Abstract
Hepatitis B virus (HBV) infection is a major cause of chronic active hepatitis, cirrhosis, and primary hepatocellular carcinoma, all of which are severe threats to human health. However, current clinical therapies for HBV are limited by potential side effects, toxicity, and drug-resistance. In this study, a cell-penetrating peptide-conjugated peptide nucleic acid (PNA), Tat-PNA-DR, was designed to target the direct repeat (DR) sequences of HBV. Tat-PNA-DR effectively inhibited HBV replication in HepG2.2.15 cells. Its anti-HBV effect relied on the binding of Tat-PNA-DR to the DR, whereby it suppressed the translation of hepatitis B e antigen (HBeAg), HBsAg, HBV core, hepatitis B virus x protein, and HBV reverse transcriptase (RT) and the reverse transcription of the HBV genome. Furthermore, Tat-PNA-DR administered by intravenous injection efficiently cleared HBeAg and HBsAg in an acute hepatitis B mouse model. Importantly, it induced an 80% decline in HBV DNA in mouse serum, which was similar to the effect of the widely used clinical drug Lamivudine (3TC). Additionally, a long-term hydrodynamics HBV mouse model also demonstrated Tat-PNA-DR's antiviral effect. Interestingly, Tat-PNA-DR displayed low cytotoxicity, low mouse acute toxicity, low immunogenicity, and high serum stability. These data indicate that Tat-PNA-DR is a unique PNA and a promising drug candidate against HBV.
Keywords: anti-HBV, cell-penetrating peptide, direct repeat sequences, dual mechanisms, low toxicity, peptide nucleic acid, serum stability
Introduction
Hepatitis B is a major health problem worldwide. Currently, more than 400 million individuals suffer from chronic hepatitis B. Persistent hepatitis B virus (HBV) infection can significantly increase the risk of chronic active hepatitis, cirrhosis, and primary hepatocellular carcinoma.1,2,3,4 More than half of liver cancer cases worldwide are attributed to hepatitis B.1,2,3 The major drugs that are currently used for clinical therapy such as interferons and nucleotide analogs are far from perfect. Interferons can cause serious side effects, such as fever, cold syndrome, and myelosuppression and can interfere with the nervous system. Nucleos(t)ide analog treatment such as lamivudine (3TC) can effectively suppress HBV replication but have the potential to induce mutations in viral DNA polymerase. Thus, new anti-HBV agents with various targets are continuously being discovered, such as NUC B1000 (a promising RNAi-based therapy)5 and cyclosporin A.6
Peptide nucleic acid (PNA) is a synthetic polymer that is commonly used in antisense therapy and clinical diagnoses.7 Its unnatural backbone, which contains repeating N-(2-aminoethyl)-glycine units, provides PNA with a high binding capacity and specificity for its complementary DNA or RNA. This unique structural feature also provides PNA with high resistance against nucleases and proteases, as well as low toxicity.7,8,9 To improve cell permeability and cellular uptake capacity, PNA molecules are usually conjugated to cell-penetrating peptide. These conjugated PNA molecules have been successfully developed as antibacterial agents,10 as well as antiviral agents against HIV, HCV, and DHBV.11,12,13 In previous studies, PNAs used were usually designed to target a single site that has a certain function in the lifecycle of the pathogenic bacteria and viruses, which elevates the risk of drug resistance to a therapeutic agent. Therefore, PNA molecules with dual or more targets in pathogenic bacteria or viruses are expected to reduce this possibility or even to improve their bioactivity. Moreover, the acute toxicity in animals, serum stability, and in vivo antiviral activity of PNA have never previously been tested. Because PNA is an important type of therapeutic agent, it is necessary to further determine the in vivo activity and safety of PNA.
HBV genome replication depends on an RNA intermediate called pregenomic RNA (pgRNA) and reverse transcriptase (RT) and is accomplished through a complex mechanism involving primer shifting that is facilitated by 3-terminal direct repeat (DR) sequence of pgRNA. DR is a crucial functional element in the synthesis of the HBV genome and it is also contained in the mRNA of the hepatitis B e antigen (HBeAg), hepatitis B core protein, hepatitis B virus x protein (HBx) and RT.3,14,15 Thus, DR sequences could be used as vital and unique targeting sequences for designing anti-HBV PNA molecules.
This study was performed to develop an effective anti-HBV PNA molecule based on the sequence and functional characteristic of DR sequences of HBV. First, we designed, synthesized, and identified an antisense PNA strand, which we have called Tat-PNA-DR, completely complementary to DR. Second, we evaluated the cytotoxicity of Tat-PNA-DR in hepatic cells and human erythrocytes and the in vitro anti-HBV activity of Tat-PNA-DR in HepG2.2.15 cells. Third, we tested the serum stability, acute toxicity and immunogenicity in mice of Tat-PNA-DR, and its antiviral activity in acute and long-term mouse models of viral hepatitis B. Finally, we tested the affinity of Tat-PNA-DR for DR sequences using an in vitro binding assay and confirmed its sequence-specific targeting by comparing its ant-HBV activity with mutated PNA molecules. Our study represents the first report to design and identify a dual-target antiviral PNA molecule in vitro and in vivo that displays an advantageous pharmacological profile and strong potential for novel antiviral drug development.
Results
Design, synthesis, purification, and identification of Tat-PNA-DR
According to PNA-synthesized literatures previously reported,16,17,18 a PNA-DR oligomer was designed and prepared using a Fmoc solid-phase synthesis protocol. The sequence of PNA-DR was MPA-GCAGAGGTGAA-Gly-NH2 (Figure 1a), complementing to HBV DR sequences (5′-UUCACCUCUGC-3′). matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis was used to measure the molecular weight (mw) of the synthesized PNA-DR oligomer. As shown in Figure 1b, the m/z value of the synthesized PNA-DR was 3,301.2, which reflects a product with mw of 3,300.2 Da. This is corresponding to the theoretical mw of the designed PNA-DR (3,300.1 Da). The MALDI-TOF spectrum therefore confirmed the successful synthesis of PNA-DR.
Figure 1.
Design, synthesis, purification, and identification of Tat-PNA-DR.(a) HBV DR sequences (5′-UUCACCUCUGC-3′) were selected to be the target for antisense PNA. The designed PNA-DR was prepared using a Fmoc solid-phase synthesis protocol and characterized using sequence determination analysis (MPA-GCAGAGGTGAA-Gly-NH2). The conjugation of PNA-DR with the Tat peptide was processed via a Michael addition reaction, and the conjugated peptide nucleic acid was named Tat-PNA-DR. Dialysis and HPLC were performed to purify the Tat-PNA-DR conjugate. (b) MALDI-TOF spectra of PNA-DR. The molecular weight of the synthesized PNA-DR was 3,300.2, which was corresponding to the theoretical molecular weight of the designed PNA-DR (3,300.1). (c) MALDI-TOF spectra of Tat-PNA-DR before HPLC purification. The observed molecular masses of 1,751.5, 3,500.4 and 5,051.5 corresponded to the excess Tat (1,751.1), Tat dimer (3,500.2) and Tat-PNA-DR (5,051.2), respectively. (d) HPLC purification of Tat-PNA-DR. The main peak from 11 to 13 minutes was manually collected and found to correspond to the conjugated Tat-PNA-DR. (e) MALDI-TOF spectra of Tat-PNA-DR after HPLC purification. The molecular weight of the purified Tat-PNA-DR was 5,051.8, which was the same as the theoretical molecular weight of the designed Tat-PNA-DR (5,051.2). (f) Purity analysis of the final product Tat-PNA-DR by analytical HPLC.
