Abstract
High–throughput technologies used to interrogate transcriptomes have been generating a great amount of publicly available gene expression data. For raw diseases that lack of clinical samples and research funding, there is a practical benefit to jointly analyze existing datasets commonly related to a specific rare disease. In this study, we collected a number of independently generated transcriptome data sets from four species: Human, Fly, Mouse and Worm. All data sets included samples with both normal and abnormal mitochondrial functions. We reprocessed each data set to standardize format, scale and gene annotation and used HomoloGene database to map genes between species. Standard procedure was also applied to compare gene expression profiles of normal and abnormal mitochondrial functions to identify differentially expressed genes. We further used meta–analysis and other integrative analyses to recognize patterns across data sets and species. Novel insights related to mitochondrial dysfunctions was revealed via these analyses, such as a group of genes consistently dysregulated by impaired mitochondrial function in multiple species. This study created a template for the study of rare diseases using genomic technologies and advanced statistical methods. All data and results generated by this study are freely available and stored at http://goo.gl/nOGWC2, to support further data mining.
Keywords: transcriptome data, mitochondrial dysfunction, integrative analysis
1. Introduction
The study of rare disease is limited by the availability of samples and other resources although it often helps researchers answer extremely important biomedical questions [1, 2, 3]. Some rare diseases can serve as models for more common one due to their close association. For example, primary mitochondrial diseases have a combined occurrence of about 1/5000 [4], but the investigation of these diseases have revealed extensive knowledge about mitochondrial dysfunction [5, 6], which has been commonly observed in diabetes [7], cancer [8] and neurodegenerative diseases [9, 10].
A deeper understanding of rare diseases can be achieved through utilization of innovative technologies, model animals, and integrative analysis of independently generated data sets. Paring modern genomic technologies with advanced statistical analysis has become an extremely valuable approach for investigating rare diseases [11, 12, 13]. This approach characterizes the overall state of one or more cellular systems, such as transcriptome, epigenome, and metabalome, and uses bioinformatic analysis to explore their interactions. It is then able to compensate for the small sample size of individual experiments with high dimension of data.
Integration of genomic data is practical challenging. Independently generated genomic data sets could be different from each other in many ways, such as technology, cell type, annotation, and data quality, which makes it critical to properly document metadata related to each experiment. Gene Expression Omnibus (GEO) [14], a public repository of transcriptome data sets, archives the metadata, raw data, and processed data of over 46, 000 data series. GEO provides an ideal platform for researchers to identify and access existing gene expression data sets related to a specific disease.
In this study, we present a semi–automated workflow to archive transcriptome data sets related to a specific rare disease, primary mitochondrial dysfunction. We have used this workflow to create Transcriptome of Mitochondrial Dysfunctions (ToMD), which currently includes 30 independent data sets and about 500 biological samples. Each of these data sets compared samples with primary mitochondrial dysfunction to those with normal or rescued mitochondrial function. Statistical methods were designed to evaluate data quality, standardize gene annotation, and perform integrative analysis. Preliminary analysis of these data sets has revealed new insights about primary mitochondrial diseases. All data sets and analysis results within archive are freely available http://goo.gl/nOGWC2.
2. Material and Methods
Below is a step-by-step description of the generation of ToMD. It is a semi-automated workflow that includes reproducible data processing and analysis steps, but also requires human decisions about data quality, sample grouping, analysis parameters, and so on.
2.1. Structure of the data archive
ToMD was structured in three tiers as described below:
Tier 1 is a collection of transcriptome data sets related to mitochondrial dysfunctions. It includes published data sets stored in GEO or other resources. All data sets were re-processed with a common procedure so they are properly normalized and annotated with unique ENTREZ gene IDs.
Tier 2 is a collection of results from pairwise comparisons. Each comparison was performed on two groups of samples within the same data set. Samples of one group usually have impaired mitochondrial functions induced by genetic or environmental factors while samples of the other group are controls with normal or rescued mitochondrial functions.
Tier 3 includes results of integrative analysis of data or results from multiple data sets. Given the high diversity of the data sets, each integrative analysis was customarily designed and documented in a separate subfolder. While it is unlikely to automate such analysis, we noticed that the availability of Tier 1 and 2 makes the integrative analysis much more efficient.
2.2. Identification of data sets
We worked closely with clinical researchers who treat mitochondrial diseases to search public repositories for transcriptome data sets related to primary mitochondrial diseases. These data sets were generated from human tissues and cell lines, as well as three model animals: C. elegans (worm), D. melanogaster (fruit fly), and M. musculus (mouse). No data generated from Sus scrofa (domestic pig) was selected due to the incompleteness of its gene annotation. Each data set includes two or more sample groups corresponding to abnormal and normal/rescued mitochondrial function. The mitochondrial dysfunction could be caused by pathogenic gene mutations in patients, knockdown/knockout of key mitochondrial genes in cultured cells or model animals, or exposure to chemicals such as Rotenone and Rapamycin.
Table 1 describes the four living species datasets. The table shows each dataset species, the dataset id that is used in this study, the dataset reference, the GEO id, the tissue, number of samples and genes, and the number of sample groups. The fly dataset compiles two different dataset sources with 12, 521 overlapped genes. The worm dataset compiles three different datasets with 16, 688 overlapped genes. The Human dataset compiles 13 different datasets with 895 overlapped genes then we excluded one of the human datasets because of too many missing values. The Mouse dataset compiles 12 different datasets with 10, 279 overlapped genes then we excluded one of the datasets because of too many missing values. Table 2 shows the four compiled species datasets, it shows the number of genes and samples for each species, in the single part of the table.
Table 1.
The four living species datasets description.
| Speciesa | DS Idb | GEO_IDc | Ref.d | Tissuee | #Samplesf | #Genesg | #Groups (classes)h |
|---|---|---|---|---|---|---|---|
| Fly | F_dataset1 | GSE10169 | [15] | Whole body | 12 | 12,522 | 4 |
| F_dataset2 | GSE32912 | [16] | S2 cell | 21 | 12,522 | 11 | |
|
| |||||||
| Worm | W_dataset1 | GSE9896 | [17] | Whole body | 10 | 16,812 | 2 |
| W_dataset2 | GSE9897 | [18] | Whole body | 10 | 16,812 | 6 | |
| W_dataset3 | GSE46051 | [19] | Whole body | 22 | 28,355 | 8 | |
|
| |||||||
| Human | H_dataset1 | GSE1462 | [20] | Skeletal muscle | 15 | 12,027 | 4 |
| H_dataset2 | GSE8648 | [21] | Fibroblast | 22 | 1,190 | 2 | |
| H_dataset3 | GSE10956 | [22] | Fibroblast | 22 | 13,031 | 2 | |
| H_dataset4 | GSE14882 | [23] | Peripheral blood | 16 | 12,027 | 2 | |
| H_dataset5 | GSE24945i | [24] | Fibroblast | 12 | 19,617 | 4 | |
| H_dataset6 | GSE26322 | [25] | Fibroblast | 12 | 19,063 | 2 | |
| H_dataset7 | GSE27041 | [26] | Fibroblast | 20 | 18,895 | 4 | |
| H_dataset8 | GSE27545 | [27] | Cybrid | 17 | 19,600 | 6 | |
| H_dataset9 | GSE28206 | [28] | Skeletal muscle | 7 | 19,600 | 2 | |
| H_dataset10 | GSE33769 | [29] | Fibroblast | 15 | 18,895 | 2 | |
| H_dataset11 | GSE33940 | [30] | Fibroblast | 20 | 19,714 | 3 | |
| H_dataset12 | GSE35642 | [31] | SK-N-MC cell | 18 | 12,027 | 6 | |
| H_dataset13 | GSE42986 | [32] | Muscle/FCL | 40 | 20,708 | 4 | |
|
| |||||||
| Mouse | M_dataset1 | GSE4330 | [33] | Myoblast (C2C12) | 21 | 14,706 | 7 |
| M_dataset2 | GSE4866 | [34] | Cochlea | 10 | 17,480 | 2 | |
| M_dataset3 | GSE5332 | [35] | MEF | 13 | 17,480 | 4 | |
| M_dataset4 | GSE5786 | [36] | Striata | 6 | 20,628 | 2 | |
| M_dataset5 | GSE6210 | [37] | Liver/Muscle | 12 | 17,480 | 4 | |
| M_dataset6 | GSE10904 | [38] | Liver | 6 | 17,480 | 2 | |
| M_dataset7 | GSE13034 | [39] | MEF | 14 | 12,279 | 4 | |
| M_dataset8 | GSE18677i | [40] | Liver | 11 | 17,480 | 8 | |
| M_dataset9 | GSE27309 | [41] | Brown adipose | 20 | 20,299 | 2 | |
| M_dataset10 | GSE27954 | [42] | Liver | 12 | 21,154 | 4 | |
| M_dataset11 | GSE30552 | [43] | Liver | 26 | 17,480 | 4 | |
| M_dataset12 | GSE34773 | [44] | Skeletal muscle | 26 | 17,480 | 4 | |
The dataset species.
