Abstract
Direct reprogramming of pancreatic nonendocrine cells into insulin-producing β-cells represents a promising approach for the treatment of insulin-dependent diabetes. However, its clinical application is limited by the potential for insertional mutagenesis associated with the viral vectors currently used for cell reprogramming. With the aim of developing a nonintegrative reprogramming strategy for derivation of insulin-producing cells, here, we evaluated a new approach utilizing synthetic messenger RNAs encoding reprogramming transcription factors. Administration of synthetic mRNAs encoding three key transcription regulators of β-cell differentiation—Pdx1, Neurogenin3, and MafA—efficiently reprogrammed the pancreatic exocrine cells into insulin-producing cells. In addition to the insulin genes expression, the synthetic mRNAs also induced the expressions of genes important for proper pancreatic β-cell function, including Sur1, Kir6.2, Pcsk1, and Pcsk2. Pretreating cells with the chromatin-modifying agent 5-Aza-2′-deoxycytidine further enhanced reprogramming efficiency, increasing the proportion of insulin-producing cells from 3.5 ± 0.9 to 14.3 ± 1.9% (n = 4). Moreover, 5-Aza-2′-deoxycytidine pretreatment enabled the reprogrammed cells to respond to glucose challenge with increased insulin secretion. In conclusion, our results support that the reprogramming of pancreatic exocrine cells into insulin-producing cells, induced by synthetic mRNAs encoding pancreatic transcription factors, represents a promising approach for cell-based diabetes therapy.
Keywords: β-cells, diabetes, insulin-producing cells, MafA, modified mRNA, Neurogenin 3, Pdx1, reprogramming, synthetic mRNA, transdifferentiation
Introduction
Reports of whole pancreas transplantations and transplantations of isolated pancreatic islets demonstrate that replacement of insulin-producing tissue can potentially cure insulin-dependent diabetes.1 However, use of this therapeutic approach is limited by a lack of suitable organ donors and the need for permanent immunosuppression. Thus, there remains a need for a safe and plentiful source of insulin-producing cells. One of the most promising methods is the differentiation of embryonic stem cells and induced pluripotent stem cells into insulin-producing cells, mainly due to its high efficiency and the high quality of derived cells.2,3 However, the clinical application of this method may be limited by the potential risk of transformation into malignant cells.4,5
Cell reprogramming has recently emerged as another promising means of generating insulin-producing cells. A terminally differentiated cell can be directly reprogrammed into the desired cell type via temporal expression of transcription factors that activate the transdifferentiation program. Specific transcription factor combinations can induce reprogramming of fibroblasts into neurons,6 cardiomyocytes,7 hepatocytes,8 and induced pluripotent stem cells.9,10 Similarly, pancreatic exocrine cells and liver bile duct epithelial cells can be transdifferentiated into insulin-producing cells through induced expression of the transcription factors Pdx1, Neurogenin3, and MafA, which participate in the natural differentiation of pancreatic β-cells.11,12,13 Insulin-producing cells derived from exocrine or liver cells by in vivo reprogramming reportedly normalize blood glucose levels in diabetic mice, demonstrating their therapeutic potential.14,15
Viral vectors are often used to introduce specific transcription factors into cells for reprogramming. However, highly efficient lentiviral and retroviral vectors can lead to the integration of viral DNA sequences into chromosomal DNA, potentially causing tumorigenic transformation.16,17 Likewise, adenoviral vectors that are considered to be nonintegrating, tend to integrate viral DNA into the host genome, although at a low frequency.18,19 Therefore, a truly integration-free reprogramming method could substantially improve the safety of the derived cells for eventual clinical application. Several integration-free techniques, utilizing episomal plasmids,20 recombinant proteins,21 Sendai RNA virus,22 miRNA,23 and synthetic mRNA have been recently reported.24 While each of these methods has both advantages and disadvantages, the most efficient method appears to be cell reprogramming using synthetic mRNAs encoding reprogramming factors.25
The present study aimed to develop a safe and integration-free method of reprogramming pancreatic exocrine cells into insulin-producing cells. For this purpose, we chose the AR42J cell line. AR42J is a rat pancreatic exocrine cell line derived from a chemically induced pancreatic tumor.26 It has been previously used as a model cell line for the analysis of pancreatic exocrine cells transdifferentiation into insulin-producing cells induced by adenoviral vectors encoding Pdx1, Neurogenin3, and MafA transcription factors.11,12 Unlike primary exocrine cells, AR42J cells possess both exocrine and neuroendocrine properties as evidenced by the expression of the neuroendocrine-specific vesicle proteins synaptophysin and S.V.2 (ref. 27). Mixed exocrine-neuroendocrine character of these cells is further evidenced by the considerable amounts of neurotransmitters glycine, glutamine, and γ-aminobutyric acid. However, AR42J cells do not express any of the islet hormones under the standard culture conditions.28 Moreover, AR42J cells have a stable phenotype upon in vitro culture and do not tend to undergo a ductal transdifferentiation under adherent culture conditions, like primary pancreatic exocrine cells do.11
Reprogramming factors were delivered into the exocrine cells in a form of synthetic mRNAs encoding the pancreatic transcription factors Pdx1, Neurogenin3, and MafA. Temporary expression of these reprogramming factors activated transdifferentiation of pancreatic exocrine cells into insulin-producing cells that expressed characteristic pancreatic β-cell markers and could process proinsulin into mature insulin and its byproduct C-peptide. The reprogrammed cells responded to glucose stimulation with limited insulin secretion, similar to that of immature β-cells.29 Our results represent the first proof that it is feasible to generate insulin-producing cells through the transdifferentiation of exocrine pancreatic cells using an integration-free protocol based on synthetic mRNAs.
Results
Induced expression of reprogramming factors upon intracellular delivery of synthetic modified mRNAs
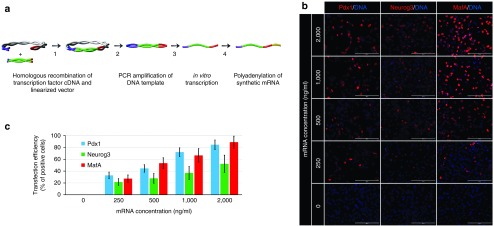
Cell reprogramming relies on ectopic expression of reprogramming transcription factors. Therefore, we first evaluated the efficiencies of transfection of each individual synthetic mRNA and expression of the encoded pancreatic transcription factors Pdx1, Neurogenin3, and MafA by the pancreatic exocrine cell line AR42J. Immunofluorescence staining revealed that transcription factor expressions were dose-dependent, with maximal expression rates achieved at a concentration exceeding 1−2 µg mRNA/ml media 20 hours post-transfection (Figure 1b,c). At a dose of 1 µg mRNA/ml media, Pdx1 was efficiently expressed by most cells (72.1 ± 7.4%, n = 5) while the expression rates of MafA (66.7 ± 11.3%, n = 5) and Neurogenin3 (36.9 ± 10.9%, n = 5) were lower and more variable as revealed by immunofluorescence staining (Figure 1b,c). Even at a higher mRNA concentration of 2 µg/ml media, variable expression was still detected, mainly for Neurogenin3 and MafA (Figure 1b,c).
Figure 1.
Scheme of DNA template construct production, in vitro transcription, and determination of efficiencies of transfection and expressions of synthetic mRNAs of the transcription factors Pdx1, Neurogenin3, and MafA by the pancreatic exocrine cell line AR42J. (a) Production of DNA template constructs and subsequent mRNA synthesis: (1) homologous recombination of transcription factor cDNA and linearized vector containing the T7 promoter, the 5′UTR (untranslated region) of the rat β-globin gene, and the 3′UTR of the human β-globin gene; (2) PCR amplification of DNA template; (3) in vitro transcription; and (4) polyadenylation of synthetic mRNA. (b, c) Dose-dependent expressions of Pdx1, Neurogenin3, and MafA upon transfection of AR42J cells with synthetic mRNAs at doses of 0, 250, 500, 1,000, and 2,000 ng/ml media as determined by immunofluorescence staining 20 hours post-transfection. Cell nuclei are stained blue with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DNA). Scale bars = 200 µm. Values are shown as mean ± standard deviation (n = 5).
