Abstract
The glycome, the full complement of glycans that cells produce, is an attractive target for molecular imaging. Imaging of the glycome in living systems has recently been enabled via bioorthogonal chemical reporter-based approaches. In this chapter, we describe two approaches to introduce bioorthogonal chemical reporters (tags) onto cell surface fucosylated glycans and glycans bearing LacNAc disaccharides, respectively. The tagged glycans can then be conjugated to imaging probes via bioorthogonal click chemistry. Similar approaches can be extended to image other sectors of the glycome in living systems.
1. Introduction
The surfaces of eukaryotic cells are covered with complex glycans that participate in a variety of physiological processes, including angiogenesis, fertilization, embryogenesis, cell adhesion, and neuronal development (Gupta et al., 2009; Ma et al., 2006; Murrey and Hsieh-Wilson, 2008; Varki et al., 2008). Inside the cell, glycans regulate transcription, translation, and protein trafficking (Gouyer et al., 2001; Hart et al., 2011). Glycoconjugates and glycoproteins are also found in certain prokaryotes, including pathogenic bacterium Campylobacter jejuni and human gut symbionts within the order Bacteroidales (Coyne et al., 2005). Studies have shown that these glycoproteins are either associated with virulence factors of clinically significant pathogens (Schmidt et al., 2003) or play critical roles in host–symbiont interactions (Comstock, 2009).
The full complement of glycans that a cell produces is collectively termed the cell's glycome. Many cellular factors, including the cell's genome, transcriptome, and proteome, and environmental cues and nutrients may have influence on the glycome (Freeze, 2006). Thus, the glycome responds to and reports on the physiological state of the cell. In humans, changes in cells’ glycome are associated with developmental disorders and defects (Varki et al., 2008), and can mark the onset of cancer and inflammation (Brooks et al., 2008; Dube and Bertozzi, 2005). The ability to visualize and characterize these changes in living systems would advance our understanding of the detailed roles of glycans in these processes and provide new clinical tools for disease diagnosis. However, glycans are assembled in a step-wise fashion by multiple enzymes, that is, monosaccharide kinase, nucleotidylyltransferase, glycosyltransferase, and thus by multiple genes. Therefore, they are not amenable to imaging techniques that rely on genetic reporters (i.e., green fluorescent protein, GFP).
Conventional methods to detect cell surface glycans rely on lectins and antibodies (Comer et al., 2001; David et al., 1992; Duijvestijn et al., 1988; Pilobello and Mahal, 2007). Lectins have been used extensively for the detection of both monosaccharides and oligosaccharides (Hirabayashi, 2008), and many of them have been commercialized. However, lectins typically have low affinity for their glycan epitopes and are often toxic (Ohba and Bakalova, 2003). Likewise, most antibodies generated against glycans are of the low-affinity IgM subtype and are tissue-impermeant. Moreover, lectin- and antibody-based imaging approaches only provide a snapshot of the labeled glycans at a particular time-point, and cannot be used for dynamic studies in a cellular environment. Glycans can also be metabolically labeled with radioisotope-bearing monosaccharides or the corresponding nucleotide sugars (Becker and Lowe, 2003; Jork et al., 1984), but the subsequent visualization of the radioisotopically labeled glycans in intact cells is not well resolved or amenable to current fluorescence microscopy techniques.
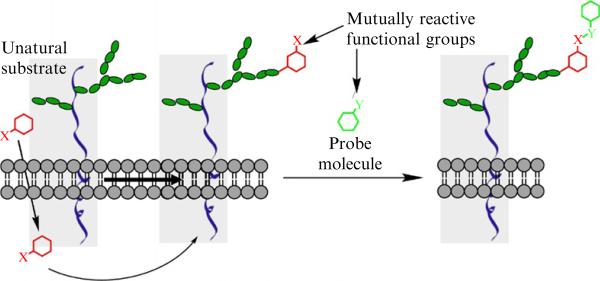
It is now possible to image the glycome in live cells or in living organisms using new tools from the emerging field of bioorthogonal click chemistry (Baskin and Bertozzi, 2007; Laughlin and Bertozzi, 2009a,b). By hijacking a cell's glycan biosynthetic machinery, a monosaccharide building block functionalized with a bioorthogonal chemical tag is incorporated into target glycoconjugates. Subsequently, a tailor-made click reaction is employed to conjugate a complementary biophysical probe, enabling visualization (Laughlin and Bertozzi, 2009a,b), or enrichment of the target glycoproteins for molecular identification (Fig. 21.1) (Hanson et al., 2007; Wang et al., 2010).
Figure 21.1.
A schematic description of metabolic labeling of cell surface glycans for fluorescence imaging. Red: an unnatural monosaccharide building block functionalized with a bioorthogonal chemical tag X; Green: a biophysical probe functionalized with a complementary chemical group Y. The covalent reaction between X and Y enables visualization or enrichment of the target glycoproteins for molecular identification.
In this chapter, we first briefly describe the most frequently used bioorthogonal click chemistry that has been applied to imaging the glycome in vivo. Next, we focus on two bioorthogonal click chemistry-based techniques developed in our laboratory for imaging fucosylated glycans and glycans bearing N-acetyl-D-lactosamine (LacNAc, Galβ1,4GlcNAc) in living systems.
2. Bioorthogonal Chemistry in Glycan Labeling: Merits and Limitations
2.1. The condensation of ketones/aldehyde with aminooxy and hydrazide reagents
The first bioorthogonal click reaction that has been exploited to label surface glycans for imaging studies is the condensation of ketones and aldehydes with aminooxy and hydrazide-bearing reagents to form stable hydrazones and oximes adducts, respectively (Fig. 21.2A). The optimal pH of these reactions is 5–6, which works for some cell lines, but is not compatible with many living systems.
