Abstract
The heterodimeric laminin receptor α6β4 integrin plays a central role in the promotion of tumor cell growth, invasion, and organotropic metastasis. As an overproduction of the integrin is often linked to a poor prognosis, the inhibition of integrin α6β4 binding to laminin is of high therapeutical interest. Here, we report on the combination of a cell-systematic evolution of ligands by exponential enrichment and a bead-based selection resulting in the first aptamer inhibiting the interaction between α6β4 integrin and laminin-332. This Integrin α6β4-specific DNA Aptamer (IDA) inhibits the adhesion of prostate cancer cells (PC-3) to laminin-332 with an IC50 value of 149 nmol/l. The Kd value concerning the aptamer's interaction with PC-3 cells amounts to 137 nmol/l. Further characterization showed specificity to α6 integrins and a half-life in murine blood plasma of 6 hours. Two truncated versions of the aptamer retained their binding capacity, but lost their ability to inhibit the interaction between laminin-332 and PC-3 cells. Confocal laser scanning microscope studies revealed that the aptamer was internalized into PC-3-cells. Therefore, in addition to the adhesion-blocking function of this aptamer, IDA could also be applied for the delivery of siRNA, microRNA or toxins to cancer cells presenting the integrin α6β4.
Keywords: aptamer, cell SELEX, integrin, laminin
Introduction
Integrins are αβ heterodimeric cell surface receptors that mediate stable adhesion between cells and their extracellular environment as well as amplifying signals from growth factor receptors and other extracellular stimuli.1,2,3 In mammalian cells, there are 18 alpha-subunits and 8 beta-subunits resulting in 24 different integrins. These bind extracellular matrix or cellular adhesion proteins, e.g., collagens, laminins or Arginin–Glycin–Aspartat (RGD)-containing proteins.2,4,5
The α6β4 integrin belongs to the group of laminin-binding integrins and is presented by epithelial cells, Schwann cells, keratinocytes, and endothelial cells.2,6 Upon binding to laminin, α6β4 integrin leads to the assembly of hemidesmosomes, which mediate stable adhesion by connecting the intracellular keratin cytoskeleton to the basement membrane.7,8 Additionally, the β4 integrin has distinctive cytoskeletal and signaling functions via its 1,017 amino-acid-long cytoplasmic domain.2,9,10 During wound healing, this domain can be phosphorylated by protein kinase C or interactions with growth factor receptors resulting in the release of the α6β4 integrin from hemidesmosomes.11,12,13,14 After relocalization from the keratin to the actin cytoskeleton, the phosphorylation of the integrin promotes the formation of filopodia and lamellae, as well as stimulating key signaling pathways facilitating migration and wound closure.2,15,16,17,18
Different types of cancer cells use this mechanism of cytoskeleton remodeling to promote invasive signaling through cooperation with growth factor receptors and alteration of the transcriptome. Under conditions where hemidesmosomes are disassembled, binding of α6β4 integrin to laminin can activate both phosphoinositide 3-OH kinase (PI3K) and RhoA, a small GTPase. Activation of these signaling pathways then facilitates tumor cell growth, invasion, and metastasis.2,14,15,18,19,20 This central role in tumor progression explains why upregulation of the α6β4 integrin gene expression is often linked to poor prognosis.8,21,22 Furthermore, Hoshino et al.23 found out, that α6β4 integrins on the surface of exosomes play a key role in the lung-specific metastasis by formation of a pre-metastatic niche. Therefore, an inhibition of the α6β4 integrin laminin interaction is of high therapeutical interest to reduce cell growth, invasion, and metastasis.
A promising strategy is the blockage of the laminin binding site at the surface of α6β4 integrin by an appropriate macromolecule. Rabinovitz and Mercurio17 showed that targeting the α6β4 integrin with a monoclonal antibody inhibited the migration of colon adenocarcinoma cells significantly. While monoclonal antibodies are commonly employed to specifically target cell surface proteins, they exhibit a number of significant drawbacks. Those include their high production cost, limited shelf life, and their high molecular weight preventing access to many biological compartments.24,25 A promising alternative to protein-based strategies for targeting cell surface markers was presented in the early 1990s in the form of small nucleic acid ligands.26,27,28 This emerging class of therapeutics, with over 900 generated species today,29 was termed aptamers. They are selected in vitro by an iterative process referred to as systematic evolution of ligands by exponential enrichment (SELEX).26,27,28 Aptamers can be selected for a wide spectrum of targets ranging from small organic molecules, dyes,26 and heavy metals to proteins30 and nanomaterials and even bacteria or eukaryotic cells.31,32 To date 11 aptamers are in clinical trials, where they are applied as therapeutics for the treatment of macular degeneration,33 coagulation,34 different forms of cancer,35 and inflammatory diseases.29,36
Here, we report on the selection of a DNA aptamer inhibiting the interaction between α6β4 integrin and laminin-332.