DS Id is the dataset id number that is used in this study.
The GEO_ID in the NCBI database.
The dataset reference.
The tissue that the sample comes from.
The number of samples in the dataset.
The number of genes in the dataset.
The number of sample groups in the dataset.
Too many missing values in this dataset.
Table 2.
Species datasets overlap and the extracted bicluster results summary.
| Overlap | Speciesa | #Genesb | #Samplesc | #Biclustersd | Avg MSRxe |
|---|---|---|---|---|---|
| Single f | F | 12,521 | 33 | 5 | 0.87 |
| M | 10,279 | 147 | 5 | 0.92 | |
| H | 895 | 233 | 6 | 0.93 | |
| W | 16,688 | 42 | 6 | 0.98 | |
|
| |||||
| Pairwise g | F,H | 443 | 256 | 8 | 0.87 |
| H,W | 377 | 265 | 6 | 0.80 | |
| M,H | 675 | 370 | 1 | 0.99 | |
| F,W | 2,724 | 75 | 9 | 0.96 j | |
| F,M | 2,944 | 180 | 7 | 0.95 j | |
| W,M | 2,240 | 189 | 8 | 0.88 | |
|
| |||||
| Triplet h | F,W,M | 1,637 | 222 | 7 | 0.92 j |
| H,F,M | 341 | 403 | 0k | – | |
| H,W,F | 307 | 298 | 0k | – | |
| H,W,M | 286 | 412 | 0k | – | |
|
| |||||
| All i | H,F,M,W | 235 | 445 | 7 | 0.85 |
F: Fly; W: Worm; M: Mouse; H: Human.
The number of genes in the dataset.
The number of samples in the dataset.
Number of biclusters with gene counts and sample counts more than or equal5.
The average mean squared residue of for the chosen biclusters with gene counts and sample counts more than or equal 5.
The overlap of the datasets in each living species.
The overlap of the species datasets in pairs.
The overlap of the species datasets in triplets.
The overlap across all species.
The best MSR results in the overlapped datasets.
All of the extracted biclusters gene counts or sample counts less than 5.
We used HomoloGene [45], a tool to build putative homology groups from the full genome of a different eukaryotic species, to construct putative orthologs for the four species. We used the putative orthologs to identified the species genes overlap in the constructed datasets as shown in table 2. Table 2 shows four groups of dataset information. The first part of the table shows the compiled datasets for each species under the heading single. The second part, pairwise; we constructed this group of datasets by identifying the overlapped genes in each pair of the species datasets. This group contains six subgroups of datasets. The third group of datasets comes from finding the overlap of three different species in one group. This group constructed from four different triplet combinations of the four species datasets. The last group is identified as the overlap between the four species datasets where we find 235 overlapped genes under 445 different samples from the dataset.
2.3. Data processing
The processing of original data sets was done independently, so they often have different scale and gene annotation. It is then necessary to re-process all data sets to make them more consistent with each other. We applied two procedures to different types of data sets. For data sets generated on Affymetrix microarrays with their raw data files available, we directly processed the raw data using RMA method [46] and the custom library files provided by BRAINARRAY [47]. The output was a data matrix annotated with non-redundant Entrez gene IDs. For data sets not generated on Affymatrix microarrays or with no raw data available, we mapped the existing annotation to Entrez gene ID and collapsed redundant IDs by taking averages. Additionally, we made sure that all data sets were properly normalized and in log-2 scale. All samples were also re-labeled to replace original sample IDs with identifiers indicative of their mitochondrial function.
Data quality was evaluated based on criteria such as number of samples, percentage of missing values, sample-sample correlation. Data sets and samples with questionable quality would be excluded from statistical analysis.
2.4. Pairwise comparison of sample groups
We repetitively performed pairwise comparisons of two groups of samples with abnormal and normal/rescued mitochondrial functions. There could be more than one comparison per data set. For example, GEO series GEO1462 includes three subtypes of mitochondrial diseases and control samples, so the pairwise comparison was performed between the controls and each subtype. Another example is series GSE42986 that includes samples collected from both muscle biopsies and fibroblast cell lines, so the comparison was performed separately for two cell types.
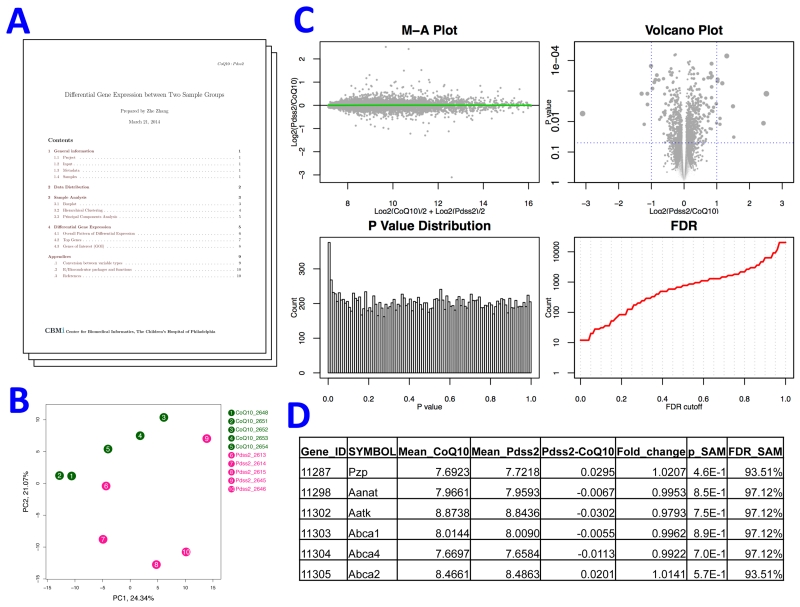
The SAM (Significance Analysis of Microarrays) method was used for all pairwise comparisons to report the folder change, p value, and false discovery rate (FDR) to indicate the differential expression of each gene between two groups. We applied our in-house workflow to generate standard outputs for each pair-wise comparison. Within the outputs, there was a PDF report that summarizes the comparison with a set of statistics, tables and plots, such as the number of differentially expressed genes under given FDR cutoffs and a plot of principal components analysis (Figure 1). Another part of the outputs is an automatically generated Excel files with the complete results of SAM analysis and the functional annotation of differentially expressed genes by DAVID.
Figure 1.
Synopsis of a pairwise comparison. The standardized outputs of a pairwise comparison of transcriptomes of two sample groups included a report file and a set of figures and tables. A) A report in PDF format that summarized data quality and comparison results. B) Unsupervised clustering of compared samples via principal components analysis. C) Visualization of comparison results. M-A plot evaluates dependence of group difference on baseline levels of gene expression. Volcano plot illustrates both magnitude (x-axis) and statistical significance (y-axis) of differential expression. P value distribution should be skewed to the left side when the comparison identifies a difference of expression profiles between two groups, and the FDR plot shows the number of genes with FDR lower than a given value. D) Full table of comparison results and DAVID functional analysis based on these results (not shown) were written to an Excel file.
2.5. Integrative analysis
A major benefit of data sets archived in ToMD is the support to efficient integrative analysis. Not only integrative analysis increases the overall statistical power by combining samples from multiple studies, but also it systematically investigates multiple biological systems and will possibly reveal knowledge that cannot be discovered from a single study. We are performing a series of such analyses based on this platform, and the results of three finished analyses are now available as part of the tier 3 in ToMD.
The first two sets of results were generated by meta–analysis. One of them used the results of 60 normal vs. abnormal pairwise comparisons and the other was performed on the results of 12 patients vs. controls comparisons generated from 8 human data sets of different cell types and disease subtypes. Fisher’s meta–analysis method [48] was used to calculate combined p values from p values from individual comparisons, and the goal was to identify genes consistently changing their expression level across comparisons.
The other analysis is an unsupervised biclustering of both samples and genes across data sets. The following sections will describe this analysis in detail.