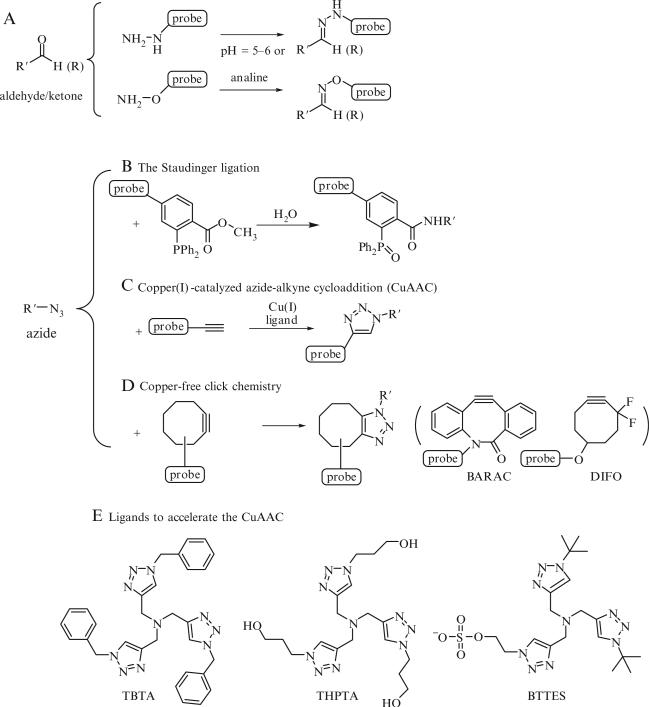
Figure 21.2.
Most frequently used bioorthogonal click chemistry for imaging studies. (A) The condensation reaction of ketones and aldehyde with aminooxy and hydrazide-bearing reagents to form stable hydrazones and oximes adducts, respectively; (B) The Staudinger ligation; (C) Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC); (D) the strain-promoted cycloaddition of azides and cyclooctynes; (E) representative examples of Cu(I)-stabilizing ligands to accelerate the CuAAC. TBTA=tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, THPTA=Tris-(hydroxypropyltriazolylmethyl)amine, BTTES=2-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl) ethyl hydrogen sulfate.
In 1997, Bertozzi and coworkers reported that an N-acetyl mannosamine analog derivatized with a levulinoyl side chain can be metabolized and incorporated into cell surface sialylated glycans in Jurkat cells. The ketone group allows subsequent reaction with a biotin hydrazide probe, and the labeled cells can then be detected using flow cytometry (Mahal et al., 1997).
To improve the biocompatibility of this reaction, Dawson and coworkers introduced aniline as a catalyst to accelerate the reaction so that the condensation can be performed at physiological conditions (Dirksen et al., 2006). The Paulson group, in a joint effort with Dawson and coworkers, used this optimized reaction to image sialylated glycans on the surface of mammalian cells (Zeng et al., 2009). In their experiment, cells were first subjected to mild periodate oxidation to selectively introduce an aldehyde at the C-7 position of sialic acid, which then underwent a condensation reaction with aminooxybiotin catalyzed by aniline (Fig. 21.2A). Subsequently, the treated cells were stained with a streptavidin conjugated with a green fluorophore. As analyzed by confocal microscope, robust labeling was achieved on the cell surface without jeopardizing membrane integrity.
2.2. Azide-associated bioorthogonal reactions
To date, azide is the most utilized bioorthogonal chemical tag for labeling glycans due to its small size and inertness to most components in a biological environment (Sletten and Bertozzi, 2009). Three bioorthogonal click reactions have been reported for labeling azide-tagged biomolecules. The Staudinger ligation covalently links the azide and an ester-functionalized triphenylphosphine via an amide bond (Fig. 21.2B) (Saxon and Bertozzi, 2000). Recently, this reaction has been successfully applied for imaging sialylated tumor cell glycans in vivo (Neves et al., 2011). As demonstrated by Brindle and coworkers, peracetylated azidoacetyl mannosamine was injected intraperitoneally to label sialic acids with an azide tag in tumor-implanted nude mice. The sialic acid-associated azides were then reacted, by Staudinger ligation, with a biotinylated phosphine probe and the biotin was detected by subsequent intravenous injection of a far-red fluorophoreor a DOTA-111In-conjugated neutravidin. At 24 h after administration of neutravidin derivatives, the mice were imaged using optical imaging or single-photon-emission computed tomography (SPECT), respectively. Positive signal resulting from the azido-dependent labeling was primarily detected in tumors. As upregulated sialylation is strongly correlated with the transformed phenotype of many cancers, this technique has the potential to be translated into a clinical setting to monitor the progression of cancer.
Though highly specific, the Staudinger ligation suffers from slow reaction kinetics and competing oxidation of the phosphine reagents (Baskin et al., 2007). By contrast, the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (Fig. 21.2C) (Rostovtsev et al., 2002; Tornoe et al., 2002), promoted by the Cu(I)-stabilizing ligand TBTA (Fig. 21.2E) (Chan et al., 2004), enjoys readily available coupling reagents and improved kinetics (Hein and Fokin, 2010; Wu and Fokin, 2007). It has been used extensively by chemical biologist for labeling glycans (Laughlin and Bertozzi, 2009a,b) and detecting glycoproteins in cell lysates (Chang et al., 2009; Hsu et al., 2007). However, the current Cu(I) catalyst formulation has two major problems: toxicity, hindering its use in living systems (Prescher and Bertozzi, 2005; Sletten and Bertozzi, 2009), and slow kinetics in aqueous solutions at micromolar substrate concentrations (Soriano del Amo et al., 2010), resulting in incomplete cycloaddition reaction (Kaltgrad et al., 2007), which hampers its use in modifying biomolecules of limited quantities.
