Abstract
The role of biotic and abiotic factors in shaping the diversity and composition of communities of plant viruses remain understudied, particularly in natural settings. In this study, we test the effects of host identity, location, and sampling year on the taxonomic composition of plant viruses in six native plant species [Ambrosia psilostachya (Asteraceae), Vernonia baldwinii (Asteraceae), Asclepias viridis (Asclepiadaceae), Ruellia humilis (Acanthaceae), Panicum virgatum (Poaceae) and Sorghastrum nutans (Poaceae)] from the Nature Conservancy’s Tallgrass Prairie Preserve in northeastern Oklahoma. We sampled over 400 specimens of the target host plants from twenty sites (plots) in the Tallgrass Prairie Preserve over 4 years and tested them for the presence of plant viruses applying virus-like particle and double-stranded RNA enrichment methods. Many of the viral sequences identified could not be readily assigned to species, either due to their novelty or the shortness of the sequence. We thus grouped our putative viruses into operational viral taxonomic units for further analysis. Partial canonical correspondence analysis revealed that the taxonomic composition of plant viruses in the target species had a significant relationship with host species (P value: 0.001) but no clear relation with sampling site or year. Variation partitioning further showed that host identity explained about 2–5 per cent of the variation in plant virus composition. We could not interpret the significant relationship between virus composition and host plants with respect to host taxonomy or ecology. Only six operational viral taxonomic units had over 5 per cent incidence over a 4-year period, while the remainder exhibited sporadic infection of the target hosts. This study is the first of its kind to document the dynamics of the entire range of viruses in multiple plant species in a natural setting.
Keywords: wild plant viruses, plant virus composition
1 Introduction
Studies to uncover patterns in the diversity and composition of viruses of wild plants have only been initiated recently (Cooper and Jones 2006; Wren et al. 2006; Melcher et al. 2008; Roossinck, Martin, and Roumagnac 2015). Understanding patterns of diversity and species composition with respect to environmental properties is a widely used approach in ecology. It is meaningful to determine how strongly a given set of environmental variables can explain the distribution of a set of species and their evolutionary relationships. For plant viruses, our current knowledge on the topic is limited to interactions of single viruses, primarily from cultivated or experimental sources. The environmental factors determining virus composition in natural plant communities across ecosystems are mostly unknown. Recent reviews emphasize the influence of both biotic and abiotic factors on the distribution of plant viruses in nature (Malmstrom, Melcher, and Bosque-Perez 2011; Roossinck 2015). Here, we focus on three attributes in determining the distribution of plant viruses: host identity; location; and time.
1.1 Host identity effects
Lovisolo, Hull, and Rosler (2003) argued that plant hosts influence the evolutionary divergence of plant viruses. In general, many plant viruses are host generalists rather than host specialists (Power and Flecker 2003). However, host ranges (defined by virologists as the set of plant species a virus can infect) of closely related plant viruses do not overlap completely (Dawson and Hilf 1992). For example, Tomato chlorosis virus and Tomato infectious chlorosis virus are closely related taxonomically and have similar influence on hosts, but they differ in host range (Wintermantle and Wisler 2006). Host identity also influences the incidence of plant viruses (Lavina, Aramburu, and Moriones 1996; Sacristán, Fraile, and García-Arenal 2004).
1.2 Location effects
A study on Barley/Cereal yellow dwarf viruses suggests a latitudinal gradient in species composition (Seabloom et al. 2010). Harrison (1981) reported biogeographic patterns in plant virus distribution. Environmental differences among sites appear to have strong influences on plant virus species composition at broad scales (Atiri, Njukeng, and Ekpo 2000). Spatial genetic structure is also reported at the local scale (Skotnicki, Mackenzie, and Gibbs 1996; Pinel et al. 2000; Hall 2006) and the regional scale (Stenger, Seifers, and French 2002; Ahmad et al. 2006).
1.3 Time effects
Many plant viruses exhibit variation in incidence over time (Coutts and Jones 2002). It might be expected that large fluctuations in plant virus incidence over time likely influence the overall virus species composition. Rates of evolutionary diversification in plant viruses may also be temporally variable (Gibbs et al. 2010).
In this study, we determined the effect of host identity, sampling site, and year on the composition of plant viruses isolated from six frequent native plants samples over a 4-year period from their natural populations in the Tallgrass Prairie Preserve (TGPP) in northeastern Oklahoma. In addition, as the preserve is a part of the intact natural tallgrass prairie ecosystem in North America that existed prior to European-American settlement (Samson, Knopf, and Ostlie 2004; Allen et al. 2009), the study also provides a unique opportunity to document the diversity of plant viruses in a natural state.
2 Materials and Methods
2.1 Field methods
Our study area is The Nature Conservancy’s TGPP, Osage County, Oklahoma (Allen et al. 2009). TGPP vegetation consists mainly of tallgrass prairie and cross timber forest that provides habitat for 763 species of vascular plants (Palmer 2007). We collected our samples within 10 m of twenty semi-randomly selected 10 m ×10 m permanent plots (Fig. 1) in tallgrass prairie vegetation (plant species composition and details of plot location are given in McGlinn, Earls, and Palmer 2010). At each sampling area, we collected at least one sample each of Ambrosia psilostachya (Asteraceae), Vernonia baldwinii (Asteraceae), Asclepias viridis (Asclepiadaceae), Ruellia humilis (Acanthaceae), Panicum virgatum (Poaceae), and Sorghastrum nutans (Poaceae). These six target plant species are readily identifiable in the vegetative state and represent some of the most frequent vascular plants of the TGPP. The samples were collected in the month of June from 2005 to 8.
