Abstract
The genetic diversity of the influenza virus hinders the use of broad spectrum antiviral drugs and favors the appearance of resistant strains. Single-stranded DNA aptamers represent an innovative approach with potential application as antiviral compounds. The mRNAs of influenza virus possess a 5′cap structure and a 3′poly(A) tail that makes them structurally indistinguishable from cellular mRNAs. However, selective translation of viral mRNAs occurs in infected cells through a discriminatory mechanism, whereby viral polymerase and NS1 interact with components of the translation initiation complex, such as the eIF4GI and PABP1 proteins. We have studied the potential of two specific aptamers that recognize PABP1 (ApPABP7 and ApPABP11) to act as anti-influenza drugs. Both aptamers reduce viral genome expression and the production of infective influenza virus particles. The interaction of viral polymerase with the eIF4GI translation initiation factor is hindered by transfection of infected cells with both PABP1 aptamers, and ApPABP11 also inhibits the association of NS1 with PABP1 and eIF4GI. These results indicate that aptamers targeting the host factors that interact with viral proteins may potentially have a broad therapeutic spectrum, reducing the appearance of escape mutants and resistant subtypes.
Keywords: aptamers, influenza virus replication, polyA-binding protein
Introduction
The absence of polymerase “proofreading” and the inability to repair errors that occur during replication enhances the genetic diversity of influenza A viruses.1,2 In addition, the segmented genome of influenza virus permits the exchange of RNA segments between genotypically different influenza viruses, promoting the generation of novel strains or subtypes.3 These two mechanisms, mutation (antigenic drift) and reassortment (antigenic shift), represent the main mechanisms of evolutionary change in influenza viruses. Three emerging influenza viruses were responsible for major pandemics in the twentieth century: the 1918 Spanish flu virus, the 1957 Asian flu virus, and the 1968 Hong Kong flu virus.4 Indeed, the 1918 Spanish flu virus was estimated to have killed 20–50 million people worldwide.5 More recently, a highly pathogenic avian virus of the H5N1 subtype has produced sporadic infections in humans and while it is associated with high rates of mortality, its poor transmission in humans prevented a more extensive spread among human populations.6 However, in 2009, a new influenza A virus of the H1N1 subtype emerged (pH1N1) that possessed high transmissibility but relatively low virulence, rapidly spreading across the entire globe and causing the first pandemic of the 21st century.7,8 Subsequently, 2013 witnessed the appearance of a new highly pathogenic avian virus of the H7N9 subtype in China.9 Thus, antigenic drift and antigenic shift appear to contribute clearly to the persistence of influenza A virus in humans, exposing human populations to a re-emerging disease. The huge impact that influenza A infections clearly have on public health underline the importance of developing efficient counter-measures and effective, long-lasting and selective drugs to treat and prevent infection by these viruses. Only in this way can the major healthcare challenge represented by the spread of influenza A be overcome.
Influenza virus and cellular mRNAs are synthesized using different pathways, but both are structurally indistinguishable containing a 5' cap structure and a 3' polyA tail.10,11,12,13 However, viral mRNA is selectively translated upon infection, while the initiation and elongation steps of cellular mRNA translation are inhibited,14 contributing to the efficient shut down of host cell protein synthesis. The majority of the host cell's mRNAs are translated in a cap-dependent manner, involving recognition of the 5'-cap structure by the eIF4F initiation factor complex.15 The eIF4F complex is comprised of three proteins: eIF4E, the cellular cap-binding factor; eIF4A, an RNA helicase responsible for the ATP-dependent elimination of secondary structures near the 5′-cap of mRNAs; and eIF4G, which serves as a scaffold for the binding of several factors.