2.6. Biclustering method
Clustering methods are used to group set of objects that shares specific features. Each group is called cluster. In the context of gene expression data under set of conditions (e.g., patients, time points), the cluster of genes is defined as a group of genes that falls in the same functional unit across the entire set of conditions in the dataset.
The process of clustering groups set of shared feature objects into groups based on only one dimension and cannot use any other dimensions simultaneously. Clustering methods aim to find groups of objects with high similarity among the same cluster members and increase the dissimilarity between different clusters.
Identifying a cluster of genes across all conditions assume similarity for all conditions which is not true in real world dataset for most cases because set of genes should be grouped based on subset of the conditions instead of all conditions.
Biclustering methods is first proposed by Cheng et al. [49] to find the biclusters in gene expression data. Biclustering methods goal is to identify biclusters that include subset of both dataset dimensions simultaneously. For example, in gene expression data where one dimension is genes list while the other is the set of samples. Biclustering methods aim to identify the subset of genes that are expressed under subset of samples simultaneously. As a result each bicluster includes a subset of genes and a subset of the samples.
In this section, we describe step-by-step procedure that we used to process and identify biclusters in each dataset.
2.6.1. Missing value recovery
The number of missing values is different across datasets. We used the R package KNN impute to identify and recover the missing values in each dataset. Two of the datasets have a large number of missing values that made it hared to recover, in one hand and in the other recovering this large amount of data will make a major change of the dataset that might not reflect the actual complexity of the dataset and affect the final results. The datasets are GSE24945 and GSE18677 with 9, 000 and 68, 000 of missing values; respectively. So we exclude them from this study.
In some datasets, some gene names were missing and hard to identify, in such cases we excluded these genes from the study. In some other cases, we have gene names duplication where the same gene occur more than once in the same dataset, we exclude the genes in such cases to eliminate any possible ambiguity in the dataset.
2.6.2. Re-normalization of data across datasets
Since the datasets come form different sources, we normalized them to the standard z – score in column-wise where the column represent samples. The z score for each value in the column is calculating using the equation:
where: x is the gene values in specific sample; μ is the column mean; and σ is the column standard deviation.
2.6.3. Mapping of gene annotation among the same species
For each species, we mapped the gene annotation of the species datasets, and then we merged them into one new dataset. The new dataset compiles the shared genes in the species datasets under all samples. This step identified four new datasets; a dataset for each species.
Table 2 shows that Worm dataset has the largest number of genes with 16, 688 across 42 samples. Human dataset has the largest number of samples compiles 233 with the smallest number of genes 895. Fly species has the smallest number of samples 33 with 12, 521 genes. Mouse dataset compiles 10, 279 genes across 147 samples.
2.6.4. Mapping of gene annotation among different species
We mapped the four species datasets that were constructed in the former step across each other to find the shared genes. The mapping included comparing the datasets in pairs, which result in identifying six new datasets, a new dataset for each pair of species. The new datasets are described in table 2. The number of shared genes across pairs of species are varies from 2, 944 shared genes between fly and mouse species datasets to 377 shared genes between human and worm species datasets.
We mapped the species dataset gene annotations in triplets. This process identified four different triplets datasets with 1, 637 genes across fly, worm and mouse to 286 across human, worm and mouse.
Mapping all of the species gene annotations results in 235 mapped genes across the four species with 445 different samples. Table 2 shows the mapping of gene annotation details.
We then processed the new datasets across the species to find the overlapped genes for each pair of species.
2.6.5. Biclustering method
Many biclustering methods are proposed in the literature for biclustering biological data. Ben-Dor et al. [50] proposed Order-Preserving Sub-Matrixes (OPSM) bicluster method for gene expression data. We used OPSM method for identifying the biclusters in the datasets. We chose OPSM for several reasons. One reason is that OPSM method proved promising results in the literature. Other reason is the availability of the method implementation.
The OPSM algorithm works as described in the following steps:
T is a set of samples and g is a gene.
The samples in T can be ordered so that the expression values are sorted in ascending order (suppose the values are all unique).
Suppose a submatrix A contains genes G and samples T .
A is a bicluster if there is an ordering (permutation) of T such that the expression values of all genes in G are sorted in ascending order.
2.6.6. Bicluster quality – Mean Squared Residue
Cheng et al. [49] proposed bicluster quality measure. The proposed measure finds the error in the bicluster expression values by calculating the difference between each expression value and both column and row means, with the bicluster mean. The proposed method is given by the following formula:
Where:
X is bicluster with |I| rows and |J| columns;
MSR(X) is the error in the bicluster;
Xij is the value in row i and column j;
is the row mean ;
is the column mean;
the overall mean;
The smaller the value of MSR(X) the better quality of the bicluster.
We modified the formula to find the coherence among the bicluster components to make it more readable by calculating it by the following formula:
In the coherence, the closer the value of MSRx to 1 is the better bicluster.
2.6.7. Software
OPSM has been implemented in Biclustering Analysis Toolbox (BicAT) [51] tool that we used to identify the biclusters from different datasets. BicAT is a JAVA-based tool that implements set of biclustering algorithms and tools for processing gene expression data. In addition to that we used R language to process, compile, analyze and plot the dataset and results.
2.6.8. Enrichment analysis
The bicluster of genes and samples assumes that the genes in the same bicluster shares common features and functionality for specific set of samples. We calculated the computational quality of the bicluster by calculating the MSR of its expression values. The computational quality measure is not enough to prove the relevance of the bicluster genes for the same group. We applied a enrichment analysis to find the biological significant of the bicluster genes and samples. The analysis test shows the related genes information.
Enrichment analysis is a computational process that decide whether an a priori defined set of genes shows statistically significant. We used for this purpose the Functional Annotation Tool from DAVID Bioinformatics Resources [52]. It is a web based tool that is used to identify the biological pathways and the enriched Gene-Ontology (GO) terms of the genes cluster. We will show the enrichment analysis result for the best biclusters MSR score, for each dataset, in section 3.
2.6.9. Statistical significant
We used Fisher’s exact test to measures the statistical significant of the bicluster of genes. It finds how likely that the enrichment of a cluster, with genes from a particular category, is to some extent greater than what is expected by chance.
We used the Benjamini correction method to correct the enrichment p–values in order to control family-wide false discovery rate under certain rate (e.g. ≤ 0.05). Benjamini correction method is one of the multiple testing correction methods that is provided by DAVID tools.
The cluster p-value range from zero to one, the closer p-value to zero the more biological significant Since the p-value is for multiple test so we used the False Discovery Rate (FDR) adjusted p–value .
For each bicluster of genes, we applied the functional annotation tool to find the Gene Ontology (GO) terms, each subset genes from the bicluster has a GO term with a p–value that shows how significant is the list of genes. The GO term describes the set of genes and what they have in common.
3. Result
3.1. Meta-analysis of pairwise comparisons
We performed 60 pairwise comparisons to identify genes differentially expressed between sample groups of data sets. These comparisons included 12, 030 to 28, 360 unique genes (mean = 17, 410) and 4 to 22 biological samples (mean = 8.4). The outputs of each comparison were standardized, including a PDF report (Figure 1), statistical results of differential gene expression, and functional analysis of differentially expressed genes via DAVID. The statistical results of each gene include both fold change (magnitude) and statistical significance (p value and FDR) of group difference. Respectively 49 and 55 comparisons identified differentially expressed genes and enriched DAVID terms with FDR = 0.05. Therefore, sample groups of most comparisons do have different gene expression profiles. Table 3 listed some of the DAVID terms identified by 10 or more comparisons.
Table 3.
The DAVID terms identified by multiple pairwise comparisons. DAVID analysis was performed on differentially expressed genes identified by each pairwise comparison. Terms enriched within these genes with FDR less than 0.05 were summarized across all 60 comparisons. This table listed some of terms most commonly identified by multiple comparisons.
| Term | ID | Num Comparisonsa |
Num Datasetb |
Num Speciesc |
|---|---|---|---|---|
| Mitochondrion | GO:0005739 | 21 | 14 | 3 |
| Oxidation reduction | GO:0055114 | 20 | 12 | 3 |
|
Generation of precursor
metabolites and energy |
GO:0006091 | 13 | 9 | 3 |
| Electron transport chain | GO:0022900 | 12 | 9 | 3 |
| Biological adhesion | GO:0022610 | 12 | 10 | 3 |
| Ribonucleoprotein complex | GO:0030529 | 10 | 9 | 3 |
| Skeletal system development | GO:0001501 | 10 | 6 | 3 |
The number of comparisons (total = 60);
Data sets (total = 28);
Species (total = 4) within which the term was identified with FDR < 0.05.