Figure 1.
Semi-randomly selected sites (solid squares) for plant virus sampling as a subset of 276 sites (open squares) at 1 km UTM grid in TGPP of northeastern Oklahoma.
At least 10 g of leaf tissue sample were collected from top leaves of each host plant. The samples were collected irrespective of any disease symptoms. Proper sanitation protocol was followed while collecting the samples to avoid contamination. Collected samples were immediately transferred to an ice-cooled chest in the field and stored at −80 °C in the laboratory before further processing for plant virus assays.
2.2 Laboratory methods
Plant virus-like particles (VLPs) and double-stranded RNA (dsRNA) were isolated from the plant samples collected for plant virus assay. VLPs were obtained by differential centrifugation as described in detail in Melcher et al. (2008). For dsRNA, total nucleic acid was obtained from 5 g of plant leaf samples by performing phenol-chloroform extraction followed by dsRNA enrichment using CF11 cellulose chromatography (Roossinck et al. 2010). The VLP method has the limitation that plant viruses that do not produce encapsidated forms and those that make unstable particles will not be captured (Melcher et al. 2008). The dsRNA method is based on the premise that uninfected plants normally do not contain detectable amount of high molecular weight dsRNA and, when present, dsRNA is considered as an indicator of presence of ssRNA, ambi-sense, or dsRNA viruses (Dodds, Morris, and Jordan 1984). Isolated VLPs were subjected to polymerase chain reaction (PCR) or reverse transcriptase-PCR (RT-PCR). Similarly, dsRNAs were subjected to RT-PCR as described (Roossinck et al. 2010). The PCR products were finally sequenced by a massively parallel sequencing technique using a Roche 454 pyrosequencer (Roossinck et al. 2010). In this study, a total of 184 and 445 samples of target species were analyzed with VLP and dsRNA methods, respectively.
The putative plant virus species were identified by performing BLAST searches of obtained nucleic acid sequences. Although the 454 sequence procedure produced virus-related reads that did not assemble into contigs within reads from many plant samples (singletons), these were disregarded for the present analysis because an occasional read could have been assigned mistakenly to the wrong plant sample. In addition, many viral sequences retained for further analysis were short, nonoverlapping, and could not regularly be assigned to virus species (whether previously known or novel). Thus, in many cases, we combined viruses into relatively broad taxonomic categories such as families or genera; i.e., the finest level of resolution possible that was consistent between samples. These finest levels (be they virus species, genera, or families) constitute our operational viral taxonomic units (OVTUs) (Dutta et al. 2014) for the remainder of the analyses. Data from this study are available on request.
2.3 Data analysis
Canonical correspondence analysis (CCA) is a direct constrained ordination method to reveal variability in a dataset related to measured environmental variables (Lepš and Šmilauer 2003). We used partial canonical correspondence analysis (pCCA) (ter Braak 1988) to factor out effects of covariables such as site, year, and host identity from each other, in our case to examine the effect of the remaining variables (site, year, or host identity) in plant virus composition. Analyses were done with the program CANOCO version 4.5 (ter Braak and Šmilauer 2002) to examine the effects of host identity, site, and year on plant virus composition in the target hosts surveyed. The plant virus composition was assayed at the OVTU level in pCCA. We performed pCCA separately for data obtained from VLP and dsRNA methods due to differences in sample size and sensitivity in detecting plant viruses by the two methods. We used the proportion of virus reads (sequenced fragments of nucleic acid) out of total reads as a measure of virus abundance; these proportions were square root transformed prior to analysis. All explanatory variables (environmental variables) are nominal (1/0). The effects of host, site, and year on virus composition against the null model of no effect were tested using a Monte Carlo permutation procedure. All randomization tests were performed with 999 iterations with permutation blocks defined by respective covariables to determine the significance of all canonical axes for all pCCA analysis under the full model. CanoDraw 4.0 (ter Braak and Šmilauer 2002) was used to generate ordination scatter plots and biplots.
Variation partitioning was applied to determine the relative importance of the host identity, site, and year on composition of viruses. The canonical equivalent of the regression coefficient of determination, R2CCA generally provides an estimate of variation. R2CCA is quantified as the ratio of the sum of all the canonical eigenvalues (explained variation) over the sum of all canonical and noncanonical eigenvalues (total variation). However, R2CCA provides a biased estimate influenced by the number of independent variables and sample size (Peres-Neto et al. 2006). Therefore, we calculated the unbiased version of this statistic (R2adj) using a permutational form of adjustment as suggested by Peres-Neto et al. (2006). It is possible that an estimate of the variation may have a negative value (Azen and Budescu 2003; Peres-Neto et al. 2006). There are two conditions possible when the variation could have a negative value using the partitioning variation approach. First, a shared variation can have a negative value when its correlation with its response variable is zero or close to zero and is correlated with another predictor variable (Azen and Budescu 2003)and second when two predictor variables are strongly correlated and their effects with the response variable are opposite to each other (Peres-Neto et al. 2006).