The third reaction, the strain-promoted cycloaddition of azides and cyclooctynes (Agard et al., 2006; Jewett et al., 2010), inherits the bio-benign characteristics of the Staudinger ligation but is further endowed with improved kinetics (Fig. 21.2D) (Baskin et al., 2007; Jewett and Bertozzi, 2010; Ning et al., 2008). Among the cyclooctynes developed by the Bertozzi and Boons group, a difluorinated cyclooctyne (DIFO) (Baskin et al., 2007), and a biarylazacyclooctynone (BARAC) (Jewett et al., 2010) showed rapid kinetics in biomolecular labeling experiments. Currently, DIFO-fluorophore conjugates are the first choice for imaging azide-tagged glycans in living systems (i.e., zebrafish embryos and Caenorhabditis elegans) (Laughlin and Bertozzi, 2009a,b; Laughlin et al., 2008). However, labeling requires a 1-h incubation time (Baskin et al., 2010; Laughlin et al., 2008). Moreover, in vivo studies revealed that cyclooctyne-based probes bind to mouse serum albumin nonspecifically, presumably via covalent reactions with cysteine residues (Chang et al., 2010). In addition, the construction of these cyclooctynes involves a multistep linear synthesis that can be technically challenging (Baskin et al., 2007; Poloukhtine et al., 2009).
Most recently, our lab discovered a new ligand for CuAAC—BTTES (Fig. 21.2E) (Soriano del Amo et al., 2010). Not only does BTTES dramatically boost the reactivity of CuAAC, it also confers the canonical CuAAC with biocompatibility. The BTTES-Cu(I) catalyst allowed, for the first time, noninvasive imaging of fucosylated glycans during zebrafish early embryogenesis. Robust labeling of the enveloping layer, the embryos’ outermost layer of cells, was achieved within 2–3 min. Using this bioorthogonal reaction, we also developed a chemoenzymatic approach for labeling cell surface glycans bearing the LacNAc disaccharide for imaging and glycomic analysis (Zheng et al., 2011).
3. Metabolic Labeling of Cell Surface Fucosylated Glycans in Zebrafish Embryos for Fluorescence Imaging
In the past 14 years, the metabolic approach combined with bioorthogonal chemistry has been successfully used for the detection and imaging of several sectors of the glycome, including mucin-type O-linked glycans (Hang et al., 2003), sialylated (Baskin et al., 2007; Chang et al., 2009; Hong et al., 2010), fucosylated glycans (Hsu et al., 2007; Rabuka et al., 2006; Soriano del Amo et al., 2010), and cytosolic O-GlcNAcylated proteins (Vocadlo et al., 2003).
In this section, we describe the detailed procedures for imaging cell surface fucosylated glycans in zebrafish embryos by microinjection of GDP-6-ethynylfucose (GDP-FucAl) followed by conjugation of azide-bearing fluorophores via the biocompatible CuAAC and imaging via confocal fluroscence microscopy.
L-fucose, the signature monosaccharide possessed by all fucosylated glycans, is a determinant of many functional glycans that play key roles in numerous physiological and pathological processes (Becker and Lowe, 2003; Lu and Stanley, 2006; Ma et al., 2006). Specific terminal glycan fucosylation confers unique properties to cell surface glycoconjugates and is often regulated in cellular differentiation and embryogenesis (Becker and Lowe, 2003). Fucosylated glycans are also critical mediators of cell–cell recognition, neurite outgrowth, and neuronal migration during central nervous system development (Brito et al., 2007). Therefore, fucosylated glycans represent an attractive target for molecular imaging. We use the zebrafish embryo as a vertebrate model system for imaging fucosylated glycans due to its rapid embryonic development, amenability to genetic and embryological manipulations, and its optical clarity.
In nature, fucosylated glycans are synthesized by fucosyltransferases, enzymes that transfer the activated fucose from GDP-fucose (GDP-Fuc) to the acceptor substrates. GDP-Fuc is biosynthesized through two independent pathways: a de novo biosynthetic pathway and a salvage pathway (Ma et al., 2006). In the salvage pathway, fucose is first activated by fucokinase to form Fuc-1-Phosphate, which is then converted to GDP-Fuc by GDP-fucose pyrophosphorylase. In vertebrates, the de novo biosynthetic pathway usually produces 90% of the total GDP-Fuc and the remaining 10% of GDP-Fuc is contributed by the salvage pathway (Becker and Lowe, 2003). Discovered by Wong and Bertozzi, the salvage pathway of cultured mammalian cells can be hijacked by unnatural fucose analogs, such as 6-azidofucose (FucAz) and 6-ethynylfucose (FucAl) (Rabuka et al., 2006); (Hsu et al., 2007).
To bypass the low yielding savage pathway, we designed a strategy to inject GDP-FucAl, directly as the metabolic precusor into the yolk sack of one-cell stage embryos (Fig. 21.3A). When GDP-FucAl diffuses into daughter cells, the alkyne-bearing fucose will be incorporated into cell surface fucosylated glycans, allowing a click reaction with a fluorophore-bound azide via the BTTES-mediated CuAAC (Soriano del Amo et al., 2010).
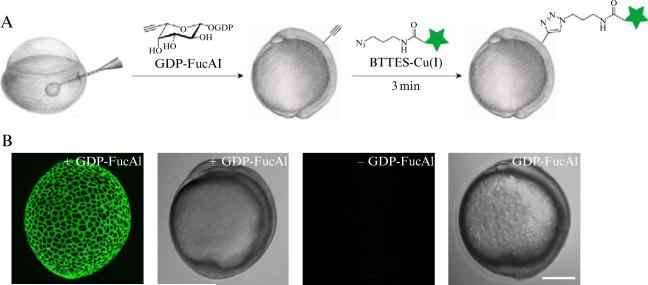
Figure 21.3.