We then performed a meta–analysis of the comparison results. To compare genes of different species (worm, fly, mouse and human), gene IDs were mapped to homolog gene clusters based on HomoloGene database. Of totally 20, 441 homolog clusters, only 1, 241 were included in all 60 comparisons while respectively 18, 459 or 15, 333 clusters were included in at least 10 or 30 comparisons. The meta-analysis identified 12 gene clusters that were ranked top 2% of differential expression by at least 8 comparisons from at least 5 data sets and 3 species (Table 4). At least 9 of these genes encode proteins located in mitochondrion. For example, ETHE1 encodes a sulfur dioxygenase within mitochondrial matrix and its mutation causes ethylmalonic encephalopathy, a metabolic disorder.
Table 4.
The homolog gene clusters identified by multiple pairwise comparisons. Genes of different species were mapped to homolog cluster for summarization across comparisons. Top 2% genes with the most significant p values were selected from each comparison. Poisson test (λ = 0.02) was performed on each gene cluster to identify those gene clusters more likely to be included in the top 2% lists. This table listed the most significant clusters identified by at least 5 independent data sets generated from at least 3 species.
| Homolog ID |
Human Gene |
Human Symbol |
Num Totala |
Num Comparisonb |
Num Datasetb |
Num Speciesb |
p Poisson |
FDR Poisson |
|---|---|---|---|---|---|---|---|---|
| 8622 | 23474 | ETHE1 | 60 | 12 | 7 | 4 | 6.17E-09 | 6.31E-05 |
| 55759 | 8140 | SLC7A5 | 52 | 11 | 5 | 3 | 1.49E-08 | 9.21E-05 |
| 3343 | 4716 | NDUFB10 | 52 | 9 | 6 | 3 | 1.55E-06 | 4.51E-03 |
| 1547 | 2617 | GARS | 60 | 9 | 5 | 3 | 4.86E-06 | 6.72E-03 |
| 2255 | 6390 | SDHB | 60 | 9 | 7 | 3 | 4.86E-06 | 6.72E-03 |
| 37514 | 513 | ATP5D | 60 | 9 | 7 | 4 | 4.86E-06 | 6.72E-03 |
| 4788 | 10667 | FARS2 | 55 | 8 | 5 | 3 | 2.01E-05 | 1.21E-02 |
| 68163 | 290 | ANPEP | 55 | 8 | 5 | 3 | 2.01E-05 | 1.21E-02 |
| 3 | 34 | ACADM | 60 | 8 | 6 | 3 | 3.70E-05 | 1.68E-02 |
| 3356 | 5106 | PCK2 | 60 | 8 | 6 | 3 | 3.70E-05 | 1.68E-02 |
| 457 | 6609 | SMPD1 | 60 | 8 | 5 | 3 | 3.70E-05 | 1.68E-02 |
| 55662 | 4967 | OGDH | 60 | 8 | 7 | 3 | 3.70E-05 | 1.68E-02 |
| 10884 | 4729 | NDUFV2 | 45 | 7 | 5 | 3 | 4.34E-05 | 1.93E-02 |
The total number of comparisons that included this gene cluster;
Num_Comparison, Num_Dataset, and Num_Species: the number of comparisons, data sets, and species that identified this cluster within the top 2% lists
Its differential expression was ranked top 2% by 12 comparisons of 7 data sets from all species. The conditions that caused differential expression of ETHE1 include: PGC-1a knockout in mouse muscle (decrease), knockdown of a cytochrome oxidase subunit in fly S2 cells (increase), treatment of worms with Rotenone (increase), muscle biopsy of MELAS patients (decrease), and so on.
Another analysis was performed on the results of pairwise comparisons to identify association between different conditions or treatments. It calculated the correlation coefficient between any two sets of fold changes of individual comparisons. Comparisons using the samples from the same data sets generally have stronger correlation to each other while significant correlation also exists between comparison results from different data sets and species (Table 5). For example, the overall transcriptomic changes in patient muscle biopsies were negatively correlated to the changes caused by treating human SK-N-MC cells with 50nM Rotenone for 4 weeks, but positively correlated to the changes caused by 5nM Rotenone for 1 week as well as Sirt3 knock out in mouse livers. Another example is the positive correlation between the changes caused by PGC-1alpha overexpression in mouse and Rotenone treatment in worm, two distantly related conditions.
Table 5.
The pairwise comparisons correlated to each other. Fold changes of two pairwise comparisons were used to calculate a correlation coefficient. This table listed pairs of the comparisons from different data sets or species having the strongest correlation to each other.
| Comparison1 | Dataset1 | Species1 | Comparison2 | Dataset2 | Species2 | Na | Corrb |
|---|---|---|---|---|---|---|---|
| 50nM Rotenone, 1 week | GSE35642 | human | Mito patients muscle | GSE42986 | human | 11472 | −0.42 |
| 5nM Rotenone, 4 weeks | GSE35642 | human | Mito patients muscle | GSE42986 | human | 11472 | 0.39 |
| Patients with CoQ10 def. | GSE33940 | human | Mito patients muscle | GSE42986 | human | 17453 | 0.37 |
| Sirt3 KO, standard diet | GSE30552 | mouse | Mito patients muscle | GSE42986 | human | 16139 | −0.37 |
| Sirt3 KO, standard diet | GSE30552 | mouse | Probucol treatment | GSE18677 | mouse | 16692 | −0.35 |
| Patients of 4977bp del | GSE1462 | human | Mito patients muscle | GSE42986 | human | 11472 | 0.34 |
| PGC1b KO in liver | GSE6210 | mouse | Rotenone, 1 day old | GSE46051 | worm | 3153 | 0.32 |
| PGC1a overexpression | GSE4330 | mouse | PGC1a KO, standard diet | GSE34773 | mouse | 12941 | −0.30 |
Number of homolog gene clusters included by both comparisons;
Pearson’s correlation coefficient between 2 sets of fold changes.
3.2. Biclustering of genes and samples across datasets
In the genes and samples biclustering part of the study, after we prepared the datasets, we applied OPSM to extracted the biclusters from each dataset. After that we ranked them based on the bicluster MSRx.
Some of the extracted biclusters includes hundreds of genes under small number of samples. In some other cases, some biclusters are including small number of genes with most of the samples. We filtered and excluded these two cases since they do not reflect a good quality clusters since one of the bicluster dimensions is very high and the other is very small.
Some cases, the number of genes or samples is less that 5. We excluded such cases from our results. Finally we included biclusters with gene counts and sample counts ≥ 5.
Table 2 summaries biclustering results for all species datasets and for all overlapped cases. In the sequel of this section we describe the biclustering results after filtering and measuring their quality.
3.2.1. Human dataset result
We identified and extracted biclusters from the human dataset and after filtering them we have 6 biclusters numbered from 3 to 8. Table 6 shows the extracted bicluster details. The highest MSRx bicluster is bicluster 3 that includes 5 genes and 13 samples.
Table 6.
Filtered human biclusters details.
| Bicluster#a | #Genesb | #Samplesc | MSRx d | Enriched%e |
|---|---|---|---|---|
| 3 | 5 | 13 | 0.9678 f | 100 |
| 4 | 7 | 11 | 0.9576 | 100 |
| 5 | 11 | 10 | 0.9388 | 100 |
| 6 | 17 | 9 | 0.9294 | 100 |
| 7 | 26 | 8 | 0.8975 | 100 |
| 8 | 49 | 7 | 0.8791 | 100 |
The bicluster serial number in this study.
The number of genes involved in the bicluster.
The number of samples involved in the bicluster.
The bicluster coherence that explained in section 2.6.6.
The percent of enriched genes from the bicluster (# enriched genes / total number of genes.)
The best MSRx score is 0.9678.
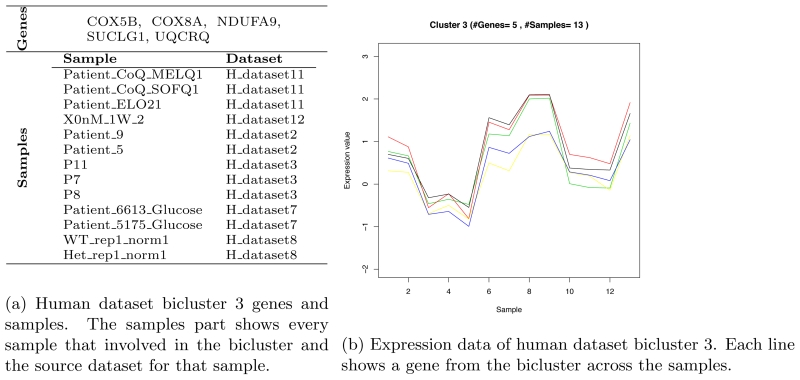
The enrichment analysis of human biclusters shows 100% genes enriched. Figure 2b shows bicluster 3 genes, samples and the bicluster plot that shows the relation between the genes and samples. Table 7 shows the enrichment analysis results for bicluster 3 using DAVID tool.