Results
Selection and identification of aptamers binding to α6β4 integrin
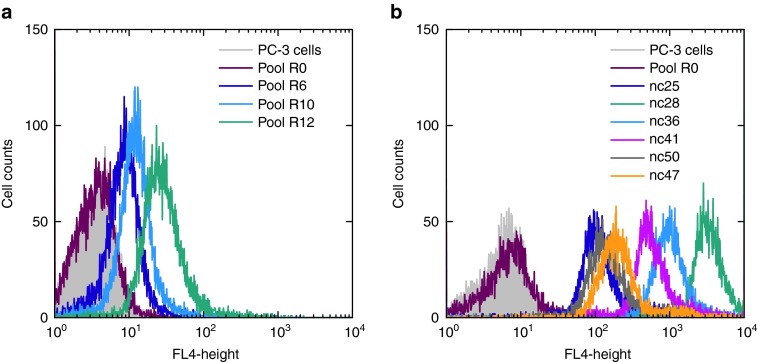
To select a DNA aptamer specific for α6β4 integrin in its native state, we started with a cell-SELEX approach using prostate cancer cells (PC-3).37,38 Cells were plated in a cell culture dish and incubated with the starting DNA library containing 1013 different species. After an enrichment of binders to PC-3-cells in the first two rounds, PC-3 β4 Integrin (ITGB4) knockdown cells were used in a preselection step to deplete aptamers specific for cell surface marker other than α6β4 integrin. However, the counter selection strategy was insufficient to prevent enrichment of nontarget specific aptamers as binding of the library to the knockdown cells could be detected via flow cytometry after selection cycle five. Therefore, in round six, we switched to a conventional SELEX with immobilized recombinant α6β4 integrin to improve the specificity. The library in round 12 showed an increased binding to the PC-3 cells via flow cytometry (Figure 1a, green) and was cloned and sequenced (see Supplementary Table S1). Twenty different nucleic acids were obtained with no apparent sequence identity shared between the individual molecules. Therefore, all nucleic acids, cyanine-5 labelled, were tested for their binding affinity using flow cytometry (Figure 1b). The highest affinity showed nucleic acid 28, henceforth termed Integrin α6β4-specific DNA Aptamer (IDA, green), followed by nc36 (blue). The starting library did not bind to PC-3-cells at all (red).
Figure 1.
Flow cytometric analyses. (a) An enrichment of binding nucleic acids in higher rounds could be detected via flow cytometry. Green: library round 12, blue: library round 10, violett: library round 6, red: starting library, grey: untreated cells. (b) FACS analysis of cloned nucleic acids. Green: nc28, blue: nc36, pink: nc41, orange: nc 47, grey: nc50, violet: nc25, red: starting library, grey: untreated cells. The nucleic acid nc28 showed the highest affinity to PC-3 cells.
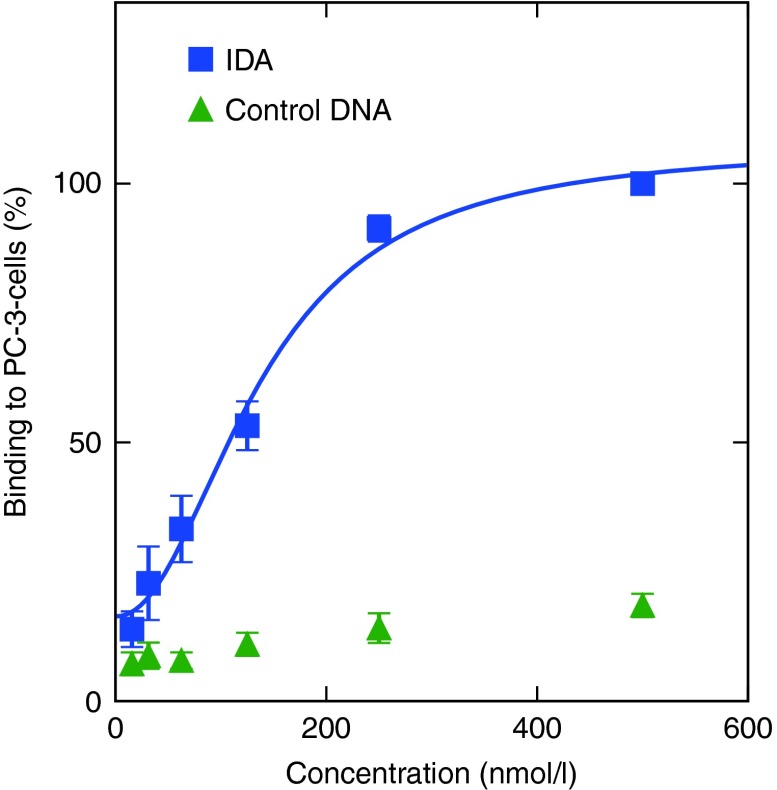
The Kd value for the interaction of IDA with α6β4 integrin in its natural environment was determined by flow cytometry. Therefore, a dilution series of fluorescently labelled IDA was incubated with a constant number of PC-3 cells. The resulting mean values were normalized to the highest binding event and plotted against the aptamer concentration with python (version 3) (Figure 2). The fitted Kd value for the binding of IDA (blue) to PC-3 cells was 137 ± 22 nmol/l. The dilution series of the control DNA (green) gave a much lower fluorescence signal compared with IDA.