A schematic description of metabolic labeling of cell surface fucosylated glycans in zebrafish embryos for fluorescence imaging. (A) workflow of metabolic labeling of fucosylated glycans followed by conjugation with fluorophores via CuAAC for imaging; (B) representative images of zebrafish embryos treated by this process: from left to right, fluorescence image of 10 hpf embryos treated with GDP-FucAl followed by a click reaction with Alexa Fluor 488-azide; the corresponding bright field image of 10 hpf zebrafish embryos treated with GDP-FucAl followed by a click reaction with Alexa Fluor 488-azide; fluorescence image of 10 hpf embryos treated without GDP-Fuc; the corresponding bright field image of 10 hpf embryos treated without GDP-Fuc followed by a click reaction with Alexa Fluor 488-azide.
3.1. Incorporation of alkyne groups to the cell surface fucosylated glycans in zebrafish embryos
3.1.1. Required materials
Microinjection apparatus
World Precision Instruments (Sarasota, FL) provides the main device (PV 820 Pneumatic PicoPump). The microinjection is done under Nikon SMZ1500 with lens Plan Apo 1 × WD70 (Nikon, Tokyo, Japan).
Microinjection needle preparation materials
Sutter Instrument Co. provides both needle glass and puller. Needle puller is flaming/brown micropipette puller, Model P-9. Needle glass is borosilicate glass with filament, OD: 1.0 mm, ID: 0.5 mm, 10 cm length, fire-polished.
Zebrafish embryos
Any wild-type zebrafish can be used in this study, for example, WIK, AB, and Tüebigen (for detailed information, see http://zebrafish.org/zirc/fish/lineAll.php). We recommend using Casper mutant (ZIRC catalog ID: ZL1714), a homozygous double mutant that lacks melanocytes and iridophores due to the mutations in both the genes of mifta and roy, because of its physical transparency. Fish lines are maintained under the recommended instruction of Zebrafish International Resource Center (ZIRC).
GDP-FucAl injection solution
20 mM GDP-FucAl in 0.2 M potassium chloride with either Alexa Fluor 594-dextran (5%, w/v) or phenol red loading dye (0.1%, w/v) as tracer. FucAl is chemically synthesized based on the reported procedure (Sawa et al., 2006) and is converted into GDP-FucAl using l-fucokinase/GDP-fucose pyrophosphorylase (FKP), a bifunctional enzyme isolated from Bacteroides fragilis 9343 (Wang et al., 2009).
Other reagents
Fish water
60 mg “Instant Ocean” per liter distilled water.
E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, pH 7.4).
Pronease E (Fisher Scientific) solution
1 mg/mL prepared with E3 embryo medium. Store at 4° C.
Agarose (Invitrogen).
Others
Fish plastic mating cage set.
35 and 100 mm tissue culture petri dish.
Fire-polished glass Pasteur pipette.
3.1.2. Single-cell zebrafish eggs preparation for microinjection
3.1.2.1. One day prior to microinjection
In the evening, prepare the mating set by inserting the mating cage with mesh bottom inside the fish tank and filling with fish water. Add the divider. Put single pair per cage.
Petri dishes for egg transfer and embryo handling have to be all coated with agarose. To make those agar-coated petri dishes, enough warm agarose (1.2%, v/w, in E3 embryo medium) is poured onto the petri dish to cover the entire surface and immediately poured out, leaving a very thin layer of agarose. Air-dry until agarose is solidified.
Microinjection dishes need to be prepared following the reported protocol (Kemp et al., 2009). Basically, a glass slide is inserted at a 45° angle across the widest part of 100 mm petri dish with three-quarters full of warm agarose (1.2%, v/w, in E3 embryo medium). Let the agarose solidify and remove the slide after complete solidification, leaving a beveled trough.
3.1.2.2. On the day of microinjection
In the morning, after the light is turned on, change the fish water and take out the divider. In this case, when fish spawn, eggs will fall through the mesh of the cage to the floor of fish tank, thus preventing the egg cannibalism by adult fish. Collect the eggs with a pipette and transfer them to a 35 mm agar-coated petri dish. Chorions have to be removed from the eggs before the microinjection by protease digestion. To achieve the complete release of the eggs, remove the water as much as possible and add 1 mg/mL pronease E in E3 embryo medium. Usually this enzymatic digestion lasts for 3–5 min, during which bubbling around chorion can be observed followed by the eggs withdrawn from the chorion. Immediately transfer those dechorioned eggs to a beaker filled with fish water by merging the petri dish into the water and gently sliding the petri dish down. Rinse the eggs with fish water for three times, then transfer the dechorionated eggs using fire-polished pipette to the microinjection dishes filled with E3 embryo medium.
3.1.3. Microinjection
Prepare the sterilized injection needle and load 2 μL GDP-FucAl injection solution. Break the tip of the needle carefully. Adjust the injection pressure and duration and inject each egg with 1 nL of injection solution. Then transfer the eggs back into the 100 mm agar-coated petri dishes filled with E3 embryo medium.
3.1.4. Zebrafish Embryo handling after microinjection
Incubate the eggs at 28 ° C in the incubator under the flow of air. Remove the unfertilized eggs within 5 hours postfertilization (hpf). Unfertilized eggs adopt abnormal cell division, if any; while fertilized eggs undergo asymmetrical divisions during this time period. Transfer the fertilized eggs to a new agar-coated petri dish filled with E3 embryo medium. The fish density should be no more than 60 per one 100 mm petri dish.