In data analysis, we reported plant virus OVTU incidence as the proportion of plant hosts in which a particular OVTU is detected. Traditionally, incidence is defined as proportion of visibly diseased plants by pathogens such as viruses (Madden and Hughes 1995). However, incidence reported in this article is based on detection of plant virus OVTUs irrespective of visual symptoms.
3 Results
We identified thirty OVTUs of plant viruses from VLP and dsRNA methods in six target plant hosts sampled over a 4-year period (Table 1). In both methods, six OVTUs were common. Out of the thirty OVTUs, only four (Cowpea chlorotic mottle virus, Cucumber mosaic virus, Maize chlorotic dwarf virus, and Oat blue dwarf virus) are previously known virus species. Some OVTUs belonged to virus families traditionally associated with fungi (Totiviridae and Chrysoviridae). We broadly grouped them with plant viruses, as we have not found any fungal association in our samples. The majority of OVTUs we identified have an RNA genome and represent fifteen viral families (Table 1). Among the thirty OVTUs (Table 1), only six (Alphaflexivirid, Asclepias asymptomatic virus [AsAV], Carmovirus, Comovirid, Partitivirid, and Totivirid) had average incidences of 5 per cent or more (range 5–53%) in one or more target hosts, while the rest were found in less than 5 per cent of samples of a host species. OVTUs with higher incidences infect plants of more than one target host species (Table 1).
Table 1.
List of plant virus OVTUs with their respective assay method, genome type, taxonomic grouping (genus and family), and host range in six target hosts.
| Putative virus OVTUs | Assay method | Genome type | Genus | Family | Host rangea |
|---|---|---|---|---|---|
| Alphaflexivirid (Alflex) | dsRNA, VNA | ssRNA | Unclassified | Alphaflexiviridae | 1,3,6 |
| Ambrosia asymptomatic virus (AmAV) | dsRNA, VNA | ssRNA | Unclassified | Alphaflexiviridae | 1 |
| Ampelovirus (Ampelo) | dsRNA | ssRNA | Ampelovirus | Closteroviridae | 4 |
| Asclepias asymptomatic virus (AsAV) | dsRNA, VNA | ssRNA | Tymovirus | Tymoviridae | 1,2,3,4,5,6 |
| Badnavirus (Badna) | dsRNA, VNA | dsDNA | Badnavirus | Caulimoviridae | 1,5,6 |
| Panicum OVTU TGP2 (Betaflex) | dsRNA | ssRNA | Unclassified | Betaflexiviridae | 3 |
| Carmovirus (Carmo) | dsRNA | ssRNA | Carmovirus | Tombusviridae | 1,3 |
| Cavemovirus (Cavemo) | VNA | dsDNA | Cavemovirus | Caulimoviridae | 1 |
| Chrysovirus (Chryso) | dsRNA | dsRNA | Chrysovirus | Chrysoviridae | 2,3,4 |
| Closterovirus (Clostero) | dsRNA | ssRNA | Closterovirus | Closteroviridae | 6 |
| Asclepias virus TGP2 (Como) | dsRNA, VNA | ssRNA | Unclassified | Secoviridae | 1,2,3,4,6 |
| Cowpea chlorotic mottle virus (CCMV) | dsRNA | ssRNA | Bromovirus | Bromoviridae | 2,4 |
| Cucumber mosaic virus (CMV) | dsRNA | ssRNA | Cucumovirus | Bromoviridae | 4 |
| Fabavirus (Faba) | dsRNA | ssRNA | Fabavirus | Secoviridae | 6 |
| Luteovirid (Luteo) | dsRNA, VNA | ssRNA | Unclassified | Luteoviridae | 1,3 |
| Maize chlorotic dwarf virus (MCDV) | dsRNA | ssRNA | Waikavirus | Secoviridae | 3 |
| Nepovirus (Nepo) | dsRNA | ssRNA | Nepovirus | Secoviridae | 6 |
| Nucleorhabdovirus (Nurhab) | dsRNA | ssRNA | Nucleorhabdovirus | Rhabdoviridae | 1 |
| Oat blue dwarf virus (OBDV) | dsRNA | ssRNA | Marafivirus | Tymoviridae | 2 |
| Oryzavirus (Oryza) | dsRNA | dsRNA | Oryzavirus | Reoviridae | 2 |
| Partitivirid (Partiti) | dsRNA | dsRNA | Unclassified | Partitiviridae | 1,2,3,4,5,6 |
| Petuvirus (Petu) | VNA | dsDNA | Petuvirus | Caulimoviridae | 2 |
| Potyvirus (Poty) | dsRNA | ssRNA | Potyvirus | Potyviridae | 4 |
| Sobemovirus (Sobemo) | dsRNA | ssRNA | Sobemovirus | Unclassified | 5 |
| Southern tomato virus (STV) | dsRNA | dsRNA | Partitivirus-like | Unclassified | 1,3,4,5 |
| Soymovirus (Soymo) | dsRNA | dsDNA | Soymovirus | Caulimoviridae | 4 |
| Passionfruit mosaic virus (Tobamo) | dsRNA | ssRNA | Tobamovirus | Virgaviridae | 6 |
| Totivirid (Toti) | dsRNA | dsRNA | Unclassified | Totiviridae | 1,2,3,4,5,6 |
| Tymovirus 2 (Tymo2) | dsRNA | dsRNA | Tymovirus | Tymoviridae | 5 |
Bold numbers represent hosts with more than 5 per cent incidence.