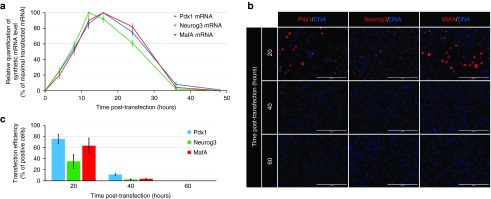
Since mRNA stability is one of the key parameters determining the gene expression rate, we also evaluated the post-transfection stability of the synthetic mRNAs. Within 4 hours, synthetic mRNA was detected in cells. The highest level of synthetic mRNA was detected between 12–16 hours post-transfection. The level of synthetic mRNA in cells substantially decreased by 24 hours post-transfection (Figure 2a), although some synthetic mRNA was detected even at 36 hours post-transfection.
Figure 2.
Stability of synthetic mRNAs of the transcription factors Pdx1, Neurogenin3, and MafA. (a) Stability of synthetic mRNAs for Pdx1, Neurogenin3, and MafA upon their transfection into AR42J cells as revealed by quantitative reverse transcription polymerase chain reaction (n = 3). (b, c) Immunofluorescence staining results showing the stability of Pdx1, Neurogenin3, and MafA at 20, 40, and 60 hours after transfection of AR42J cells with the corresponding synthetic mRNAs at a dose of 1 µg mRNA/ml media. Cell nuclei (DNA) are stained blue by 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride. Scale bars = 200 µm. Values are shown as mean ± standard deviation (n = 5).
At the protein level, expression of the encoded pancreatic transcription factors was most intense at 20 hours after transfection of cells with synthetic mRNAs at a dose of 1 µg mRNA/ml media (Figure 2b,c). Positive staining was detected even 40 hours post-transfection, although the staining intensity and the number of positive cells significantly declined. All positive cells disappeared within 60 hours post-transfection.
Simultaneous coexpression of reprogramming transcription factors upon intracellular delivery of synthetic modified mRNAs
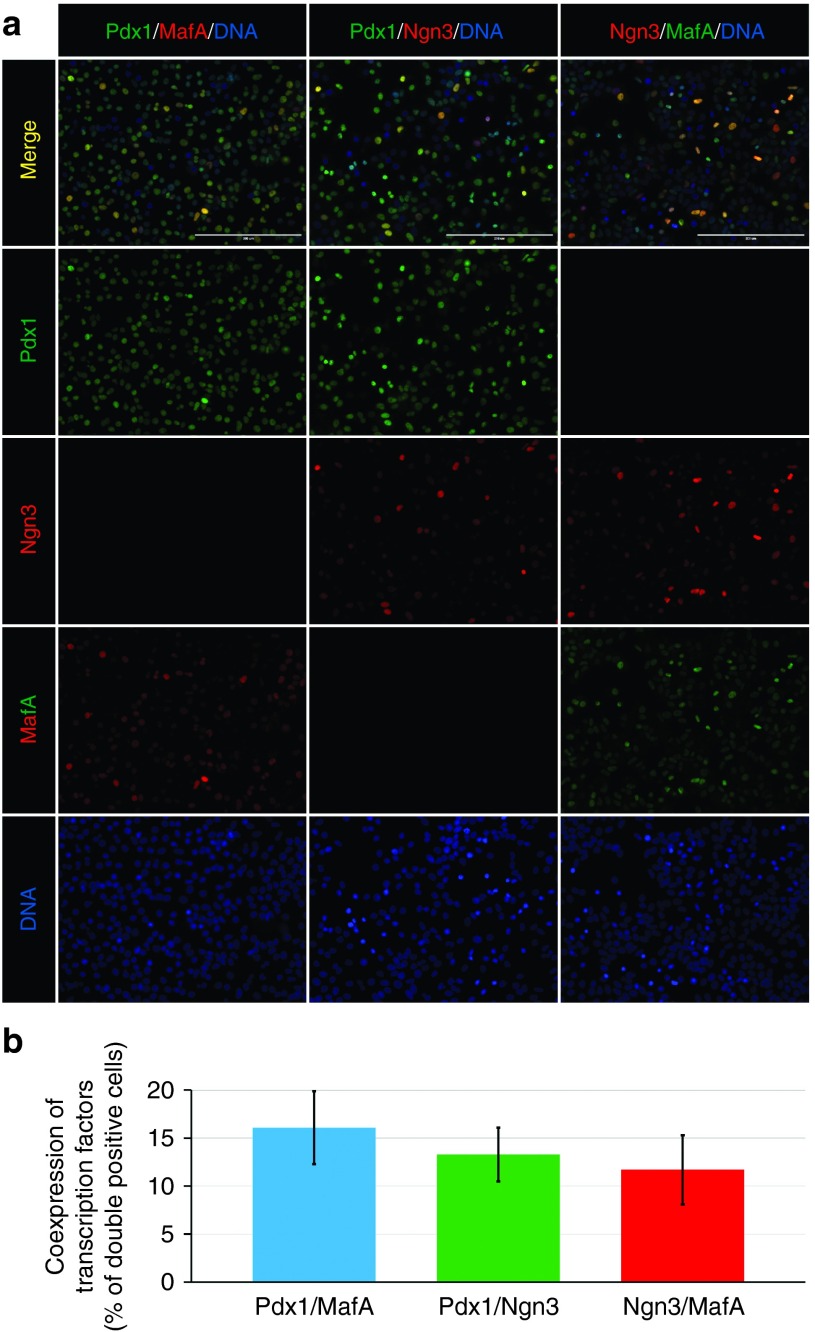
Efficient cell reprogramming requires simultaneous expression of transcription factors. Therefore, we evaluated the coexpression of the reprogramming transcription factors upon simultaneous transfection of cells with all three synthetic mRNAs (Pdx1, Neurogenin3, and MafA) at a dose of 500 ng of each mRNA/ml media (Figure 3a). Transcription factor coexpression was mainly limited by the expression rates of Neurogenin3 and MafA, since Pdx1 was expressed by most of the Neurogenin3- and MafA-positive cells. The rates of double-positive cells were 16.1 ± 3.8% (n = 4) for Pdx1 and MafA, 13.3 ± 2.8% (n = 4) for Pdx1 and Neurogenin3, and 11.7 ± 3.6% (n = 4) for MafA and Neurogenin3 (Figure 3b).
Figure 3.
Transcription factors coexpression. (a, b) Immunofluorescence staining results showing coexpression of the transcription factors Pdx1, Neurogenin3 (Ngn3), and MafA following simultaneous transfection of AR42J cells with all three synthetic mRNAs at a dose of 500 ng of each mRNA/ml media. Double-positive cells are indicated by yellow color in the upper row. Cell nuclei (DNA) are stained blue with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride. Scale bars = 200 µm. Values are shown as mean ± standard deviation (n = 4).