3.2. Conjugation of Azide-bound fluorophores by click chemistry
3.2.1. Required materials
Reagents for click reaction
Alexa Fluor-488 azide (Invitrogen)
2.5 mM stock solution in water. Store at 4 ° C in dark.
The CuSO4-BTTES catalyst
[BTTES]:[CuSO4] = 6:1, [CuSO4] = 2.5 mM, in water. BTTES is synthesized and purified based on our published work (Soriano del Amo et al., 2010) and adjusted to pH 7.4 by 1 M NaOH before mixing with CuSO4 solution. BTTES stock should be stored at 4 ° C. Copper sulfate solution can be prepared from dissolving commercially available copper sulfate pentahydrate. Copper sulfate stock solution can be stored at room temperature and last for months. Seal the stock solution bottle with parafilm to avoid water evaporation and check the copper concentration by UV–vis spectrometry if necessary.
Tip
We recommend using fresh prepared catalyst if possible, but premixed CuSO4–BTTES catalyst can last for 1–3 weeks when stored at 4 ° C.
Sodium ascorbate
100 mM stock in water. Sodium ascorbate is a strong reductant used to generate Cu(I) from Cu(II) in situ. In aqueous solutions, sodium ascorbate is easily oxidized so the stock solution has to be freshly prepared every time right before the experiment.
Bathocuproine sulphonate (BCS)
50 mM stock in water. BCS is a biocompatible copper chelator used to quench the CuAAC. It is sensitive to light and the stock solution should be stored at 4 ° C in dark.
Additional reagents
E3 embryo medium
Agarose
Disposable
96-well plate
Fire-polished glass Pasteur pipette
Glass petri dish
3.2.2. Click chemistry performance
Coat the base of 96-well plate with agarose before transferring the embryos as described in Section 3.1.2. Add 92 μL E3 embryo medium to each well followed by addition of 4 μL Alexa Fluor-488 azide stock solution and 2 μL CuSO4–BTTES catalyst 1:6 complex. Gently shake the plate to mix. Then carefully transfer the embryos at desired developmental stages (e.g., late gastrula, tissue segmentation, and early larva, etc.) into the well using a fire-polished glass Pasteur pipette. Each well should contain less than five embryos. To initiate the click reaction, add 2.5 μL freshly prepared sodium ascorbate stock and shake gently.
Tip
The final concentration for each reagent in click reaction is: Alexa Fluor-488 azide: 100 μM; CuSO4: 50 μM; BTTES: 300 μM; sodium ascorbate: 2.5 mM.
After 3 min, add 2 μL BCS (final concentration: 1 mM) to quench the reaction then dilute the reaction system immediately by adding 100 μL E3 embryo medium. Transfer the embryos to a glass petri dish and wash the treated embryos two times with 15 mL E3 embryo medium.
3.3. Imaging
3.3.1. Required apparatus
Confocal fluorescence microscope: Leica SP5 Confocal fluorescent microscope is used in our studies to obtain the images following the manual instruction.
Additional reagents
Ultralow melting point agarose (Invitrogen)
E3 embryo medium
Disposable
MatTek glass bottom microwell dish
3.3.2. Procedure
Prepare ultralow melting point agarose in E3 embryo medium at the concentration of 1.2% (w/v). Place a drop of the agarose solution on a MatTek glass bottom microwell dish. Use a fire-polished glass Pasteur pipette to transfer an embryo into the agarose drop. Position the embryos based on your experiment design (e.g., dorsally or laterally). Set the microwell dish on ice for 5 min to solidify the agarose drop, then add E3 embryo medium gently to the dish until it covers the agarose drop. Place the microwell dish onto the microscope workstation and start to acquire images. Fluorescence and bright field images are acquired sequentially using a 5 μm step interval. Composite figures are prepared using ImageJ (Collins, 2007) (Fig. 21.3B).
4. Labeling Cell Surface Glycans Bearing the LacNAc Disaccharide in Chinese Hamster Ovary(CHO) Cells for Fluorescence Imaging
Although the metabolic glycan labeling strategy we described in Section 3 gives us promising results in vivo, there are two downsides to this approach: (1) certain unnatural sugars, such as 6-azidofucose (Rabuka et al., 2006) and Ac4ManNAz impart different levels of toxicity to live cells and organisms; (2) the metabolic incorporation of a monosaccharide derivative cannot specifically label a oligosaccharide of defined composition due to the prevalence of the monosaccharide in various oligosaccharides on the cell surface.
In this section, we describe an alternative method to label cell surface oligosaccharide glycans in live mammalian cells using a chemoenzymatic strategy instead of hijacking the glycan biosynthetic pathways with unnatural substrates. The sector of the glycome we target here is glycans bearing N-acetyllactosamine (LacNAc, Galβ1,4GlcNAc). This protocol utilizes a recombinant Helicobacter pylori α-(1,3)-fucosyltransferase C168S mutant (α-1,3-FucTM) to transfer a C-6 azide- or alkyne-tagged fucose residue to the 3-OH of N-acetylglucosamine of the LacNAc disaccharide. The bioorthogonal tag can then be selectively derivatized with probes via CuAAC or the copper-free click chemistry as described in Section 3. for imaging (Fig. 21.4A).
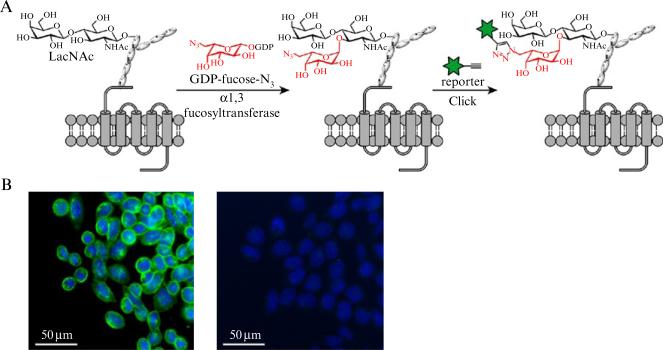
Figure 21.4.
