Abstract
Epidermolytic palmoplantar keratoderma (EPPK) is a relatively common autosomal-dominant skin disorder caused by mutations in the keratin 9 gene (KRT9), with few therapeutic options for the affected so far. Here, we report a knock-in transgenic mouse model that carried a small insertion–deletion (indel) mutant of Krt9, c.434delAinsGGCT (p.Tyr144delinsTrpLeu), corresponding to the human mutation KRT9/c.500delAinsGGCT (p.Tyr167delinsTrpLeu), which resulted in a human EPPK-like phenotype in the weight-stress areas of the fore- and hind-paws of both Krt9+/mut and Krt9mut/mut mice. The phenotype confirmed that EPPK is a dominant-negative condition, such that mice heterozygotic for the K9-mutant allele (Krt9+/mut) showed a clear EPPK-like phenotype. Then, we developed a mutant-specific short hairpin RNA (shRNA) therapy for EPPK mice. Mutant-specific shRNAs were systematically identified in vitro using a luciferase reporter gene assay and delivered into Krt9+/mut mice. shRNA-mediated knockdown of mutant protein resulted in almost normal morphology and functions of the skin, whereas the same shRNA had a negligible effect in wild-type K9 mice. Our results suggest that EPPK can be treated by gene therapy, and this has significant implications for future clinical application.
Keywords: epidermolytic palmoplantar keratoderma, indel, knock-in, Krt9 gene (mouse), shRNA therapy
Introduction
Keratins are a superfamily of cytoskeletal filament-forming proteins that provide structural stability and flexibility in the skin and other epithelia. They play a critical role in maintaining the shape of keratinocytes, building the intermediate filament (IF, 10 nm in diameter) networks, and providing stability to the tissue.1,2,3 Among the keratins, keratin 9 (K9), a type-I IF protein, is prominently expressed in the suprabasal cells of the palms and soles, and the thick “primary epidermal ridges” form a specialized tissue designed to withstand more mechanical trauma than other glabrous skin.1,4 Mutations in the K9 gene (KRT9) can cause epidermolytic palmoplantar keratoderma (EPPK, OMIM: 144200), a relatively common autosomal-dominant skin disorder characterized by diffuse yellowish thickening of the skin on the palms and soles with distinct erythematous borders, with or without knuckle pads, digital mutilation, and camptodactyly, during the first weeks or months after birth.5,6,7 Previous studies of the Krt9-null mouse indicated that deficiency of K9 causes calluses marked by hyperpigmentation exclusively localized to the stress-bearing footpads.8
None of the keratin defects have effective treatments so far.1,2,3,4 Therefore, it has been proposed that mutation-specific small interfering RNA (siRNA) can be used to reduce the synthesis of abnormal protein in dominant keratin disorders with little or no effect on the wild-type allele, leading to a more functional cytoskeleton and thus greater tissue integrity.8,9 Studies of a transgenic mouse model of epidermolysis bullosa simplex have demonstrated that even 50% knockdown of the mutant allele prevents the development of epidermal blistering.10 The expression of mutant K6a, K14, K5, and K9 alleles is effectively downregulated by mutation-specific siRNAs.9,11,12,13,14 Development of this approach has now progressed to an efficacious small-scale human clinical trial showing significant reduction in plantar keratoderma.15 Thus, siRNA-mediated downregulation of mutant keratins may present new and specific targets in the treatment of genodermatoses.
EPPK provides a better model than other keratin disorders for RNAi therapy because (i) it involves only one disease-causing gene, KRT9, or KRT1 (with mild symptoms), (ii) K9 forms a cytoskeleton together with other keratins, and its expression is restricted to the upper strata of the palmoplantar epidermis, and (iii) most KRT9 mutations occur within codons 163 (p.Arg163) and 157 (p.Met157) (www.interfil.org). Notably, the plantar epidermis of knockout heterozygous Krt9-null (Krt9+/-) mice display a normal phenotype without disease, demonstrating that a single copy of K9 is sufficient to maintain healthy skin in vivo, and mutation-specific siRNA therapy may be feasible.8,9