Supplementation with vaccinia virus-derived type I interferon receptor B18R prevents cell death during repeated transfection of synthetic modified mRNAs
Efficient cell reprogramming also requires that transcription factor expression continue over a sufficient time period. Therefore, synthetic mRNA was delivered in the form of lipid complexes, allowing repeated transfection. However, repeated daily transfection with synthetic mRNAs at a dose exceeding 1 µg/ml led to the induction of apoptosis and substantial cell loss over the 3-day period (Supplementary Figure S1). This may have been due to activation of the cellular innate immune response, which serves as an antiviral defense mechanism against DNA and RNA viruses30 and is characterized by inflammatory cytokine production, protein synthesis inhibition, and apoptosis induction.31 Innate immune response activation by exogenous mRNA can be limited by incorporating modified nucleotide bases into the synthetic mRNA24,30,32 and by dephosphorylation of 5′ triphosphates via phosphatase treatment.24,33 However, using the modified nucleotides pseudouridine and 5-methylcytidine in our mRNA synthesis and phosphatase treatment were not sufficient to prevent cell loss caused by the repeated transfection. Therefore, we further tested the use of the recombinant protein B18R—a soluble receptor of type I interferons—which has previously been used during the highly efficient synthetic mRNA-induced reprogramming of skin fibroblasts into induced pluripotent stem cells.24
Supplementation of culture media with B18R, significantly improved cell survival and attenuated the cell apoptosis induced by repeated transfection of synthetic mRNAs (Supplementary Figure S1). Therefore, in our further experiments, we used B18R supplementation with repeated daily transfection. This addition allowed us to achieve prolonged expressions of Pdx1, Neurogenin3, and MafA for at least 10 days (Supplementary Figure S2a). However, in order to limit the potential activation of innate immune response by the increased amount of synthetic mRNA exceeding 2 µg/ml, we used only 500 ng/ml of each mRNA (1,500 ng/ml of all three mRNAs) for repeated daily cotransfection, during the 10-day reprogramming period.
The coexpression of Pdx1, Neurogenin3, and MafA transcription factors was slightly increased following 10 days repeated daily cotransfection of all three synthetic mRNAs, in comparison with a single simultaneous transfection (Supplementary Figure S2). The rates of double-positive cells following 10 days repeated daily cotransfection were 20.5 ± 3.2% (n = 4) for Pdx1 and MafA, 17.8 ± 3.4% (n = 4) for Pdx1 and Neurogenin3, and 15.1 ± 5.1% (n = 4) for MafA and Neurogenin3 (Supplementary Figure S2b).
Reprogramming of pancreatic exocrine cells into insulin-producing cells using synthetic modified mRNAs encoding Pdx1, Neurogenin3, and MafA
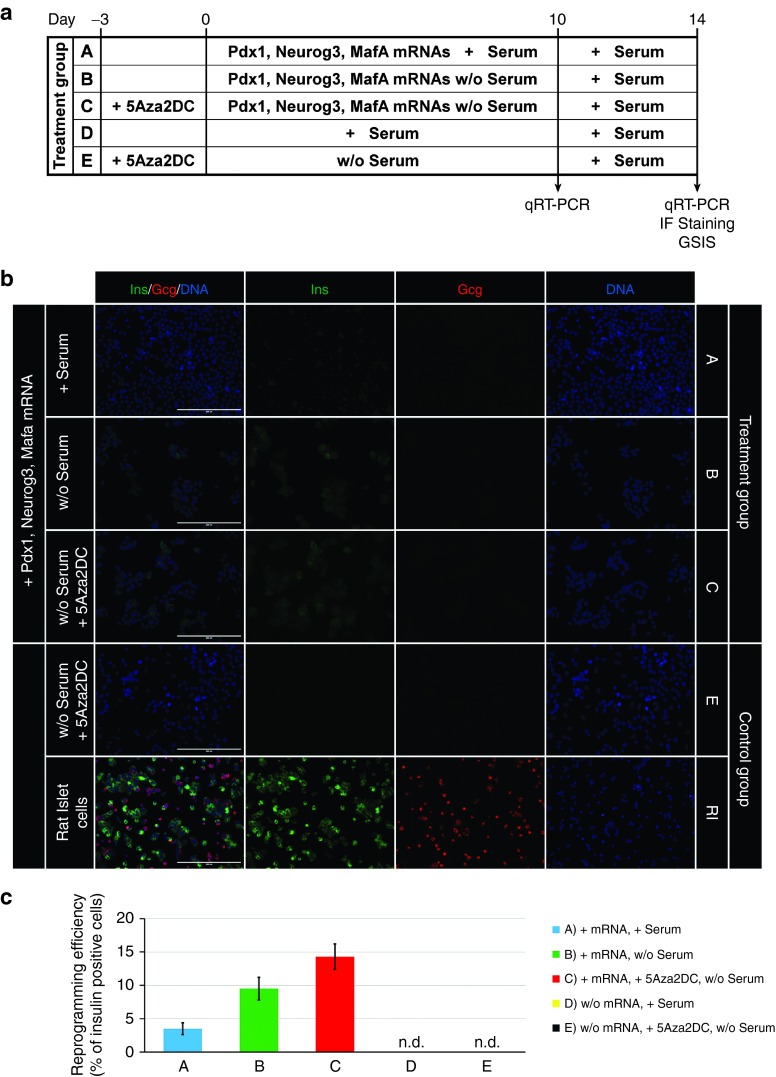
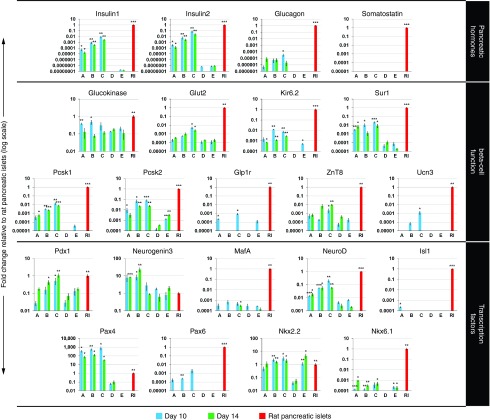
We next investigated the potential of the synthetic modified mRNAs encoding Pdx1, Neurogenin3, and MafA for reprogramming pancreatic exocrine cells into insulin-producing cells. AR42J cells were transfected daily for 10 days with a combination of all three synthetic mRNAs, at doses of 500 ng/ml each, and cultured in serum-containing medium (denoted as a treatment group A) (Figure 4a). During the reprogramming period, cells began to express pancreatic hormones insulin and glucagon. However, the reprogramming efficiency was very low, with immunofluorescence staining showing only 3.5 ± 0.9% (n = 4) insulin-positive cells (Figure 4b). While the insulin expression was detected at the mRNA and protein levels, the expression of glucagon was detectable only at the mRNA level (Figure 5). The results of quantitative reverse transcription polymerase chain reaction further showed that repeated daily transfection with the synthetic mRNAs led to upregulation or induction of genes important for pancreatic β-cell differentiation (Pax4 and Nkx2.2) and function (Kir6.2, Sur1, Pcsk1, Pcsk2, and Glp1r) (Figure 5). However, some transcription factors (Isl1, Ngn3, Nkx6.1, and Pax6) and genes important for proper function (Glut2 and ZnT8) were upregulated only slightly or not at all (Figure 5). Detection of C-peptide by immunofluorescence staining (Figure 6a) revealed proper processing of prohormone peptide proinsulin into mature insulin and its byproduct C-peptide by the neuroendocrine endoproteases Pcsk1 and Pcsk2.
Figure 4.
Scheme of the experimental design and evaluation of reprogramming efficiency. (a) Overview of the reprogramming protocol and subsequent analyses. Cell samples were divided into five groups based on culture conditions and the administration of all three reprogramming transcription factors (Pdx1, Neurogenin3, and MafA) for 10 days in the form of synthetic mRNAs at a dose of 500 ng of each mRNA/ml media. Cells were either cultured in serum-containing medium with mRNA transfection (treatment group A), cultured in serum-free medium with mRNA transfection (treatment group B), or pretreated for 3 days with 5-Aza-2′-deoxycytidine and cultured in serum-free medium with mRNA transfection (treatment group C). The expression profiles were compared with those of non-transfected AR42J cells that were either cultured in serum-containing medium (control group D), or pretreated for 3 days with 5-Aza-2′-deoxycytidine and cultured in serum-free medium (control group E) and of native rat pancreatic islets (control group RI). (b, c) Evaluation of reprogramming efficiency by immunofluorescence staining for the β-cell marker insulin (Ins) and the α-cell marker glucagon (Gcg). Insulin and glucagon expression was compared with non-transfected AR42J cells that were pretreated for 3 days with 5-Aza-2′-deoxycytidine and cultured in serum-free medium (control group E) and native rat pancreatic islet cells (control group RI). Cell nuclei (DNA) are stained blue with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride. Scale bars = 200 μm. Values are shown as mean ± standard deviation (n = 4). n.d., not detected.