Indeed, in addition to eIF4E and eIF4A, eIF4GI binds the eIF3 factor that allows translational machinery to be recruited to the mRNA associated with the eIF4F complex, and it also binds the poly(A)-binding protein (PABP1) that can be considered as part of the translation initiation complex. Sequence analysis of the gene encoding PABP1 from several organisms reveals four conserved RNA recognition motifs that are responsible for poly(A) binding16,17 and that support the interaction with eIF4GI.18 The PABP1-eIF4G interaction provokes the circularization of the mRNA,18,19 which stimulates the translation of mRNAs containing cap structure and a poly-A tail.20,21
We previously described interactions between several influenza virus proteins and components of the cell's translational complexes. NS1 interacts directly with eIF4GI and PABP1, both in vivo and in vitro.22,23 In addition, we provided evidence that the viral polymerase, an heterotrimer cap-binding complex composed of PA, PB1, and PB2 subunits, interacts with translation initiation complexes, specifically with eIF4GI independently of NS1.24 These interactions play a significant role in the control of influenza virus mRNA translation, which is independent of the cellular eIF4E cap-binding factor24 but fully dependent on eIF4A and eIF4GI.25,26
Aptamers are structured polynucleotide sequences that are isolated from randomized oligonucleotide libraries through the systematic evolution of ligands by exponential enrichment (SELEX), and they selectively bind target molecules with high affinity and specificity.27,28 Aptamers can form stable and specific complexes with their targets, with dissociation constants in the nanomolar range. Due to their chemical and biological characteristics (small size, high stability, lack of immunogenicity, ease of chemical synthesis, adaptable, etc), aptamers have been used as agents for targeted therapeutics and molecular diagnostics. Thus, we assessed the use of DNA aptamers targeting the Leishmania infantum PABP (LiPABP),29 which also efficiently recognize human PABP1, as a new approach to combat influenza virus. These aptamers may interfere with the interactions between influenza virus proteins and components of the cell's translation apparatus, reducing viral multiplication and possibly compromising viral variability. Two aptamers have been studied and they both reduced viral titers, representing potentially novel tools to control influenza virus infection.
Results
Characterization of aptamers recognizing Leishmania and human PABP1 protein
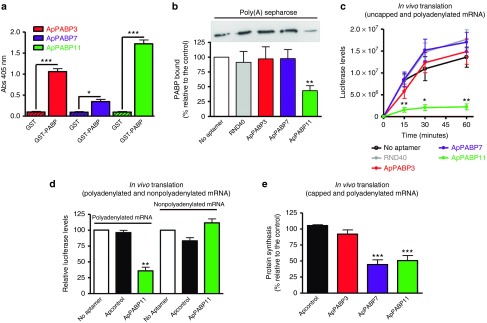
We recently selected and characterized three aptamers that recognize PABP1 from Leishmania infantum (LiPABP) and that bind to it with high affinity.29 This LiPABP contains the main domains present in PABPs from other organisms, including humans,30 conserving the four characteristic RNA recognition motifs responsible for poly(A) binding, as well as the highly conserved RNP1 and RNP2 motifs. The C-terminal KITGMLLE motif of human PABP1 involved in the interaction with regulatory proteins and translation factors is also conserved. Thus, we performed enzyme-linked oligonucleotide assay (ELONA), to study whether these aptamers recognize recombinant human GST-PABP1 protein.23 All three aptamers, ApPABP3, ApPABP7, and ApPABP11 bind to human GST-PABP1 with significantly higher affinity than to the GST protein (Figure 1a). Hence, the potential effect of the three aptamers on the human PABP1–poly(A) interaction was investigated in a competition assay and compared with that of naive random ssDNA oligonucleotides that contained a central randomized region of 40 nucleotides flanked by two conserved 18-nucleotides regions in each end (RND40). Like LiPABP,29 only ApPABP11 reduced the amount of human PABP1 bound to poly(A)-sepharose (62%) while the ApPABP3 and ApPABP7 aptamers, and RND40 had no effect on this interaction (Figure 1b).
Figure 1.