A schematic description of chemoenzymatic labeling cell surface glycans bearing LacNAc in Lec2 CHO cells for fluorescence imaging. (A) workflow of the chemoenzymatic labeling LacNAc by GDP-FucAz followed by conjugation of fluorophores for imaging; (B) Fluorescence images Lec2 CHO cells labeled by this method: left: fluorescence image of Lec2 CHO cells treated with GDP-FucAz followed by a click reaction with Alexa Fluor 488-alkyne; right: fluorescence image of Lec2 CHO cells treated with GDP-Fuc followed by a click reaction with Alexa Fluor 488-alkyne. Green: cell surface labeled by Alexa Fluor 488; Blue: nucleus stained by Hoechst 33342 dye.
Since LacNAc-bearing glycans are developmentally regulated and the upregulation of LacNAc is correlated with malignant phenotypes, such as colorectal cancer (Ichikawa et al., 1999), this method for imaging LacNAc may serve as a powerful tool for tracing the changes in cell surface LacNA-cylation level during the development and has the potential to be transferred to a clinical setting for disease diagnosis.
4.1. Incorporation of alkyne groups into the CHO cell surface glycans bearing the LacNAc disaccharide via a chemoenzymatic approach
We chose a mutant CHO cell line—Lec2 CHO cells as a model system for labeling glycans bearing the LacNAc disaccharide. The major glycan epitope on the surface of Lec2 CHO cells are polyLacNAc on N-linked glycans (North et al., 2010). H. pylori α-(1,3)-fucosyltransferase C168S mutant (α-1,3-FucTM) is used to incorporate unnatural fucose analogs to LacNAc residues for the second step click modification. C168S mutation minimizes the dimerization of the parent enzymes while maintaining its activity.
4.1.1. Required materials
Cell line and media
Lec2 CHO mutant is established by Patnaik and Stanley (2006). This cell line can grow both in suspension and in monolayer. Incubation condition is 5.0% carbon dioxide and water-saturated at 37 ° C.
Growing medium
α-Minimum essential medium (α-MEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS).
α-1,3-Fucosyltransferase-M (a-1,3-FucMT)
α-1,3-FucTM can be generated using our established protocol (Zheng et al., 2011). The enzyme can be store at high concentration (>5 mg/mL) with 10% glycerol at 4 ° C and is stable for 3 months. The specific enzymatic activity of α-1,3-FucTM is ~6.0 U/mg protein (one unit is defined as the amount of enzyme that is required to convert 1 μM of GDP-fucose per min at 37 ° C).
Other reagents
GDP-FucAl stock solution
5 mM, in water. Store at 4 ° C (see Section 3.1.1).
GDP-FucAz stock solution
5 mM, in water. Store at 4 ° C. 6-azidofucose (FucAz) is chemically synthesized based on the reported procedure (Zheng et al., 2011) and is converted into GDP-FucAz using FKP by the same method as described for the synthesis of GDP-FucAl in Section 3.1.1.
Fucosylation buffer
HBSS buffer supplemented with 20 mM MgSO4, 3 mM HEPES and 1% FBS. Store at 4 ° C.
PBS + 1% FBS
Ca2+ and Mg2+ free supplemented with 1% FBS (v/v).
Store at 4 ° C.
Disposables
96-well round-bottom plates
Regular and multiple-channel pipette and pipette tips (Rainin Instrument)
Tissue Culture tubes
TC-treated, 16 × 125 mm, screw cap (Cornings Corp.)
4.1.2. Enzymatic incorporation of alkyne fucose derivatives on to LacNAc residues of the glycans on CHO cell surface
For flow cytometry
We grow Lec2 CHO cells in 15 mL screw-capped cell culture tube and harvest them when the cell density reaches ~1.0 × 106 cells/mL. Cultured cells are washed three times with PBS + 1% FBS (200 μL each time, for three times, centrifuge at 300 × g for 3 min) and resuspended in fucosylation buffer at the concentration of 5.0 × 106 cells/mL. Place 90 μL cell suspension in a 96-well plate (final density: ~0.45 × 106 cells per well). Treat the cells with 100 μM of GDP-FucAl and 30 mU of α-(1,3)FucT-M at 37 ° C for 10 min. After the reaction, wash the treated cells three times with PBS + 1% FBS then resuspend the cells in 100 μL of PBS + 1% FBS.
For imaging
Seed the Lec2 CHO cells at a density of 3000 cells per well in an eight-well LabTek II chambered cover glass with 100 μL α-MEM media (10% FBS). Allow the cells to grow for 72 h followed by washing for three times with PBS + 1% FBS (200 μL each time, for three times, centrifuge at 300 × g for 3 min). Then treat the cells with 200 μM GDP-FucAz in the presence of 30 mU α-(1,3)FucT-M in fucosylation buffer for 10 min at 37 ° C. After the incubation wash the cells three times with α-MEM medium.
4.2. Conjugation of azide-bound probes by click chemistry
4.2.1. Required materials
Biotin-azide (Click Chemistry Tools)
2.5 mM stock solution in water. Store at −20 ° C.
Streptavidin-Alexa Fluor 488 (Invitrogen)
100 μg/mL in water. Store at 4 ° C in dark.
Alexa Fluor-488 alkyne (Invitrogen)
10 mM in water. Store at 4 ° C in dark. CuSO4-BTTES catalyst, Sodium ascorbate and BCS are all prepared as described in Section 3.2.1.
Hoechst 33342 dye
1 mg/mL in DMSO. Store at −20 ° C.
α-MEM medium
Store at 4 ° C.