Figure 5.
Gene expression profiles of reprogrammed cells were analyzed by quantitative reverse transcription polymerase chain reaction at the end of reprogramming (day 10—blue bars) and at 4 days after the last transfection with synthetic mRNAs (day 14—green bars). AR42J cells were treated with all three synthetic mRNAs (Pdx1, Neurogenin3, and MafA) for 10 days at a dose of 500 ng of each mRNA/ml media. Cells were either cultured in serum-containing medium with mRNA transfection (treatment group A), cultured in serum-free medium with mRNA transfection (treatment group B), or pretreated for 3 days with 5-Aza-2′-deoxycytidine and cultured in serum-free medium with mRNA transfection (treatment group C). The gene expression profiles were compared with those of native rat pancreatic islets (control group RI) and of nontransfected AR42J cells that were either cultured in serum-containing medium (control group D), or pretreated for 3 days with 5-Aza-2′-deoxycytidine and cultured in serum-free medium (control group E). Endogenous expressions of Pdx1, Neurogenin3, and MafA genes were determined using reverse primers specific for the 3′UTR (untranslated region) of each particular gene, which were not specific for synthetic mRNAs. The expression levels are presented relative to gene expression of rat pancreatic islets (normalized to 1). Values are shown as mean ± standard deviation (n = 5). Statistical analysis was performed using a two-tailed unpaired Student's t-test with Holm–Bonferroni correction. Samples were compared with nontransfected AR42J cells cultured in serum-containing medium (control group D). Asterisks indicate statistical significance: *P < 0.05, **P < 0.01, ***P < 0.001.
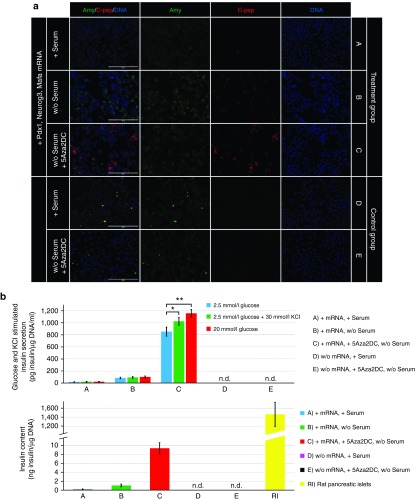
Figure 6.
Reprogramming efficiency and determination of insulin secretion capacity and insulin content. (a) Reprogramming efficiency was evaluated by immunofluorescence staining for the exocrine marker amylase (Amy) and the β-cell marker C-peptide (C-pep). Cell nuclei (DNA) are stained blue with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride. Scale bars = 200 μm. (b) Glucose-stimulated insulin secretion of cell samples was determined by sequential 60-minute incubations at low (2.5 mmol/l) and high (20 mmol/l) glucose concentrations. The effect of depolarizing agent KCl on insulin secretion was determined by sequential 60-minute incubations at low (2.5 mmol/l) glucose concentration followed by low (2.5 mmol/l) glucose concentration with 30 mmol/l KCl. Insulin content in cell lysates was determined following KCl stimulated insulin secretion capacity test. Values are shown as mean ± standard deviation (n = 5). n.d., not detected. Statistical analysis was performed using a two-tailed unpaired Student's t-test. Asterisks indicate statistical significance: *P < 0.05, **P < 0.01.
Serum exclusion from culture medium enhances reprogramming efficiency
We next attempted to improve the reprogramming efficiency by optimizing the culture conditions. It has previously been shown that exclusion of serum from culture medium can significantly improve reprogramming efficiency.34 Indeed, our results showed that replacing fetal bovine serum with human serum albumin (denoted as a treatment group B) (Figure 4a) led to more efficient reprogramming, characterized by a greater proportion of insulin-positive cells (9.5 ± 1.7%, n = 4) and a higher insulin and C-peptide expression rates (Figures 4b and 6a). These results were confirmed by quantitative reverse transcription polymerase chain reaction (Figure 5), which revealed further upregulation of genes important for a proper pancreatic β-cell function, including Glut2, Kir6.2, Pcsk1, and Pcsk2. However, the reprogrammed cells were not glucose-responsive as detected by inefficient glucose-stimulated insulin secretion (88 ± 12 versus 101 ± 15 pg insulin/µg DNA/ml) (n = 5) upon exposure to high glucose concentration (2.5 versus 20 mmol/l glucose) (Figure 6b).
Effect of DNA demethylation on cell reprogramming
Cell reprogramming efficiency depends on both the ectopic expression of reprogramming factors and the induction of endogenous genes. Thus, we further evaluated the effect of 5-Aza-2′-deoxycytidine on cell reprogramming and endogenous transcription factor expression. The chromatin-modifying agent 5-Aza-2′-deoxycytidine inhibits DNA methyltransferase activity, resulting in DNA demethylation, chromatin structure remodeling, and subsequently increased accessibility of genes for transcription factors—which is a necessary condition for gene expression activation.
Pretreatment of cells with 5-Aza-2′-deoxycytidine, followed by transfection with the synthetic mRNAs (denoted as a treatment group C) (Figure 4a) further improved reprogramming efficiency, as revealed by the increased proportion of insulin-producing cells (14.3 ± 1.9%, n = 4, Figure 4b); greater insulin gene expression; and upregulation of the functional genes Glut2 and Pcsk1, the transcription factors NeuroD and Pax6, and the maturation marker Urocortin3 (Figure 5).35 Moreover, only the reprogramming protocol that included 5-Aza-2′-deoxycytidine pretreatment induced glucose-responsive reprogrammed cells, and led to glucose-stimulated insulin secretion (842 ± 72 versus 1,157 ± 58 pg insulin/µg DNA/ml) (n = 5) upon exposure to high glucose concentration (2.5 vs. 20 mmol/l glucose) (Figure 6b). Insulin release under the basal glucose level (2.5 mmol/l glucose) was also induced by depolarizing agent potassium chloride (863 ± 78 versus 1,025 ± 66 pg insulin/µg DNA/ml) (n = 5), albeit at a lower extent than by high glucose concentration (Figure 6b).
However, in spite of improved reprogramming efficiency promoted by DNA demethylation, incomplete reprogramming of AR42J exocrine cells was also revealed by significantly lower insulin content (9.3 ± 1.3 ng insulin/µg DNA) (n = 5) in comparison with rat pancreatic islets (1,460.7 ± 268.1 ng insulin/µg DNA) (Figure 6b). Moreover, endogenous expression of Pdx1, Neurogenin3 and MafA transcription factors at protein level was not detected at the end of reprogramming period (day 14) (Supplementary Figure S3).
Discussion
Here, we report that pancreatic cells of exocrine origin can be transdifferentiated into insulin-producing cells using synthetic mRNAs encoding key transcription regulators of β-cell differentiation. To our knowledge, this is the first demonstration of direct reprogramming of pancreatic exocrine cells into insulin-producing cells using a nonintegrative approach involving intracellular delivery of synthetic mRNAs. Although the reprogrammed cells were not fully equivalent to primary β-cells, they shared important similarities. The reprogrammed cells produced mature insulin and its byproduct C-peptide by using the neuroendocrine endoproteases Pcsk1 and Pcsk2 that process the prohormone peptide proinsulin. Moreover, the reprogrammed cells expressed key elements of glucose-sensing mechanisms—including the glycolytic enzyme glucokinase, glucose transporter isoform-2 (Glut2), and the ATP-sensitive potassium channel subunits Sur1 and Kir6.2—which are required to properly sense blood glucose level and for subsequent insulin secretion. Finally, the reprogrammed cells responded to glucose challenge with increased insulin secretion, although at a lower rate than the primary β-cells. However, the reprogrammed cells were not fully equivalent to primary β-cells, as shown by the low stimulatory index and the inability to increase insulin secretion upon membrane depolarization by KCl. Moreover, insulin content of reprogrammed cells was significantly lower in comparison with rat pancreatic islets. The immature phenotype of reprogrammed cells can be explained by insufficient expression of the transcription factors and of the genes responsible for the complex β-cell-specific expression program.