a1, A. psilostachya; 2, As. viridis; 3, P. virgatum; 4, R. humilis; 5, S. nutans; 6, V. baldwinii.
3.1 Effects of plant hosts on plant virus composition
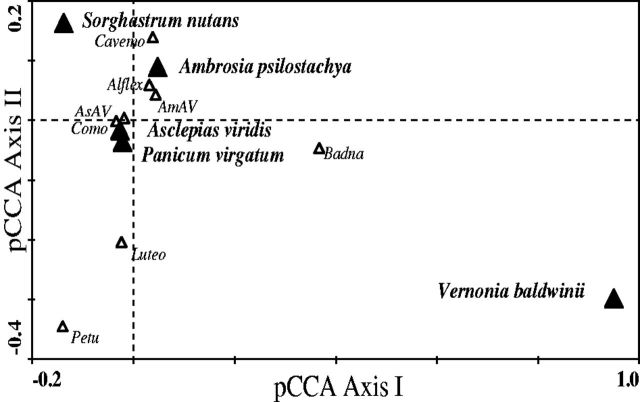
The pCCA on data from both dsRNA and VLP methods with sites and years as covariables showed that host identity had a significant effect on the composition of plant virus (P < 0.001). Variation partitioning on dsRNA and VLP data indicated about 5 per cent (R2adj = 0.049) and 2 per cent (R2adj = 0.016) of variation in OVTU composition, respectively, is significantly explained by the identity of target host plants. The distribution of OVTUs on the pCCA biplot indicates their relative proportion of abundance in the target host plants as detected by dsRNA (Fig. 2) and VLP (Fig. 3) methods. OVTUs common to both methods such as Alphaflexivirid, Comovirid, and AsAV show relatively similar positions in relation to the plant hosts in the ordination space.
Figure 2.
Species-environment biplot of pCCA using dsRNA data with sites and year as covariables. Host plants are represented as nominal variables (solid triangles). The eigenvalue for the first and the second axes are 0.514 and 0.312, respectively. For clarity, only plant virus OVTUs (open triangles) with abundances over 1 per cent of the maximum abundance are displayed. The values of pCCA axis I and II were generated in ordination matrices using nominal values of environmental variables (here, host identity after factoring out site and year effects) and virus composition, respectively. Refer to Table 1 for the full names of the plant virus OVTUs.
Figure 3.
Species-environment biplot of pCCA using VLP data with sites and year as covariables. Host plants are represented as nominal variables (solid triangles) and plant virus OVTUs as open triangles. The eigenvalue for the first and the second axes are 0.389 and 0.155, respectively. Note that since no plant virus was identified from R. humilis from VLP method, the host does not appear in the biplot. The values of pCCA axis I and II were generated in ordination matrices using nominal values of environmental variables (here, host identity after factoring out site and year effects) and virus composition, respectively. Refer to Table 1 for the full names of the plant virus OVTUs.
Despite the high level of statistical significance, we could not interpret the biplots of pCCA (Figs 2 and 3) with respect to host plant taxonomy. For example, A. psilostachya and V. baldwinii are both members of Asteraceae but their positions appear almost opposite in the second pCCA axis in the biplots constructed from both dsRNA and VLP data. Similarly, the centroids of the two grasses (P. virgatum and S. nutans) are spaced apart in the ordination diagrams (Figs 2 and 3) implying a difference in their taxonomic compositions of viruses.
Likewise, we cannot readily interpret the pCCA biplots with respect to host plant ecology. There is an intriguing suggestion that the second axis of the dsRNA pCCA (Fig. 2) is associated with pollination mode—i.e., the three species (As. viridis, R. humilis, and V. baldwinii) at the bottom have showy, insect pollinated flowers, while the remainders are wind pollinated. However, we are not confident of this interpretation as the second axis (eigenvalue = 0.155) has a much weaker eigen value than the first (eigenvalue = 0.389).
A large proportion of OVTUs (∼70% sampled) were detected from only one or two species of the target hosts and their incidence was less than 5 per cent in those host species (Table 1). Thus, the apparent specialization may be in part an artifact of low frequency. Only six OVTUs isolated had incidences over 5 per cent in at least one host but their magnitude of incidence varied with the target hosts (Table 2). For example, AsAV incidence varied from 6 to 41 per cent in the target hosts with highest incidence (41%) in As. viridis and the lowest (6%) in P. virgatum (Table 2).
Table 2.