Figure 2.
Determination of the Kd value for the interaction of IDA with PC-3 cells (blue); control DNA (green). A dilution series from 500 to 15.6 nmol/l was incubated with a constant amount of cells and analyzed by flow cytometry. Bound DNA was normalized to 500 nmol/l as 100%.
Inhibition assay
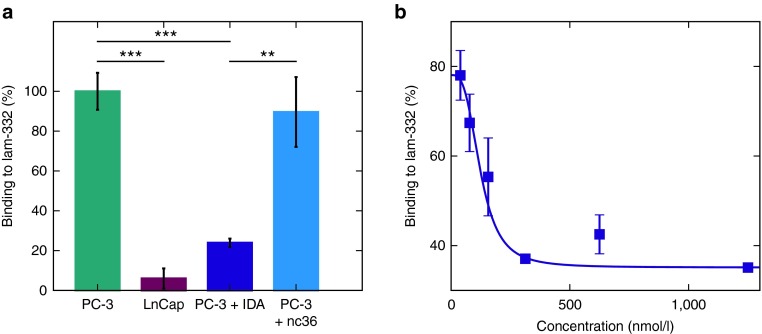
To test the ability of IDA and nc36 to block the interaction between α6β4 integrin and laminin-332, we performed an inhibition assay. α6β4 integrin presenting PC3-cells and primarily α6β1 presenting LnCap cells (see Supplementary Figure S1) were pre-incubated with the aptamers IDA or nc36 and then transferred to 96 well plates precoated with laminin-332. After washing, attached cells were counted with the CellTiter-Glo® Assay (Promega).
IDA (violet) inhibits the binding of PC-3 cells to laminin-332 with a P value < 0.001 compared with untreated cells (green) and a value <0.05 compared with aptamer nc36 (blue). LnCapcells (red) did not show any binding to laminin-332 (Figure 3a). Next, we wanted to identify the IC50 value of IDA in a dilution series of the aptamer. The resulting luminescence signal was normalized to bound PC-3 cells as 100% (Figure 3b). The apparent Ki of IDA was 123 ± 44 nmol/l. The IC50 value was determined to be ~149 nmol/l.
Figure 3.
IDA inhibits the interaction between PC-3 cells and laminin-332. (a) PC-3 cells were incubated with IDA (violet), nc36 (blue) or without DNA (green) in a laminin-coated plate. Only IDA showed a reduced binding of PC-3 cells. LnCap cells not presenting the integrin showed no adhesion (red). (b) Dilution series of IDA (1.25 µmol/l–39 nmol/l) to determine the IC50 value and the Ki.
Specificity
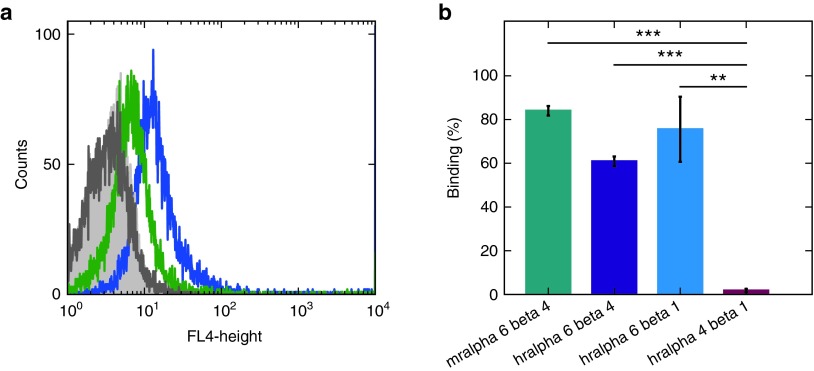
To analyze the specificity of IDA, it was incubated at 37 °C with PC-3 cells and PC-3 integrin β4 knockdown cells. A clear fluorescence shift could be detected for PC-3 cells presenting the integrin after incubation with IDA (Figure 4a, blue). Although there appears to be moderate binding of IDA to β4-deficient cells, the fluorescence shift is much less pronounced (green). As the reduced fluorescence shift is in accordance with a reduction of α6 integrin surface levels (see Supplementary Figure S2), this indicates an α6 integrin specificity. To assure that IDA is specific for α6 integrins, we performed an electrophoretic mobility shift assay with human recombinant integrins α6β4, α6β1, rα4β1 and murine α6β4, respectively (Figure 4b). IDA bound to human α6β4 (violet) and α6β1 (blue) as well as to murine α6β4 (green), but not to α4β1 (red). Raw data are provided in the supplementary material section (see Supplementary Figure S3)
Figure 4.