The first indel mutation within exon 1 of KRT9, c.500delAinsGGCT (p.Tyr167delinsTrpLeu), was identified in our laboratory.16 Here, we established a Krt9/c.434delAinsGGCT (p.Tyr144delinsTrpLeu) knockin transgenic mouse model which had a phenotype similar to the clinical symptoms of human EPPK and evaluated the feasibility and efficiency of mutant-specific short hairpin RNA (shRNA) therapy in vivo.
Results
Krt9-mutant knockin mice develop an EPPK-like phenotype
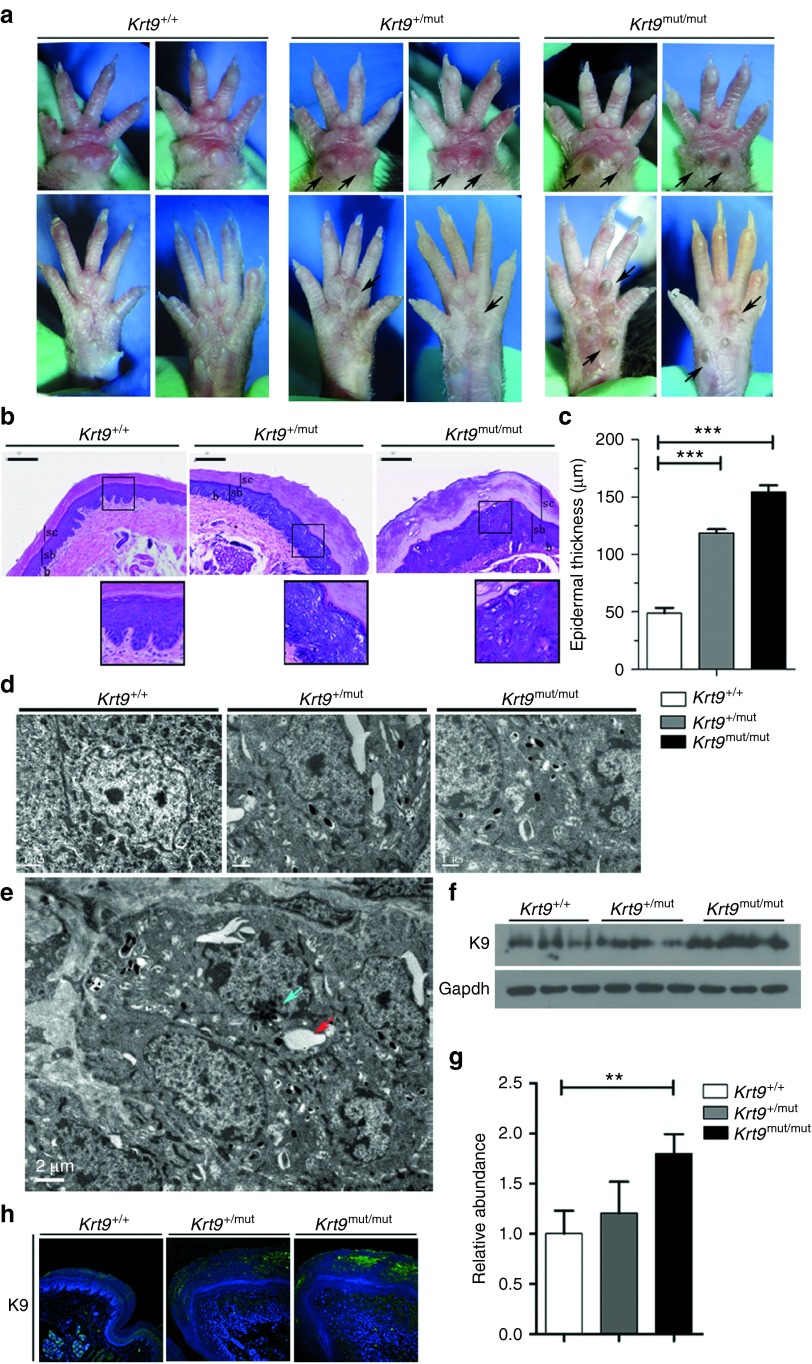
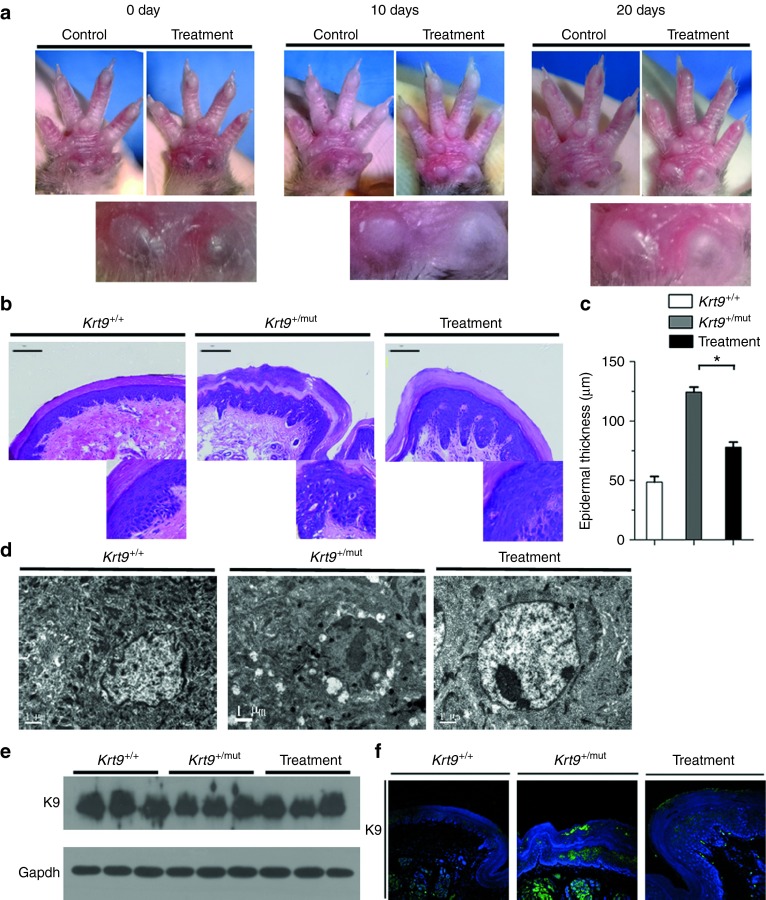
Krt9/c.434delAinsGGCT (p.Tyr144delinsTrpLeu) homozygous mutant (Krt9mut/mut), heterozygous mutant (Krt9+/mut), and wild-type (wt; Krt9+/+) mice were maintained. Hyperpigmented calluses appeared on the weight-stress areas of the fore- and hind-paws of Krt9mut/mut mice at 3–4 weeks and of Krt9+/mut mice at 5–6 weeks. A clear phenotype in Krt9+/mut mice was not observed until 11–12 weeks, almost 4 weeks later than in Krt9mut/mut mice (data not shown). Hyperpigmented calluses and acanthosis in the hind-paws were as severe as those in the fore-paws of adult Krt9-mutant mice (Figure 1a). Hematoxylin and eosin (H & E) staining showed remarkable footpad epidermal thickening, expansion of the epidermis and massive hyperkeratosis, papillomatous hyperplasia, marked hypergranulosis and acanthosis, and vacuolated cells in the upper portion of the epidermis with atypical shapes (Figure 1b). The thickness of footpads in Krt9+/mut mice was greater than twofold that in Krt9+/+ mice, while in Krt9mut/mut mice it was ~3-fold thicker (Figure 1c). Ultrastructural examination showed cytolysis of keratinocytes and abnormal aggregation of tonofilaments in the suprabasal layers of the epidermis (Figure 1d,e). Both western blotting and semi-quantitative western blotting analysis revealed that the expression of K9 was slightly higher in Krt9+/mut mice and significantly higher in Krt9mut/mut mice than that in Krt9+/+ mice (Figure 1f,g). Immunofluorescent staining for K9 in Krt9+/+, Krt9+/mut, and Krt9mut/mut mice showed that all the expressed K9 in the suprabasal layers of the palmoplantar epidermis (Figure 1h). The distribution of K9 in Krt9+/+ was smooth and uniform, while abnormal accumulations occurred in Krt9+/mut and Krt9mut/mut mice.
Figure 1.
EPPK-like phenotype in Krt9-mutant (Krt9+/mut and Krt9mut/mut) mice. (a) Gross appearance of the fore-paws (upper panels) and hind-paws (lower panels) of wild-type (Krt9+/+), heterozygous (Krt9+/mut), and homozygous (Krt9mut/mut) 12-week-old mice. Arrows indicate hyperpigmented calluses on the major stress-bearing footpads of adult Krt9+/mut and Krt9mut/mut mice. (b) H & E-stained cross-sections of the footpads of the fore-paws of Krt9+/+, Krt9+/mut, and Krt9mut/mu littermates (n = 4). The boxed areas indicate vacuolated cells with atypical shapes. sc, stratum corneum; sb, suprabasal layer; b, basal layer; bars = 100 μm. (c) Thickness of epidermis in H & E-stained footpads. The histogram depicts the mean thickness of each region calculated from 10 independent measurements taken over the length of a representative section from each genotype (n = 3), and the error bars represent the standard error of the mean (***P < 0.001). (d) Transmission electron microscopy images of the footpads of the fore-paws from Krt9+/mut and Krt9mut/mut mice showing increased numbers of melanin cells. (e) The cytoskeleton of cells in Krt9+/mut mice was disrupted by cell lysis (green arrow), and abnormal aggregation of keratin filaments or proteins (red arrow). (f–h) Immunoblotting (f) in the footpads of fore-paws from Krt9+/+, Krt9+/mut, and Krt9mut/mut mice (n = 3) with K9, and semi-quantitative immunoblotting analyses relative to (g) Gapdh (**P < 0.01), together with (h) immunofluorescent staining.