To enhance the delivery of PNA-DR into the cytoplasm and nuclei of hepatic cells, it was fused to Tat, a cell-penetrating peptide that was derived from the HIV.19,20,21 The Tat and PNA-DR conjugation was synthesized via a Michael addition reaction between the maleimide group of PNA and the cysteine residue of the Tat peptide (Figure 1a). The conjugated reaction was evaluated by MALDI-TOF analysis, which was used to analyze the reaction product, as shown in Figure 1c. The observed m/z of 1,752.5, 3,501.4, and 5,052.5 corresponded to the excess Tat (theoretical mw: 1,751.1), Tat dimer (3,500.2) and Tat-PNA-DR (5,051.2), respectively. These signals indicated the successful synthesis of Tat-PNA-DR. The reaction products were then dialyzed and the conjugated Tat-PNA-DR was purified using RP-high-performance liquid chromatography (HPLC) (Figure 1d). The MALDI-TOF spectrum for the purified Tat-PNA-DR indicates a signal with m/z of 5,052.8, corresponding to the theoretical molecular mass (5,051.2 Da) of Tat-PNA-DR (Figure 1e). The analytical HPLC confirmed that the purity of the final Tat-PNA-DR product was >95% (Figure 1f).
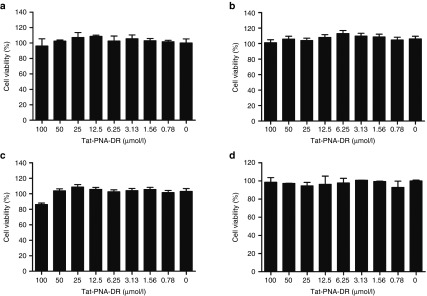
Cytotoxicity of Tat-PNA-DR
The cytotoxicity of Tat-PNA-DR in HepG2.2.15, HepG2, and L-02 cells was tested using MTT assays. At a concentration lower than 100 μmol/l, Tat-PNA-DR did not significantly affect cell proliferation or survival in any of the three cell lines. The viability of HepG2.2.15 cells (Figure 2a) and HepG2 cells (Figure 2b) that were treated with 100 μmol/l Tat-PNA-DR was greater than 95%. The viability of L-02 cells treated with 100 μmol/l Tat-PNA-DR was higher than 80% (Figure 2c), indicating that 100 μmol/l or less Tat-PNA-DR was minimally cytotoxic to cells. Hemolysis assay showed that the viability of erythrocytes was more than 95% when the concentration of Tat-PNA-DR was lower than 100 μmol/l (Figure 2d), indicating a low hemolytic effect on erythrocytes. These data showed that the oligomer Tat-PNA-DR has low cytotoxity in vitro.
Figure 2.
Cytotoxicity of Tat-PNA-DR. The cytotoxicity of Tat-PNA-DR was measured in HepG2.2.15 (a), HepG2 (b), and L-02 (c) cells using MTT assays. The cytotoxicity of Tat-PNA-DR was evaluated in human erythrocytes (d) using hemolytic assays. The concentrations ranged from 0 to 100 μmol/l.
Anti-HBV activity of Tat-PNA-DR in vitro
To investigate the effect of Tat-PNA-DR on HBV replication in vitro, HepG2.2.15 cells were cultured in the presence of 10-fold serial dilutions of Tat-PNA-DR. HBeAg and HBsAg levels were measure in HepG2.2.15 cell supernatants using enzyme-linked immunosorbent assay (ELISA), and the amount of HBV progeny DNA was determined using real-time PCR. As shown in Figure 3a,b, Tat-PNA-DR potently inhibited the cellular supernatant levels of HBeAg and HBsAg in a dose-dependent manner. Real-time PCR data indicated that HBV progeny DNA in HepG2.2.15 media was also inhibited by Tat-PNA-DR in a dose-dependent manner (Figure 3c). Furthermore, western blot analysis showed that Tat-PNA-DR efficiently reduced intracellular HBV protein levels in HepG2.2.15 cells. As shown in Figure 3d–g, HBsAg, HBV core protein, x protein, and RT expression levels were all reduced in a dose-dependent manner. Because it is a drug that specifically inhibits the reverse transcription of HBV, 3TC did not affect the expression of any HBV viral proteins.
Figure 3.
Anti-HBV activity of Tat-PNA-DR in HepG2.2.15 cells. (a, b) The inhibitory effect of Tat-PNA-DR against HBeAg and HBsAg in HepG2.2.15 culture medium was measured using ELISA. (c) The inhibitory effect of Tat-PNA-DR against HBV DNA in HepG2.2.15 culture medium was measured using real-time PCR. Values represent the mean ± SEM of five independent samples. (d–g) The inhibitory effect of Tat-PNA-DR against the HBsAg, HBV core, HBx, and HBV RT proteins in HepG2.2.15 cell lysates was determined using western blot analysis. The drug 3TC was used as a control. (h) The inhibitory effect of Tat-PNA-DR against HBV DNA in HepG2.2.15 cells was determined using southern blot analysis. The drug 3TC, which is a nucleoside reverse transcriptase inhibitor, was used as a control. (i) The inhibitory effect of Tat-PNA-DR against HBV RNA in HepG2.2.15 cells was determined using northern blot analysis. The drug 3TC was used as a control.
Southern and northern blot analysis were used to measure the levels of intracellular HBV DNA (Figure 3h) and RNA (Figure 3i) in HepG2.2.15 cells. After treatment with Tat-PNA-DR for 48 hours, both HBV DNA and RNA were significantly reduced in a dose-dependent manner. As a positive control, 3TC clearly inhibited the synthesis of HBV DNA but not HBV RNA. These results suggest that Tat-PNA-DR can inhibit the expression of HBV proteins and the synthesis of both HBV DNA and RNA, revealing that it has multiple modes of action as an anti-HBV agent.
Serum stability, acute toxicity and immunogenicity in mice of Tat-PNA-DR
To apply Tat-PNA-DR in an in vivo antiviral study, we tested the serum stability, acute toxicity, and immunogenicity in mice of Tat-PNA-DR. After incubation in 10% serum for less than 24 hours, the inhibitory rate of Tat-PNA-DR against HBeAg was still 68.11% (Figure 4a), while the inhibitory rate of Tat-PNA-DR against HBsAg was 62.98% (Figure 4b), which was similar to the rate in the untreated Tat-PNA-DR. These results suggest that Tat-PNA-DR has high serum stability.