Average incidence (%) of six plant virus OVTUsa over 4 years in the target hosts.
| Plant host | Alflex | AsAV | Carmo | Como | Partiti | Toti |
|---|---|---|---|---|---|---|
| A. psilostachya | 6 | 15 | 6 | 0 | 9 | 8 |
| As. viridis | 0 | 41 | 0 | 15 | 13 | 13 |
| P. virgatum | 1 | 6 | 1 | 4 | 3 | 10 |
| R. humilis | 0 | 13 | 0 | 2 | 38 | 53 |
| S. nutans | 0 | 9 | 0 | 0 | 7 | 16 |
| V. baldwinii | 1 | 14 | 0 | 4 | 6 | 10 |
aAlflex, Alphaflexivirid; AsAV, Asclepias asymptomatic virus; Carmo, Carmovirus; Como, Comovirin; Partiti, Partitivirid; Toti, Totivirid.
3.2 Effects of sites and year on plant virus composition
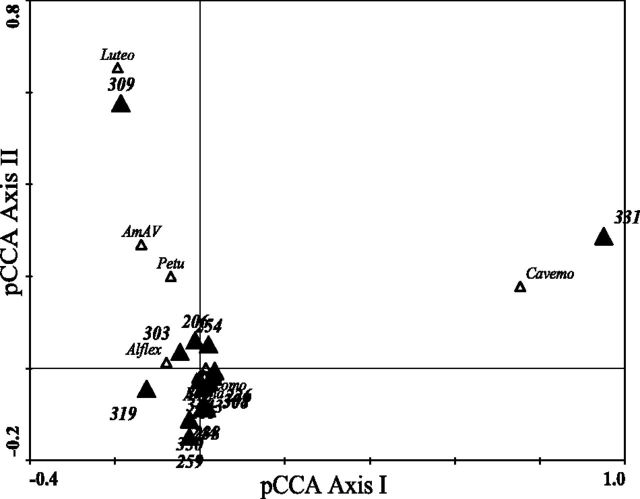
The effect of sites on plant virus composition was not consistent between the two datasets (dsRNA and VLP). A pCCA from dsRNA data showed no significant effect of sites on OVTU composition (P value: 0.98; ordination figure not shown). The finding was also supported by a negligible adjusted fraction (R2adj = −0.01) of variation explained by variation partitioning analysis. In the case of VLP data, pCCA showed a significant effect of sites on OVTU composition (P value: 0.003). However, variation explained after calculating correction by variation partitioning analysis with VLP data was negligible (R2adj = −0.03) similar to dsRNA. The biplot from VLP data indicates that the major signature for significance seems to be influenced by Luteovirid’s (Luteo) association with plot 309 and Cavemovirus (Cavemo) with plot 331 that occupy extreme positions in the ordination space (Fig. 4). Besides Luteo and Cavemo, the centroids of the rest of the plots as nominal variables and plant virus OVTUs aggregate close to the origin in the ordination space implying low spatial variation in virus composition with the exception of the two outlying plots (Fig. 4). Our data also show that Luteo was isolated from multiple target hosts in same year (2005) from plot 309 and Cavemo was found in multiple years in A. psilostachya from plot 331. Moreover, plots 309 and 331 represent the northernmost and southernmost plots, respectively, in our sampling and are likely to be most ‘different’ of any plots, as they (especially plot 331) are highly disturbed and close to cultivated areas.
Figure 4.
Plant virus OVTUs and site biplot of pCCA using VLP data with year and host identity as covariables. Note that most of site and plant virus OVTUs’ centroids are aggregated close to the origin of the ordination axes except Luteo and Cavemo associated with plots 309 and 331, respectively. Sites are represented as nominal variables and depicted as solid triangles with their three digit numerical codes in the ordination space. The plant virus OVTUs are symbolized with open triangles and their abbreviated names. The eigenvalue for the first and the second axes are 0.722 and 0.52, respectively. The values of pCCA axis I and II were generated in ordination matrices using nominal values of environmental variables (here, sites after factoring out host identity and year effects) and virus composition, respectively. Refer to Table 1 for the full names of the plant virus OVTUs.
The effect of year on plant virus composition was not significant with either dsRNA (P value: 0.47) or VLP (P value: 0.38) data. Variation partitioning also indicated less than 0.1 per cent variation in plant virus OVTUs composition explained by dsRNA (R2adj = 0.001) and VLP (R2adj = −0.073) data.
4 Discussion
Despite the growing interest in plant viruses in natural plant communities, the composition of plant viruses in native hosts is rarely studied. In this study, we investigated the plant viruses in six frequent target plant hosts (A. psilostachya, V. baldwinii, As. viridis, R. humilis, P. virgatum, and S. nutans) in the TGPP and tested the effects of sites, years, and host identity on the plant virus composition. Moreover, we reported diversity of plant viruses in the six target host plants and compared the incidence of some frequently occurring plant viruses with the rest from the total pool of plant viruses studied.