Dominant-negative mutation of K9 induces an imbalance in a subset of palmoplantar keratins, irregular proliferation and differentiation, and impaired cell mobility
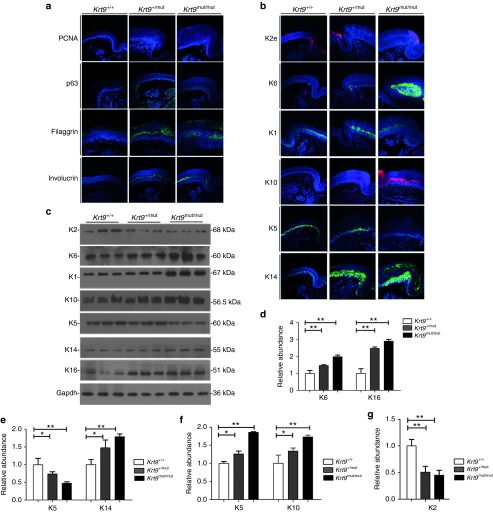
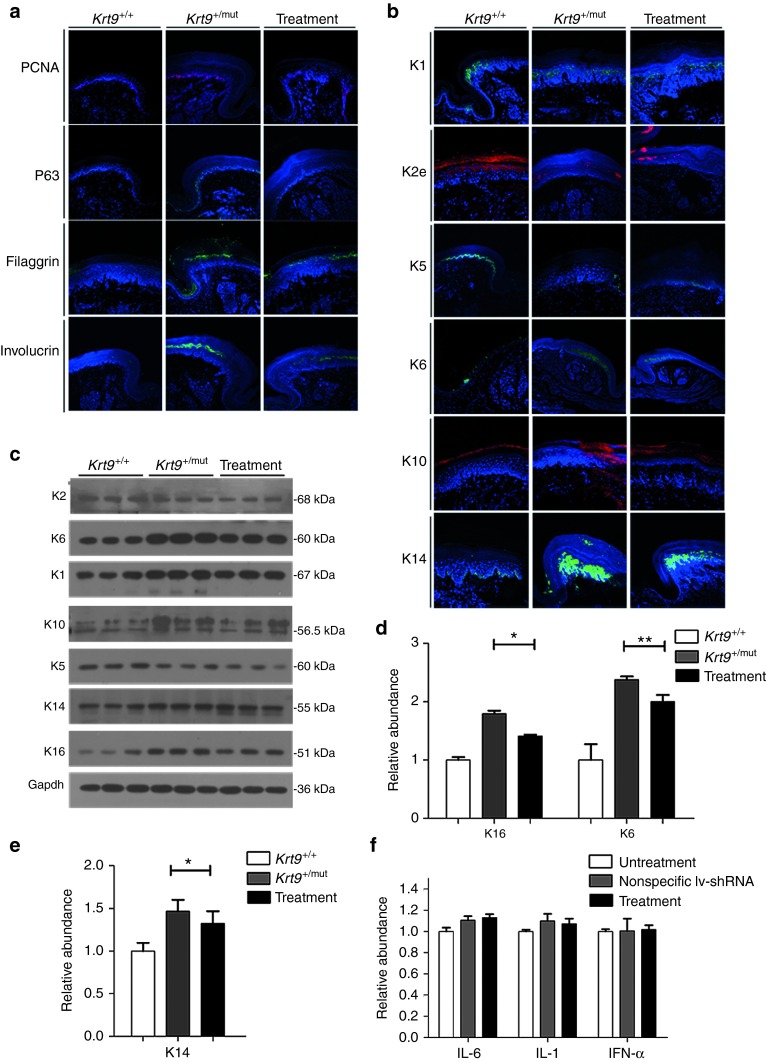
Immunofluorescent staining for the proliferation marker proliferating cell nuclear antigen (PCNA) and the transcription factor p63, a key regulatory element of basal keratinocyte proliferation, differentiation, and survival, revealed an increase in the number and staining intensity of PCNA- and p63-positive cells in the footpads of K9-mutant mice (Figure 2a).17 The involucrin staining intensity increased significantly in the suprabasal layer. In addition, filaggrin, the stratum granulosum marker, was more intermittent and disorganized in the footpads of Krt9+/mut and Krt9mut/mut mice (Figure 2a).
Figure 2.
Dominant-negative mutation in K9 induces an imbalance in a subset of palmoplantar keratins, and disrupts proliferation, differentiation, and migration. (a and b) Immunofluorescent labeling (a) of the proliferation markers PCNA and p63, and the differentiation markers filaggrin and involucrin, together with (b) the palmoplantar keratins K5, K14, K1, K10, K2, and K6 in the footpads of fore-paws from Krt9+/+, Krt9+/mut, and Krt9mut/mut littermates (n = 3). (c) Immunoblotting of the footpads of fore-paws against K5, K14, K1, K10, K2, K6, K16, and Gapdh antibodies, and semi-quantitative immunoblotting analyses relative to (d–g) Gapdh. Mean relative abundances (n = 3) and error bars (mean ± SEM) are shown (*P < 0.05; **P < 0.01). PCNA, proliferating cell nuclear antigen.
The expression of the stress-response and wounding-healing keratins, K6 and K16, was significantly higher in Krt9+/mut and Krt9mut/mut mice than that in wt mice (Figures 2b–d). The localization of K14 in the footpads of Krt9+/mut and Krt9mut/mut mice extended into the suprabasal epidermis and as far as the uppermost granular layer, with abnormal aggregation (Figure 2b). Although both K5 and K14 were appropriately localized to the basal layer, semi-quantitative western blotting analysis showed an increase in K14 but a decrease in K5 (Figure 2b,c,e). These results suggested that the synthesis of various epidermal keratins is regulated individually. The suprabasal keratins K1 and K10 both increased significantly in response to dysfunction of K9 in the footpads (Figure 2c,f), with local expansion and abnormal aggregation (Figure 2b). Semi-quantitative western blotting analysis of K2 revealed a 60% decrease in the footpads of Krt9+/mut and Krt9mut/mut mice compared with normal epidermis (Figure 2g).
Dominant-negative mutation of K9 does not impair the epidermal barrier
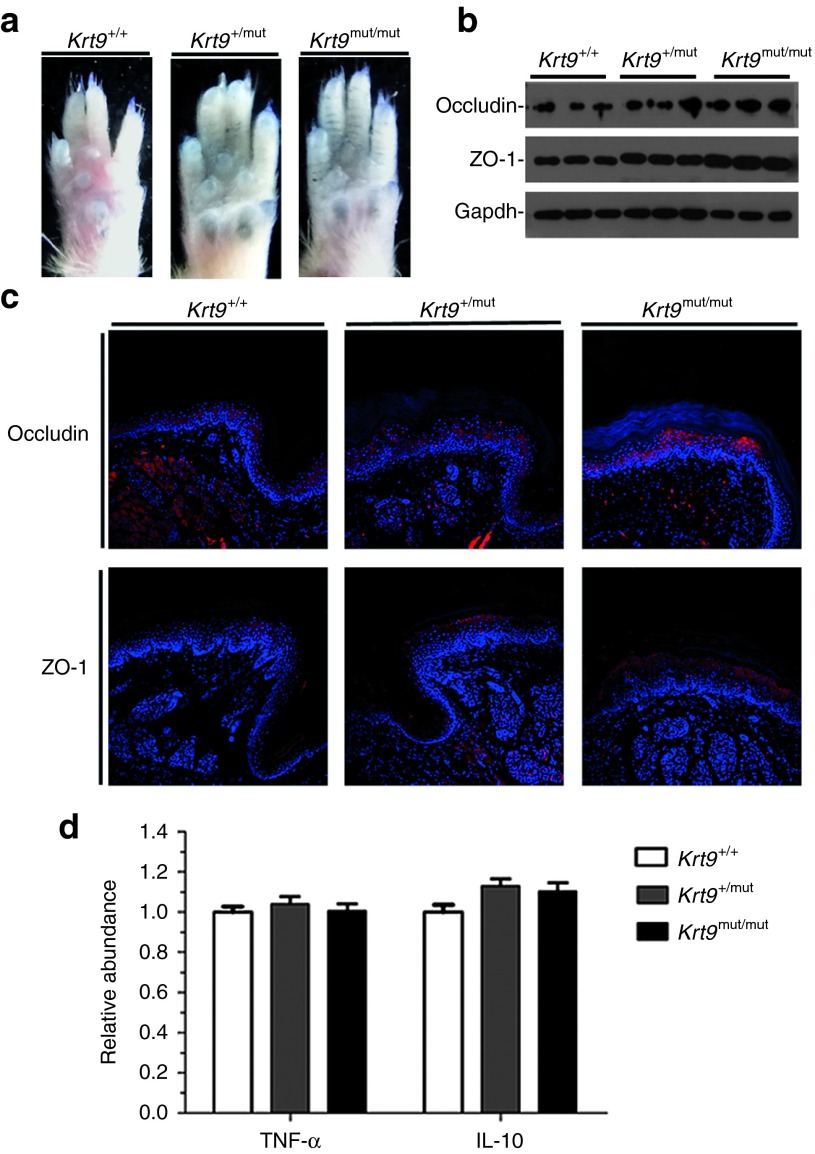
The dye-penetration assay is the gold standard for evaluating outside-in epidermal barrier function.18 In this method, toluidine blue staining is carried out in the paws of adult animals.19 Stained areas did not appear on the fore- and hind-paws of wt and Krt9-mutant mice, suggesting that K9 dysfunction does not impair the epidermal barrier (Figure 3a).