Cell transdifferentiation is characterized by suppression of the original expression program and induction of a newly acquired one,36 which requires the expression of key regulatory transcription factors. Thus, reprogramming efficiency could potentially be improved by inducing the expressions of additional transcription factors. We propose that the transcription factors Nkx6.1, Pax6, and Isl1 are the most promising candidates for improving reprogramming efficiency, since their expressions were greatly limited or absent in our transdifferentiated cells. Each of these three transcription factors is active during the later phase of β-cell differentiation, and in mature β-cells.37,38,39 Nkx6.1, Pax6, and Isl1 reportedly have positive effects on expressions of the insulin gene itself and of several key regulators of glucose-stimulated insulin secretion.40,41,42
Induction of the Nkx6.1 transcription factor may have been limited by possible repression by the exocrine cell-specific transcription factors Ptf1a and RbpJ.43,44 Although, we only rarely observed amylase and C-peptide double-positive cells, we cannot exclude that the reprogrammed cells may have persistently expressed Ptf1a and RbpJ. On the other hand, Pax6 and Isl1, are downstream targets of the Neurogenin3 transcription factor.39,45 Thus, it seems that the ectopic Neurogenin3 expression by reprogrammed cells was insufficient to induce endogenous expressions of Pax6 and Isl1. The limited Pax6 expression by our reprogrammed cells is in agreement with previous findings in insulin-secreting cells derived from human pancreatic ductal cells.46 That study also revealed insufficient induction of endogenous Pax6 expression upon reprogramming induced by Pdx1, Neurogenin3, and MafA, and reported that ectopic Pax6 expression was required to enhance the expressions of insulin and other β-cell functional genes.
The epigenetic status of the transdifferentiated cells may have also influenced the induction of endogenous gene expression. Activation of gene expression during cellular differentiation requires remodeling of the gene-specific DNA chromatin structure from transcriptionally inactive heterochromatin into active euchromatin.47 Thus, inappropriate chromatin remodeling can lead to insufficient induction of endogenous gene expression. Our results showed that the chromatin modifying agent 5-Aza-2′-deoxycytidine positively impacted cellular reprogramming and upregulation of gene expression. However, 5-Aza-2′-deoxycytidine only modulates DNA methylation status, not any other possible epigenetic modifications.
Gene expression can also be limited by repressive modifications of histone proteins that substantially impact chromatin structure. For example, the trimethylation of lysine 27 at histone H3 (H3K27) induces formation of an inactive heterochromatin structure.47,48 A recent study comparing histone modifications between pancreatic exocrine cells and β-cells demonstrated one such H3K27 repressive modification of the Nkx6.1, Pax6, and Isl1 genes in pancreatic exocrine cells.48 Furthermore, this same repressive H3K27 modification was detected in genes important for β-cell function following the in vitro differentiation of embryonic stem cells into insulin-producing cells.49 The Glp1r and Urocortin3 genes were among those marked with a repressive modification, and were also inefficiently induced in our reprogrammed cells. The same previous work demonstrated the importance of proper chromatin modifications on gene expression, by showing the effect of an in vivo terminal differentiation in embryonic stem cell-derived cells. The in vitro terminally differentiated insulin-producing cells were associated with repressive histone modifications and with insufficient induction of genes important for β-cell function. On the other hand, in vivo terminal differentiation of embryonic stem cell-derived cells into insulin-producing cells induced permissive histone modifications and significantly higher expressions of the β-cell functional genes.49
Our reprogrammed insulin-producing cells did not resemble the so-called polyhormonal cells that produce insulin along with the other pancreatic hormones glucagon and somatostatin.29 Therefore, we assume that our reprogramming protocol induced transdifferentiation directly to the β-like cell phenotype. During reprogramming, we observed substantial induction of the Pax4 transcription factor, which is transiently overexpressed during the early phase of pancreatic endocrine cells differentiation.50 Pax4 restricts endocrine cell differentiation into the β- and δ-cell lineages via repression of the α-cell-specific transcription factor Arx.51 Moreover, Pax4 and the transcription factor Nkx2.2 further specify the differentiation of endocrine progenitors into the β-cell phenotype.52 While the endogenous expression of Pax4 transcription factor was induced following the reprogramming period, Nkx2.2 is naturally expressed by AR42J cells, and its expression was only slightly upregulated by the reprogramming factors. Further specification of AR42J cells into β-like cell phenotype could be promoted by the Pdx1 transcription factor that activates genes essential for β-cell identity and represses those associated with α-cell identity. Therefore, the ectopic overexpression of Pdx1 transcription factor, that is also naturally expressed by AR42J cells, can further stimulate the reprogramming of AR42J cells into β-like cell lineage. In addition to the transcription factors that are active during the early phase of β-cell differentiation, we also observed slight induction of the β-cell maturation marker Urocortin3 (ref. 35) at the end of the reprogramming period. Therefore, we assume that our reprogrammed insulin-producing cells resemble partially differentiated immature β-cells. This immature phenotype could be caused by insufficient induction of additional transcription factors such as MafA, that are responsible for the final maturation and proper function of pancreatic β-cells. Limited endogenous expression of MafA, which was significantly under-expressed in comparison with the native β-cells, can be explained by insufficient induction of Nkx6.1, Pax6, and Isl1 transcription factors that all positively regulate the MafA expression.33,36,37,38
Previous studies have reported the reprogramming of exocrine cells into insulin-producing cells using adenoviral vectors.11,12,13,14,15,36 Although adenoviral vectors are highly efficient in the delivery and expression of introduced genes, the application of this process is limited by the potential for insertional mutagenesis,18,19 and by the prolonged persistence in infected cells that does not allow modulation of the reprogramming process.46 The presently described mRNA-based reprogramming resolves all of these issues. The mRNA chemical structure eliminates the risk of insertional mutagenesis or any other effect on cellular DNA. Moreover, the intracellular stability of mRNA is limited by permanent endogenous degradation, such that synthetic mRNA establishes only transient expression of the encoded gene. The use of synthetic mRNAs to induce temporal and sequential expression of different combinations of reprogramming factors allows to mimic the natural cellular differentiation process, in which some transcription factors are expressed only transiently while others are expressed over longer period of time.39,44,45 Moreover, appropriate transcription factor stoichiometry can be achieved at different stages of cellular reprogramming.
On the other hand, disadvantages of mRNA-based reprogramming may include the need for repeated transfection and the potential cytotoxic effects of the synthetic mRNA. However, these issues could potentially be overcome by mRNA sequence optimization to improve the stability and translation efficiency, consequently reducing the required dose of mRNA.53 Elimination of the cytotoxic effects of synthetic mRNA may also be promoted by high-performance liquid chromatography purification. These cytotoxic effects are mainly caused by aberrant byproducts formed during in vitro mRNA synthesis.54,55 Highly efficient high-performance liquid chromatography purification can substantially reduce the amount of such byproducts in the final mRNA preparation, consequently eliminating cytotoxic effects and activation of the innate immune response by transfected cells.55 It is worth noting that we did not test any of these possible improvements of synthetic mRNA in our present study. We only used the vaccinia virus B18R receptor of type I interferons to eliminate innate immune response activation. B18R application allowed us to achieve long-term repeated transfection of synthetic mRNA and to eliminate its cytotoxic effects. However, the addition of sequence optimization and high-performance liquid chromatography purification to our protocol would likely further improve the reprogramming efficiency and reduce the negative side effects.