Figure 4.
Serum stability, acute toxicity, and immunogenicity in mice of Tat-PNA-DR. (a, b) Serum stability of Tat-PNA-DR. Tat-PNA-DR was incubated in culture medium containing 10% FBS for 0, 4, 8, 12, and 24 hours at 37 °C. At the indicated timepoints, preincubated Tat-PNA-DR aliquots were used for the antiviral assay. The levels of HBeAg and HBsAg in HepG2.2.15 culture medium were measured using ELISA. Values represent the mean ± SEM of five independent samples. (c, d) The acute toxicity of Tat-PNA-DR in BALB/c mice. Tat-PNA-DR was injected into mice intravenously at a dose of 50.5 mg/kg. The survival rate and the average body weights of the treated mice were calculated each day after injection for 7 days (n = 5). (e, f) Mice sera were collected each day, and then ALT and AST levels were measured by blood biochemistry tests (n = 5). Normal saline was used as the negative control. (g, h) Mice sera were collected each day. IgG and IgM levels were measured by ELISA (n = 5). Values represent the mean ± SEM of five independent samples.
The acute toxicity in mice of Tat-PNA-DR was identified in 6- to 7-week-old BALB/c mice. Tat-PNA-DR was administered via intravenous injection at a concentration of 50.5 mg/kg (a 10-fold experimental treatment dose), and then mice were housed and fed in a SPF laboratory animal room for 7 days. As shown in Figure 4c, the survival rate in Tat-PNA-DR-treated mice did not vary in the control mice treated with normal saline each day throughout the treatment period. However, the average body weight of the Tat-PNA-DR-treated mice increased from 19.23 to 21.67 g, which was similar to the body growth observed in the negative control group, which grew from 19.82 to 22.01 g (Figure 4d). As shown in Figure 4e,f, serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels from Tat-PNA-DR treated mice did not show a significant difference with those of normal saline-treated mice during the whole 7 days treatment, which suggested that Tat-PNA-DR may have low hepatic toxicity. At a dose of 50.5 mg/kg, Tat-PNA-DR therefore did not induce any fatal or apparent toxic symptoms in mice. We further investigated the immunogenicity of Tat-PNA-DR in mice. As shown in Figure 4g,h, no detectable IgG or IgM antibodies were elicited in mouse sera by Tat-PNA-DR, similar to normal saline treatment. These results indicate that Tat-PNA-DR has low acute toxicity and low immunogenicity in mice.
Inhibitory activity of Tat-PNA-DR against HBV in vivo
To further investigate the anti-HBV function of Tat-PNA-DR in vivo, we used a mouse model of acute hepatitis B, as described in previous studies.22,23 Viral antigens were measured in mouse blood 48 hours after a hydrodynamic injection of pUC18-HBV1.3 plasmids. As shown in Figure 5a,b, the average concentration of HBeAg and HBsAg in the serum of the Tat-PNA-DR-treated mice was 4.55 and 15.40 PEIU/ml, respectively, which was 27 and 44%, respectively, of concentration observed in the untreated acute hepatitis B mice (16.27 and 34.63 PEIU/ml, respectively). These data indicate the efficient clearance of HBV antigens. As a negative control, neither HBeAg nor HBsAg was detected in the blood of the mice that were not administered pUC18-HBV1.3. The average level of HBV DNA in serum was determined using real-time PCR. As shown in Figure 5c, the average HBV DNA concentration in the serum of Tat-PNA-DR-treated mice was 1.4 × 104 copies/ml, which was 20% of the number of copies observed in the untreated acute hepatitis B mice (6.9 × 104 copies/ml), indicating the efficient elimination of HBV DNA. In the 3TC-treated mice, the concentration of HBV DNA decreased to 1.2 × 104 copies/ml, which was similar to the reduction observed following treatment with Tat-PNA-DR. The organ coefficients of the livers were measured by weighting the liver and the whole mouse. As shown in Figure 5d, the average organ coefficient of livers from the untreated acute hepatitis B mice was 5.17%, while it was 4.56% in the Tat-PNA-DR-treated mice. The average organ coefficient was 4.39% in the blank control mice. There was also no significant reduction in the organ coefficient in the 3TC-treated mice.
Figure 5.
Anti-HBV activity of Tat-PNA-DR in hepatitis B mouse models. An acute hepatitis B mouse model (a–f) and a long-term hydrodynamics HBV mouse model (g–i) were both used to measure anti-HBV activity of Tat-PNA-DR in vivo. (a, b) HBeAg and HBsAg levels were measured in the sera of mice (n = 5) using ELISA. ***Denotes P < 0.001. (c) HBV DNA levels were measured in the sera of mice (n = 5) using real-time PCR. ***Denotes P < 0.001. (d) Organ coefficients of mouse livers (n = 5). The organ coefficients were measured by weighing the liver and the whole mouse. The average organ coefficient of the livers of the mice that were injected with pUC18-HBV1.3 was higher than that of the BLANK-group mice. This increase was reversed by the injection of Tat-PNA-DR. There was also no significant reduction in the organ coefficient in the 3TC-treated mice. **Denotes P < 0.01. (e, f) The inhibitory activity of Tat-PNA-DR against HBV core in the hepatocytes of HBV-infected mice, measured using Quantum Dots Fluorescence Immunoassay. The frequency of HBV core-positive hepatocytes was significantly lower in the Tat-PNA-DR-treated mice than in the untreated group. The clearance of HBV core-positive hepatocytes was 63%. **Denotes P < 0.01. (g, h) HBeAg and HBsAg levels were measured in the sera of mice by ELISA every 3 days. “PEIU” is an international standard unit which was put forward by Paul Ehrlich Institut, which is an Agency of German Federal Ministry of Health. ***Denotes P < 0.001 and **Denotes P < 0.01. (i) HBV DNA levels were also quantitated in the sera of mice every 3 days using real-time PCR. **Denotes P < 0.01.
To evaluate the abundance of HBV protein expression in mouse liver tissues, we used Quantum Dots Fluorescence Immunoassays to detect the HBV core protein in mouse livers after they were fixed in a 4% paraformaldehyde solution. As shown in Figure 5e,f, the frequency of HBV core-positive hepatocytes was 4.46% in the untreated acute hepatitis B mice, which decreased to 1.67% in the Tat-PNA-DR-treated mice. The inhibition of the expression of the HBV core protein in mouse livers was therefore consistent with its effect on HBeAg and HBV DNA in mouse blood. These results indicate that Tat-PNA-DR inhibits HBV replication in mouse livers and that it reduces the secretion of HBV antigens and HBV DNA into mouse blood.