Figure 3.
Dominant-negative mutation of K9 does not impair the epidermal barrier. (a) Toluidine blue penetration in the footpads of fore-paws from adult Krt9+/+, Krt9+/mut, and Krt9mut/mut littermates. (b) Immunoblotting in the footpads of fore-paws from adult Krt9+/+, Krt9+/mut, and Krt9mut/mut littermates (n = 3) with markers of the TJ barrier, occludin, and ZO-1. (c) Immunofluorescent labeling of the footpads of fore-paws against occludin and ZO-1. (d) qPCR analysis of markers associated with inflammation, TNF-α, and anti-inflammatory IL-10.
The tight-junction (TJ) barrier is located in the granular layer of the epidermis. Occludin is important in TJ stability and barrier function,20 while zonula occludens-1 (ZO-1), located on the cytoplasmic membrane surface of intercellular TJs, is involved in signal transduction at the cell–cell junctions.21 TJ proteins are strongly regulated by inflammatory factors such as TNF-α and the anti-inflammatory cytokine IL-10. To evaluate the barrier functions of TJs in K9-mutant mice, we assessed the expression of occludin and ZO-1, together with TNF-α and IL-10. Western blotting analysis revealed an increase in occludin and ZO-1, consistent with the results of immunofluorescent staining (Figure 3b,c). But TNF-α and IL-10 showed no significant changes in Krt9+/+, Krt9 +/mut, and Krt9mut/mut mice by real-time qPCR (Figure 3d).
Mutant-specific siRNA knockdown of the Krt9 mutant allele is specific and effective in vitro
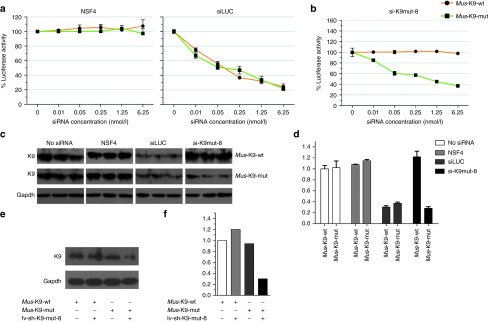
A firefly luciferase reporter assay was developed to screen all 16 possible allele-specific siRNAs in order to identify one that inhibited the Krt9 mutant allele specifically and effectively but had little or no effect on the wt allele, as required by a realistic model of continuous Krt9 expression. Stably-transfected pfLUC-Mus-ex1Krt9-c.434delAinsGGCT/wt (pfLUC-ex1Krt9-mutant/wt) and pfLUC-Homo-ex1KRT9/c.500delAinsGGCT/wt (pfLUC-ex1KRT9-mutant/wt) 293T cell lines were used for selection. A positive control siRNA targeting firefly luciferase (siLUC) and a nonspecific control siRNA (NSC4) were used to define the expression levels of pfLUC-ex1Krt9-mutant/wt and pfLUC-ex1KRT9-mutant/wt and detect potential knockdown. The results showed that NSC4 did not affect the expression of either pfLUC-ex1Krt9-mutant/wt or pfLUC-ex1KRT9-mutant/wt, whereas siLUC inhibited them (Figure 4a and Supplementary Figure S1a). In each experiment, the siRNA concentration was varied from 6.25 nmol/l to 10 pmol/l in fivefold decrements in order to measure the potency separately from the specificity. Among the 16 designed siRNAs tested for Krt9/c.434delAinsGGCT and KRT9/c.500delAinsGGC, the optimal inhibitor for further development was si-K9mut-8, with 70–80% knockdown of expression of the mutant allele at the highest siRNA concentration of 6.25 nmol/l. Expression of both the human and mouse mutant alleles was knocked down, while the wt allele was little affected (Figure 4b and Supplementary Figure S1b). Complete siRNA data sets are shown in Supplementary Figures S2a and S3a.
Figure 4.
RNA interference (RNAi)-mediated independent mutant-specific inhibition of the K9 dominant-negative allele. (a and b) Dominant-negative allele, wild-type K9 (K9-WT), or mutant luciferase construct 293T with pfLUC-ex1Krt9 WT/434delAinsGGCT stable cells were transfected with 0–6.25 nmol/l of (a) the control siRNAs (siLUC or NSC4) and (b) si-K9mut-8, the most promising mutant-specific inhibitor. (c and d) Semi-quantitative analyses of immunoblotting, relative to Gapdh, were used to quantify the specificity and potency of siRNA inhibition (5 nmol/l). (e and f) lv-sh-K9mut-8 affects 293T cells with pfLUC-ex1Krt9WT/434delAinsGGCT. Semi-quantitative analyses of immunoblotting, relative to Gapdh, were used to quantify the specificity and potency of shRNA inhibition. Results are the mean ± SEM.
Western blotting was analyzed using an antibody against keratin 9 to validate the allele-specific siRNA. si-K9mut-8 siRNA was transfected at 5 nmol/l into stable pfLUC-ex1Krt9-mutant/wt and pfLUC-ex1KRT9-mutant/wt 293T cells, and successfully inhibited the expression of mutant K9, with no discernible effect on the wt allele (Figure 4c and Supplementary Figure S1c). Therefore, si-K9mut-8 was chosen for further experiments.
Based on the above data, the lentiviral shRNA vector lv-sh-K9mut-8, which selectively expressed si-K9mut-8, was synthesized (Genechem, Shanghai, China). Stable pfLUC-ex1Krt9-mutant/wt cells were treated directly with lv-sh-K9mut-8, and a notable knockdown was found, reducing expression of the mutant allele by almost 80% (Figure 4e,f). To investigate off-target effects, HaCat cells were treated with lv-sh-K9mut-8. Fluorescence microscopy showed that the infection rate was close to 100%. The main related keratins were assessed as well. No evident reduction in any keratins was found compared with untreated HaCat cells (Supplementary Figure S1e). These data suggested that lv-sh-K9mut-8 is highly specific.
