Abstract
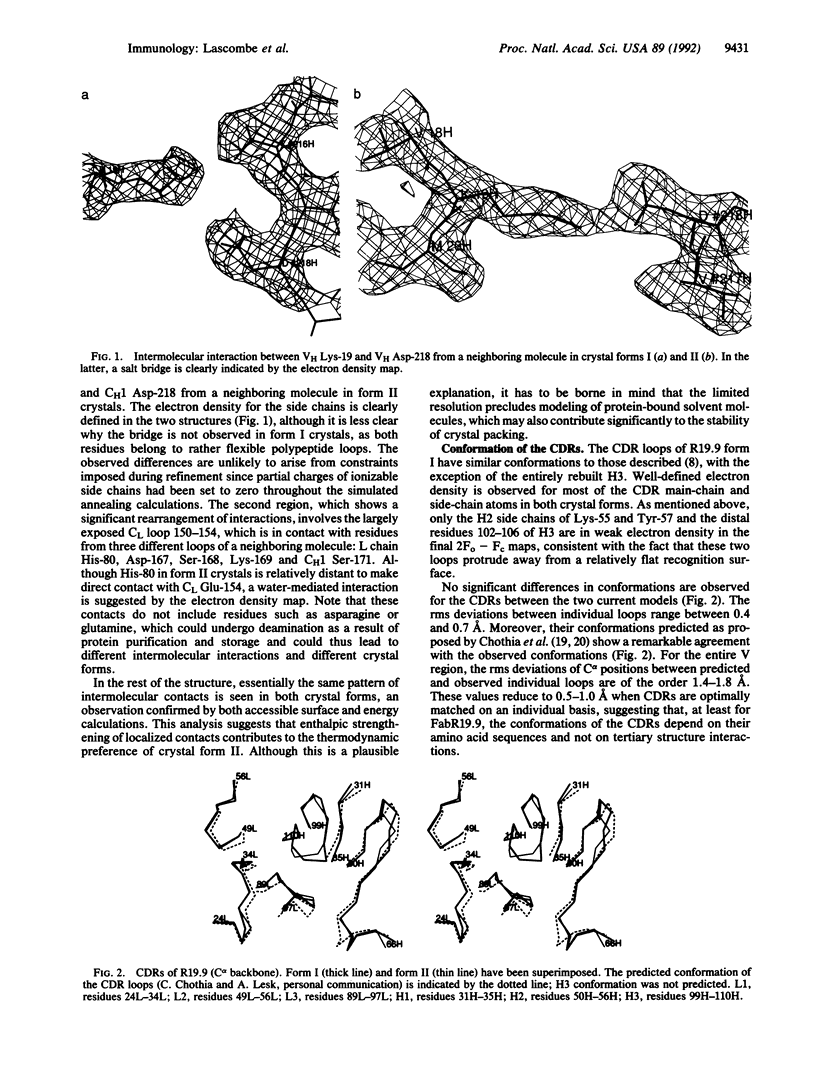
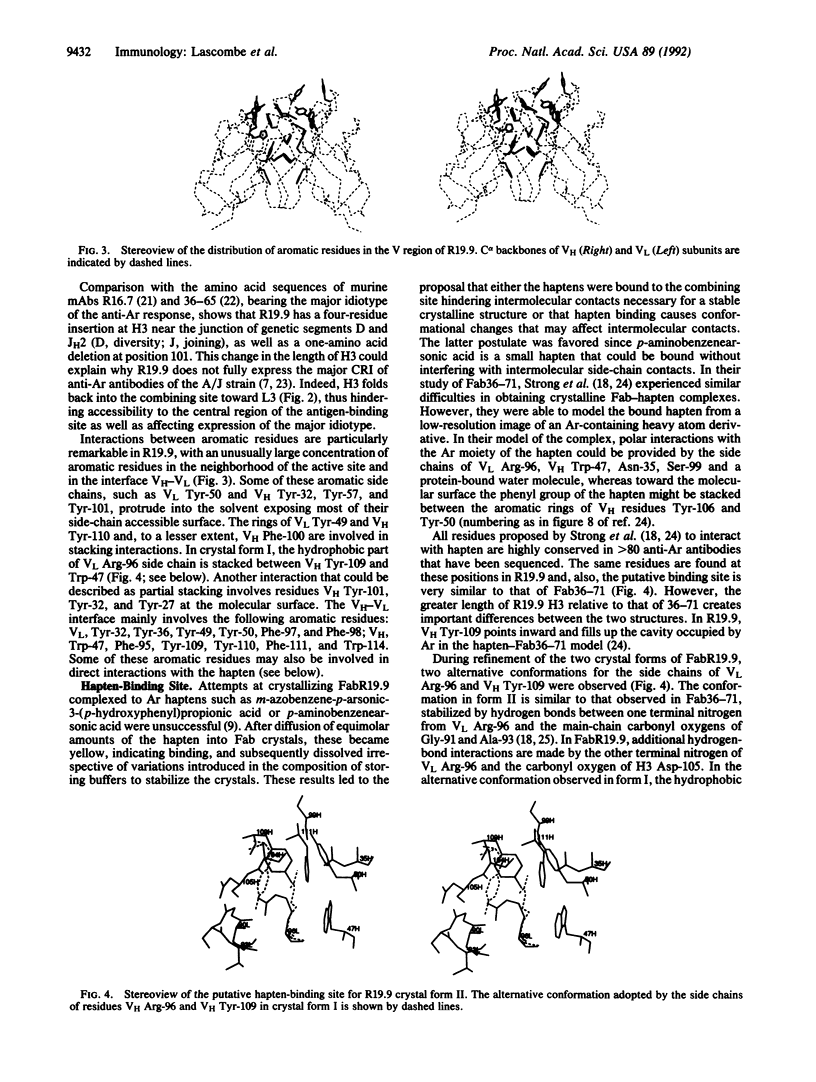
The three-dimensional structure of FabR19.9 from a well-characterized anti-p-azobenzenearsonate monoclonal antibody has been determined by x-ray diffraction techniques in two crystalline forms (I and II) to a resolution of 2.8 and 2.7 A, respectively. Essentially the same tertiary and quaternary structure of the Fab is observed in the two forms. The major difference resides in the intermolecular contacts, which are interpreted to favor an irreversible transition from the metastable form I to the more stable form II. The third complementarity-determining region of the heavy chain (H3) folds back over the combining site and requires rearrangement for hapten binding. This dynamic requirement on H3 is consistent with its mobility in the structure and can explain hapten binding to an otherwise inaccessible antibody combining site.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bentley G. A., Boulot G., Riottot M. M., Poljak R. J. Three-dimensional structure of an idiotope-anti-idiotope complex. Nature. 1990 Nov 15;348(6298):254–257. doi: 10.1038/348254a0. [DOI] [PubMed] [Google Scholar]
- Brown A. R., Lamoyi E., Nisonoff A. Relationship of idiotypes of the anti-p-azophenylarsonate antibodies of A/J and BALB/c mice. J Immunol. 1981 Apr;126(4):1268–1273. [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987 Aug 20;196(4):901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M., Tramontano A., Levitt M., Smith-Gill S. J., Air G., Sheriff S., Padlan E. A., Davies D., Tulip W. R. Conformations of immunoglobulin hypervariable regions. Nature. 1989 Dec 21;342(6252):877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- Edmundson A. B., Ely K. R., Herron J. N. A search for site-filling ligands in the Mcg Bence-Jones dimer: crystal binding studies of fluorescent compounds. Mol Immunol. 1984 Jul;21(7):561–576. doi: 10.1016/0161-5890(84)90041-5. [DOI] [PubMed] [Google Scholar]
- Gill-Pazaris L. A., Lamoyi E., Brown A. R., Nisonoff A. Properties of a minor cross-reactive idiotype associated with anti-p-azophenylarsonate antibodies of A/J mice. J Immunol. 1981 Jan;126(1):75–79. [PubMed] [Google Scholar]
- Greene M. I., Nelles M. J., Sy M. S., Nisonoff A. Regulation of immunity to the azobenzenearsonate hapten. Adv Immunol. 1982;32:253–300. doi: 10.1016/s0065-2776(08)60723-3. [DOI] [PubMed] [Google Scholar]