Determination of the specificity of IDA. (a) Flow cytometry analysis. IDA binds to PC-3 cells (blue) with a higher affinity than to β4 integrin knockdown cells (green). The auto fluorescence of knockdown cells is shown in dark grey, the fluorescence of PC-3 cells in light grey. (b) Electrophoretic mobility shift assay. The aptamer binds to murine (green) and humane (violet) recombinant α6β4 integrin, as well as humane α6β1 (blue), but not to recombinant α4β1 (red).
Internalization of IDA into PC-3 cells
The internalization of IDA into PC-3 cells was analyzed with a confocal laser scanning microscope. Therefore, PC-3 cells were incubated with either cyanine 5 (cy5)-labelled IDA (IDA-cy5) or control DNA (coDNA-cy5). IDA was able to bind the cell surface at 4 °C (see Supplementary Figure S4a) and was internalized into the cells at 37 °C (Figure 5a), whereas the control coDNA-cy5 did not appear to even bind to the cells (Figure 5b, Supplementary Figure S4b). To determine, if IDA is able to induce internalization of the α6β4 integrin, cells surface levels after aptamer incubation were analyzed by flow cytometry (see Supplementary Figure S5). No significant decrease of these levels could be detected after 5, 10 or 15 minutes of aptamer incubation at 37 °C. This indicates that IDA is internalized upon natural receptor recycling.
Figure 5.
Confocal laser scanning microscopy analyses. (a) IDA is internalized by PC-3 cells. (b) The nonbinding DNA showed no fluorescence signal.
Truncation of IDA
In order to truncate IDA, secondary structure predictions were performed using the RNA structure webserver (see Supplementary Figure S6a).39 This software predicted a long unpaired region consisting of the forward primer and three small stem loops. The last loop was imparted into the reverse primer. Two smaller constructs were designed, named IDA-A and IDA-B. IDA-A was shortened to 44 nucleotides including all three stem loops, whereas IDA-B had 36 nt including only the two stem loops in the former randomized part (see Supplementary Figure S6b,c). Both constructs were able to bind to PC-3 cells as analyzed by flow cytometry (see Supplementary Figure S7). However, they lost the ability to block the integrin-laminin-binding as confirmed by an inhibition assay (see Supplementary Figure S8).
Determining the stability of IDA
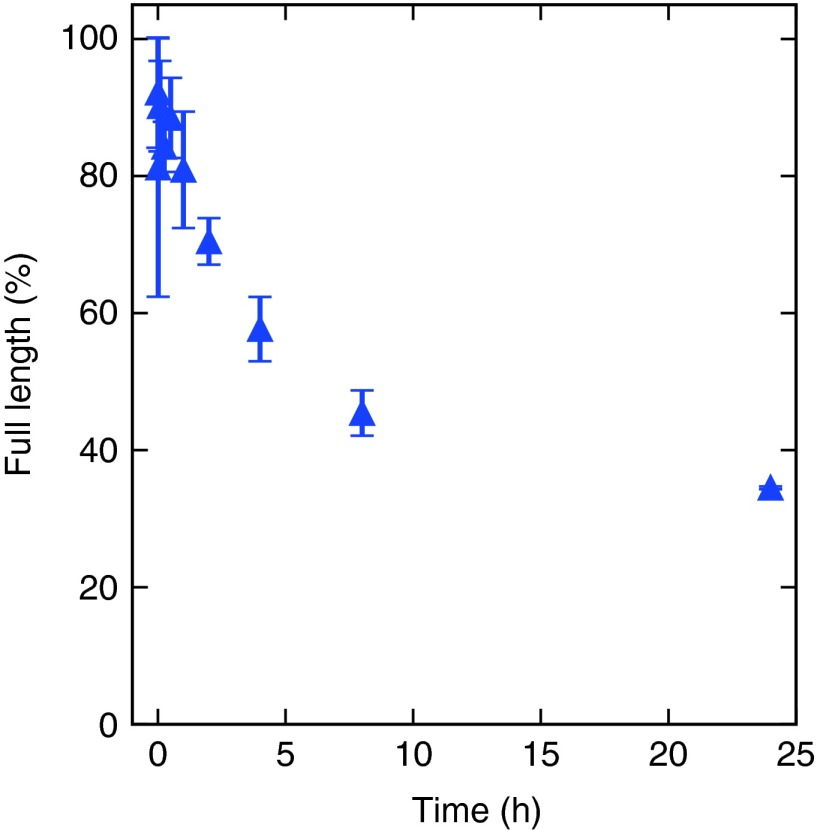
The stability of IDA was analyzed in murine blood plasma over a period of 24 hours (Figure 6). IDA had a half-life time around 6 hours. Raw data are provided in the supplementary material (see Supplementary Figure S9)
Figure 6.
Stability assay of IDA in murine plasma. The half-life turned out to be around 6 hours.