In general, the plant virus composition in six target plant hosts is significantly related to host plant identity but not related to year. We found weak site effects on the plant virus composition in VLP data that is most likely due to association of two viruses with two extreme plots. A recent large-scale study of the composition of four Barley/Cereal yellow dwarf viruses (B/CYDVs) along a latitudinal gradient by Seabloom et al. (2010) found a decline in turnover of B/CYDVs (β diversity) among sites along the lower to higher latitudinal gradient. In another study, three different subpopulations of Kennedya yellow mosaic virus isolates are found according to latitude along the eastern coast of Australia (Skotnicki, Mackenzie, and Gibbs 1996). The data from our study showed no influence of locations on taxonomic composition of the plant viruses, which may be attributable to low environmental variability or short spatial extent (the maximum inter-site distance was ∼19 km). It is also possible that spatial variation is more prominent at lower taxonomic levels such as among different isolates or strains of the same virus species. In this study, little variation in virus composition among sites also implies few constraints in transmission of plant viruses in our study area. Low nucleotide sequence variation in a widely distributed novel tymovirus, AsAV in TGPP was reported earlier by our group (Min et al. 2012).
No significant difference in plant virus composition was noted over a 4-year period in the natural plant community of TGPP. Although temporal dynamics in plant virus incidence due to shift in climatic and management regimes have been reported in the literature (Coutts and Jones 2002; Cadle-Davidson and Bergstrom 2004), at the taxonomic level of the plant virus, we studied here no such influence over years on plant virus composition is evident.
Identity of host is the only factor found in this study that significantly explains variation in plant virus composition from both VLP and dsRNA data. The site shows a significant relationship with the plant virus composition in the case of VLP data. The variation explained in plant viruses by host identity is a much smaller percentage of explained variance (∼2–5%) but still contributes a significant amount of explained variation at an alpha level of 0.05 and is biologically important. It is clear that plant virus composition is not clearly related to host taxonomy at the family level nor to ecological attributes of the plants. Dawson and Hilf (1992) also found inconsistent relationships of plant viruses with the taxonomy of their hosts. However, the explanation for the inconsistency is not known. It is interesting that although the grass family (Poaceae) hosts a number of specialized viruses in cultivated systems (Hull 2001; Lapierre 2004), we found that viruses from our two grass species were not substantially different from those of the dicotyledonous hosts sampled.
Another interesting observation in our data was that a large proportion of viruses infecting the target hosts include plant viruses with sporadic incidences. We predict that such sporadic infecting plant viruses may represent colonization efforts of dispersing plant viruses as they belong to diverse taxa of plant viruses. Influence of migrating populations of archeal viruses in local pools on species composition have been reported in the hot springs of the Yellowstone National Park (Snyder et al. 2007).
Most of the plant viruses identified in the target host plants in our study include viruses that were not previously reported. This observation is consistent with viruses isolated from other natural habitats such as terrestrial (Roossinck 2012), aquatic (López-Bueno et al. 2009; Rohwer and Thurber 2009) and soil (Kimura et al. 2008) environments. Previously undescribed plant viruses as reported here could open a new avenue for studying plant viruses and their ecology in natural systems. This article is a part of the Plant Virus Biodiversity and Ecology project, which documented many novel plant viruses from the TGPP (Melcher et al. 2008; Muthukumar et al. 2009; Shah 2010; Thapa et al. 2012). The large number of previously unreported plant viruses and the importance of plant host on their species composition open up new avenues for investigation. Indeed the characterization of several newly identified TGPP viruses recently reported provides anecdotal evidence supporting the importance of host species as a determinant of what viruses are likely to be found in them. The dynamics of viruses in a complex natural setting are likely to yield insights into the evolution of specialization and the emergence of diseases.
Acknowledgements
We thank the Nature Conservancy for allowing us to carry out fieldwork in the TGPP, Oklahoma. In particular, we acknowledge Bob Hamilton, Director of the TGPP, for his support and coordination during the fieldwork. We thank Vijay Muthukumar and Tao Ding for preparation of VLP and Graham B. Wiley and Bruce A. Roe for pyrosequencing. For assistance in plant sampling, we thank Wyatt Sharber, Kelly Derennaux, Fumiko Shirakura, Matthew Allen, Kiyoshi Sasaki, Olga Blinkova, Tracy Feldman, Rick Nelson, Byoung Eun Min, Guoan Shen, Veenita Grover, Peter Earls, and Will Lowry. The study under the Plant Virus Biodiversity and Ecology (PVBE) Project was supported by NSF-EPSCOR (grant EPS-0447262) and this work was supported by the Oklahoma Agricultural Experiment Station to U.M.
Conflict of interest. None declared.