Figure 2.
Human dataset bicluster 3.
Table 7.
Sample of the enrichment analysis results. Enrichment analysis results for the highest human dataset MSRx bicluster, bicluster 3, using DAVID tools.
| Categorya | Termb | Genesc | %d | P–Value | Fisher Exact |
|---|---|---|---|---|---|
| SP_PIR_KEYWORDS | respiratory chain | NDUFA9, COX8A, UQCRQ, COX5B |
80 | 1.90E-07 | 8.50E-10 |
| GOTERM_CC_FAT | mitochondrial inner membrane | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 3.20E-07 | 7.60E-09 |
| GOTERM_CC_FAT | organelle inner membrane | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 4.30E-07 | 1.10E-08 |
| GOTERM_CC_FAT | mitochondrial membrane | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 8.90E-07 | 2.70E-08 |
| GOTERM_CC_FAT | mitochondrial envelope | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 1.10E-06 | 3.70E-08 |
| SP_PIR_KEYWORDS | mitochondrion | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 3.50E-06 | 1.50E-07 |
| GOTERM_CC_FAT | mitochondrial part | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 4.70E-06 | 2.20E-07 |
| GOTERM_CC_FAT | organelle envelope | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 5.50E-06 | 2.60E-07 |
| GOTERM_CC_FAT | envelope | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 5.60E-06 | 2.70E-07 |
| GOTERM_BP_FAT | generation of precursor metabo- lites and energy |
NDUFA9, SUCLG1, COX8A, UQCRQ |
80 | 4.80E-05 | 1.40E-06 |
| GOTERM_CC_FAT | mitochondrion | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 5.20E-05 | 4.40E-06 |
| GOTERM_CC_FAT | organelle membrane | NDUFA9, SUCLG1, COX8A, UQCRQ, COX5B |
100 | 5.40E-05 | 4.60E-06 |
| UP_SEQ_FEATURE | transit peptide:Mitochondrion | NDUFA9, SUCLG1, COX8A, COX5B |
80 | 5.70E-05 | 1.70E-06 |
| SP_PIR_KEYWORDS | transit peptide | NDUFA9, SUCLG1, COX8A, COX5B |
80 | 5.90E-05 | 1.80E-06 |
| KEGG_PATHWAY | Parkinson’s disease | NDUFA9, COX8A, UQCRQ, COX5B |
80 | 6.10E-05 | 1.90E-06 |
| KEGG_PATHWAY | Oxidative phosphorylation | NDUFA9, COX8A, UQCRQ, COX5B |
80 | 6.40E-05 | 2.00E-06 |
| KEGG_PATHWAY | Alzheimer’s disease | NDUFA9, COX8A, UQCRQ, COX5B |
80 | 1.30E-04 | 5.00E-06 |
| KEGG_PATHWAY | Huntington’s disease | NDUFA9, COX8A, UQCRQ, COX5B |
80 | 1.70E-04 | 7.40E-06 |
| GOTERM_MF_FAT | hydrogen ion transmembrane transporter activity |
COX8A, UQCRQ, COX5B |
60 | 2.80E-04 | 3.20E-06 |
| GOTERM_MF_FAT | monovalent inorganic cation trans- membrane transporter activity |
COX8A, UQCRQ, COX5B |
60 | 3.80E-04 | 4.90E-06 |
| SP_PIR_KEYWORDS | mitochondrion inner membrane | COX8A, UQCRQ, COX5B |
60 | 5.90E-04 | 9.80E-06 |
| GOTERM_MF_FAT | inorganic cation transmembrane transporter activity |
COX8A, UQCRQ, COX5B |
60 | 7.90E-04 | 1.50E-05 |
| KEGG_PATHWAY | Cardiac muscle contraction | COX8A, UQCRQ, COX5B |
60 | 1.40E-03 | 3.40E-05 |
| SP_PIR_KEYWORDS | mitochondrial inner membrane | COX8A, COX5B | 40 | 3.90E-03 | 9.20E-06 |
| SP_PIR_KEYWORDS | oxidoreductase | NDUFA9, COX8A, COX5B |
60 | 4.90E-03 | 2.40E-04 |
| SP_PIR_KEYWORDS | membrane-associated complex | COX8A, COX5B | 40 | 6.60E-03 | 2.70E-05 |
| SP_PIR_KEYWORDS | oxidative phosphorylation | COX8A, COX5B | 40 | 7.30E-03 | 3.20E-05 |
| GOTERM_MF_FAT | oxidoreductase activity, acting on heme group of donors |
COX8A, COX5B | 40 | 8.60E-03 | 4.50E-05 |
| GOTERM_MF_FAT | cytochrome-c oxidase activity | COX8A, COX5B | 40 | 8.60E-03 | 4.50E-05 |
| GOTERM_MF_FAT | heme-copper terminal oxidase activity | COX8A, COX5B | 40 | 8.60E-03 | 4.50E-05 |
| GOTERM_MF_FAT | oxidoreductase activity, acting on heme group of donors, oxygen as acceptor |
COX8A, COX5B | 40 | 8.60E-03 | 4.50E-05 |
| SP_PIR_KEYWORDS | electron transfer | COX8A, COX5B | 40 | 1.00E-02 | 6.60E-05 |
| SP_PIR_KEYWORDS | electron transport | NDUFA9, UQCRQ | 40 | 2.10E-02 | 2.60E-04 |
| GOTERM_CC_FAT | respiratory chain | NDUFA9, UQCRQ | 40 | 2.30E-02 | 3.40E-04 |
| GOTERM_BP_FAT | cellular respiration | NDUFA9, SUCLG1 | 40 | 2.80E-02 | 5.00E-04 |
| GOTERM_BP_FAT | electron transport chain | NDUFA9, UQCRQ | 40 | 3.30E-02 | 6.90E-04 |
| INTERPRO | NAD(P)-binding domain | NDUFA9, SUCLG1 | 40 | 3.60E-02 | 7.90E-04 |
| UP_SEQ_FEATURE | cross-link:Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin) |
SUCLG1, UQCRQ | 40 | 4.10E-02 | 1.10E-03 |
| GOTERM_BP_FAT | energy derivation by oxidation of organic compounds |
NDUFA9, SUCLG1 | 40 | 4.20E-02 | 1.10E-03 |
| SP_PIR_KEYWORDS | isopeptide bond | SUCLG1, UQCRQ | 40 | 6.50E-02 | 2.70E-03 |
| GOTERM_CC_FAT | mitochondrial lumen | NDUFA9, SUCLG1 | 40 | 6.90E-02 | 3.00E-03 |
| GOTERM_CC_FAT | mitochondrial matrix | NDUFA9, SUCLG1 | 40 | 6.90E-02 | 3.00E-03 |
| SP_PIR_KEYWORDS | acetylation | NDUFA9, SUCLG1, COX5B |
60 | 9.30E-02 | 2.10E-02 |
The original source (i.e., database) of the term.
The enrichment terms associated with the gene list.
The list of genes from the bicluster that is involved in the term.
The percentage of the bicluster genes that involved in this term (Involved genes/total genes).
3.2.2. Mouse dataset result
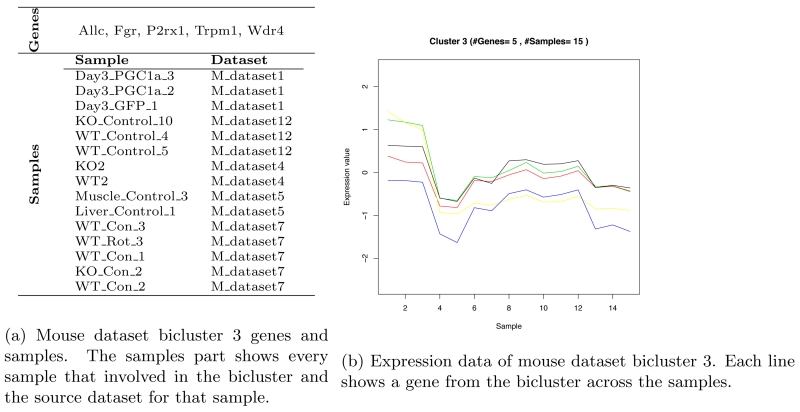
We identified and extracted five biclusters from the mouse dataset after filtering. Table 8 shows the extracted bicluster details. Bicluster 3 has the highest MSRx score. It includes 5 genes and 15 samples. Figure 3b shows bicluster 3 genes, samples and the bicluster plot that shows the genes, samples and the relation between them.