In addition, we used a long-term hydrodynamics HBV mouse model to evaluate the long-acting anti-HBV activity of Tat-PNA-DR, as described in previous studies.22,24 Viral antigens were measured in mouse blood each 3 days after the first injection of Tat-PNA-DR at day 0. As shown in Figure 5g,h, the average concentration of HBeAg and HBsAg in the serum of the Tat-PNA-DR-treated hepatitis B mice was significantly decreased at day 3, compared with that of the untreated, 3TC treated or Tat-PNA-DRm (a two-site mutant of Tat-PNA-DR)-treated hepatitis B mice. After the second injection at day 7, its inhibition effect on HBeAg and HBsAg maintained until the end of experiment at day 15. These data indicate the efficient and long-acting clearance of HBV antigens by Tat-PNA-DR. As a negative control, neither HBeAg nor HBsAg was detected in the blood of the mice that were not administered pAAV/HBV1.2. As shown in Figure 5i, the average HBV DNA concentration in the serum of the Tat-PNA-DR-treated hepatitis B mice was also reduced, compared with the untreated or the Tat-PNA-DRm-treated hepatitis B mice, and its inhibitory activity against HBV DNA was reinforced by the second injection. As an inhibitor of HBV reverse transcription, 3TC reduced the concentration of HBV DNA significantly during the whole treating period. Tat-PNA-DRm had no obvious inhibition effect on HBV DNA.
Tat-PNA-DR interferes with HBV gene translation by targeting DR sequences
To determine whether the inhibitory activity of Tat-PNA-DR against HBV proteins was due to the interruption of translational processes, HepG2 cells were transiently transfected with constructs encoding HBeAg, HBsAg, HBV core protein, x protein, and RT. After cells were treated with Tat-PNA-DR for 48 hours, ELISA and western blot analysis showed that HBeAg (Figure 6e), HBV core protein (Figure 6b), x protein (Figure 6c) and RT (Figure 6d) were significantly reduced. However, Tat-PNA-DR did not affect the expression of HBsAg in the transient transfection system (Figure 6a), contrast to its effect in HepG2.2.15 cells, which could be explained by the previous results that knockdown of HBx could downregulate the expression of HBsAg in HepG2.2.15 cells.25 Furthermore, we found that the overexpression of HBx resulted in a significant decrease of inhibitory effect of Tat-PNA-DR against HBsAg in HepG2.2.15 cells (Figure 6k), which provided a solid evidence that HBx could regulate the expression of HBsAg.
Figure 6.
Tat-PNA-DR interferes with HBV gene translation by targeting DR sequences. Different constructs encoding the HBeAg, HBsAg, HBV core, HBV x, and RT proteins were transfected into HepG2 cells. After 24 hours, the transfected cells were treated with Tat-PNA-DR and incubated for another 48 hours. The protein levels of HBsAg (a), HBV core (b), HBV x (c) and RT (d) were measured using western blot assays, and HBeAg (e) was measured using ELISA. The mRNA levels of HBeAg (f), HBsAg (g), HBV core (h), HBx (i) and RT (j) in the transiently transfected HepG2 cells were analyzed using real-time PCR analysis. (k) HepG2.2.15 cells were transiently transfected with a CMV-promoter-driven plasmid of HBx, named pcDNA3.1-HBx. The concentration of HBsAg in cell culture medium was measured by ELISA after a 48-hour incubation with Tat-PNA-DR. Values represent the mean ± SEM of five independent samples.
Meanwhile, total RNA was extracted for each group and measured using real-time PCR. The levels of HBeAg (Figure 6f), HBsAg (Figure 6g), HBV core protein (Figure 6h), HBx protein (Figure 6i), and HBV RT (Figure 6j) mRNA were not affected by treatment with Tat-PNA-DR. Clearly, Tat-PNA-DR does not immediately downregulate the transcription of HBV viral proteins. This could have been the result of a difference in binding efficiency of Tat-PNA-DR to the double-stranded DNA and single-stranded RNA. Thus, Tat-PNA-DR interferes with HBV genes' translation, but not transcription.
Tat-PNA-DR inhibits HBV replication by specifically binding to DR sequences
To confirm its anti-HBV mechanism, a two-site mutant of Tat-PNA-DR (Tat-PNA-DRm) was prepared to investigate the sequence specificity of its anti-HBV activity. HepG2.2.15 cells were cultured in the presence of Tat-PNA-DR or Tat-PNA-DRm for 48 hours. An ELISA was used to measure the levels of extracellular HBeAg (Figure 7a) and HBsAg (Figure 7b). Tat-PNA-DR inhibited the expression of HBeAg and HBsAg, while Tat-PNA-DRm displayed no obvious inhibitory activity. Extracellular HBV DNA was measured by real-time PCR (Figure 7c). Tat-PNA-DRm also hardly decreased HBV DNA. Western blot assay of HBV viral proteins presented similar results. HBsAg (Figure 7d), HBV core protein (Figure 7e), HBx protein (Figure 7f), and HBV RT (Figure 7g) were all reduced by treatment of Tat-PNA-DR, but not significantly affected by Tat-PNA-DRm. The comparison of effects between Tat-PNA-DR and Tat-PNA-DRm on HBV RNA was also carried out by northern blot. The result showed that inhibitory activity of Tat-PNA-DRm against HBV RNA was extremely lower than that of Tat-PNA-DR (Figure 7h).
Figure 7.
Tat-PNA-DR inhibits HBV replication by binding DR in a sequence-specific manner. (a, b) Inhibitory activity of Tat-PNA-DR and Tat-PNA-DRm (mutant of Tat-PNA-DR) against the expression of HBeAg and HBsAg in HepG2.2.15 culture medium. HepG2.2.15 cells were incubated with Tat-PNA-DR or Tat-PNA-DRm for 48 hours. The levels of HBeAg and HBsAg in the culture medium were measured using ELISA. (c) Comparison of inhibitory activity on extracellular HBV DNA of Tat-PNA-DR and Tat-PNA-DRm was performed by a real-time PCR. The data above are shown as the mean ± SEM of five independent samples. (d–g) Western blot assay was used to measure the effects of Tat-PNA-DR and Tat-PNA-DRm on intracellular HBV proteins. (h) Inhibitory activity of Tat-PNA-DR and Tat-PNA-DRm on HBV RNA was measured by northern blot with a DIG-labeled probe. (i) In vitro binding assay of Tat-PNA-DR and HBV DR. Native and mutated RNA oligonucleotides corresponding to the HBV DR were synthesized and labeled with biotin. The labeled RNA oligonucleotides were used for in vitro binding assays. Shifted bands indicate the binding between conjugates of Tat-PNA-DR and RNA oligonucleotides.
To determine the target selectivity of Tat-PNA-DR, RNA oligonucleotides corresponding to DR sequences were synthesized and biotin labeled. Probes were incubated with Tat-PNA-DR solution or ddH2O to determine whether Tat-PNA-DR can efficiently bind to DR. The result showed that there appeared to be remarkable shift bands in the lanes of Tat-PNA-DR and DR RNA oligonucleotides interacting groups. To measure the specificity of the binding between Tat-PNA-DR and DR sequence, a mutant probe with two random mutation (5′-UGCACCUAUGC-3′) sites was designed and prepared. The result showed that Tat-PNA-DR was unable to bind the mutant probe (Figure 7i). These data indicate that Tat-PNA-DR can bind HBV DR sequences in a sequence-specific manner. Thus, Tat-PNA-DR can specifically bind to DR, and this binding is necessary for its anti-HBV activity.