Lentivirus-delivered mutant-specific shRNA relieves the EPPK-like phenotype effectively and selectively in vivo
To determine whether lv-sh-K9mut-8 affects the process of hyperkeratosis in vivo, 12-week-old Krt9+/mut mice (n = 4) received an injection of 50 μl lv-sh-K9mut-8 into the right fore-paw every 10 days to deliver a viral dose of 1 × 108 infectious units /ml and imaged before and after each injection. Hyperpigmented calluses in the right fore-paw pad regressed after the first injection (day 10) and were sloughed off after the second injection (day 20) (Figure 5a). The controls, Krt9+/mut mice injected with lentivirus containing nonsilencing shRNA, showed no phenotype change (Supplementary Figure S4a), suggesting that stimulation by neither the injection nor the lentivirus vector had an impact on the EPPK-like phenotype. The results indicated that the lv-sh-K9mut-8 has potential efficacy and specificity in the in vivo environment.
Figure 5.
Hyperpigmented calluses on the forepaws are partly rescued by injection of lentivirus-delivered mutant-specific shRNA. (a) Right fore-paws of Krt9+/mut were injected with shRNA every 10 days for 20 days (n = 4); the left fore-paws served as controls. (b and c) H & E-stained cross-sections of the footpads of fore-paws (b) and (c) the thickness of the epidermis (n = 3). Results are the mean ± SEM. **P < 0.05. (d) Ultrastructural analysis showed reducing of disrupted cytoskeletal integrity and abnormal keratin filament assembly after treatment. (e and f) Immunoblotting (e) and immunofluorescent staining against K9 (f) in the footpads of right fore-paws from Krt9+/mut mice on injection day 20.
H & E staining on day 20 (after the second injection) showed that the epidermal thickness of the footpad decreased, with less expansion of the epidermis and less massive hyperkeratosis. Marked improvement in the arrangement of the suprabasal cells in the upper portion of the epidermis was also observed (Figure 5b,c).
Immunofluorescent staining showed that the abnormal accumulation of K9 in heterozygous mice returned to a smooth distribution after shRNA treatment (Figure 5f). Immunofluorescent staining for PCNA and p63 revealed fewer positive cells in the footpads of treated Krt9 mutant mice than in controls (Figure 6a). Significant decreases in involucrin and filaggrin were also observed, and the expression of K6, K16, and K14 also decreased after treatment (Figure 6b–e).
Figure 6.
Mutant-specific shRNA therapy improves the abnormality of other palmoplantar keratins. (a and b) On injection day 20, immunofluorescent labeling of proliferation and differentiation (a) together with other palmoplantar keratins (b) in the footpads of fore-paws from the treatment group (n = 3). (c) Immunoblotting of the footpads of fore-paws against K5, K14, K1, K10, K2, K6, K16, and Gapdh antibodies. (d and e) Semi-quantitative immunoblotting analyses relative to Gapdh. Mean relative abundances (n = 3) and error bars (*P < 0.05; **P < 0.01) are shown. (f) No increase of IL-6, IL-1, and IFN-α was demonstrated by qPCR.
To investigate the immunostimulatory effect of lv-sh-K9mut-8 therapy, markers associated with the anti-inflammatory gene programs of Toll-like receptor 3 (TLR3) (IL-6, IL-1) and TLR7 (IFN-α), the main mediators of the immune response, were assessed by q-PCR after injection, and no evident change was found (Figure 6f).
Discussion
The indel is a relatively rare type of mutation causing human genetic disease, comprising 1.5% of the mutations encountered in the human genome.22 Several indel hotspots such as “GTAAGT” (and its complement “ACTTAC”) have been noted in 211 different indels underlying genetic diseases.22 Here, the indel hotspot motif of KRT9/c.500delAinsGGCT is “TCTTAC.”16 Our Krt9/c.434delAinsGGCT (p.Tyr144delinsTrpLeu) mutant knock-in mouse model exhibited a human EPPK-like phenotype, including hyperkeratosis calluses on the footpads of both the fore- and hind-paws (Figure 1a). Pathologic analysis showed significant hypergranulosis and acanthosis, vacuolated cells, abnormal clumps of tonofilaments, and keratohyaline granules (Figure 1b,d,e). The EPPK-like phenotype in Krt9mut/mut mice was more severe than that in Krt9+/mut mice, confirming that EPPK is a dominant-negative disease and the heterozygous K9-mutant allele (Krt9+/mut) contributes to EPPK-like pathogenesis. Our studies of heterozygotic and Krt9-null mice reconfirmed that K9 is required for normal palmoplantar physiology and that its dysfunction or loss-of-function causes pathology in vivo.1,2,3,4,8
Stabilization of the epidermis is due to a regulated balance between proliferation, differentiation, and migration, so keratin mutations in humans give rise to epithelial cells displaying perturbations in IF architecture and mechanical fragility.1,2,3,4 In the development of epithelia, p63 functions as a key regulator of cellular proliferation, differentiation, and survival in both physiological and pathological contexts.17 Upregulation of PCNA and p63 indicates that proliferation is stimulated by an imbalance in the expression of keratin. We found that increased expression of involucrin and the appearance of filaggrin in terminal differentiation were also clearly disrupted in the footpad epidermis of Krt9-mutant mice (Figure 2a). Upon commitment to differentiation, specific switches of keratin gene expression have been highly conserved throughout evolution and are paralleled by striking changes in the density and organization of keratin IFs in keratinocytes. Upregulation of the basal epidermal keratins such as K14 in Krt9-mutant mice implied that they complement K9 in the formation of IFs. In addition, K14 did not remain restricted to the basal epidermis and showed abnormal aggregation in the mutant mice (Figure 2b). Increased expression of wound healing-associated keratin K6 in the footpad epidermis of Krt9-mutant mice suggested stressful epithelial stimulation such as chronic inflammation.23 Loss of the normal IF cytoskeleton could lead to increased mechanical fragility of the suprabasal keratinocytes and corneocytes. Hyperproliferation and abnormal differentiation disrupt the process of cornification, impair epidermal homoeostasis, and increase epidermal turnover and thickening, including hyperkeratosis.24
Several mice with keratin modifications, such as Krt1-, Krt10-, or Krt16-null mice, have epidermal barrier dysfunction, so we speculated that the extremely thickened stratum corneum in Krt9-mutant mice may also impair the skin barrier.25,26,27 However, to our surprise, the expression of occludin and ZO-1 increased in these mice (Figure 3b,c). This may be due to the fact that occludin and ZO-1 were located in the granular layer, which was thickened in the footpads of the fore- and hind-paws. No stained area was present within the toluidine blue region in the forepaws of wt and Krt9-mutant adult mice, suggested that the outside-in epidermal barrier had been acquired. These results indicated that the dominant-negative mutation of K9 causes compact hyperkeratosis and does not damage the epidermal barrier.