Characterization of the aptamers recognizing the Leishmania and human PABP1 proteins. (a) Recombinant GST-PABP protein or GST were plated at 500 ng/well and incubated with the digoxigenin-labeled aptamers (10 nmol/l). Anti-digoxigenin-POD antibodies were added for 1 hour and revealed with an 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) solution at 405 nm. All the experiments were performed in triplicate and the average of three different experiments is shown: *P < 0.05, ***P < 0.001. (b) Top, a representative western blot showing the amount of endogenous PABP1 from HEK293T cells lysates bound to poly(A)-Sepharose in the presence or absence of RND40 or the PABP aptamers. Bottom, the protein bands were quantified and the values normalized relative to the PABP1 bound in the absence of aptamers. The bars represent the mean ± standard error of the mean (SEM) of four different experiments and the statistical significance is relative to the controls/no aptamer: **P < 0.01. (c) In vitro translation was performed using a Luciferase mRNA uncapped and polyadenylated provided with the Rabbit Reticulocyte Lysate kit (Promega) in the presence or absence (no aptamer) of 4 µmol/l aptamers or RND40. Luciferase activity was measured after different incubation times and the data represent the mean ± SEM of four independent experiments. Statistical significance relative to control/no aptamer: *P < 0.05; **P < 0.0l. (d) Transcription-translation assay using a Luciferase mRNA from a plasmid with or without a poly(A) tail as a template in the presence or absence (no aptamer) of 0.8 µmol/l ApPABP11 or the ApControl. Luciferase levels were measured after a 60-minute incubation. The data are expressed as the percentage of activity relative to the control/no aptamer and represent the mean ± SEM of four different experiments. **P < 0.01. (e) The rate of protein synthesis was measured as described in the Materials and Methods. The results are expressed as the activity relative to the control (no aptamer) and they represent the mean ± SEM of four independent experiments: statistical significance relative to the ApControl. ***P < 0.001.
The effect of PABP1 aptamers on in vitro translation
We assessed the effect of these aptamers on the in vitro translation of different mRNAs, first assaying the translation of uncapped and polyadenylated luciferase mRNA in the presence or absence of each aptamer (4 µmol/l), or RND40 (Figure 1c). While ApPABP3 and ApPABP7 slightly reduced the amount of luciferase produced, ApPABP11 strongly inhibited luciferase translation in a concentration-dependent manner (data not shown). To evaluate the role of the polyA tail in this inhibition, additional experiments were performed using a transcription/translation system and uncapped luciferase mRNAs with or without the poly(A) tail. An unstructured ssDNA aptamer with 38xAG (ApControl) was added as a further control as this aptamer fails to acquire any type of secondary structure. While ApPABP11 strongly inhibited the translation of polyadenylated luciferase mRNA, the translation of nonpolyadenylated luciferase mRNA was not affected by this aptamer (Figure 1d). Hence, the effect of ApPABP11 appears to require the presence of poly(A) at the 3'end of the mRNA.
The effect of PABP1 aptamers on translation in vivo
The effect of aptamers on in vivo translation was also analyzed in HEK293T cells transfected with control or PABP1-specific aptamers (60 nmol/l) for 4 hours, and measuring 3H-Methionine incorporation into proteins after 1 hour (see Materials and Methods). The ApPABP7 and ApPABP11 aptamers both inhibited protein synthesis in these assays (55 and 50% inhibition, respectively), whereas ApPABP3 and ApControl did not affect mRNA translation in vivo (Figure 1e). These results indicate that the ApPABP7 and ApPABP11 aptamers bind to human PABP1 and inhibit the translation of cellular mRNAs in vivo, encouraging us to characterize their potential activity against influenza virus.