Table 8.
Filtered mouse biclusters details.
| Bicluster#a | #Genesb | #Samplesc | MSRx d | Enriched%e |
|---|---|---|---|---|
| 3 | 5 | 15 | 0.9471 f | 40 |
| 4 | 6 | 13 | 0.9228 | 33.3 |
| 5 | 10 | 12 | 0.9362 | 70 |
| 6 | 19 | 11 | 0.8810 | 63.2 |
| 7 | 43 | 10 | 0.9070 | 86 |
The bicluster serial number in this study.
The number of genes involved in the bicluster.
The number of samples involved in the bicluster.
The bicluster coherence that explained in section 2.6.6.
The percent of enriched genes from the bicluster (# enriched genes / total number of genes.)
The best MSRx score is 0.9471.
Figure 3.
Mouse dataset bicluster 3.
The enrichment analysis of bicluster 3 of mouse dataset shows that 40% of the genes are enriched. Bicluster 7 has the highest enrichment genes percent. The following genes are part of the extracted biclusters form the mouse dataset and none of them is enriched:
Fgr, Wdr4, Allc, Icos, Pigu, Polr2a, Tat, Rab25, Cecr5, Dntt, Gtse1, Gstm3, Llgl2, Srrm1, Pih1d1, Tspo2, Rdh7
Table 8 describes mouse dataset biclusters details with enrichment analysis result.
3.2.3. Worm dataset result
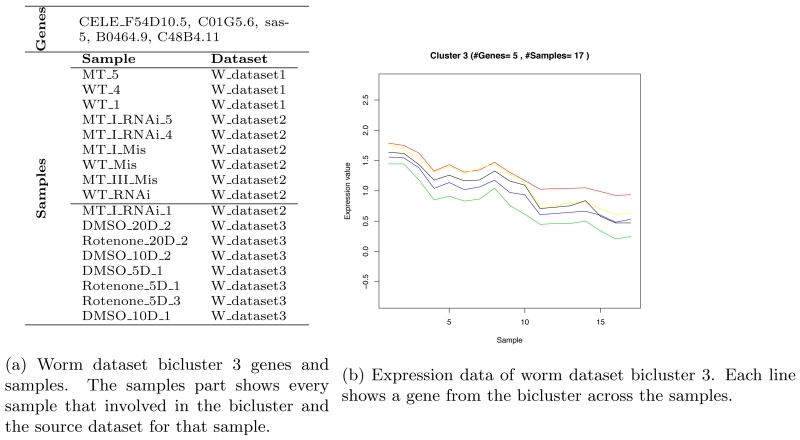
We applied OPSM to the worm dataset. The method identified 6 different biclusters, after filtering, with different MSRx scores. Bicluser 3 shows the highest MSRx score; however, none of its genes were enriched. Bicluster 6 shows the highest enrichment gene percent. Table 9 shows the extracted bicluster details. Figure 4b shows bicluster 3 genes, samples and the bicluster plot that shows the correlation between the bicluster genes for the specified set of samples.
Table 9.
Filtered worm biclusters details.
| Bicluster#a | #Genesb | #Samplesc | MSRx d | Enriched%e |
|---|---|---|---|---|
| 3a | 5 | 17 | 0.9969 f | 0 |
| 4a | 9 | 16 | 0.9820 | 0 |
| 5 | 15 | 15 | 0.9804 | 46.7 |
| 6 | 37 | 14 | 0.9767 | 64.9 |
| 7 | 43 | 13 | 0.9693 | 58.1 |
| 8 | 112 | 12 | 0.9736 | 64.3 |
The bicluster serial number in this study.
The number of genes involved in the bicluster.
The number of samples involved in the bicluster.
The bicluster coherence that explained in section 2.6.6.
The percent of enriched genes from the bicluster (# enriched genes / total number of genes.)
The best MSRx score is 0.9969.
Figure 4.
Worm dataset bicluster 3.
The following genes are part of the extracted biclusters form the worm dataset and none of them is enriched:
C01G5.6, F54D10.5, B0464.9, sas-5, C13F10.7, C15H11.8, C18E3.9, K03H1.7, syn-16, C02F5.13, C48B4.11, Y57A10A.16, cpn-1, F18A1.8, ZK1248.11, cids-1, C39E9.12, D1007.8, T02C12.3, C04G6.4, C28A5.1, fbxa-210, F40F8.11, F49C12.9, F55A11.7, K06H6.2, lgg-3, T06D8.9, ZK1248.15, ztf-9, B0261.7, B0491.1, clec-127, C01G10.7, C01G8.1, C08F8.3, C14B1.2, C24D10.4, C42C1.12, C43E11.12, C43H8.2, F17A9.2, dhs-22, egg-2, fsn-1, F21D5.6, F41G3.6, F54A3.6, inx-22, K05C4.7, mvb-12, W01D2.5, pak-2, C35D10.10, Y54G11A.9, ZK1067.3
Table 9 shows worm dataset biclusters details with enrichment analysis results.
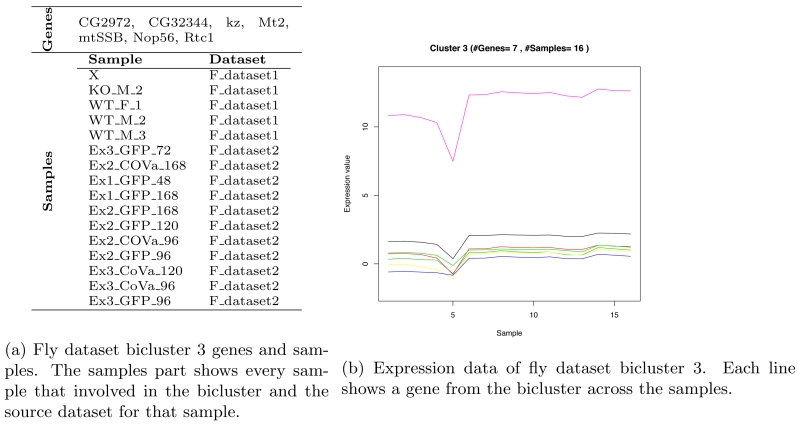
3.2.4. Fly dataset result
OPSM was able to extract, after filtering, 5 biclusters from fly dataset. The highest bicluster MSRx score is bicluster 3. It involves 7 genes and 16 samples. Table 10 shows the extracted biclusters details.
Table 10.
Filtered fly biclusters details.
| Bicluster#a | #Genesb | #Samplesc | MSRx d | Enriched%e |
|---|---|---|---|---|
| 3 | 7 | 16 | 0.8875 f | 85 |
| 4 | 9 | 15 | 0.8634 | 77.8 |
| 5 | 20 | 14 | 0.8619 | 75 |
| 6 | 33 | 13 | 0.8801 | 78.8 |
| 7 | 70 | 12 | 0.8691 | 72.9 |
The bicluster serial number in this study.
The number of genes involved in the bicluster.
The number of samples involved in the bicluster.
The bicluster coherence that explained in section 2.6.6.
The percent of enriched genes from the bicluster (# enriched genes / total number of genes.)
The best MSRx score is 0.8875.
Figure 5b shows bicluster 3 genes, samples and the bicluster plot that shows the bicluster genes, samples and the relation between them. Bicluster 3 shows the best enrichment percent.
Figure 5.
Fly dataset bicluster 3.