4.2.2. Click chemistry performance
For flow cytometry
After in situ fucosylation, add 2.5 μL biotin-azide and 2 μL CuSO4-BTTES catalyst to the Lec2 CHO cells suspension (final concentration: [biotin-azide] = 100 μM [CuSO4] = 50 μM, [BTTES]:[CuSO4] = 6:1) and ascorbate (final concentration = 2.5 mM). Mix well immediately after sodium ascorbate is added. Wait for 3 min and Quench the click reaction by the addition of 2 μL BCS. Wash the cells three times and resuspend the cells in 100 μL PBS + 1% FBS with streptavidin-Alexa Fluor 488 (final concentration = 1 μg/mL). Incubate in dark at 4 ° C for 30 min. Wash the cells three times with PBS + 1% FBS.
For imaging
The labeling of Lec2 CHO cells for fluorescence imaging is similar to what described above with the replacement of GDP-FucAl with GDP-FucAz, and biotin-azide with an alkyne-bearing fluorophore—Alexa Fluor-488 alkyne (this step can also be accomplished using GDP-FucAl and an azide-bearing fluorophore). In the cell suspension, add Alexa Fluor-488 alkyne (final concentration 1 μM), CuSO4–BTTES catalyst ([CuSO4] = 50 μM, [BTTES]:[CuSO4] = 6:1) and sodium ascorbate (final concentration = 2.5 mM). Mix well. After 3 min, quench the reaction with BCS (final concentration = 1 mM). Wash the cells with the α-MEM medium followed by treatment with Hoechst 33342 dye to stain the nucleus (1:1000 dilution in medium of the stock). Incubate the cells at room temperature in dark for 3 min and wash for three times with α-MEM medium (10% FBS), and image in the presence of 100 μL of the medium.
4.3. Labeling, characterization and imaging
4.3.1. Required materials
Device
Flow cytometry experiments are performed on a Becton Dickinson FACScan analog bench top analyzer using a 488 nm argon laser.
Reagents
FACS buffer: HBSS buffer, pH 7.4, 1% bovine serum albumin, 2 μg/mL 7-AAD, 0.2% NaN3. Store at 4 ° C in dark.
4.3.2. Labeling efficiency characterization by flow cytometry
After Lec2 CHO cells are treated as described in Section 4.2.2, they can be resuspended in 400 μL of FACS buffer for flow cytometric analysis. At least 18,000 cells are recorded for each sample. Flow cytometry data were analyzed using Flowjo (TriStar Inc.). Mean fluorescence intensity (MFI) is calculated for live cells. Cell viability is ascertained by gating the sample on the basis of forward scatter (to sort by size) and FL3 (to sort by 7-AAD negative).
4.3.3. Imaging
Image the cells prepared in Section 4.2.2 using a Zeiss Axio Observer. The channels imaged are DAPI (for nucleus staining) and FITC (for membrane staining). Images acquired were processed using Axiovision (Carl Zeiss MicroImaging). Composite figures are prepared using ImageJ (download at: http://rsbweb.nih.gov/ij/) and Photoshop CS2 (Adobe) (Fig. 21.4B).
ACKNOWLEDGMENTS
Research in the authors’ laboratory was supported by the National Institutes of Health (GM080585 and GM093282), the Mizutani Foundation for Glycoscience, and DuPont (DuPont Young Professor Award).
REFERENCES
- Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- Baskin JM, Bertozzi CR. Bioorthogonal click chemistry: Covalent labeling in living systems. QSAR Comb. Sci. 2007;26:1211–1219. [Google Scholar]
- Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, Lowe JB. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- Brito C, Escrevente C, Reis CA, Lee VM, Trojanowski JQ, Costa J. Increased levels of fucosyltransferase IX and carbohydrate Lewis(x) adhesion determinant in human NT2N neurons. J. Neurosci. Res. 2007;85:1260–1270. doi: 10.1002/jnr.21230. [DOI] [PubMed] [Google Scholar]
- Brooks SA, Carter TM, Royle L, Harvey DJ, Fry SA, Kinch C, Dwek RA, Rudd PM. Altered glycosylation of proteins in cancer: What is the potential for new anti-tumour strategies. Anticancer Agents Med. Chem. 2008;8:2–21. doi: 10.2174/187152008783330860. [DOI] [PubMed] [Google Scholar]
- Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper (I)-stabilizing ligands in catalysis. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew. Chem. Int. Ed. 2009;48:4030–4043. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. USA. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- Comstock LE. Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe. 2009;5:522–526. doi: 10.1016/j.chom.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- David G, Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H. Developmental changes in heparan sulfate expression: In situ detection with mAbs. J. Cell Biol. 1992;119:961–975. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen A, Dirksen S, Hackeng TM, Dawson PE. Nucleophilic catalysis of hydrazone formation and transimination: Implications for dynamic covalent chemistry. J. Am. Chem. Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation–potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Duijvestijn AM, Horst E, Pals ST, Rouse BN, Steere AC, Picker LJ, Meijer CJ, Butcher EC. High endothelial differentiation in human lymphoid and inflammatory tissues defined by monoclonal antibody HECA-452. Am. J. Pathol. 1988;130:147–155. [PMC free article] [PubMed] [Google Scholar]