The EPPK-like phenotype of the Krt9-indel mice was slightly different from that reported in Krt9-null mice.8,9 K1/K10 filaments are believed to be important for epidermal formation because they are highly cross-linked with the cornified envelope. Due to the high degree of sequence homology shared by K10 and K9, not only in the highly-conserved rod domain but also in large parts of the head and tail domains, and their overlapping suprabasal palmoplantar expression patterns, it has been suggested that K10 and K9 have functions in common.1,28 Moreover, it has long been postulated that K9 also interacts with K1 in the palmoplantar epidermis.1,2,3,4 Consequently, it is reasonable to assume that K1 and K10 could be upregulated to compensate for the K9 deficiency in Krt9-null mice. However, no changes in the quantity or distribution of K1/K10 were found in Krt9-null mice.8,9 Interestingly, significant upregulation of both K1 and K10 was found in our Krt9-indel mice (Figure 2b). Considering these two different phenomena, the assumption that increasing K1 and K10 compensates for dysfunction of K9 is not convincing. We suggest that a small in-frame deletion/insertion affecting the rod domain results in conformational changes and the formation of aberrant heteropolymers, which prevents proteins from normal degradation and the differentiation or migration of K1 and K10. The phenotype of intensified hyperpigmentation, increased callus size, and sloughing off every 3 weeks in Krt9-null mice was not observed in our study (Supplementary Figure S4b,c),8,9 and may be due to the different mutation sites. Collectively, these findings suggested that the localized acanthosis and hyperkeratosis phenotype is triggered by an increase in the proliferation state of basal keratinocytes and a more rapid transition towards terminal differentiation, which ultimately impairs terminal differentiation and cornification.
The application of RNAi can be mediated in two ways: chemically synthesized double-stranded small interfering RNA (siRNA), and vector-based short hairpin RNA (shRNA). RNAi therapeutics have shown potential in many animal models, allowing for transition to the clinic, and at least 10 RNAi-based drugs are currently in early-phase clinical trials for cancer and HIV.29,30 shRNA for the treatment of hepatitis B has been approved for clinical trials by the US Food and Drug Administration. These herald a new age of gene therapy for the correction of inherited genetic disorders.31
The skin, as the largest organ, mainly functions as a physical barrier to prevent the entry of foreign pathogens. It forms very thick keratoderma, especially in keratosis, making the delivery of siRNA/shRNA a great challenge.15,32 In addition, the activity of siRNA peaks rapidly, ~24 hours postdelivery, and diminishes within 48 hours. In a study of pachyonychia congenita, siRNAs were injected twice a week, and the keratosis partly degraded in the treated site after 14 weeks of treatment. However, the changes rapidly returned to baseline after the siRNA was discontinued.32 shRNA has several advantages over siRNA: shRNA constructs have high potency and sustained effects using low copy-numbers, resulting in less up- and down-regulation of off-target transcripts than a corresponding siRNA, with activation for longer periods, which reduces the number of injections.33 Thus, we decided to introduce an shRNA lentiviral vector to treat the EPPK mouse. Hyperkeratosis and acanthosis were significantly relieved after injection, suggesting that the EPPK-like phenotype can be rescued by treatment with lentiviral-driven shRNA which inhibits the generation of abnormal K9, decreases the proliferation and terminal differentiation, and enables the partial return to normal physiological processes, without major adverse side-effects (Figure 6).
All RNAi-based therapeutics may have off-target effects as well as immune responses and toxicity associated with the RNAi construct itself or its delivery vehicle.34 Some studies have shown that naked siRNA activates TLR3 on the surface of vascular endothelial cells and triggers nonspecific anti-angiogenic effects in vivo.34,35 The vector-driven shRNA approach to RNAi does not permit specific chemical modification of the silencing construct,36 but it is reasonable to believe that shRNA is less likely to induce an inflammatory response through cytoplasmic dsRNA receptors in vivo because shRNA is spliced by endogenous mechanisms.37 However, both adenoviral- and AAV-based vectors can trigger downstream immune responses, thereby potentially limiting their effectiveness in certain therapeutic settings.36 The use of unmethylated CpG-free plasmid or minimal plasmid vector attenuates this potentially toxic effect.38
Using the endogenous processing machinery, optimized shRNA constructs allow for highly potent and sustainable effects using low copy-numbers resulting in fewer off-target effects and thereby ensuring greater safety.36 When integrated with an effective delivery system, there are compelling data to support the implementation of shRNA as a next-generation therapeutic strategy for keratin disease.31,32 However, a notable difficulty is the intense pain associated with injection, which limits further use of this type of treatment in its current form. Excitingly, a study with a motorized microneedle array device has shown that functional self-delivery of siRNA can be effective with little or no pain; this predicts successful clinical translation.37
In conclusion, we re-confirmed the view that EPPK is an autosomal-dominant inherited disorder, a dominant-negative disease in which heterozygosity for the Krt9-mutant allele (Krt9+/mut) contributes to a compact hyperkeratosis phenotype. We present the first reported ‘‘knock-in'' organotypic model for a dominant type-I keratin mutation. Our data substantiate that shRNA therapy is able to reduce the expression of the K9 indel mutation specifically and potently, preventing the keratosis phenotype. These specific shRNA molecules hold great promise for personalized medicine.
Materials and methods
Animals. Krt9/c.434delAinsGGCT mutant mice were generated using Krt9/c.434delAinsGGCT C57Bl/6NTac embryonic stem cells through homologous recombination (from the Model Animal Research Center of Nanjing University, Nanjing, China).
Mice were on a C57BL/6 genetic background and housed in groups with a 12-hours dark/light cycle and free access to food and water in accordance with the Regulations on Mouse Welfare and Ethics of Nanjing University, China. All procedures were conducted with the approval of the relevant authority. All experiments were conducted on mice between 12 and 16 weeks of age.
Histopathology and immunofluorescence. The fore-paw footpad epidermis from Krt9+/+, Krt9+/mut, and Krt9mut/mut littermates (n = 3/genotype) was dissected and fixed for 4 hours in 4% paraformaldehyde/phosphate-buffered saline at 4 °C. Then tissues were dehydrated, paraffin embedded, and cut at 5 μm. For histopathological analyses, sections were deparaffinized and stained with H & E according to standard protocols. Epidermal thickness was measured using Photoshop software. For immunofluorescence analysis, rehydrated paraffin-embedded sections were washed in phosphate-buffered saline, blocked in 5% normal goat serum for 1 hour at room temperature, incubated with primary antibody (in 2.5% normal goat serum) for 24 hours at 4 °C overnight, washed three times in phosphate-buffered saline, incubated with secondary antibody (in 2.5% normal goat serum) for 1 hour, counterstained with 4′,6-diamidino-2-phenylindole, mounted, and imaged.
Transmission electron microscopy and barrier function assays. For transmission electron microscopy, tissues from the footpads of fore-paws were fixed in 2.5% glutaraldehyde in cacodylate buffer for 24 hours at 4 °C. After rinsing twice with cacodylate buffer, samples were post-fixed in aqueous osmium tetroxide for 2 hours. Then, samples were incubated in uranyl acetate and dehydrated through a graded series of ethanol concentrations. Ultra-thin sections were infiltrated ′with complete embedding resin and stained with uranyl acetate and lead citrate and examined on a transmission electron microscopy (Gatan Erlangshen ES500W; Pleasanton, CA).
To assess outside-in barrier function, toluidine blue staining was done on the paws of adult Krt9+/+, Krt9+/mut, and Krt9mut/mut mice as described.19
Immunoblotting. Proteins were extracted from cells or the footpad tissues of fore- and hind-paws dissected from Krt9+/+, Krt9+/mut, and Krt9mut/mut littermates (n = 3/genotype) and snap-frozen in liquid nitrogen. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto PVDF membranes at 100 V for 70 minutes. The membranes were blocked, washed, and incubated with the primary antibodies overnight at 4 °C. The next day, the membranes were washed and incubated with horseradish peroxidase-conjugated IgG (1:10,000) at room temperature for 1 hour. Signals were detected by enhanced chemiluminescence.