The following genes are part of the extracted bicluster form fly dataset and none of them is enriched:
CG2972, CG14270, CG11788, CG1749, sec13, CG10424, CG11875, CG13016, CG17249, CG18004, CG31223, mRpS23, CG32554, CG33213, CG3353, CG6617, CG7484, CG8090, Rae1, CG9947, CG12975, CG14286, CG6073
3.2.5. Different species mapping result
We mapped the four species gene annotations among the four species to find the overlapped genes among them. The overlapped datasets are falls in one of the following three categories based on the number of overlapped datasets:
Species pairs: In this category, we mapped the species datasets in pairs. Table 2 shows 6 overlapped pairs of datasets. The overlapped dataset of fly with worm result in 2, 724 genes across 75 samples produced highest number of biclusters 9 in this category. The biclusters extracted from this dataset shows the best average MSRx results that means these bicluster are with high quality 0.96, that means the set of genes and samples in each bicluster share common features. The next best average biclusters quality is fly and mouse dataset with average MSRx of 0.95. This show that fly dataset has common features with both worm and mouse datasets whereas the result biclusters of the fly and human overlap shows relatively small number of overlapped genes with average quality of biclusters. The overlapped dataset of human and worm shows smallest number of shared genes and the smallest average MSRx. We extracted one bicluster form the mouse and human overlapped dataset, this bicluster was with MSRx = 0.99; however, all of this bicluster samples are from mouse samples and the human samples are not included.
Species triplets: In this category, we mapped the species datasets three species at a time to find the overlapped genes. The result is four different datasets with 1, 637 overlapped genes between fly, mouse and worm. We extracted 7 biclusters from this dataset with average MSRx of 0.92, three biclusters of them included samples from the three species and the other four biclusters included samples from only two species. The other three overlapped datasets have an average of 300 overlapped genes among the species. The biclusters that extracted from these three datasets are of either number of genes less than 5 or number of samples less than 5 and they excluded from the list of biclusters.
All four species: The overlap among the four species produced a dataset of 235 genes and 445 samples, 7 biclusters extracted from this dataset with average MRSx = 0.85. However, none of the extracted biclusters involved samples from all species. Three biclusters involved samples from human, fly and worm datasets. Two biclusters involved human and mouse dataset samples. One bicluster involoved human, fly and mouse and the last bicluster involved human fly and mouse samples.
4. Discussion
We developed a solution to overcome some of the difficulties in studying rare diseases. Genomic data sets seemingly divergent from each other, but commonly related to mitochondrial dysfunctions, were collected to generate a centralized platform of data mining, ToMD. Although many of the data processing and data analysis steps can be automated, we noticed that the key to the success of such a platform is the close control of data quality, sample grouping, and result interpretation by investigators with different domain knowledge.
Biclustering methods identify biclusters by involving both dimensions of dataset simultaneously, genes and samples in this study. Each bicluster includes a group of genes that falls in the same functional group under a subset of samples. We applied a robust biclustering method, OPSM, to identify co–regulated genes and samples across the different datasets. The method construct a bicluster if there is a permutation of the bicluster samples such that the expression values of the bicluster genes are sorted in ascending order.
Integrative analysis ToMD data sets have revealed new insights about mitochondrial diseases. For example, meta-analysis identified genes consistently having differential expression when mitochondrial functions were impaired across a variety of cell types and species. Follow-up study of the roles played by these genes in mitochondrial diseases will improve our understanding about these diseases and mitochondrial function in general.
The currents contents of ToMD are mostly static files. One of our future plans is to create a web interface that allows users to query, filter and analyze the data, as well as generate dynamic results and plots, which will further facilitate the process of knowledge discovery. Once the value of ToMD is further confirmed, we will apply the same principal to generate similar resources for other types of rare diseases, such as childhood cancers and congenital heart diseases.
5. Conclusion
In conclusion, we developed a practical strategy to reuse public data for the study of rare diseases whose low occurrence makes it difficult to collect large sample cohorts and obtain sufficient research funding. We proved that it is possible to integrate transcriptome data sets sharing a common link to a rare disease, but generated from different platforms, cell types, and even species.
Acknowledgments
The work was supported in part by US National Science Foundation award (IIS-0644366) to Xuewen Chen, and by the P30-HD026979 grant from the National Institute of Health. We thank Dr. Marni J. Falk for her help to identify data sets related to mitochondrial diseases.
References
- [1].Fishman MC. Author response to comment on “power of rare diseases: Found in translation”. Science translational medicine. 2014;6(228):228lr1–228lr1. doi: 10.1126/scitranslmed.3008809. [DOI] [PubMed] [Google Scholar]
- [2].van der Meer JW, Simon A, Dinarello CA. Comment on “power of rare diseases: Found in translation”. Science translational medicine. 2014;6(219):219le1–219le1. doi: 10.1126/scitranslmed.3007911. [DOI] [PubMed] [Google Scholar]
- [3].Fishman MC. Power of rare diseases: Found in translation. Science translational medicine. 2013;5(201):201ps11–201ps11. doi: 10.1126/scitranslmed.3006800. [DOI] [PubMed] [Google Scholar]
- [4].Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders—past, present and future. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2004;1659(2):115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- [5].Rossignol R, et al. The expanding universe of mitochondrial research. The international journal of biochemistry & cell biology. 2013;45(1):2–3. doi: 10.1016/j.biocel.2012.10.015. [DOI] [PubMed] [Google Scholar]
- [6].Wallace DC, et al. A mitochondrial bioenergetic etiology of disease. J Clin Invest. 123(4) doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dela F, Helge JW. Insulin resistance and mitochondrial function in skeletal muscle. The international journal of biochemistry & cell biology. 2013;45(1):11–15. doi: 10.1016/j.biocel.2012.09.019. [DOI] [PubMed] [Google Scholar]
- [8].Iommarini L, Calvaruso MA, Kurelac I, Gasparre G, Porcelli AM. Complex i impairment in mitochondrial diseases and cancer: Parallel roads leading to different outcomes. The international journal of biochemistry & cell biology. 2013;45(1):47–63. doi: 10.1016/j.biocel.2012.05.016. [DOI] [PubMed] [Google Scholar]
- [9].Oliveira J, Jekabsons MB, Chen S, Lin A, Rego AC, Gonçalves J, Ellerby LM, Nicholls DG. Mitochondrial dysfunction in huntington’s disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. Journal of neurochemistry. 2007;101(1):241–249. doi: 10.1111/j.1471-4159.2006.04361.x. [DOI] [PubMed] [Google Scholar]
- [10].Eckmann J, Eckert SH, Leuner K, Muller WE, Eckert GP. Mitochondria: mitochondrial membranes in brain ageing and neurodegeneration. The international journal of biochemistry & cell biology. 2013;45(1):76–80. doi: 10.1016/j.biocel.2012.06.009. [DOI] [PubMed] [Google Scholar]
- [11].Zhang Z, Gasser DL, Rappaport EF, Falk MJ. Cross-platform expression microarray performance in a mouse model of mitochondrial disease therapy. Molecular genetics and metabolism. 2010;99(3):309–318. doi: 10.1016/j.ymgme.2009.10.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, et al. Transcriptional dysregulation in nipbl and cohesin mutant human cells. PLoS biology. 2009;7(5):e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Z, Chen D, Fenstermacher DA. Integrated analysis of independent gene expression microarray datasets improves the predictability of breast cancer outcome. BMC genomics. 2007;8(1):331. doi: 10.1186/1471-2164-8-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. Ncbi geo: archive for functional genomics data sets—update. Nucleic acids research. 2013;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fernández-Ayala DJ, Chen S, Kemppainen E, MC O’Dell K, Jacobs HT. Gene expression in a drosophila model of mitochondrial disease. PLoS One. 2010;5(1):e8549. doi: 10.1371/journal.pone.0008549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Freije WA, Mandal S, Banerjee U. Expression profiling of attenuated mitochondrial function identifies retrograde signals in drosophila. G3: Genes— Genomes— Genetics. 2012;2(8):843–851. doi: 10.1534/g3.112.002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kayser E-B, Morgan PG, Hoppel CL, Sedensky MM. Mitochondrial expression and function of gas-1 in caenorhabditis elegans. Journal of Biological Chemistry. 2001;276(23):20551–20558. doi: 10.1074/jbc.M011066200. [DOI] [PubMed] [Google Scholar]