Discussion
Aptamers are an emerging class of therapeutics as up to date 11 aptamers are under clinical trials.29 To increase this number, a variety of different SELEX approaches were developed, including the selection of aptamers targeting recombinant proteins as well as whole cells.40 Even complete in vivo selections have been performed.41
As cancer cells use the α6β4 integrin/laminin-332 interaction to activate signaling pathways promoting tumor cell growth, invasion and metastasis, the inhibition of this interaction is of high therapeutic interest.2,14,15,18,19,20,42 In order to select an aptamer with the ability to block this interaction, we combined a cell-SELEX, which had the advantage for proteins being in their natural state, with a bead-based selection to further enhance the specificity of the selected library. This selection strategy resulted in the identification of 20 different clones with no sequence identity. Only the best binder, termed IDA, was also able to inhibit the adhesion of PC-3 cells to laminin-332 with a resulting IC50 value of 149 nmol/l. Binding of the aptamer to PC-3 cells was shown to depend on α6 integrins and an electrophoretic mobility shift assay further verified specificity to human recombinant integrins α6β4 and α6β1, respectively. No binding could be detected to α4β1, but the aptamer recognizes murine α6β4. This data indicate that the aptamer binds to α6 integrins, which opens the possibility to block the interaction of α6β1 with other laminins as well. As both α6 integrins presented on exosomes play a key role in lung-tropic metastasis, the inhibition of both integrins is beneficial.23 Additionally to IDA's function as an inhibitor, the aptamer could be used as a tumor marker for cancer cells highly presenting α6 integrins, e.g., squamous or thyroid carcinomas or for the detection of these exosomes.
The internalization of the aptamer opens the possibility to deliver drugs directly to cancer cells overexpressing the α6β4 integrin. Several aptamer-drug conjugates have been published so far including siRNA, microRNA, toxins, and functionalized nanoparticles.43,44,45
Truncated IDA was still able to bind to PC-3 cells in an α6β4 integrin-dependent manner. Blocking of the cell surface protein for laminin, however, was impaired after shortening of the aptamer. This observation indicates that the binding site of the aptamer is near but not identical to the binding pocket of the α6β4 integrin. Therefore, the inhibition could be caused by a region of the aptamer that is not essential for binding to its target protein but rather extending from the binding side, masking the laminin binding pocket of the α6β4 integrin.
For an in vivo application, the stability of the aptamer is of great importance. We studied the stability of IDA in blood plasma, resulting in a half-life time around 6 hours. For a further application, modifications of the aptamer need to be performed to increase the IC50 value as well as the stability. This could be done using modified nucleotides or by combination with a nucleic acid binding to a different binding site of the integrin, like nc36.
In summary, we selected a DNA aptamer inhibiting the integrin–laminin interaction, which is also internalized into target cells. Future studies need to reveal, if the aptamer has an influence on the signaling pathways promoted by the integrin.
Materials and methods
Chemicals. All chemicals were purchased from Sigma-Aldrich (Hamburg, Germany), unless otherwise stated. Buffers were prepared using deionized water obtained from a water purification system (Millipore, Billerica, MA).
Oligonucleotides. All ssDNAs were synthesized, modified, and purified by Sigma-Aldrich (Hamburg, Germany).
Proteins. All integrins were purchased from R&D Systems (Abingdon, UK) and laminin-332 from Biolamina AB (Stockholm, Sweden).
Cell culture. Cell culture of PC-3 cells was carried out using RPMI (PAN Biotech) with 10% FCS (PAA/GE Healthcare, K41-001), 60 mg/mL penicillin (PAA/GE Healthcare, P11-010) and 100 mg/mL streptomycin (PAA/GE Healthcare, P11-010) at 37 °C and in water-saturated atmosphere containing 5% CO2.
Selection of aptamers for integrin β4. The selection of aptamers for integrin β4 was accomplished by a combination of cell-SELEX and conventional SELEX using the recombinant α6β4 integrin. The starting ssDNA library (GCCTGTTGTGAGCCTCCTAAC-N39-CATGCTTATTCTTGTCTCCC) was purchased from Sigma Aldrich HPLC-purified using the primers described by Mayer et al.46 The forward primer contained a 5′ cyanine 5 label and the reverse primer a 5′ phosphate needed for strand displacement. Cells, cultured in a cell culture dish (100 × 20 mm), were incubated with 500 pmol of the starting library at 37 °C for 30 minutes in selection buffer (1× PBS, 3 mmol/l MgCl2, 1 mmol/l CaCl2, 1 mg/ml BSA and 1 mg/ml salmon sperm DNA). In the first round, cells were washed three times with the selection buffer. Cells were detached with a cell scraper and binding ssDNAs were eluted with 50 µl preheated water at 80 °C for 5 minutes. After centrifugation at 400×g for 5 minutes to remove the cells, eluted nucleic acids were amplified using PCR amplification (30 seconds at 95 °C, 1 minute at 60 °C, 1 minute at 72 °C) for an appropriate number of PCR cycles. The strand displacement was done by Lambda Exonuclease digestion for 30 minutes at 37 °C.47 In round three, ITGB4 knockdown cells were used in a preselection step. Therefore, the pool was incubated with knockdown cells for 30 minutes at 37 °C. The supernatant was then used in the selection step.