Discussion
In recent years, PNA has been used in antisense therapies and for clinical diagnoses, which strongly suggested its potential applications.11,12,13 PNA has been used as a resource for antiviral drug development.11,12,13 For example, PNA molecules have been designed as inhibitors of HIV-1 tat-dependent trans-activation, which then exhibited dose-dependent inhibitory activity against tat-dependent trans-activation in a HeLa cell assay.13 PNAs targeting the bulge and upper stem of the pgRNA of DHBV had been reported to efficiently inhibit DHBV reverse transcription.12 An antisense PNA targeting the X-RNA of HCV has also been reported to interfere with HCV replication.11 In this study, we rationally designed, synthesized, and identified a cell-penetrating peptide-conjugated PNA (Tat-PNA-DR) that targets DR sequences of the HBV RNA. Tat-PNA-DR effectively inhibited the replication of HBV in vitro and in vivo. Our work represents the first report to develop a PNA molecule that is an inhibitor of the human hepatitis B virus.
DR sequences are contained within the precore mRNA and mRNA for the HBV x protein, which encodes the HBV core protein, e antigen, x protein, and RT. In the unique life cycle of HBV, the normal function of the DR sequence located at the 3′ end of the HBV pgRNA (DR1*) is essential for the translocation of HBV RT during reverse transcription of the HBV genome. The process of reverse transcription relies on the presence of HBV RT and RT translocation. Therefore, DR sequences are potential antiviral targets for blocking the synthesis of the HBV core protein, e antigen, RT and x protein, and its reverse transcription. In this study, we designed and synthesized an 11-nt PNA strand that is completely complementary to HBV DR in consideration for the double and essential roles of DR sequences in HBV replication.
Because Tat-PNA-DR may bind with DR sequences in the mRNA of HBV viral proteins, it may therefore disturb the ribosome scanning and ribosomal translation of HBV viral proteins. Our western blot analysis and real-time PCR data indicate that the translation of the HBeAg, HBV core protein, x protein, and RT can be directly inhibited by Tat-PNA-DR, which targets DR. However, intracellular HBsAg was also significantly reduced. Because DR sequences are not contained in the mRNA of HBsAg, we assumed that this was caused by the regulatory functions of other HBV viral proteins during the life cycle of HBV. This assumption was supported by previously published studies showing that HBx protein is crutial for the transcriptional activation of HBV genes and the expression of HBsAg in HepG2.2.15 cells was upregulated by HBV x protein.25,26,27,28 To further provide hard experimental evidence, HepG2.2.15 cells were transiently transfected with a CMV-promoter-driven plasmid of HBx, pcDNA3.1-HBx. HBsAg in cell culture medium was measured by ELISA after incubation with Tat-PNA-DR for 48 hours (Figure 6k). The expression of HBsAg was decreased after the treatment of Tat-PNA-DR in the wild-type HepG2.2.15 cells, consistent with the above data. Overexpression of HBx in HepG2.2.15 cells appeared to increase the expression of HBsAg. Importantly, the inhibitory rate of Tat-PNA-DR against HBsAg was significantly reduced in the HBx overexpressed HepG2.2.15 cells. Therefore, our experimental data confirmed that HBsAg reduction treated by Tat-PNA-DR in HepG2.2.15 cells resulted from the decreasing of HBx with Tat-PNA-DR treatment.
Since it forms a steady complimentary pair with Tat-PNA-DR, DR sequence located at the 3′-terminal of HBV pgRNA (DR1*) could therefore no longer serve as the destination for the translocation of the short nascent minus strand DNA. As a consequence of inhibiting the expression and translocation of RT, Tat-PNA-DR blocked the reverse transcription of the HBV genome. Our in vitro binding experiment demonstrated that Tat-PNA-DR can bind to HBV DR in a sequence-specific manner and that this binding is necessary for its anti-HBV activity in HepG2.2.15 cells. Most studies that have reported the discovery or creation of new antisense agents were focused on a single functional target that plays a role in the lifecycle of the pathogen, but our study may open a new avenue through which researchers can focus on targets such as nucleotide sequences, proteins, peptides, and even physiological processes that exert multiple biological functions during the life process of these pathogens. Thus, it is possible to block the lifecycle of a pathogen in several ways using one agent. Tat-PNA-DR inhibited the replication of HBV in vitro and in vivo via a dual mechanism: it targeted DR sequences to prevent both the reverse transcription of HBV genome and the translation of the HBV proteins. Tat-PNA-DR makes this kind of academic intention possible. Our work will help to accelerate the discovery of efficient drug leads.
We found that Tat-PNA-DR exhibited a dose-dependent anti-HBV activity in HepG2.2.15 cells. However, HBeAg, HBsAg, and HBV DNA were significantly reduced after treatment with Tat-PNA-DR in a mouse model of acute hepatitis B. Tat-PNA-DR cleared ~ 73% of HBeAg and 80% of HBV DNA. The amount of HBV DNA eliminated by treatment with Tat-PNA-DR was similar to that eliminated by treatment with the clinical drug 3TC. Viral hepatitis B may also cause liver edema. As a consequence of its inhibitory effect on HBV replication, Tat-PNA-DR also possessed the ability to reduce liver edema. In a long-term hydrodynamics HBV mouse model, Tat-PNA-DR also exerted significantly and long-acting anti-HBV activity by an injection every other 7 days.
In this investigation, we wanted to design a PNA sequence targeting HBV DR sequences (named as PNA-DR), which possibly interfered multiple aspects of HBV life cycle. On the other hand, PNA-DR was conjugated with a cellular membrane penetrating peptide (Tat) to enhance its bioavailability (Supplementary Figure S1). In fact, the coinjection of NAG-MLP with potent chol-siRNAs targeting conserved HBV sequences resulted in multilog repression of viral RNA, proteins, and viral DNA with long duration of effect in hydrodynamics HBV-persistence mouse model and HBV-transgenic mouse models.29 Therefore, a hepatocyte-specific PNA-DR conjugate will be developed in the follow-up study, which may improve the in vivo anti-HBV activity.
Tat-PNA-DR did not have an obvious influence on cell proliferation and survival in cultured hepatic cells at a concentration of 100 μmol/l. Similar to the results of tests of its cytotoxicity, 100 μmol/l or less Tat-PNA-DR did not result in detectable hemolysis in human erythrocytes. Meanwhile, after treatment with 10% serum for 24 hours or less, Tat-PNA-DR retained its inhibitory activity against HBV replication, indicating that it has good serum stability. PNA molecules have such a feature that they are neither nucleic acids due to the pseudopeptide skeleton, nor peptides because their monomers are not amino acids. May be the reason for strong molecular stability of Tat-PNA-DR, PNA molecules are hardly recognized and degraded by nucleases and proteases. After intravenous injection of a high concentration of Tat-PNA-DR (50.5 mg/kg), there was no increase in mortality, and the mice displayed body growth was similar to that of the negative control group. Alanine aminotransferase and aspartate aminotransferase levels in mice sera also did not show significant difference between the Tat-PNA-DR-treated and the negative control groups. Many clinical applications and developed drugs/agents are limited by their side effects, which can include toxicity. Its low in vitro and in vivo toxicity suggests that Tat-PNA-DR is a potential anti-HBV agent. It is certainly necessary for Tat-PNA-DR to be comprehensively tested for its safety in future research in preclinical studies or clinical trials.