- [18].Falk M, Zhang Z, Rosenjack J, Nissim I, Daikhin E, Nissim I, Sedensky M, Yudkoff M, Morgan P. Metabolic pathway profiling of mitochondrial respiratory chain mutants in¡ i¿ c. elegans¡/i¿. Molecular genetics and metabolism. 2008;93(4):388–397. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, Platzer M, Ristow M. Neuronal ros signaling rather than ampk/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Molecular metabolism. 2013;2(2):92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Crimi M, Bordoni A, Menozzi G, Riva L, Fortunato F, Galbiati S, Del Bo R, Pozzoli U, Bresolin N, Comi GP. Skeletal muscle gene expression profiling in mitochondrial disorders. The FASEB journal. 2005;19(7):866–868. doi: 10.1096/fj.04-3045fje. [DOI] [PubMed] [Google Scholar]
- [21].Cížková A, Stráneckỳ V, Ivánek R, Hartmannová H, Nosková L, Piherová L, Tesařová M, Hansíková H, Honzík T, Zeman J, et al. Development of a human mitochondrial oligonucleotide microarray (h-mitoarray) and gene expression analysis of fibroblast cell lines from 13 patients with isolated f1fo atp synthase deficiency. BMC genomics. 2008;9(1):38. doi: 10.1186/1471-2164-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Čížková A, Stráneckỳ V, Mayr JA, Tesařová M, Havlíčková V, Paul J, Ivánek R, Kuss AW, Hansíková H, Kaplanová V, et al. Tmem70 mutations cause isolated atp synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nature genetics. 2008;40(11):1288–1290. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- [23].Mende S, Royer L, Herr A, Schmiedel J, Deschauer M, Klopstock T, Kostic VS, Schroeder M, Reichmann H, Storch A. Whole blood genome-wide expression profiling and network analysis suggest melas master regulators. Neurological research. 2011;33(6):638–655. doi: 10.1179/1743132810Y.0000000016. [DOI] [PubMed] [Google Scholar]
- [24].Ivanov VN, Ghandhi SA, Zhou H, Huang SX, Chai Y, Amundson SA, Hei TK. Radiation response and regulation of apoptosis induced by a combination of trail and chx in cells lacking mitochondrial dna: A role for nf-κb–stat3-directed gene expression. Experimental cell research. 2011;317(11):1548–1566. doi: 10.1016/j.yexcr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kovářová N, Čížková Vrbacká A, Pecina P, Stráneckỳ V, Pronicka E, Kmoch S, Houštěk J. Adaptation of respiratory chain biogenesis to cytochrome¡ i¿ c¡/i¿ oxidase deficiency caused by¡ i¿ surf1¡/i¿ gene mutations. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2012;1822(7):1114–1124. doi: 10.1016/j.bbadis.2012.03.007. [DOI] [PubMed] [Google Scholar]
- [26].Voets A, Huigsloot M, Lindsey P, Leenders A, Koopman W, Willems P, Rodenburg R, Smeitink J, Smeets H. Transcriptional changes in oxphos complex i deficiency are related to anti-oxidant pathways and could explain the disturbed calcium homeostasis. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2012;1822(7):1161–1168. doi: 10.1016/j.bbadis.2011.10.009. [DOI] [PubMed] [Google Scholar]
- [27].Chae S, Ahn BY, Byun K, Cho YM, Yu M-H, Lee B, Hwang D, Park KS. A systems approach for decoding mitochondrial retrograde signaling pathways. Science signaling. 2013;6(264):rs4. doi: 10.1126/scisignal.2003266. [DOI] [PubMed] [Google Scholar]
- [28].Mormeneo E, Jimenez-Mallebrera C, Palomer X, De Nigris V, Vázquez-Carrera M, Orozco A, Nascimento A, Colomer J, Lerín C, Gómez-Foix AM. Pgc-1α induces mitochondrial and myokine transcriptional programs and lipid droplet and glycogen accumulation in cultured human skeletal muscle cells. PloS one. 2012;7(1):e29985. doi: 10.1371/journal.pone.0029985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fernández-Ayala DJ, Guerra I, Jiménez-Gancedo S, Cascajo MV, Gavilán A, DiMauro S, Hirano M, Briones P, Artuch R, De Cabo R, et al. Survival transcriptome in the coenzyme q10 deficiency syndrome is acquired by epigenetic modifications: a modelling study for human coenzyme q10 deficiencies. BMJ open. 3(3) doi: 10.1136/bmjopen-2012-002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fernández-Ayala DJ, Guerra I, Jiménez-Gancedo S, Cascajo MV, Gavilán A, DiMauro S, Hirano M, Briones P, Artuch R, De Cabo R, et al. Survival transcriptome in the coenzyme q10 deficiency syndrome is acquired by epigenetic modifications: a modelling study for human coenzyme q10 deficiencies. BMJ open. 3(3) doi: 10.1136/bmjopen-2012-002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cabeza-Arvelaiz Y, Schiestl RH. Transcriptome analysis of a rotenone model of parkinsonism reveals complex i-tied and-untied toxicity mechanisms common to neurodegenerative diseases. PloS one. 2012;7(9):e44700. doi: 10.1371/journal.pone.0044700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Falk M, Zhang Z, Rosenjack J, Nissim I, Daikhin E, Nissim I, Sedensky M, Yudkoff M, Morgan P. Metabolic pathway profiling of mitochondrial respiratory chain mutants in¡ i¿ c. elegans¡/i¿. Molecular genetics and metabolism. 2008;93(4):388–397. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nature genetics. 2006;38(5):576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- [34].Someya S, Yamasoba T, Kujoth GC, Pugh TD, Weindruch R, Tanokura M, Prolla TA. The role of mtdna mutations in the pathogenesis of age-related hearing loss in mice carrying a mutator dna polymerase γ. Neurobiology of aging. 2008;29(7):1080–1092. doi: 10.1016/j.neurobiolaging.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mtor controls mitochondrial oxidative function through a yy1–pgc-1&agr; transcriptional complex. nature. 2007;450(7170):736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- [36].Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of pgc-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127(1):59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- [37].Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim Y-B, Cinti S, et al. Hypomorphic mutation of pgc-1β causes mitochondrial dysfunction and liver insulin resistance. Cell metabolism. 2006;4(6):453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peng M, Falk MJ, Haase VH, King R, Polyak E, Selak M, Yudkoff M, Hancock WW, Meade R, Saiki R, et al. Primary coenzyme q deficiency in pdss2 mutant mice causes isolated renal disease. PLoS genetics. 2008;4(4):e1000061. doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, Plun-Favreau H, Edwards R, Teismann P, Esposti M, et al. Mitochondrial dysfunction triggered by loss of htra2 results in the activation of a brain-specific transcriptional stress response. Cell Death & Differentiation. 2009;16(3):449–464. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- [40].Zhang Z, Gasser DL, Rappaport EF, Falk MJ. Cross-platform expression microarray performance in a mouse model of mitochondrial disease therapy. Molecular genetics and metabolism. 2010;99(3):309–318. doi: 10.1016/j.ymgme.2009.10.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. Sirt3 opposes reprogramming of cancer cell metabolism through hif1α destabilization. Cancer cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Falk MJ, Polyak E, Zhang Z, Peng M, King R, Maltzman JS, Okwuego E, Horyn O, Nakamaru-Ogiso E, Ostrovsky J, et al. Probucol ameliorates renal and metabolic sequelae of primary coq deficiency in pdss2 mutant mice. EMBO molecular medicine. 2011;3(7):410–427. doi: 10.1002/emmm.201100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stančáková A, Goetzman E, Lam MM, Schwer B, et al. Sirt3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular cell. 2011;44(2):177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Finley LW, Lee J, Souza A, Desquiret-Dumas V, Bullock K, Rowe GC, Procaccio V, Clish CB, Arany Z, Haigis MC. Skeletal muscle transcriptional coactivator pgc-1α mediates mitochondrial, but not metabolic, changes during calorie restriction. Proceedings of the National Academy of Sciences. 2012;109(8):2931–2936. doi: 10.1073/pnas.1115813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the national center for biotechnology information. Nucleic acids research. 2007;35(suppl 1):D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix genechip probe level data. Nucleic acids research. 2003;31(4):e15–e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of genechip data. Nucleic acids research. 2005;33(20):e175–e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fisher RA. Statistical methods for research workers. Genesis Publishing Pvt Ltd; 1925. [Google Scholar]
- [49].Cheng Y, Church GM. Biclustering of expression data. Ismb. 2000;8:93–103. [PubMed] [Google Scholar]
- [50].Ben-Dor A, Chor B, Karp R, Yakhini Z. Discovering local structure in gene expression data: the order-preserving submatrix problem. Journal of computational biology. 2003;10(3-4):373–384. doi: 10.1089/10665270360688075. [DOI] [PubMed] [Google Scholar]
- [51].Barkow S, Bleuler S, Prelić A, Zimmermann P, Zitzler E. Bicat: a biclustering analysis toolbox. Bioinformatics. 2006;22(10):1282–1283. doi: 10.1093/bioinformatics/btl099. [DOI] [PubMed] [Google Scholar]
- [52].Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA, et al. David: database for annotation, visualization, and integrated discovery. Genome biol. 2003;4(5):P3. [PubMed] [Google Scholar]