Beginning with round six, the procedure was changed to a conventional SELEX. Therefore, the human recombinant α6 (X1) β4 Integrin (100 pmol) was coupled to magnetic Beads (SIMAG-Carboxyl) with EDC-NHS and incubated with the pool in 100 µl selection buffer at 37 °C for 30 minutes. The beads were washed three times, with increasing washing steps for later rounds, and eluted with 50 µl preheated water at 80 °C for 5 minutes. Afterwards, an amplification step was done as described above. Altogether 12 SELEX rounds were done.
Cell inhibition assay. For analyzing the inhibition capacity of the integrin β4 aptamer, a cell inhibition assay was done. Therefore, a black 96-well plate was coated with 0.5 µg/ml laminin-332 (Biolamina) in 60 µl RPMI overnight at 4 °C. Cells were detached with accutase (PAN Biotech). 150,000 cells were either left untreated or incubated with relevant nucleic acids for 30 minutes at RT. For measuring the IC50 value, a dilution series of IDA was done beforehand. After washing, the 96-well plate twice with RPMI, the corresponding samples were added to the wells. After incubation at RT for 30 minutes, the plate was washed eight times with 60 µl 1×PBS. Attached and viable cells were detected via CellTiter-Glo® Assay (Promega). The data were plotted with python (version 3) and fitted with a Hill1-Fit. Measurements were done in triplicate.
Flow cytometry. The binding affinity of IDA to PC-3 cells as well as the specificity of the aptamer was analyzed via flow cytometry. Therefore, cells were detached with accutase for 2 minutes at 37 °C. In each case, 200,000 cells were washed with 100 µl RPMI and incubated with relevant ssDNAs for 20 minutes at RT.
For determining the Kd value, a dilution series was done beforehand. After two washing steps with 200 µl 1×PBS, the fluorescence intensities of the cells were determined with a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA) counting 10,000 events. The fluorescence was then evaluated using the BD CellQuest Pro software (Version 3.2.1).
For determining if IDA induces internalization, cells were incubated with 5 µmol/l IDA or a control nucleic acid for 5, 10 or 15 minutes at 37 °C prior to a β4 integrin antibody staining.
Electrophoretic mobility shift assay. The specificity of IDA was analyzed using an electrophoretic mobility shift assay. A constant amount of radioactively labelled ssDNA (>1 nmol/l) was incubated with 750 nmol/l protein for 20 minutes at 37 °C. After addition of 1 µl 6× DNA loading dye, the samples were loaded on a 5% non-denaturing polyacrylamide gel (acrylamide/bisacrylamide 37.5:1). The separation was done at 60 V and 4°C for 2 hours. The gel was frozen at −20 °C and the bands were detected via autoradiography. The amount of bound DNA was calculated by dividing the overall intensity per lane with bound fractions. Measurements were done in duplicate and plotted with python (version 3).
Laser scanning microscopy. Cells were sowed into a Nunc™ Glass Bottom Dish and cultivated for 24 hours to let them reach 80% confluence. Then 100 nmol/l ssDNA was added to the cells. After incubation at 4 °C or 37 °C for 20 minutes, cells were washed twice with RPMI without phenolred. Results were observed with an Olympus IX-81 microscope.
Stability assay. To determine the stability of IDA, a constant amount of radioactively labelled ssDNA was incubated in murine blood plasma at 37 °C for 24 hours. After 0, 1, 5, 15, and 30 minutes and 1, 2, 4, 8 and 24 hours, respectively, samples were taken, mixed with RNA loading dye and frozen in liquid nitrogen. The samples were analyzed by a 10% denaturing PAGE and bands were detected via autoradiography. Band intensities were determined with Quantity One and the amount of full length DNA in each lane was calculated with the 0 minute sample as 100%. Measurements were done in duplicate and plotted with python (version 3).