Conclusion
We rationally designed, synthesized, and identified an antisense cell-penetrating peptide-conjugated PNA strand, which we called Tat-PNA-DR, which was completely complementary to HBV DR sequences. Tat-PNA-DR exerted considerable inhibitory effects against HBV replication in vitro and in vivo via a unique dual mechanism. Moreover, Tat-PNA-DR also exhibited low hemolysis and cytotoxicity, low acute toxicity in mice, and good serum stability. To the best of our knowledge, Tat-PNA-DR is the first PNA to display anti-HBV activity and low toxicity in thorough in vitro and in vivo evaluations. Our results show that Tat-PNA-DR is a unique PNA with a specific dual mechanism and a new lead with clinical potential against viral hepatitis B.
Materials and methods
Reagents. Fmoc/Bhoc-protected PNA monomers (Fmoc-PNA-A(Bhoc)-OH, Fmoc-PNA-C(Bhoc)-OH, Fmoc-PNA-G(Bhoc)-OH, and Fmoc-PNA-T-OH) were purchased from Panagene (South Korea). Rink Amide-AM resin (100–200 mesh, loading: 0.52 mmol/g), glycine, N-methylmorpholine, and 2-(7-azo-benzotriazole)-N,N,N′,N′-tetramethyluronium hexafluorophosphate were purchased from GL Biochem (Shanghai, China). Maleimidopropionic acid was obtained from J&K Scientific (Beijing, China). Trifluoroacetic acid, N,N′-dimethylformamide (DMF), and N-methylpyrrolidone were of analytical grade and were distilled before use. All other organic reagents and solvents were acquired from Shanghai Reagent Chemical (China) and used directly.
The positive drug control lamivudine (3TC), the penicillin, and streptomycin used in cell cultures and the dimethylsulfoxide (DMSO) used for MTT analysis were all purchased from Sigma (Sigma, St. Louis, MO). The dose of 3TC used for the experiments of in vitro and in vivo anti-HBV activity was 20 μmol/l and 0.5 mg/kg body weight, respectively. Real-time PCR primers and RNA oligonucleotides were synthesized by Sangon Biological Engineering Technology Company (Shanghai, China).
Synthesis of maleimidopropionic acid terminated the PNA oligomer PNA-DR. The HBV DR sequences (5′-UUCACCUCUGC-3′) were used as the targeting sequence for PNA design. The designed oligomer was named PNA-DR. The PNA-DR oligomer was synthesized manually according to the solid phase synthesis protocol16,17,18 using Fmoc/Bhoc-protected PNA monomers, as shown in Figure 1. The PNA chain was grown on Rink Amide-AM resin in a stepwise manner, which was downloaded to approximate 0.12 mmol/g in the first building block by capping the remaining sites. Typically, the PNA was synthesized by repeating the steps including Fmoc removal and monomer coupling and capping using a 12 μmol scale. Piperidine in DMF (20%, v/v) was used to remove the Fmoc-protecting group, while PNA coupling was performed using a N-methylpyrrolidone solution containing PNA monomer (4 equiv), 2-(7-azo-benzotriazole)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (3.6 equiv) and N-methylmorpholine (8 equiv) for 90 minutes. A mixture of acetic anhydride and 2, 6-lutidine in DMF (5:6:89, v/v/v) was used as the capping reagent. After every step, the resin was washed four times with DMF to remove the remaining reactants. The maleimidopropionic acid group was conjugated at the end following the same method. After all couplings, the resin was washed with DMF, methanol, and dichloromethane three times each and then dried in vacuum. The PNA was recovered by full deprotection and cleavage from the resin by employing a cocktail containing trifluoroacetic acid, m-cresol, and water (95:3:2, v/v/v) for 2 hours. The reaction product was filtered, concentrated by rotary evaporation and then precipitated using excess cold ether. The precipitate was collected using centrifugation and then dried under vacuum. The product was confirmed by MALDI-TOF mass spectrum on an Axima TOF2 mass spectrometer (Shimadzu, Kyoto, Japan).
Conjugation, purification, and identification of Tat-PNA-DR. The Tat peptide (NH2-GRKKRRQRRRPPGC-COOH) was obtained from GL Biochem (Shanghai, China). The cysteine in the C-terminus was designed to react with the maleimide group of PNA-DR to form the Tat-PNA-DR maleimide conjugate,30,31 as shown in Figure 1a. Briefly, PNA-DR and Tat were dissolved in distilled water at the mole ratio of 1:2 and then stirred for 2 days under a steady flow of N2 to ensure anaerobic conditions. The solution was purified primarily via dialysis in a dialysis bag (mw cut-off = 2,000) against distilled water and then freeze dried. The obtained Tat-PNA product was analyzed using MALDI-TOF mass spectrometry and further purified using HPLC. HPLC was performed using a preparative C18 reversed phase column with a 0.1% trifluoroacetic acid/H2O/acetonitrile gradient (10–30%) at 1.0 ml/minute for 45 minutes on a Gilson FAST Prep HPLC system. Every major peak was collected and verified by MALDI-TOF. The correct fraction was selected and freeze dried to obtain the purified final product, the purity of which was measured using analytical HPLC. Similarly, a two-site mutant of Tat-PNA-DR (Tat-PNA-DRm, targeting to RNA oligonucleotides 5′-UGCACCUAUGC-3′) was synthesized, conjugated, and identified according to the above protocols.
Cell culture. The HepG2.2.15, HepG2, and L-02 cells used in this study were cultured in Dulbecco's modified Eagle's medium (Invitrogen, CA) supplemented with 10% (vol/vol) fetal bovine serum (Gibco), 50 units/ml penicillin and 50 μg/ml streptomycin at 37 °C in a 5% carbon dioxide incubator.
MTT assay. HepG2.2.15, HepG2 and L-02 cells were seeded in 96-well plates (104 cells per well) and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum in 5% CO2 at 37 °C for 24 hours. Then, Tat-PNA-DR was added to the medium in a series of concentrations. After 48 hours of incubation, 20 μl of MTT solution (5 mg/ml in PBS buffer; Invitrogen) was added to the medium in each well, and the plate was incubated in 5% CO2 at 37 °C for 4 hours. Then, the medium was replaced by 100 μl DMSO. The plate was then gently shaken for 10 minutes at room temperature to completely dissolve the precipitated crystal purple formazan. The absorbance was measured using a microplate reader at a wavelength of 570 nm.