References
- Ahmad Y. A., et al. (2006) ‘Geographical Distribution of Four Sugarcane Yellow Leaf Virus Genotypes’, Plant Disease, 90: 1156–60. [DOI] [PubMed] [Google Scholar]
- Allen M. S., et al. (2009) Lessons from the Prairie: Research at the Nature Conservancy’s Tallgrass Prairie Preserve. Stillwater, OK: Oklahoma Academy of Sciences. [Google Scholar]
- Atiri G. I., Njukeng A. P., Ekpo E. J. A. (2000) ‘Climate in Relation to Plant Virus Epidemiology and Sustainable Disease Management in West Africa’, Journal of Sustainable Agriculture, 16: 17–30. [Google Scholar]
- Azen R., Budescu D. V. (2003) ‘The Dominance Analysis Approach for Comparing Predictors in Multiple Regression’, Psychological Methods, 8: 129–48. [DOI] [PubMed] [Google Scholar]
- Cadle-Davidson L., Bergstrom G. C. (2004) ‘The Effects of Postplanting Environment on the Incidence of Soilborne Viral Diseases in Winter Cereals’, Phytopathology, 94: 527–34. [DOI] [PubMed] [Google Scholar]
- Cooper J., Jones R. A. C. (2006) ‘Wild Plants and Viruses: Under-Investigated Ecosystems’, Advances in Virus Research, 67: 2–32. [DOI] [PubMed] [Google Scholar]
- Coutts B. A., Jones .R. A. C. (2002) ‘Temporal Dynamics of Spread of Four Viruses within Mixed Species Perennial Pastures’, Annals of Applied Biology, 140: 37–52. [Google Scholar]
- Dawson W. O., Hilf M. E. (1992) ‘Host-Range Determinants of Plant Viruses’, Annual Review of Plant Physiology and Plant Molecular Biology, 43: 527–55. [Google Scholar]
- Dodds J. A., Morris T. J., Jordan R. L. (1984) ‘Plant Viral Doublestranded RNA’, Annual Review of Phytopathology, 22: 151–68. [Google Scholar]
- Dutta M., et al. (2014) ‘Genomics Characterization of Ambrosia Asymptomatic Virus 1 and Evidence of Other Tymovirales Members in the Oklahoma Tallgrass Prairie Revealed by Sequence Analysis’, Archives of Virology, 159: 1755–64. [DOI] [PubMed] [Google Scholar]
- Gibbs A. J., et al. (2010) ‘Time—The Emerging Dimension of Plant Virus Studies’, Journal of General Virology, 91: 13–22. [DOI] [PubMed] [Google Scholar]
- Hall G. (2006) ‘Selective Constraint and Genetic Differentiation in Geographically Distinct Barley Yellow Dwarf Virus Populations’, Journal of General Virology, 87: 3067–75. [DOI] [PubMed] [Google Scholar]
- Harrison B. D. (1981) ‘Plant Virus Ecology: Ingredients, Interactions and Environmental Influences’, Annuals of Applied Biology, 99: 195–209. [Google Scholar]
- Hull R. (2001) Matthews Plant Virology. San Diego, CA: Academic Press. [Google Scholar]
- Kimura M., et al. (2008) ‘Ecology of Viruses in Soils: Past, Present and Future Perspectives’, Soil Science & Plant Nutrition, 54: 1–32. [Google Scholar]
- Lapierre H. (2004) ‘Host range and position of viruses of Poaceae in the virus taxonomy’, in Lapierre H., Signoret P. A. (eds) Viruses and Virus diseases of Poaceae (Gramineae). Paris: Institut national De La Recherche Agronomique, pp. 274–278. [Google Scholar]
- Lavina A., Aramburu J., Moriones E. (1996) ‘Occurrence of Tomato Spotted Wilt and Cucumber Mosaic Virus in Field-Grown Tomato Crops and Associated Weeds in Northeastern Spain’, Plant Phytopathology, 4: 25–38. [Google Scholar]
- Lepš J., Šmilauer P. (2003) Multivariate Analysis of Ecological Data Using CANOCO. Cambridge, UK: Cambridge University Press. [Google Scholar]
- López-Bueno A., et al. (2009) ‘High Diversity of the Viral Community from an Antarctic Lake’, Science, 326: 858–61. [DOI] [PubMed] [Google Scholar]
- Lovisolo O., Hull R., Rosler O. (2003) ‘Coevolution of Viruses with Hosts and Vectors and Possible Paleontology’, Advances in Virus Research, 62: 325–79. [DOI] [PubMed] [Google Scholar]
- Madden L. V., Hughes G. (1995) ‘Plant Disease Incidence: Distributions, Heterogeneity, and Temporal Analysis’, Annual Review of Phytopathology, 33: 529–64. [DOI] [PubMed] [Google Scholar]
- Malmstrom C. M., Melcher U., Bosque-Perez N. A. (2011) ‘The Expanding Field of Plant Virus Ecology: Historical Foundations, Knowledge Gaps, and Research Directions’, Virus Research, 159: 84–94. [DOI] [PubMed] [Google Scholar]
- McGlinn D. J., Earls P. G., Palmer M. W. (2010) ‘A Twelve-Year Study on the Scaling of Vascular Plant Composition in an Oklahoma Tallgrass Prairie’, Ecology, 91: 1872. [Google Scholar]
- Melcher U., et al. (2008) ‘Evidence for Novel Viruses by Analysis of Nucleic Acids in Virus-Like Particle Fractions from Ambrosia psilostachya’, Journal of Virological Methods, 152: 49–55. [DOI] [PubMed] [Google Scholar]