Our present results demonstrate that using synthetic mRNAs encoding pancreatic transcription factors to reprogram pancreatic exocrine cells into insulin-producing cells, could represent a safe and promising approach for cell-based diabetes therapy. However, there remains a need for further optimization of the synthetic mRNAs, the culture conditions, and the combination of transcription factors to achieve efficient reprogramming into insulin-producing cells that are functionally equivalent to the native β-cells.
Materials and Methods
Construction of DNA Templates. Figure 1a shows the scheme for the production of DNA template constructs and subsequent RNA synthesis. All oligonucleotides were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). The Supplementary Note S1 includes the oligonucleotide sequences used for DNA template construction. The Pdx1, Neurogenin3, and MafA coding regions were derived by reverse transcription of mRNA isolated from primary rat pancreatic islet cells, using gene-specific primers (Supplementary Table S1) and the AccuScript High-Fidelity 1st Strand cDNA Synthesis Kit (Agilent, Santa Clara, CA), following the manufacturer's instructions. Polymerase chain reaction (PCR) amplification of cDNA was performed using the same gene-specific primers and Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA), following the manufacturer's instructions. DNA template constructs were prepared using the pAcGFP1-N3 vector (Clontech, Mountain View, CA) along with the gBlock gene fragment (IDT) that contains sequences encoding the T7 RNA polymerase promoter site, the 5′ untranslated region (UTR) of the rat β-globin gene, two PstI cloning sites, and the 3′UTR of the human β-globin gene. The gBlock gene fragment was inserted into the BamHI and NheI (New England Biolabs) sites of the linearized pAcGFP1-N3 vector by homologous recombination, using the In-Fusion PCR cloning kit (Clontech), following the manufacturer's instructions. Then, the pAcGFP1-N3 vector with the integrated gBlock gene fragment was further linearized using the PstI restriction enzyme (New England Biolabs). The In-Fusion PCR cloning kit was then used to insert cDNA of each transcription factor coding region into the PstI-linearized vector. To verify the DNA sequence of the prepared vectors, we used the BigDye Terminator v3.1 Cycle Sequencing Kit with a 3130 Genetic Analyzer (Life Technologies, Grand Island, NY).
The NheI restriction enzyme (New England Biolabs) was used to excise a DNA template encoding the T7 RNA polymerase promoter site, the 5′UTR of the rat β-globin gene, the transcription factor open reading frame, and the 3′UTR of the human β-globin gene from the vector. This excised fragment was purified by agarose gel electrophoresis and the QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA). Isolated DNA fragment was PCR amplified with DNA template-specific primers (Supplementary Table S2) and Q5 High-Fidelity DNA Polymerase (New England Biolabs), following the manufacturer's instructions. The final PCR product was purified as described above and quantified by Qubit fluorometer (Life Technologies).
Synthesis of mRNA. RNA was synthesized using a T7 mScript Standard mRNA Production System (CELLSCRIPT, Madison, WI), with 20-μl reactions containing 2 μg of purified DNA template. We used a custom ribonucleotide blend comprising 3′-0-Me-m7G(5′)ppp(5′)G ARCA cap analog, pseudouridine triphosphate, 5-methylcytidine triphosphate (TriLink Biotechnologies, San Diego, CA), adenosine triphosphate, and guanosine triphosphate (New England Biolabs). The final reaction mixture contained 6 mmol/l ARCA cap analog, 3.0 mmol/l adenosine triphosphate, and 1.5 mmol/l of each the other nucleotides. Reactions were incubated for 1 hour at 37 °C and treated with DNase following the manufacturer's instructions. Next, RNA was purified via ammonium acetate precipitation, and treated with Antarctic phosphatase (New England Biolabs) for 2 hours at 37 °C to remove residual 5′-triphosphates. Treated RNA was again purified by ammonium acetate precipitation and polyadenylated for 2 hours at 37 °C using the Poly(A) Polymerase, Yeast (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. Finally, the polyadenylated RNA was purified with the MEGAclear Transcription Clean-Up Kit (Life Technologies), diluted with RNAsecure Resuspension Solution (Life Technologies), and quantified by Qubit fluorometer. Synthetic mRNA quality was assessed using the Agilent RNA 6000 Nano Kit with an Agilent 2100 Bioanalyzer (Agilent).
Cell culture. The rat pancreatic exocrine cell line AR42J (Sigma-Aldrich) was cultured in Ham's F-12K medium (Life Technologies) containing 15% fetal bovine serum (Sigma-Aldrich) and 1% GlutaMAX supplement (Life Technologies). Cells were plated at 1 × 104 cells per well in 96-well-culture tissue dishes (Greiner Bio-One, Frickenhausen, Germany) on an extracellular matrix derived from the human bladder carcinoma cell line HTB-9, which was prepared as previously reported with a slight modification:
HTB-9 cells (American Type Culture Collection, Manassas, VA) were cultured in 96-well plates with Roswell Park Memorial Institute medium (Sigma-Aldrich) containing 10% fetal bovine serum and 1% GlutaMAX supplement. Cells were grown to confluence and cultured for an additional 3 days to allow extracellular matrix deposition. To decellularize the culture wells while leaving the intact extracellular matrix attached to the well surface, media was aspirated and each well was incubated for 5 minutes at 37 °C with 100 μl distilled water containing 20 mmol/l NH4OH and 0.1% Triton X-100 (Sigma-Aldrich). The NH4OH solution was then triturated four times and aspirated. Plates were inspected under microscope to ensure cell removal, and were washed five times with 37 °C phosphate-buffered saline (PBS) prior to seeding of AR42J cells.
During cell reprogramming, the AR42J cells were cultured in either serum-containing or serum-free Ham's F-12K medium (Figure 4a). Serum-free Ham's F-12K medium was supplemented with 0.5% human serum albumin, 1% insulin-transferrin-selenium, 1% Eagle's Minimum Essential Medium (MEM) nonessential amino acids (Life Technologies), 50 ng/ml epidermal growth factor, 10 ng/ml fibroblast growth factor 2, and 80 ng/ml insulin-like growth factor (PeproTech, Rocky Hill, NJ). Cell samples pretreated with 5-Aza-2′-deoxycytidine were cultured in serum-containing Ham's F-12K medium supplemented with 500 nmol/l 5-Aza-2′-deoxycytidine diluted in dimethyl sulfoxide (Sigma-Aldrich) for 3 days prior to reprogramming.
RNA transfection. RNA transfection was carried out using Lipofectamine MessengerMAX mRNA Transfection Reagent (Life Technologies). With Opti-MEM basal media (Life Technologies), synthetic mRNA was diluted to a concentration of 20 ng/μl and Lipofectamine MessengerMAX mRNA Transfection Reagent was diluted 33×. Diluted mRNA and transfection reagent were pooled 1:1 and incubated at room temperature for 5 minutes before being dispensed to the culture media. RNA transfections were performed in either serum-containing or serum-free Ham's F-12K medium, both supplemented with 200 ng/ml B18R interferon inhibitor (eBioscience, San Diego, CA).
Immunostaining. Cells were washed in Hank's Balanced Salt Solution (Sigma-Aldrich) and fixed in 4% paraformaldehyde for 15 minutes. Fixed cells were washed with PBS and blocked/permeabilized by a 30-minute incubation at room temperature with PBS containing 5% donkey serum (Sigma-Aldrich) and 0.3% Triton X-100 (Sigma-Aldrich). The cells were then stained in blocking buffer with primary antibodies for 30 minutes at 37 °C, washed, and then stained with secondary antibodies for 30 minutes at 37 °C with protection from light. Cell nuclei were stained for 15 minutes at room temperature with NucBlue Fixed Cell ReadyProbes Reagent (Life Technologies) diluted 1:10 in PBS. The following primary antibodies were used: rabbit anti-Pdx1 (1:200), rabbit anti-MafA (1:200), rabbit anti-insulin (1:300), mouse anti-C-peptide (1:100), mouse anti-glucagon (1:200) (Abcam, Cambridge, United Kingdom), mouse anti-Neurogenin3 (1:800), mouse anti-Pdx1 (1:400) (Developmental Studies Hybridoma Bank, Iowa City, IA), and rabbit anti-α-amylase (1:200) (Sigma-Aldrich). The secondary antibodies were donkey anti-mouse or donkey anti-rabbit IgG Alexa Fluor 555 and/or Alexa Fluor 647 (Life Technologies) at a 1:400 dilution. Images were acquired with the EVOS FL Auto Cell Imaging System (Life Technologies). Positive cells was quantified from at least ten visual fields (with 100× magnification) using the EVOS FL automatic cell counting tool.