Quantification of HBeAg, HBsAg, and HBV DNA in the culture medium. HepG2.2.15 cells were seeded in 24-well plates at 5 × 104 cells per well and incubated at 37 °C for 12 hours. Then, we treated the cells with various concentrations of Tat-PNA-DR for 48 hours. HBV DNA in the culture medium was extracted and measured using real-time PCR (QIAGEN, Valencia, CA). HBeAg and HBsAg levels were quantitated using an ELISA kit.
Western blot analysis. Cells were lysed in radioimmunoprecipitation assay buffer (50 mmol/l Tris-HCl [pH 7.5], 150 mmol/l sodium chloride, 1% Nonidet P40, and 0.5% sodium deoxycholate) with phenylmethylsulfonyl fluoride. Cell lysates were electrophoresed in sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose membranes (Millipore, Bedford, MA). The antibodies used for probing were as follows: mouse monoclonal anti-HBsAg (1:200 dilution, Abcam, Cambridge, UK), rabbit polyclonal anti-HBV core (1:500 dilution, Dako-Cytomation, Carpinteria, CA), mouse monoclonal anti-Hep B xAg (1:200 dilution, Santa Cruz Biotechnology, CA), mouse monoclonal anti-Hep B Pol (1:200 dilution, Santa Cruz Biotechnology, CA), and anti-β-actin (1:5,000 dilution, Santa Cruz Biotechnology, CA) were used as primary antibodies, and the secondary antibodies were conjugated to horseradish peroxidase.
Southern and northern blot analyses. HBV capsid DNA was collected from cytoplasm using a viral DNA kit (OMEGA bio-tek, GA, USA). The samples were analyzed using a standard southern blot procedure. HBV DNA was then detected using a DIG-labeled probe that was generated using a PCR DIG Probe Synthesis Kit (Roche, Basel, Switzerland). HepG2.2.15 cell total RNA was extracted using TRIzol reagent (Invitrogen, CA, USA) following the manufacturer's instructions and then electrophoresed in a 1.5% formaldehyde agarose gel. HBV RNAs were detected using the same DIG probe that was used for southern blot analysis. GAPDH was also detected using a DIG probe as an internal control.
Hemolysis assay. Erythrocytes were obtained from fresh human blood that was centrifuged for 10 minutes at 1,000×g and washed three times using HEPES buffer. Then, the cells were resuspended in normal saline and seeded in 96-well plates (107 cells per well). Tat-PNA-DR was added in a series of concentrations, normal saline was used as the negative control and 0.1% triton-X100 was used as the positive control. Plates were incubated at 37 °C for 1 hour. The supernatants were collected after centrifugation for 10 minutes at 1,000×g, and the absorbance of hemoglobin was measured at a wavelength of 570 nm.
Serum stability testing. Tat-PNA-DR was incubated with 10% serum or ddH2O at 37 °C for 4, 8, 12 and 24 hours. Then, Tat-PNA-DR was added to HepG2.2.15 cells at 10 μmol/l and the cells were incubated at 37 °C in 5% CO2 for 48 hours. The concentrations of HBeAg and HBsAg were detected using ELISA, and the antiviral activity was then evaluated.
Acute toxicity and immunogenicity in mice. All animal studies were approved by the Institutional Animal Care and Use Committee at Wuhan University. BALB/c mice were treated with Tat-PNA-DR in a series of concentrations via tail intravenous injections. An equal volume of normal saline was used as the negative control. All of the mice were fed in a SPF laboratory animal room for another 7 days. The mortality and body weights of the mice were recorded every day. Mice sera was collected each day, and then alanine aminotransferase and aspartate aminotransferase levels were measured by blood biochemistry tests. To test the antigenicity of Tat-PNA-DR, IgG and IgM levels from mice sera were measured by ELISA.
Anti-HBV assay in an acute hepatitis B mouse model. The in vivo anti-HBV activity of Tat-PNA-DR was analyzed using a mouse model of acute hepatitis B. Male BALB/c mice (6- to 7-week-old) were injected with 20 μg of pUC18-HBV1.3 in a volume of normal saline equivalent to 8% of the body mass of each mouse. All of the plasmid solution was delivered within 5 seconds. After the mice were fed under the same conditions for 24 hours, Tat-PNA-DR was injected into the mice via the tail vein at a dose of 5.05 mg/kg. Serum was collected after another 24 hours, and livers were obtained by dissection. The mass of the livers was weighed. The amount of HBeAg, HBsAg, and HBV DNA in the serum was quantitated using the same methods mentioned for the HepG2.2.15 cell culture medium assays. The distribution and density of HBV core proteins in liver tissues were analyzed using Quantum Dots Fluorescence Immunoassays.
Anti-HBV assay in a long-term hydrodynamics HBV mouse model. A long-term hydrodynamics HBV mouse model was used to measure the long-acting anti-HBV activity of Tat-PNA-DR. pAAV/HBV1.2 plasmid was used to construct the model. Similar to the acute hepatitis B mouse model, male BALB/c mice (6- to 7-week-old) were injected with 20 μg of pAAV/HBV1.2 in a volume of normal saline equivalent to 8% of the body mass of each mouse. All of the plasmid solution was delivered within 5 seconds. 24 hours after the injection of plasmid (day 0), 5.05 mg/kg Tat-PNA-DR or Tat-PNA-DRm was injected by tail vein. A second injection was performed at day 7. Mice blood was collected every 3 days. The amount of HBeAg, HBsAg, and HBV DNA in the mouse serum was quantitated as described before.
Real-time PCR assay of HBV gene mRNA. HepG2 cells were seeded in six-well plates at 5 × 105 per well. After 12 hours of incubation, cells were transiently transfected with constructs encoding HBeAg, HBsAg, HBV core protein, x protein or RT using Lipofectamine 2000 (Invitrogen). The Tat-PNA-DR treatment was administered 24 hours after transfection. Normal saline was used as the negative control. After incubating the cells at 37 °C in 5% CO2 for 48 hours, the cells were harvested. Total RNA was extracted using TRIzol reagent and quantified using a SYBR Green PCR assay kit and an ABI 7500 system after it was reverse transcribed into cDNA using a First-Strand Synthesis Supermix kit (Invitrogen, CA, USA). The relative quantity of mRNA of the HBV genes was calculated and normalized to that of GAPDH.
In vitro binding assay. Tat-PNA-DR was incubated with biotin-labeled oligonucleotides and competitors in gel-shift binding buffer for 30 minutes at room temperature. Competitors were incubated with Tat-PNA-DR for 5 minutes before the addition of the biotin-labeled oligonucleotides for competition assays. Samples were then electrophoresed in 5% native polyacrylamide gels and transferred to nitrocellulose membranes (Millipore, MA). The signals were detected using a Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher scientific, MA). The sequences of the oligonucleotides were the same as that of DR (5′-UUCACCUCUGC-3′). Two random bases (5′-UGCACCUAUGC-3′) were mutated to analyze the specificity of Tat-PNA-DR binding.