SUPPLEMENTARY MATERIAL Table S1. Name and sequence of used nucleic acids and primers. Figure S1. Determining the surface level of α6 (A) and β4 (B) integrin on LnCap cells via flow cytometry. Grey: untreated Lncap cells, green: Integrin specific antibody, red: Isotype control. Figure S2. Determining the surface level of α6 integrin on PC-3 cells via flow cytometry. Black: PC-3 cells with an isotype control, dark green: PC-3 cells with an α6 integrin specific antibody, grey: PC-3 integrin β4 knockdown cells with an isotype control, dark green: PC-3 integrin β4 knockdown cells with an α6 integrin specific antibody. Figure S3. Determining the specificity of IDA by electrophoretic mobility shift assay. The aptamer binds to murine (lane 2) and humane (lane 3) recombinant α6β4 Integrin, as well as humane α6β1 (lane 4), but not to recombinant α4β1 (lane 5). Lane 1 and 6 show the aptamer migration in the absence of proteins. Figure S4. Confocal laser scanning microscopy analyses. (A) IDA binds to the surface of PC-3 cells at 4°C. The nonbinding DNA showed no fluorescence signal (B). Figure S5. IDA does not induce internalization of α6β4 integrin determined by flow cytometry. Cell surface levels of α6β4 Integrin on PC-3 cells were analyzed with an ITGB4 antibody after preincubation of cells with 5 µM IDA at 37°C for 5 (violet), 10 (blue) or 15 min (green) or with a control DNA (red). No significant change can be seen. An antibody isotype control showed no binding to PC-3 cells (yellow). Figure S6. Structure analysis of IDA (A) and resulted Truncations. IDA-A (B) includes all three stem loops, whereas IDA-B (C) contains only two of them. Figure S7. FACS Analysis of truncated IDA-constructs. Grey: untreated PC-3 cells, violet: IDA-cy5, orange: IDA-A-cy5, pink: IDA-B. Both truncated nucleic acids are still binding. Figure S8. The truncations are not able to inhibit the binding of PC-3 cells to lam-332. Violett: IDA, orange: IDA-A, pink: IDA-B, blue: nc36. Figure S9. 10% denat. PAA-gel for detecting the stability of IDA in murine blood plasma. Samples taken after 0 (lane 1), 1(lane 2), 5 (lane 3), 15 (lane 4), 30 min (lane 5) and 1 (lane 6), 2 (lane 7), 4 (lane 8), 8 (lane 9) und 24 h (lane 10) of incubation at 37 °C.
Supplementary Material
References
- Hynes, RO (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687. [DOI] [PubMed] [Google Scholar]
- Stewart, RL and O'Connor, KL (2015). Clinical significance of the integrin α6β4 in human malignancies. Lab Invest 95: 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson, KD, Maddula, VS, Seligmann, BE, Shearstone, JR, Khan, A and Mercurio, AM (2012). Effects of β4 integrin expression on microRNA patterns in breast cancer. Biol Open 1: 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant, C, Monsuur, HN, Norman, JC and Sonnenberg, A (2011). Mechanisms of integrin activation and trafficking. Curr Opin Cell Biol 23: 607–614. [DOI] [PubMed] [Google Scholar]
- Campbell, ID and Humphries, MJ (2011). Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol 3: a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio, AM, Rabinovitz, I and Shaw, LM (2001). The alpha 6 beta 4 integrin and epithelial cell migration. Curr Opin Cell Biol 13: 541–545. [DOI] [PubMed] [Google Scholar]
- Litjens, SH, de Pereda, JM and Sonnenberg, A (2006). Current insights into the formation and breakdown of hemidesmosomes. Trends Cell Biol 16: 376–383. [DOI] [PubMed] [Google Scholar]
- Giancotti, FG (2007). Targeting integrin beta4 for cancer and anti-angiogenic therapy. Trends Pharmacol Sci 28: 506–511. [DOI] [PubMed] [Google Scholar]
- de Pereda, JM, Wiche, G and Liddington, RC (1999). Crystal structure of a tandem pair of fibronectin type III domains from the cytoplasmic tail of integrin alpha6beta4. EMBO J 18: 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio, AM and Rabinovitz, I (2001). Towards a mechanistic understanding of tumor invasion–lessons from the alpha6beta 4 integrin. Semin Cancer Biol 11: 129–141. [DOI] [PubMed] [Google Scholar]
- Rabinovitz, I, Toker, A and Mercurio, AM (1999). Protein kinase C-dependent mobilization of the alpha6beta4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J Cell Biol 146: 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant, C, Frijns, E, Wilhelmsen, K and Sonnenberg, A (2008). Regulation of hemidesmosome disassembly by growth factor receptors. Curr Opin Cell Biol 20: 589–596. [DOI] [PubMed] [Google Scholar]
- Tsuruta, D, Hashimoto, T, Hamill, KJ and Jones, JCR (2011). Hemidesmosomes and focal contact proteins: functions and cross-talk in keratinocytes, bullous diseases and wound healing. J Dermatol Sci 121: 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti, A, Kedeshian, PA, Dans, M, Curatola, AM, Gagnoux-Palacios, L and Giancotti, FG (2001). EGF-R signaling through Fyn kinase disrupts the function of integrin alpha6beta4 at hemidesmosomes: role in epithelial cell migration and carcinoma invasion. J Cell Biol 155: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, LM, Rabinovitz, I, Wang, HH, Toker, A and Mercurio, AM (1997). Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell 91: 949–960. [DOI] [PubMed] [Google Scholar]