Cell culture and infection of 293T cells with lentivirus. Human 293FT (ATCC, Manassas, VA; #CRL-3249) and 293T (ATCC; #CRL-3216) embryonic kidney cells, and HaCaT (The Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China; #141198) human keratinocytes were grown in DMEM (Gibco, Uxbridge, UK; #1885084) supplemented with 10% fetal bovine serum (Gibco, Uxbridge, UK; #16000–044). Cells were incubated at 37 °C in a humidified atmosphere of 5% CO2 and passaged following standard laboratory procedures. Lentivirus was allowed to directly infect the 293T cells for 24 hours, and puromycin was used for 72 hours. Then, the medium was replaced with new medium to obtain stable cell lines.
Construction of plasmids and shRNAs. pfLUC-Mus-ex1Krt9-c.434delAinsGGCT/wt (pfLUC-ex1Krt9-mutant/wt) and pfLUC-Homo-ex1KRT9/c.500delAinsGGCT/wt (pfLUC-ex1KRT9-mutant/wt) including the 5′ and exon 1 regions were PCR-amplified from genomic DNA isolated from EPPK mice and patients harboring the KRT9 mutations. The following oligonucleotides were used for these PCR amplifications: Mus-Krt9-F: CGGGATCCCTGAGACCGCTTCCTGGCTA; Mus-Krt9-R: ATAAGAATGCGGCCGCTGGCAGTGTTGCTCTCCTCC; Homo-KRT9-F: CGGGATCCGCACTTGGGAGCCGGTAGCACT; Homo-KRT9-R: ATAAGAATGCGGCCGCCTGGTCCTTGAGATCATCAA. Following amplification, the fragments were cloned into the Not I and BamH I restriction sites of the Luciferase reporter plasmid by pCDH-luc (a kind gift from Nanjing University).
Lentiviruses containing pfLUC-ex1Krt9-mutant/wt and pfLUC-ex1KRT9-mutant/wt were produced as follows: 293FT cells were seeded and, at 24 hours after plating, co-transfected using Lipofectamine 2000 (Invitrogen; #11668-019) with 1.0 μg of pfLUC-ex1Krt9/WT, pfLUC-ex1Krt9/434delAinsGGCT, pfLUC-ex1KRT9/WT, or pfLUC-ex1KRT9/500delAinsGGCT plasmid, 0.75 μg psPAX2 (YouBio, Hunan, China; #VT1444), and 0.25 μg pMD2.G (YouBio, Hunan, China; #VT1443). After 48 hours, the medium was replaced with fresh medium. The supernatants were collected, and re-constructed lentiviruses were harvested by centrifugation and then stored at −80 °C for further experiments.
Construction of lentiviral Krt9mut-8-shRNA vector (lv-sh-K9mut-8): The vector was GV248. shRNA template forward strand: 5′-RRM-siRNA sense strand (GGCCTCTTGGCTCATGGAG) + loop (CTCGAG) + RRM-siRNA antisense strand (CTCCATGAGCCAAGAGGCC) + polymerase III terminator (TTTTTTg) was synthesized by Genechem (Shanghai, China).
siRNA design. siRNAs are generally used in a double-stranded 19 + 2 format. Thus, to target the mutant, there are 16 possible positions within the siRNA molecule where the mutant base can be positioned. Whole siRNA sequence walks for K9 mutations are shown in Supplementary Figures S2a and S3a. A positive control siRNA (siLuc) targeting the firefly luciferase gene and a nonspecific control siRNA (NSC4) were synthesized; they had no specific effect and inhibited the expression of luciferase (NSC4: 5′-UAGCGACUAAACACAUCAAUU-3′; siLUC: 5′-GUGCGUUGCUAGUACCAACUU-3′). All siRNAs were synthesized by Ruibo Biotech (Guangzhou, China).
Luciferase reporter assay. A luciferase reporter assay (Promega, Southampton, UK; #E1910) was used to measure the effect of siRNA on the luciferase expression of stable 293T cells with the firefly luciferase reporter constructs (pfLUC-ex1Krt9/WT, pfLUC-ex1Krt9/434delAinsGGCT, pfLUC-ex1KRT9/WT, and pfLUC-ex1KRT9/500delAinsGGCT). Briefly, at 24 hours after plating, stable 293T cells were transfected with 0–6.25 nmol/l of the indicated siRNA using Lipofectamine 2000 (Invitrogen). The luciferase reporter assay (Promega) was performed 24 hours after transfection following the manufacturer's instructions. Luciferase was measured sequentially using a luminometer (Turner Biosystems, CA; #2030-000).
shRNA injections and imaging. A total volume of 50 μl lv-sh-K9mut-8 was injected into the footpads of Krt9+/mut mice (12 weeks) intradermally with a needle, once every 10 days. The injections were administered twice, with each treatment group containing four animals. Images were captured before and after injection. As a control, an equivalent quantity of nonspecific shRNA (lv-shRNA) was injected into the control group.
Quantitative RT-PCR. Total RNA was extracted from the footpads of fore- and hind-paws using RNAIso Plus (Takara, Shiga, Japan; #9109) and purified using the manufacturer's protocol. RNA was reverse-transcribed into cDNA using the PrimeScriptTM RT reagent kit with gDNA Eraser (Takara, Shiga, Japan; #RR047A). Quantitative RT-PCR was used to amplify the target genes with SYBR Premix Ex TaqTM (Takara; #DRR420A). Relative gene expression was calculated using the comparative Ct (2−ΔΔCT) method and normalized to the housekeeping gene Gapdh.
SUPPLEMENTARY MATERIAL Figure S1. RNA interference (RNAi)-mediated independent mutant-specific inhibition of the K9 dominant-negative allele. Figure S2. Complete siRNA sequence walks for K9 mutation Krt9-c.434delAinsGGCT and measurements of luciferase activities. Figure S3. Complete siRNA sequence walks for K9 mutation KRT9/c.500delAinsGGCT and measurements of luciferase activities. Figure S4. EPPK-like lesions were persistent in K9-mutant mice.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (30972644, 81350018, and 81450065) and the Ningbo Natural Science Foundation (2015A610187 and 2012A610192). The authors have no conflict of interest to declare.