SUPPLEMENTARY MATERIAL Figure S1. Cellular localization and uptake of Tat-PNA-DR in HepG2.2.15 cells. HepG2.2.15 cells were incubated with FITC-labeled Tat-PNA-DR for 4 h, 24 h and 48 h. At the indicated timepoints, cells were washed and analyzed using confocal microscopy.
Acknowledgments
This work was supported by grants from the National Science Fund of China for Excellent Young Scholar (No. 31422049), National Natural Science Fund in China (No. 31572289), Hubei Science Fund for Excellent Scholars (No. 2015CFA042), Fundamental Research Funds for the Central Universities in China (Nos. 2042014kf0205 and 2042015kf0255), and China Scholarship Council (No. 201308420306). We thank Ying Zhu (College of Life Sciences, Wuhan University, Wuhan, China) for plasmids: pUC18-HBV1.3, pcDNA-HBeAg, pcDNA-HBsAg, pcDNA-HBV core, pcDNA-HBx, and pcDNA-HBV RT.
Supplementary Material
References
- Beasley, RP (1988). Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61: 1942–1956. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, D and Thio, CL (2010). Review of hepatitis B therapeutics. Clin Infect Dis 51: 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, TM, Guo, H and Guo, JT (2007). Molecular virology of hepatitis B virus for clinicians. Clin Liver Dis 11: 685–706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, WL and Chung, RT (2010). Viral hepatocarcinogenesis. Oncogene 29: 2309–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish, RG, Satishchandran, C, Young, M and Pachuk, C (2011). RNA interference and its potential applications to chronic HBV treatment: results of a Phase I safety and tolerability study. Antivir Ther 16: 547–554. [DOI] [PubMed] [Google Scholar]
- Nkongolo, S, Ni, Y, Lempp, FA, Kaufman, C, Lindner, T, Esser-Nobis, K et al. (2014). Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol 60: 723–731. [DOI] [PubMed] [Google Scholar]
- Ray, A and Nordén, B (2000). Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J 14: 1041–1060. [DOI] [PubMed] [Google Scholar]
- Nielsen, PE and Egholm, M (1999). An introduction to peptide nucleic acid. Curr Issues Mol Biol 1: 89–104. [PubMed] [Google Scholar]
- Pellestor, F and Paulasova, P (2004). The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur J Hum Genet 12: 694–700. [DOI] [PubMed] [Google Scholar]
- Bai, H, You, Y, Yan, H, Meng, J, Xue, X, Hou, Z et al. (2012). Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials 33: 659–667. [DOI] [PubMed] [Google Scholar]
- Ahn, DG, Shim, SB, Moon, JE, Kim, JH, Kim, SJ and Oh, JW (2011). Interference of hepatitis C virus replication in cell culture by antisense peptide nucleic acids targeting the X-RNA. J Viral Hepat 18: e298–e306. [DOI] [PubMed] [Google Scholar]
- Robaczewska, M, Narayan, R, Seigneres, B, Schorr, O, Thermet, A, Podhajska, AJ et al. (2005). Sequence-specific inhibition of duck hepatitis B virus reverse transcription by peptide nucleic acids (PNA). J Hepatol 42: 180–187. [DOI] [PubMed] [Google Scholar]
- Turner, JJ, Ivanova, GD, Verbeure, B, Williams, D, Arzumanov, AA, Abes, S et al. (2005). Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res 33: 6837–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger, A and Nassal, M (1996). Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J Virol 70: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal, M and Schaller, H (1996). Hepatitis B virus replication—an update. J Viral Hepat 3: 217–226. [DOI] [PubMed] [Google Scholar]
- Beck, F (2002). Solid phase synthesis of PNA oligomers. Methods Mol Biol 208: 29–41. [DOI] [PubMed] [Google Scholar]
- Bendifallah, N, Rasmussen, FW, Zachar, V, Ebbesen, P, Nielsen, PE and Koppelhus, U (2006). Evaluation of cell-penetrating peptides (CPPs) as vehicles for intracellular delivery of antisense peptide nucleic acid (PNA). Bioconjug Chem 17: 750–758. [DOI] [PubMed] [Google Scholar]
- Liu LH, Li ZY, Rong L, Qin SY, Lei Q, Cheng H et al. (2014). Self-assembly of hybridized peptide nucleic acid amphiphiles. Acs Macro Lett 3: 467–471. [DOI] [PubMed] [Google Scholar]
- Eguchi, A, Akuta, T, Okuyama, H, Senda, T, Yokoi, H, Inokuchi, H et al. (2001). Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J Biol Chem 276: 26204–26210. [DOI] [PubMed] [Google Scholar]
- Fittipaldi, A, Ferrari, A, Zoppé, M, Arcangeli, C, Pellegrini, V, Beltram, F et al. (2003). Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J Biol Chem 278: 34141–34149. [DOI] [PubMed] [Google Scholar]
- Wadia, JS, Stan, RV and Dowdy, SF (2004). Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med 10: 310–315. [DOI] [PubMed] [Google Scholar]
- Yang, PL, Althage, A, Chung, J and Chisari, FV (2002). Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA 99: 13825–13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z, Hong, W, Zeng, Z, Wu, Y, Hu, K, Tian, X et al. (2012). Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4α in vitro and in vivo. J Biol Chem 287: 30181–30190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, LR, Wu, HL, Chen, PJ and Chen, DS (2006). An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci USA 103: 17862–17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, HY, Cheng, J, Xing, CY, Wang, JJ, Su, R, Wei, XY et al. (2011). Evaluation of hepatitis B viral replication and proteomic analysis of HepG2.2.15 cell line after knockdown of HBx. Hepatobiliary Pancreat Dis Int 10: 295–302. [DOI] [PubMed] [Google Scholar]
- Lucifora, J, Arzberger, S, Durantel, D, Belloni, L, Strubin, M, Levrero, M et al. (2011). Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 55: 996–1003. [DOI] [PubMed] [Google Scholar]
- Benhenda, S, Cougot, D, Buendia, MA and Neuveut, C (2009). Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res 103: 75–109. [DOI] [PubMed] [Google Scholar]
- Xie, Q, Zhang, S, Wang, W, Li, YM, Du, T, Su, XL et al. (2012). Inhibition of hepatitis B virus gene expression by small interfering RNAs targeting cccDNA and X antigen. Acta Virol 56: 49–55. [DOI] [PubMed] [Google Scholar]
- Wooddell, CI, Rozema, DB, Hossbach, M, John, M, Hamilton, HL, Chu, Q et al. (2013). Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol Ther 21: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi, SK and Nielsen, PE (2002). Synthesis of PNA-peptide conjugates. Methods Mol Biol 208: 43–52. [DOI] [PubMed] [Google Scholar]
- Hoyle, CE, Lowe, AB and Bowman, CN (2010). Thiol-click chemistry: a multifaceted toolbox for small molecule and polymer synthesis. Chem Soc Rev 39: 1355–1387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.