- Freeze HH. Genetic defects in the human glycome. Nat. Rev. Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- Gouyer V, Leteurtre E, Zanetta JP, Lesuffleur T, Delannoy P, Huet G. Inhibition of the glycosylation and alteration in the intracellular trafficking of mucins and other glycoproteins by GalNAcalpha-O-bn in mucosal cell lines: An effect mediated through the intracellular synthesis of complex GalNAcalpha-O-bn oligosaccharides. Front. Biosci. 2001;6:D1235–D1244. doi: 10.2741/gouyer. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Bansal P, Ganguly A, Bhandari B, Chakrabarti K. Human zona pellucida glycoproteins: Functional relevance during fertilization. J. Reprod. Immunol. 2009;83:50–55. doi: 10.1016/j.jri.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. Tailored glycoproteomics and glycan site mapping using saccharide-selective bioorthogonal probes. J. Am. Chem. Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein JE, Fokin VV. Copper-catalyzed azide-alkyne cycloaddition (CuAAC) and beyond: New reactivity of copper(I) acetylides. Chem. Soc. Rev. 2010;39:1302–1315. doi: 10.1039/b904091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi J. Concept, strategy and realization of lectin-based glycan profiling. J. Biochem. (Tokyo) 2008;144:139–147. doi: 10.1093/jb/mvn043. [DOI] [PubMed] [Google Scholar]
- Hong V, Steinmetz NF, Manchester M, Finn MG. Labeling live cells by copper-catalyzed alkyne–azide click chemistry. Bioconjug. Chem. 2010;21:1912–1916. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc. Natl. Acad. Sci. USA. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Nakayama J, Sakura N, Hashimoto T, Fukuda M, Fukuda MN, Taki T. Expression of N-acetyllactosamine and beta1,4-galactosyltransferase (beta4GalT-I) during adenoma-carcinoma sequence in the human colorectum. J. Histochem. Cytochem. 1999;47:1593–1602. doi: 10.1177/002215549904701211. [DOI] [PubMed] [Google Scholar]
- Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jork R, Schmitt M, Lossner B, Matthies H. Dopamine stimulated L-fucose incorporation into brain proteins is related to an increase in fucokinase activity. Biomed. Biochim. Acta. 1984;43:261–270. [PubMed] [Google Scholar]
- Kaltgrad E, Sen Gupta S, Punna S, Huang CY, Chang A, Wong CH, Finn MG, Blixt O. Anti-carbohydrate antibodies elicited by polyvalent display on a viral scaffold. Chem. Bio. Chem. 2007;8:1455–1462. doi: 10.1002/cbic.200700225. [DOI] [PubMed] [Google Scholar]
- Kemp HA, Carmany-Rampey A, Moens C. Generating chimeric zebrafish embryos by transplantation. J. Vis. Exp. 2009;29 doi: 10.3791/1394. 10.3791/1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. In vivo imaging of Caenorhabditis elegans glycans. ACS Chem. Biol. 2009a;4:1068–1072. doi: 10.1021/cb900254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. Imaging the glycome. Proc. Natl. Acad. Sci. USA. 2009b;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Stanley P. Roles of O-fucose glycans in notch signaling revealed by mutant mice. Methods Enzymol. 2006;417:127–136. doi: 10.1016/S0076-6879(06)17010-X. [DOI] [PubMed] [Google Scholar]
- Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- Murrey HE, Hsieh-Wilson LC. The chemical neurobiology of carbohydrates. Chem. Rev. 2008;108:1708–1731. doi: 10.1021/cr078215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves AA, Stockmann H, Harmston RR, Pryor HJ, Alam IS, Ireland-Zecchini H, Lewis DY, Lyons SK, Leeper FJ, Brindle KM. Imaging sialylated tumor cell glycans in vivo. FASEB J. 2011;25:2528–2537. doi: 10.1096/fj.10-178590. 10.1096/fj.10-178590. [DOI] [PubMed] [Google Scholar]
- Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem. Int. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North SJ, Huang HH, Sundaram S, Jang-Lee J, Etienne AT, Trollope A, Chalabi S, Dell A, Stanley P, Haslam SM. Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 2010;285:5759–5775. doi: 10.1074/jbc.M109.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba H, Bakalova R. Relationships between degree of binding, cytotoxicity and cytoagglutinating activity of plant-derived agglutinins in normal lymphocytes and cultured leukemic cell lines. Cancer Chemother. Pharmacol. 2003;51:451–458. doi: 10.1007/s00280-003-0607-y. [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Stanley P. Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- Pilobello KT, Mahal LK. Lectin microarrays for glycoprotein analysis. Methods Mol. Biol. 2007;385:193–203. doi: 10.1007/978-1-59745-426-1_14. [DOI] [PubMed] [Google Scholar]
- Poloukhtine AA, Mbua NE, Wolfert MA, Boons GJ, Popik VV. Selective labeling of living cells by a photo-triggered click reaction. J. Am. Chem. Soc. 2009;131:15769–15776. doi: 10.1021/ja9054096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Bertozzi CR. Chemistry in living systems. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. A chemical reporter strategy to probe glycoprotein fucosylation. J. Am. Chem. Soc. 2006;128:12078–12079. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Riley LW, Benz I. Sweet new world: Glycoproteins in bacterial pathogens. Trends Microbiol. 2003;11:554–561. doi: 10.1016/j.tim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Sletten EM, Bertozzi CR. Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan A, Levy M, Liu Y, Marlow FL, Wu P. Biocompatible copper(I) catalysts for in vivo imaging of glycans. J. Am. Chem. Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. Cold Spring Harbor, New York: 2008. [PubMed] [Google Scholar]
- Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD, 3rd, Wu P. Chemoenzymatic synthesis of GDP-L-fucose and the Lewis X glycan derivatives. Proc. Natl. Acad. Sci. USA. 2009;106:16096–17101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Fokin VV. Catalytic azide-alkyne cycloaddition: Reactivity and applications. Aldrichimica Acta. 2007;40:7–17. [Google Scholar]
- Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Meth. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Jiang H, Gros M, Soriano Del Amo D, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, Wu P. Tracking N-acetyllactosamine on cell-surface glycans in vivo. Angew. Chem. Int. Ed. 2011;50:4113–4118. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]