- Min B. E., et al. (2012) ‘Molecular Characterization, Ecology, and Epidemiology of a Novel Tymovirus in Asclepias viridis from Oklahoma’, Phytopathology, 102: 166–76. [DOI] [PubMed] [Google Scholar]
- Muthukumar V., et al. (2009) ‘Non-Cultivated Plants of the Tallgrass Prairie Preserve of Northeastern Oklahoma Frequently Contain Virus-Like Sequences in Particulate Fractions’, Virus Research, 141: 169–73. [DOI] [PubMed] [Google Scholar]
- Palmer M. W. (2007) ‘The Vascular Flora of the Tallgrass Prairie Preserve, Osage County, Oklahoma’, Castanea, 72: 235–46. [Google Scholar]
- Peres-Neto P. R., et al. (2006) ‘Variation Partitioning of Species Data Matrices: Estimation and Comparison of Fractions’, Ecology, 87: 2614–25. [DOI] [PubMed] [Google Scholar]
- Pinel A., et al. (2000) ‘Molecular Variability of Geographically Distinct Isolates of Rice yellow mottle virus in Africa’, Archives of Virology, 145: 1621–38. [DOI] [PubMed] [Google Scholar]
- Power A. G., Flecker A. S. (2003) ‘Virus specificity in disease systems: are species redundant?’, in Kareiva P., Lavin S. A. (eds) The Importance of Species: Perspectives on Expandability and Triage. Princeton, NJ, Princeton University Press, pp. 330–346. [Google Scholar]
- Rohwer F., Thurber R. V. (2009) ‘Viruses Manipulate the Marine Environment’, Nature, 459: 207–12. [DOI] [PubMed] [Google Scholar]
- Roossinck M. J. (2012) ‘Plant Virus Metagenomics: Biodiversity and Ecology’, The Annual Review of Genetics, 46: 359–69. [DOI] [PubMed] [Google Scholar]
- Roossinck M. J. (2015) ‘Plants, Viruses and the Environment: Ecology and Mutualism’, Virology, 479-80: 271–7. [DOI] [PubMed] [Google Scholar]
- Roossinck M. J., Martin D. P., Roumagnac P. (2015) ‘Plant Virus Metagenomics: Advances in Virus Discovery’, Phytopathology, 105: 716–27. [DOI] [PubMed] [Google Scholar]
- Roossinck M. J., et al. (2010) ‘Ecogenomics: Using Massively Parallel Pyrosequencing to Understand Virus Ecology’, Molecular Ecology, 19: 81–8. [DOI] [PubMed] [Google Scholar]
- Sacristán S., Fraile A., García-Arenal F. (2004) ‘Population Dynamics of Cucumber Mosaic Virus in Melon Crops and in Weeds in Central Spain’, Phytopathology, 94: 992–8. [DOI] [PubMed] [Google Scholar]
- Samson F. B., Knopf F. L., Ostlie W. R. (2004) ‘Great Plains Ecosystems: Past, Present, and Future’, Wildlife Society Bulletin, 32: 6–15. [Google Scholar]
- Seabloom E. W., et al. (2010) ‘Viral Diversity and Prevalence Gradients in North American Pacific Coast Grasslands’, Ecology, 91: 721–32. [DOI] [PubMed] [Google Scholar]
- Shah V. G. (2010) ‘Development of Oligonucleotide-Based Microarrays for the Detection of Plant Viruses and the First Characterization of a Plant Virus Belonging to the Family Totiviridae’, PhD thesis, Oklahoma State University, Stillwater, Oklahoma. [Google Scholar]
- Skotnicki M. L., Mackenzie A. M., Gibbs A. J. (1996) ‘Genetic Variation in Populations of Kennedya Yellow Mosaic Tymovirus’, Archives of Virology, 141: 99–110. [DOI] [PubMed] [Google Scholar]
- Snyder J. C., et al. (2007) ‘Virus Movement Maintains Local Virus Population Diversity,’ Proceedings of the National Academy of Sciences United States of America, 104: 19102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D. C., Seifers D. L., French R. (2002) ‘Patterns of Polymorphism in Wheat Streak Mosaic Virus: Sequence Space Explored by a Clade of Closely Related Viral Genotypes Rivals that between the Most Divergent Strains’, Virology, 302: 58–70. [DOI] [PubMed] [Google Scholar]
- ter Braak C. J. F. (1988) ‘Partial canonical correspondence analysis,’ in Bock H. H. (ed) Classification and Related Methods of Data Analysis. Amsterdam: North-Holland, pp. 551–8. [Google Scholar]
- ter Braak C. J. F., Šmilauer P. (2002) CANOCO Reference Manual and CanoDraw for Windows User’s Guide: Software for Canonical Community Ordination (Version 4.5). Ithaca, NY: Microcomputer Power. [Google Scholar]
- Thapa V., et al. (2012) ‘Detection of Members of the Secoviridae in the Tallgrass Prairie Preserve, Osage County, Oklahoma, USA’, Virus Research, 167: 34–42. [DOI] [PubMed] [Google Scholar]
- Wintermantel W. M., Wisler G. C. (2006) ‘Vector Specificity, Host Range, and Genetic Diversity of Tomato Chlorosis Virus’, Plant Disease, 90: 814–9. [DOI] [PubMed] [Google Scholar]
- Wren J. D., et al. (2006) ‘Plant Virus Biodiversity and Ecology’, PLoS Biology, 4: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]