition, we demonstrate that there is not a statistically significant interaction between PTEN and ERG with respect to RFS.
Acknowledgments
Funding/Support and role of the sponsor: Funding for this research was provided in part by the Canary Foundation, a Prostate Cancer Foundation Young Investigator Award (T.L.L.), and a generous gift from Mr. David H. Koch (T.L.L). The Canary Foundation played a role in data collection.
Footnotes
Author contributions: Tamara L. Lotan had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Lotan, Brooks, Wei, Morais, Hawley, Fazli, Hurtado-Coll, Troyer, McKenney, Simko, Carroll, Gleave, Lance, Lin, Nelson, Thompson, True, Feng .
Acquisition of data: Lotan, Morais.
Analysis and interpretation of data: Lotan, Wei.
Drafting of the manuscript: Lotan, Brooks, Wei, Troyer.
Critical revision of the manuscript for important intellectual content: Lotan, Brooks, Wei.
Statistical analysis: Wei, Feng.
Obtaining funding: Lotan, Brooks, Troyer, McKenney, Simko, Carroll, Gleave, Lin, Nelson, Thompson, True, Feng.
Administrative, technical, or material support: None.
Supervision: Lotan, Brooks.
Other: None.
Financial disclosures: Tamara L. Lotan certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23. 3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 7.Cairns P, Okami K, Halachmi S, et al. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 8.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–6. [PubMed] [Google Scholar]
- 9.Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9:1474–9. [PubMed] [Google Scholar]
- 10.Verhagen PC, van Duijn PW, Hermans KG, et al. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol. 2006;208:699–707. doi: 10.1002/path.1929. [DOI] [PubMed] [Google Scholar]
- 11.Bedolla R, Prihoda TJ, Kreisberg JI, et al. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13:3860–7. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz M, Grignard G, Margue C, et al. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int J Cancer. 2007;120:1284–92. doi: 10.1002/ijc.22359. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sircar K, Yoshimoto M, Monzon FA, et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;218:505–13. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 15.Han B, Mehra R, Lonigro RJ, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reid AH, Attard G, Ambroisine L, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–84. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krohn A, Diedler T, Burkhardt L, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181:401–12. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 18.Lotan TL, Gurel B, Sutcliffe S, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–73. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonarakis ES, Keizman D, Zhang Z, et al. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012;118:6063–71. doi: 10.1002/cncr.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaux A, Peskoe SB, Gonzalez-Roibon N, et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25:1543–9. doi: 10.1038/modpathol.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimoto M, Joshua AM, Cunha IW, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 22.Troyer DA, Jamaspishvili T, Wei W, et al. A multicenter study shows PTEN deletion is strongly associated with seminal vesicle involvement and extracapsular extension in localized prostate cancer. Prostate. 2015;75:1206–15. doi: 10.1002/pros.23003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krohn A, Freudenthaler F, Harasimowicz S, et al. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014;27:1612–20. doi: 10.1038/modpathol.2014.70. [DOI] [PubMed] [Google Scholar]
- 24.Gumuskaya B, Gurel B, Fedor H, et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis. 2013;16:209–15. doi: 10.1038/pcan.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bismar TA, Yoshimoto M, Duan Q, Liu S, Sircar K, Squire JA. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. Histopathology. 2012;60:645–52. doi: 10.1111/j.1365-2559.2011.04116.x. [DOI] [PubMed] [Google Scholar]
- 26.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson A, Graff RE, Bauer SR, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersson A, Lis RT, Meisner A, et al. Modification of the association between obesity and lethal prostate cancer by TMPRSS2:ERG. J Natl Cancer Inst. 2013;105:1881–90. doi: 10.1093/jnci/djt332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bismar TA, Yoshimoto M, Vollmer RT, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–85. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 32.Reid AH, Attard G, Brewer D, et al. Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol. 2012;25:902–10. doi: 10.1038/modpathol.2011.207. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Chi P, Rockowitz S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawley S, Fazli L, McKenney JK, et al. A model for the design and construction of a resource for the validation of prognostic prostate cancer biomarkers: the Canary Prostate Cancer Tissue Microarray. Adv Anat Pathol. 2013;20:39–44. doi: 10.1097/PAP.0b013e31827b665b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaux A, Albadine R, Toubaji A, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–20. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–8. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lotan T, Morais C, Wei W, et al. PTEN status determination in prostate cancer: comparison of IHC and FISH in a large multi-center cohort. Mod Pathol. 2015;28:241A. [Google Scholar]
- 38.Heselmeyer-Haddad KM, Berroa Garcia LY, Bradley A, et al. Single-cell genetic analysis reveals insights into clonal development of prostate cancers and indicates loss of PTEN as a marker of poor prognosis. Am J Pathol. 2014;184:2671–86. doi: 10.1016/j.ajpath.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steurer S, Mayer PS, Adam M, et al. TMPRSS2-ERG fusions are strongly linked to young patient age in low-grade prostate cancer. Eur Urol. 2014;66:978–81. doi: 10.1016/j.eururo.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 40.Leinonen KA, Saramaki OR, Furusato B, et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:2333–44. doi: 10.1158/1055-9965.EPI-13-0333-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontugne J, Lee D, Cantaloni C, et al. Recurrent prostate cancer genomic alterations predict response to brachytherapy treatment. Cancer Epidemiol Biomarkers Prev. 2014;23:594–600. doi: 10.1158/1055-9965.EPI-13-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuzick J, Yang ZH, Fisher G, et al. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer. 2013;108:2582–9. doi: 10.1038/bjc.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, Karnes RJ. combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol. 2015;67:326–33. doi: 10.1016/j.eururo.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.