Mapping aptamer recognition within human PABP1
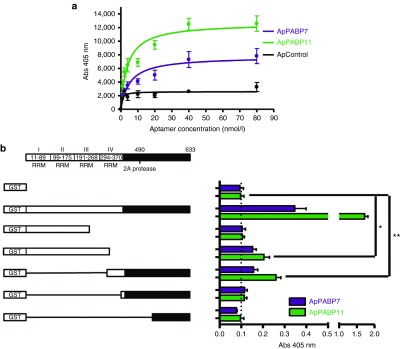
The binding affinities and the region where the aptamers ApPABP7 and ApPABP11 interact with PABP1 were analyzed. The binding capacity was studied by ELONA assays in which the GST-PABP1 protein was incubated with increasing concentrations of digoxigenin-labeled aptamers (0–80 nmol/l). Both aptamers detected the PABP1 protein in a concentration-dependent manner, showing ApPABP11 a higher affinity for PABP1 than ApPABP7 (Figure 2a).
Figure 2.
Affinity of the aptamers for human PABP1 and mapping analysis. (a) Enzyme-linked oligonucleotide assays for hPABP1-aptamer binding. GST-PABP1 was plated at 0.5 µg/well and then incubated with digoxigenin-labeled aptamers at 0–80 nmol/l. The bars represent the mean ± standard error of the mean (SEM) of three independent experiments. (b) Scheme of the PABP1 functional domains and the GST–PABP1 deletion mutants used (left panel). GST or GST-PABP1 and deletion mutants were plated at 2.5 pmol/well, and incubated with Dig-ApPABP7 and Dig-ApPABP11 (10 nmol/l). The bars represent the mean ± SEM of three independent experiments: *P < 0.05; **P < 0.01.
ELONA experiments were also performed to analyze the region of PABP1 recognized by aptamers 7 and 11 using a set of GST–PABP1 deletion mutants as the targets (ref. 23; Figure 2b). A PABP1 mutant lacking the first 307 N-terminal amino acids was recognized by both aptamers, whereas the aptamers did not bind to PABP1 deletion mutants lacking the 365 or 535 N-terminal amino acids. Both aptamers were unable to bind to the PABP1 mutant containing only the first 234 amino acids (GST–PABP1 234), yet they recognized a PABP1 mutant containing the first 319 amino acids (GST–PABP1 319). These results strongly suggest that ApPABP7 and ApPABP11 interact with a domain that resides between amino acids 307 and 364 of PABP1, a region within the RRM4 domain.
The effect of aptamers on viral translation
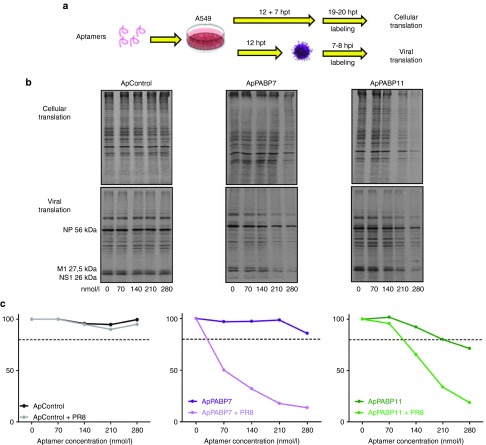
We tested how the aptamers affected the translation of cellular and influenza virus mRNAs in vivo. A549 cells were transfected with increasing amounts of the control aptamer or the specific ApPABP7 and ApPABP11 aptamers, and 12 hours post-transfection (hpt) the cells were infected with the A/PR8/8/34 (PR8) strain of influenza virus at a high multiplicity of infection (MOI) or they were left uninfected. Protein translation was evaluated 7 hours later by metabolically labeling the cells for 1 hour with 35S-Methionine and then quantifying its incorporation using a phosphorimager (a scheme of the experimental conditions is depicted in Figure 3a). As expected, the control aptamer did not inhibit cellular mRNA translation, whereas aptamers 7 and 11 reduced mRNA translation in the cells by 20% at 280 and 210 nmol/l, respectively (Figure 3b,c). This reduction was weaker than that observed in HEK293T cells 4 hours after aptamer transfection (Figure 1e), which may reflect different transfection efficiencies, differences in cell susceptibility to the aptamers, intracellular degradation of aptamers at longer times post-transfection (see below) or a combination of these factors. While the control aptamer did not inhibit the translation of viral mRNAs, even at the highest concentration (280 nmol/l), an important dose-dependent decrease was provoked by the specific aptamers with aptamer 7 being particularly effective. These results indicate that aptamers targeting PABP1 can impair the translation of cellular mRNAs but that they are much more effective in inhibiting the translation of influenza virus mRNAs.