- Kresina T. F., Rosen S. M., Nisonoff A. Degree of heterogeneity of binding specificities of antibodies to the phenylarsonate group that share a common idiotype. Mol Immunol. 1982 Nov;19(11):1433–1439. doi: 10.1016/0161-5890(82)90190-0. [DOI] [PubMed] [Google Scholar]
- Kuettner M. G., Wang A. L., Nisonoff A. Quantitative investigations of idiotypic antibodies. VI. Idiotypic specificity as a potential genetic marker for the variable regions of mouse immunoglobulin polypeptide chains. J Exp Med. 1972 Mar 1;135(3):579–595. doi: 10.1084/jem.135.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoyi E., Estess P., Capra J. D., Nisonoff A. Presence of highly conserved idiotypic determinants in a family of antibodies that constitute an intrastrain cross-reactive idiotype. J Exp Med. 1980 Sep 1;152(3):703–711. doi: 10.1084/jem.152.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascombe M. B., Alzari P. M., Boulot G., Saludjian P., Tougard P., Berek C., Haba S., Rosen E. M., Nisonoff A., Poljak R. J. Three-dimensional structure of Fab R19.9, a monoclonal murine antibody specific for the p-azobenzenearsonate group. Proc Natl Acad Sci U S A. 1989 Jan;86(2):607–611. doi: 10.1073/pnas.86.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser T., Wysocki L. J., Margolies M. N., Gefter M. L. Evolution of antibody variable region structure during the immune response. Immunol Rev. 1987 Apr;96:141–162. doi: 10.1111/j.1600-065x.1987.tb00513.x. [DOI] [PubMed] [Google Scholar]
- Mariuzza R. A., Amit A. G., Boulot G., Saludjian P., Saul F. A., Tougard P., Poljak R. J., Conger J., Lamoyi E., Nisonoff A. Crystallization of the fab fragments of monoclonal anti-p-azophenylarsonate antibodies and their complexes with haptens. J Biol Chem. 1984 May 10;259(9):5954–5958. [PubMed] [Google Scholar]
- Meek K., Jeske D., Slaoui M., Leo O., Urbain J., Capra J. D. Complete amino acid sequence of heavy chain variable regions derived from two monoclonal anti-p-azophenylarsonate antibodies of BALB/c mice expressing the major cross-reactive idiotype of the A/J strain. J Exp Med. 1984 Oct 1;160(4):1070–1086. doi: 10.1084/jem.160.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun G., Sanz I., Meek K., Tucker P., Capra J. D. The molecular genetics of the arsonate idiotypic system of A/J mice. Adv Immunol. 1988;42:95–164. doi: 10.1016/s0065-2776(08)60843-3. [DOI] [PubMed] [Google Scholar]