Gene expression analysis. Total RNA was isolated using the RNeasy Mini Plus kit (Qiagen, Valencia, CA). The RNA was then treated for 1 hour at 37 °C with Turbo DNase (Life Technologies), repurified using RNA Clean & Concentrator-5 (Zymo Research, Irvine, CA), and quantitated using a Qubit fluorometer. Next, 500 ng of isolated RNA was reverse-transcribed at 50 °C for 60 minutes with random hexamer and anchored oligo dT primers (5:1 ratio) using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Rotkreuz, Switzerland) following the manufacturer's instructions. The generated cDNAs were analyzed by PCR using FastStart Universal SYBR Green Master Rox (Roche) with gene-specific primers (Integrated DNA Technologies) for each detected mRNA (Supplementary Table S3). PCR started with 10 minutes at 95 °C, which was followed by 40 cycles of 15 seconds at 95 °C (denaturation) and 1 minute at 62 °C (annealing/extension). Reactions and data analysis were carried out using a ViiA 7 Real-Time PCR System (Life Technologies). All samples were assayed in triplicates. Fold-changes in gene expression were determined using the ΔΔCT method, with normalization to β-actin expression.
Apoptosis assay. To test the synthetic mRNA for cytotoxic effects we used CellEvent Caspase-3/7 Green ReadyProbes Reagent (Life Technologies), following the manufacturer's instructions. AR42J cells were cultured in a 96-well plate, and transfected twice for 2 days with a mixture of Pdx1, Ngn3, and MafA synthetic mRNAs (1:1:1 ratio) at a total dose of 1–2 μg/ml. On the third day, we analyzed induction of apoptosis by the synthetic mRNA. CellEvent Caspase-3/7 Green Reagent and NucBlue Live ReadyProbes Reagent (Life Technologies) were added to each well and incubated at 37 °C for 30 minutes in a CO2 incubator. Next, the cell samples were washed three times with PBS, and images were acquired using an EVOS FL Auto Cell Imaging System (Life Technologies). The number of apoptotic cells and total cell number were determined from at least 10 visual fields (at 100× magnification) using the EVOS FL automatic cell counting tool.
Glucose-stimulated insulin secretion assay. Cell samples were cultured in 24-well plates and then washed three times with 0.5 ml Krebs solution (128 mmol/l NaCl, 5 mmol/l KCl, 2.7 mmol/l CaCl2, 1.2 mmol/l MgCl2, 1 mmol/l Na2HPO4, 1.2 mmol/l KH2PO4, 5 mmol/l NaHCO3, and 10 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) containing 0.1% human serum albumin and 2.5 mmol/l glucose (low-glucose solution). To normalize insulin secretion, the cell samples were then preincubated for 1 hour in the low-glucose solution. Then the low-glucose solution was refreshed and the cell samples were again incubated for 1 hour. A 250-µl sample of the low-glucose solution supernatant was aspired, centrifuged at 10,000 g for 5 minutes at 4 °C, and then immediately frozen and stored at −80 °C until analysis. The cell samples were washed three times with 0.5 ml of Krebs-Ringer solution containing 20 mmol/l glucose (high-glucose solution) or 2.5 mmol/l glucose and 30 mmol/l KCl (high KCl solution), and incubated for an additional hour. A 250-µl sample of the high-glucose solution or high KCl solution supernatant was aspired, centrifuged at 10,000 g for 5 minutes at 4 °C, and immediately frozen and stored at −80 °C until analysis.
For analysis, the cells were lysed in 0.3 ml RIPA buffer (Sigma-Aldrich) and DNA content was determined using a Qubit fluorometer. In samples from the glucose-stimulated insulin secretion assay and cell lysates, insulin content was determined using the Insulin 125I RIA kit (MP Biomedicals, Orangeburg, NY) according to the manufacturer's instructions. All incubation steps were performed at 37 °C in a CO2 incubator, and all solutions were equilibrated to 37 °C prior to use.
Statistical analysis. Statistical analyses were performed using a two-tailed unpaired Student's t-test with Holm–Bonferroni correction using GraphPad software. P values of <0.05 were considered to indicate statistically significant differences. The numbers of independent experiments performed are indicated in the text. Mean values are presented with standard deviations in the format (mean ± standard deviation).
SUPPLEMENTARY MATERIAL Figure S1. Evaluation of the effect of recombinant B18R interferon inhibitor on prevention of apoptosis upon repeated transfection of exocrine cells with synthetic mRNAs. Figure S2. Transcription factors co-expression after repeated daily transfections. Figure S3. Endogenous transcription factors co-expression after repeated daily transfections. Table S1. Primers used for reverse-transcription and PCR amplification of transcription factors cDNAs and PCR amplification of DNA templates used for in vitro transcription. Table S2. Primers used for PCR amplification of DNA templates for in vitro transcription. Table S3. Primers used for qRT-PCR gene expression analysis. Note S1. Oligonucleotide sequences of DNA constructs and templates used for in vitro transcription. Supplementary Information
Acknowledgments
T.K. managed the project, designed the experiments, executed the experiments, analyzed and interpreted the data, and wrote the paper. I.L. and S.L. executed the experiments and analyzed the data. L.K. executed the experiments. F.S. interpreted the data and wrote the paper. This work was supported by an IGA grant (project reference number NT12190-5/2011) from the Ministry of Health, Czech Republic. We thank Milan Jirsa for discussions and critical reading of this manuscript and Jan Kriz for providing samples of rat pancreatic islets. The authors declare no conflict of interest.