- O'Connor, KL, Chen, M and Towers, LN (2012). Integrin α6β4 cooperates with LPA signaling to stimulate Rac through AKAP-Lbc-mediated RhoA activation. Am J Physiol Cell Physiol 302: C605–C614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz, I and Mercurio, AM (1997). The integrin alpha6beta4 functions in carcinoma cell migration on laminin-1 by mediating the formation and stabilization of actin-containing motility structures. J Cell Biol 139: 1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, KL, Nguyen, BK and Mercurio, AM (2000). RhoA function in lamellae formation and migration is regulated by the alpha6beta4 integrin and cAMP metabolism. J Cell Biol 148: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb, EA and Mercurio, AM (2005). Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev 24: 413–423. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos, SN, Blaikie, P, Yoshioka, T, Guo, W and Giancotti, FG (2004). Integrin beta4 signaling promotes tumor angiogenesis. Cancer Cell 6: 471–483. [DOI] [PubMed] [Google Scholar]
- Kusuma, N, Denoyer, D, Eble, JA, Redvers, RP, Parker, BS, Pelzer, R et al. (2012). Integrin-dependent response to laminin-511 regulates breast tumor cell invasion and metastasis. Int J Cancer 130: 555–566. [DOI] [PubMed] [Google Scholar]
- Cruz-Monserrate, Z, Qiu, S, Evers, BM and O'Connor, KL (2007). Upregulation and redistribution of integrin alpha6beta4 expression occurs at an early stage in pancreatic adenocarcinoma progression. Mod Pathol 20: 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A, Costa-Silva, B, Shen, TL, Rodrigues, G, Hashimoto, A, Tesic Mark, M et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe, AD, Pai, S and Ellington, A (2010). Aptamers as therapeutics. Nat Rev Drug Discov 9: 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, D, Zheng, C, Zhou, SF, Qiao, S, Tran, PH, Pu, C et al. (2015). Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics 5: 1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Tuerk, C and Gold, L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Robertson, DL and Joyce, GF (1990). Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344: 467–468. [DOI] [PubMed] [Google Scholar]
- Lao, YH, Phua, KK and Leong, KW (2015). Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano 9: 2235–2254. [DOI] [PubMed] [Google Scholar]
- Mittelberger, F, Meyer, C, Waetzig, GH, Zacharias, M, Valentini, E, Svergun, DI et al. (2015). RAID3—an interleukin-6 receptor-binding aptamer with post-selective modification-resistant affinity. RNA Biol 12: 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupold, SE, Hicke, BJ, Lin, Y and Coffey, DS (2002). Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 62: 4029–4033. [PubMed] [Google Scholar]
- Meyer, C, Hahn, U and Rentmeister, A (2011). Cell-specific aptamers as emerging therapeutics. J Nucleic Acids 2011: 904750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, EW, Shima, DT, Calias, P, Cunningham, ET Jr., Guyer, DR and Adamis, AP (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5: 123–132. [DOI] [PubMed] [Google Scholar]
- Rusconi, CP, Scardino, E, Layzer, J, Pitoc, GA, Ortel, TL, Monroe, D et al. (2002). RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419: 90–94. [DOI] [PubMed] [Google Scholar]
- Hoellenriegel, J, Zboralski, D, Maasch, C, Rosin, NY, Wierda, WG, Keating, MJ et al. (2014). The spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 123: 1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, O, Pawar, RD, Purschke, W, Eulberg, D, Selve, N, Buchner, K et al. (2007). Spiegelmer inhibition of CCL2/MCP-1 ameliorates lupus nephritis in MRL-(Fas)lpr mice. J Am Soc Nephrol 18: 2350–2358. [DOI] [PubMed] [Google Scholar]
- Mayer, G, Ahmed, MS, Dolf, A, Endl, E, Knolle, PA and Famulok, M (2010). Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc 5: 1993–2004. [DOI] [PubMed] [Google Scholar]
- Sefah, K, Shangguan, D, Xiong, X, O'Donoghue, MB and Tan, W (2010). Development of DNA aptamers using Cell-SELEX. Nat Protoc 5: 1169–1185. [DOI] [PubMed] [Google Scholar]
- Reuter, JS and Mathews, DH (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmostuk, M, Rimpelova, S, Gbelcova, H and Ruml, T (2015). Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv 33: 1141–1161. [DOI] [PubMed] [Google Scholar]
- Ozer, A, Pagano, JM and Lis, JT (2014). New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol Ther Nucleic Acids 3: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta, D, Kobayashi, H, Imanishi, H, Sugawara, K, Ishii, M and Jones, JC (2008). Laminin-332-integrin interaction: a target for cancer therapy? Curr Med Chem 15: 1968–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J and Rossi, JJ (2014). Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol Ther Nucleic Acids 3: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruspe, S, Meyer, C and Hahn, U (2014). Chlorin e6 conjugated interleukin-6 receptor aptamers selectively kill target cells upon irradiation. Mol Ther Nucleic Acids 3: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruspe, S, Mittelberger, F, Szameit, K and Hahn, U (2014). Aptamers as drug delivery vehicles. Chem Med Chem 9: 1998–2011. [DOI] [PubMed] [Google Scholar]
- Mayer, G, Ahmed, MS, Dolf, A, Endl, E, Knolle, PA and Famulok, M (2010). Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc 5: 1993–2004. [DOI] [PubMed] [Google Scholar]
- Avci-Adali, M, Paul, A, Wilhelm, N, Ziemer, G and Wendel, HP (2010). Upgrading SELEX technology by using lambda exonuclease digestion for single-stranded DNA generation. Molecules 15: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.