- Rini J. M., Schulze-Gahmen U., Wilson I. A. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992 Feb 21;255(5047):959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- Rose D. R., Strong R. K., Margolies M. N., Gefter M. L., Petsko G. A. Crystal structure of the antigen-binding fragment of the murine anti-arsonate monoclonal antibody 36-71 at 2.9-A resolution. Proc Natl Acad Sci U S A. 1990 Jan;87(1):338–342. doi: 10.1073/pnas.87.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein T. L., Gefter M. L. Affinity analysis of idiotype-positive and idiotype-negative Ars-binding hybridoma proteins and Ars-immune sera. Mol Immunol. 1983 Feb;20(2):161–168. doi: 10.1016/0161-5890(83)90127-x. [DOI] [PubMed] [Google Scholar]
- Siegelman M., Capra J. D. Complete amino acid sequence of light chain variable regions derived from five monoclonal anti-p-azophenylarsonate antibodies differing with respect to a crossreactive idiotype. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7679–7683. doi: 10.1073/pnas.78.12.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz M., Gefter M. L., Brodeur P., Riblet R., Marshak-Rothstein A. The genetic basis of antibody production: the dominant anti-arsonate idiotype response of the strain A mouse. Eur J Immunol. 1982 Dec;12(12):1023–1032. doi: 10.1002/eji.1830121208. [DOI] [PubMed] [Google Scholar]
- Stanfield R. L., Fieser T. M., Lerner R. A., Wilson I. A. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science. 1990 May 11;248(4956):712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- Strong R. K., Campbell R., Rose D. R., Petsko G. A., Sharon J., Margolies M. N. Three-dimensional structure of murine anti-p-azophenylarsonate Fab 36-71. 1. X-ray crystallography, site-directed mutagenesis, and modeling of the complex with hapten. Biochemistry. 1991 Apr 16;30(15):3739–3748. doi: 10.1021/bi00229a022. [DOI] [PubMed] [Google Scholar]
- Strong R. K., Petsko G. A., Sharon J., Margolies M. N. Three-dimensional structure of murine anti-p-azophenylarsonate Fab 36-71. 2. Structural basis of hapten binding and idiotypy. Biochemistry. 1991 Apr 16;30(15):3749–3757. doi: 10.1021/bi00229a023. [DOI] [PubMed] [Google Scholar]