Supplementary Material
References
- Fuchs, E and Cleveland, DW (1998). A structural scaffolding of intermediate filaments in health and disease. Science 279: 514–519. [DOI] [PubMed] [Google Scholar]
- Herrmann, H, Strelkov, SV, Burkhard, P and Aebi, U (2009). Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest 119: 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza, V (2011). Keratins in health and cancer: more than mere epithelial cell markers. Oncogene 30: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary, MB, Coulombe, PA and McLean, WH (2004). Intermediate filament proteins and their associated diseases. N Engl J Med 351: 2087–2100. [DOI] [PubMed] [Google Scholar]
- Reis, A, Hennies, HC, Langbein, L, Digweed, M, Mischke, D, Drechsler, M et al. (1994). Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK). Nat Genet 6: 174–179. [DOI] [PubMed] [Google Scholar]
- Du, ZF, Wei, W, Wang, YF, Chen, XL, Chen, CY, Liu, WT et al. (2011). A novel mutation within the 2B rod domain of keratin 9 in a Chinese pedigree with epidermolytic palmoplantar keratoderma combined with knuckle pads and camptodactyly. Eur J Dermatol 21: 675–679. [DOI] [PubMed] [Google Scholar]
- Ke, HP, Jiang, HL, Lv, YS, Huang, YZ, Liu, RR, Chen, XL et al. (2014). KRT9 gene mutation as a reliable indicator in the prenatal molecular diagnosis of epidermolytic palmoplantar keratoderma. Gene 546: 124–128. [DOI] [PubMed] [Google Scholar]
- Fu, DJ, Thomson, C, Lunny, DP, Dopping-Hepenstal, PJ, McGrath, JA, Smith, FJ et al. (2014). Keratin 9 is required for the structural integrity and terminal differentiation of the palmoplantar epidermis. J Invest Dermatol 134: 754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie Pedrioli, DM, Fu, DJ, Gonzalez-Gonzalez, E, Contag, CH, Kaspar, RL, Smith, FJ et al. (2012). Generic and personalized RNAi-based therapeutics for a dominant-negative epidermal fragility disorder. J Invest Dermatol 132: 1627–1635. [DOI] [PubMed] [Google Scholar]
- Cao, T, Longley, MA, Wang, XJ and Roop, DR (2001). An inducible mouse model for epidermolysis bullosa simplex: implications for gene therapy. J Cell Biol 152: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson, RP, Smith, FJ, McLean, WH, Landthaler, M, Leube, RE and Kaspar, RL (2006). SiRNA-mediated selective inhibition of mutant keratin mRNAs responsible for the skin disorder pachyonychia congenita. Ann NY Acad Sci 1082: 56–61. [DOI] [PubMed] [Google Scholar]
- Werner, NS, Windoffer, R, Strnad, P, Grund, C, Leube, RE and Magin, TM (2004). Epidermolysis bullosa simplex-type mutations alter the dynamics of the keratin cytoskeleton and reveal a contribution of actin to the transport of keratin subunits. Mol Biol Cell 15: 990–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson, RP, Smith, FJ, Reeves, RE, Contag, CH, Leake, D, Leachman, SA et al. (2008). Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J Invest Dermatol 128: 594–605. [DOI] [PubMed] [Google Scholar]
- Atkinson, SD, McGilligan, VE, Liao, H, Szeverenyi, I, Smith, FJ, Moore, CB et al. (2011). Development of allele-specific therapeutic siRNA for keratin 5 mutations in epidermolysis bullosa simplex. J Invest Dermatol 131: 2079–2086. [DOI] [PubMed] [Google Scholar]
- Leachman, SA, Hickerson, RP, Schwartz, ME, Bullough, EE, Hutcherson, SL, Boucher, KM et al. (2010). First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol Ther 18: 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, XH, Zhang, XN, Mao, W, Chen, HP, Xu, LR, Chen, H et al. (2004). A novel mutation of keratin 9 in a large Chinese family with epidermolytic palmoplantar keratoderma. Br J Dermatol 150: 647–651. [DOI] [PubMed] [Google Scholar]
- Candi, E, Cipollone, R, Rivetti di Val Cervo, P, Gonfloni, S, Melino, G and Knight, R (2008). p63 in epithelial development. Cell Mol Life Sci 65: 3126–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsuga, K (2014). Epidermal barriers. Cold Spring Harb Perspect Med 4: a018218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman, MJ, Sisi, P, Banbury, DN and Byrne, C (1998). Patterned acquisition of skin barrier function during development. Development 125: 1541–1552. [DOI] [PubMed] [Google Scholar]
- Saitou, M, Furuse, M, Sasaki, H, Schulzke, JD, Fromm, M, Takano, H et al. (2000). Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell 11: 4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas, TK, Chen, XN, Rowe, LB, Birkenmeier, EH, Fanning, AS, Anderson, JM et al. (1995). Localization of the tight junction protein gene TJP1 to human chromosome 15q13, distal to the Prader-Willi/Angelman region, and to mouse chromosome 7. Genomics 30: 594–597. [DOI] [PubMed] [Google Scholar]
- Chuzhanova, NA, Anassis, EJ, Ball, EV, Krawczak, M and Cooper, DN (2003). Meta-analysis of indels causing human genetic disease: mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat 21: 28–44. [DOI] [PubMed] [Google Scholar]
- Paladini, RD, Takahashi, K, Bravo, NS and Coulombe, PA (1996). Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J Cell Biol 132: 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt, J and Magin, TM (2002). Hyperproliferation, induction of c-Myc and 14-3-3sigma, but no cell fragility in keratin-10-null mice. J Cell Sci 115: 2639–2650. [DOI] [PubMed] [Google Scholar]
- Reichelt, J, Büssow, H, Grund, C and Magin, TM (2001). Formation of a normal epidermis supported by increased stability of keratins 5 and 14 in keratin 10 null mice. Mol Biol Cell 12: 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, W, Kumar, V, Beer, HD, Richter, M, Wohlenberg, C, Reuter, U et al. (2012). Keratin 1 maintains skin integrity and participates in an inflammatory network in skin through interleukin-18. J Cell Sci 125: 5269–5279. [DOI] [PubMed] [Google Scholar]
- Lessard, JC and Coulombe, PA (2012). Keratin 16-null mice develop palmoplantar keratoderma, a hallmark feature of pachyonychia congenita and related disorders. J Invest Dermatol 132: 1384–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbein, L, Heid, HW, Moll, I and Franke, WW (1993). Molecular characterization of the body site-specific human epidermal cytokeratin 9: cDNA cloning, amino acid sequence, and tissue specificity of gene expression. Differentiation 55: 57–71. [DOI] [PubMed] [Google Scholar]
- Heidel, JD, Liu, JY, Yen, Y, Zhou, B, Heale, BS, Rossi, JJ et al. (2007). Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res 13: 2207–2215. [DOI] [PubMed] [Google Scholar]
- Almaça, J, Faria, D, Sousa, M, Uliyakina, I, Conrad, C, Sirianant, L et al. (2013). High-content siRNA screen reveals global ENaC regulators and potential cystic fibrosis therapy targets. Cell 154: 1390–1400. [DOI] [PubMed] [Google Scholar]
- Rao, DD, Vorhies, JS, Senzer, N and Nemunaitis, J (2009). siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev 61: 746–759. [DOI] [PubMed] [Google Scholar]
- McLean, WH and Moore, CB (2011). Keratin disorders: from gene to therapy. Hum Mol Genet 20(R2): R189–R197. [DOI] [PubMed] [Google Scholar]
- Rao, DD, Senzer, N, Cleary, MA and Nemunaitis, J (2009). Comparative assessment of siRNA and shRNA off target effects: what is slowing clinical development. Cancer Gene Ther 16: 807–809. [DOI] [PubMed] [Google Scholar]
- Jackson, AL and Linsley, PS (2010). Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov 9: 57–67. [DOI] [PubMed] [Google Scholar]
- Takanashi, M, Sudo, K, Ueda, S, Ohno, S, Yamada, Y, Osakabe, Y et al. (2015). Novel types of small RNA exhibit sequence- and target-dependent angiogenesis suppression without activation of toll-like receptor 3 in an age-related macular degeneration (AMD) mouse model. Mol Ther Nucleic Acids 4: e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, DH and Rossi, JJ (2007). Strategies for silencing human disease using RNA interference. Nat Rev Genet 8: 173–184. [DOI] [PubMed] [Google Scholar]
- Hickerson, RP, Wey, WC, Rimm, DL, Speaker, T, Suh, S, Flores, MA et al. (2013). Gene silencing in skin after deposition of self-delivery siRNA with a motorized microneedle array device. Mol Ther Nucleic Acids 2: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew, NS and Scheule, RK (2005). Toxicity of cationic lipid-DNA complexes. Adv Genet 53: 189–214. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.