Supplementary Material
References
- Shapiro, AM, Ricordi, C, Hering, BJ, Auchincloss, H, Lindblad, R, Robertson, RP et al. (2006). International trial of the Edmonton protocol for islet transplantation. N Engl J Med 355: 1318–1330. [DOI] [PubMed] [Google Scholar]
- Kroon, E, Martinson, LA, Kadoya, K, Bang, AG, Kelly, OG, Eliazer, S et al. (2008). Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 26: 443–452. [DOI] [PubMed] [Google Scholar]
- Pagliuca, FW, Millman, JR, Gürtler, M, Segel, M, Van Dervort, A, Ryu, JH et al. (2014). Generation of functional human pancreatic β cells in vitro. Cell 159: 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David, U, Arad, G, Weissbein, U, Mandefro, B, Maimon, A, Golan-Lev, T et al. (2014). Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun 5: 4825. [DOI] [PubMed] [Google Scholar]
- Peterson, SE and Loring, JF (2014). Genomic instability in pluripotent stem cells: implications for clinical applications. J Biol Chem 289: 4578–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen, T, Ostermeier, A, Pang, ZP, Kokubu, Y, Südhof, TC and Wernig, M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K, Nam, YJ, Luo, X, Qi, X, Tan, W, Huang, GN et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya, S and Suzuki, A (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475: 390–393. [DOI] [PubMed] [Google Scholar]
- Takahashi, K, Tanabe, K, Ohnuki, M, Narita, M, Ichisaka, T, Tomoda, K et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Takahashi, K and Yamanaka, S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Akinci, E, Banga, A, Greder, LV, Dutton, JR and Slack, JM (2012). Reprogramming of pancreatic exocrine cells towards a beta (β) cell character using Pdx1, Ngn3 and MafA. Biochem J 442: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, MJ, Docherty, HM, Chen, Y and Docherty, K (2012). Efficient differentiation of AR42J cells towards insulin-producing cells using pancreatic transcription factors in combination with growth factors. Mol Cell Endocrinol 358: 69–80. [DOI] [PubMed] [Google Scholar]
- Banga, A, Akinci, E, Greder, LV, Dutton, JR and Slack, JM (2012). In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc Natl Acad Sci USA 109: 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q, Brown, J, Kanarek, A, Rajagopal, J and Melton, DA (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455: 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga, A, Greder, LV, Dutton, JR and Slack, JM (2014). Stable insulin-secreting ducts formed by reprogramming of cells in the liver using a three-gene cocktail and a PPAR agonist. Gene Ther 21: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo, M, Payen, E, Negre, O, Wang, G, Hehir, K, Fusil, F et al. (2010). Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 467: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina, S, Von Kalle, C, Schmidt, M, McCormack, MP, Wulffraat, N, Leboulch, P et al. (2003). LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302: 415–419. [DOI] [PubMed] [Google Scholar]
- Stephen, SL, Montini, E, Sivanandam, VG, Al-Dhalimy, M, Kestler, HA, Finegold, M et al. (2010). Chromosomal integration of adenoviral vector DNA in vivo. J Virol 84: 9987–9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen, SL, Sivanandam, VG and Kochanek, S (2008). Homologous and heterologous recombination between adenovirus vector DNA and chromosomal DNA. J Gene Med 10: 1176–1189. [DOI] [PubMed] [Google Scholar]
- Yu, J, Hu, K, Smuga-Otto, K, Tian, S, Stewart, R, Slukvin, II et al. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324: 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H, Wu, S, Joo, JY, Zhu, S, Han, DW, Lin, T et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4: 381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, T, Yuasa, S, Oda, M, Egashira, T, Yae, K, Kusumoto, D et al. (2010). Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 7: 11–14. [DOI] [PubMed] [Google Scholar]
- Miyoshi, N, Ishii, H, Nagano, H, Haraguchi, N, Dewi, DL, Kano, Y et al. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8: 633–638. [DOI] [PubMed] [Google Scholar]
- Warren, L, Manos, PD, Ahfeldt, T, Loh, YH, Li, H, Lau, F et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger, TM, Daheron, L, Brickler, TR, Entwisle, S, Chan, K, Cianci, A et al. (2015). A comparison of non-integrating reprogramming methods. Nat Biotechnol 33: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker, DS, Lilja, HS, French, J, Kuhlmann, E and Noll, W (1979). Transplantation of azaserine-induced carcinomas of pancreas in rats. Cancer Lett 7: 197–202. [DOI] [PubMed] [Google Scholar]
- Rosewicz, S, Riecken, EO and Wiedenmann, B (1992). The amphicrine pancreatic cell line AR42J: a model system for combined studies on exocrine and endocrine secretion. Clin Investig 70: 205–209. [DOI] [PubMed] [Google Scholar]
- Zhou, J, Wang, X, Pineyro, MA and Egan, JM (1999). Glucagon-like peptide 1 and exendin-4 convert pancreatic AR42J cells into glucagon- and insulin-producing cells. Diabetes 48: 2358–2366. [DOI] [PubMed] [Google Scholar]
- Bruin, JE, Erener, S, Vela, J, Hu, X, Johnson, JD, Kurata, HT et al. (2014). Characterization of polyhormonal insulin-producing cells derived in vitro from human embryonic stem cells. Stem Cell Res 12: 194–208. [DOI] [PubMed] [Google Scholar]
- Karikó, K and Weissman, D (2007). Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel 10: 523–532. [PubMed] [Google Scholar]
- Barber, GN (2001). Host defense, viruses and apoptosis. Cell Death Differ 8: 113–126. [DOI] [PubMed] [Google Scholar]
- Kormann, MS, Hasenpusch, G, Aneja, MK, Nica, G, Flemmer, AW, Herber-Jonat, S et al. (2011). Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29: 154–157. [DOI] [PubMed] [Google Scholar]
- Nallagatla, SR, Toroney, R and Bevilacqua, PC (2008). A brilliant disguise for self RNA: 5'-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol 5: 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, MJ, Muir, KR, Docherty, HM, Drummond, R, McGowan, NW, Forbes, S et al. (2013). Suppression of epithelial-to-mesenchymal transitioning enhances ex vivo reprogramming of human exocrine pancreatic tissue toward functional insulin-producing β-like cells. Diabetes 62: 2821–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, B, Hrvatin, SS, Schuetz, C, Bonal, C, Rezania, A and Melton, DA (2012). Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol 30: 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W, Nakanishi, M, Zumsteg, A, Shear, M, Wright, C, Melton, DA et al. (2014). In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. Elife 3: e01846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, AE, Taylor, BL, Benthuysen, JR, Liu, J, Thorel, F, Yuan, W et al. (2013). Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet 9: e1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, M, Neubüser, A, Kalamaras, J, Ee, HC, Martin, GR and German, MS (1997). Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 11: 1662–1673. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel, VM, Scheel, DW, Conners, JR, Kalamaras, J, Lee, JE, Anderson, DJ et al. (2000). Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127: 3533–3542. [DOI] [PubMed] [Google Scholar]
- Taylor, BL, Liu, FF and Sander, M (2013). Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep 4: 1262–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmain, Y, Katz, LS, Masson, MH, Cheyssac, C, Poisson, C and Philippe, J (2012). Pax6 is crucial for β-cell function, insulin biosynthesis, and glucose-induced insulin secretion. Mol Endocrinol 26: 696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediger, BN, Du, A, Liu, J, Hunter, CS, Walp, ER, Schug, J et al. (2014). Islet-1 Is essential for pancreatic β-cell function. Diabetes 63: 4206–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, AE, Freude, KK, Nelson, SB and Sander, M (2010). Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell 18: 1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda, HE, Benitez, CM and Kim, SK (2013). Gene regulatory networks governing pancreas development. Dev Cell 25: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, P, May, CL, Lamounier, RN, Brestelli, JE and Kaestner, KH (2008). Defining pancreatic endocrine precursors and their descendants. Diabetes 57: 654–668. [DOI] [PubMed] [Google Scholar]
- Lee, J, Sugiyama, T, Liu, Y, Wang, J, Gu, X, Lei, J et al. (2013). Expansion and conversion of human pancreatic ductal cells into insulin-secreting endocrine cells. Elife 2: e00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T and Dent, SY (2014). Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15: 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arensbergen, J, García-Hurtado, J, Moran, I, Maestro, MA, Xu, X, Van de Casteele, M et al. (2010). Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res 20: 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, R, Everett, LJ, Lim, HW, Patel, NA, Schug, J, Kroon, E et al. (2013). Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell 12: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda, B, Chowdhury, K, Torres, M, Oliver, G and Gruss, P (1997). The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature 386: 399–402. [DOI] [PubMed] [Google Scholar]
- Collombat, P, Mansouri, A, Hecksher-Sorensen, J, Serup, P, Krull, J, Gradwohl, G et al. (2003). Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 17: 2591–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J, Elghazi, L, Parker, SE, Kizilocak, H, Asano, M, Sussel, L et al. (2004). The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell differentiation. Dev Biol 266: 178–189. [DOI] [PubMed] [Google Scholar]
- Thess, A, Grund, S, Mui, BL, Hope, MJ, Baumhof, P, Fotin-Mleczek, M et al. (2015). Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol Ther 23: 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso, FJ, Dabrowski, M, Wadzack, J and Nierhaus, KH (1995). Self-coded 3'-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem 270: 6298–6307. [DOI] [PubMed] [Google Scholar]
- Karikó, K, Muramatsu, H, Ludwig, J and Weissman, D (2011). Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 39: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.