Figure 3.
Effect of the PABP1 aptamers on cellular and viral mRNA translation. (a) A549 cells were transfected with the corresponding aptamers at increasing concentrations and 12 hpt, the aptamers were washed and the cells were left uninfected or they were infected with A/PR8/8/34 strain at 2 plaque forming units/cell. At 7 hours after mock-infection or infection with influenza virus, the cells were metabolically labeled with 35S-Met for 1 hour and the cell extracts were obtained. (b,c) Cell extracts of A549 cells transfected with control (ApControl) or specific aptamers (ApPABP7 and ApPABP11), uninfected or infected from part (a), were resolved in sodium dodecyl sulfate-denaturing gels and the label quantified in a phosphorimager. The experiment was performed twice and a representative image is shown. Quantificatin of nucleoprotein protein was performed to analyze viral protein synthesis. Entire lines were quantified in the noninfected cells.
Effect of aptamers on cellular metabolism
To examine the effect of PABP1 aptamers on influenza virus infection, we first evaluated the potential toxicity of aptamers at different concentrations in uninfected cells. A549 cells were transfected with the aptamers and their toxicity was determined by evaluating the metabolic activity of the cell biomass using the (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell viability assay at 18 hours post-transfection (hpt).31 As can be seen, the highest toxicity was obtained with 280 nmol/l of each aptamer although they did not reach the CC50's doses (Supplementary Figure S1a). To further evaluate aptamer toxicity, we performed metabolic labeling in A549 cells transfected with increasing concentrations of each aptamer. Eighteen hours postransfection, the cells were metabolically labeled for 1 hour with 35S-Methionine and label incorporated into proteins were then quantitated using a phosphorimager. As can be seen, an important inhibition of cellular protein synthesis was obtained with 380 nmol/l of ApPABP7 and ApPABP11 aptamers compared with the control aptamer (Supplementary Figure S1b).
The effect of aptamers on viral infection
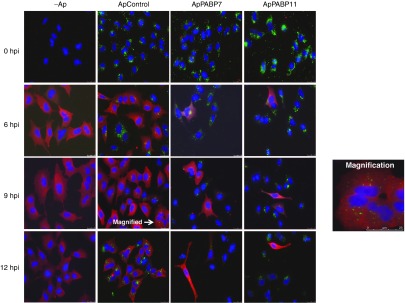
Since specific PABP1 aptamers efficiently inhibit viral mRNA translation, we examined their effect on viral infection. To that aim, we used a concentration of aptamers of 140 nmol/l, a concentration at which the PABP1 aptamers only poorly affect cellular translation but at which they inhibit viral translation (Figure 3). We evaluated the consequence of aptamer transfection on the expression and distribution of influenza virus proteins at high MOI. A549 cells were transfected with 140 nmol/l of the alexa-fluor 488 aptamers, and then the cells were infected at 12 hpt with the A/Victoria/3/75 (VIC) strain of influenza virus at 2 plaque forming units (PFU)/cell and the distribution of viral proteins was analyzed by immunofluorescence at different hpi. ApPABP7 and ApPABP11 transfection was associated with an important reduction in the number of influenza-infected cells relative to the untransfected cells or those transfected with the control aptamer (Figure 4).
Figure 4.
The effect of the PABP1 aptamers on influenza virus protein expression and distribution. A549 cells were transfected with 140 nmol/l of the indicated Alexa-488-labeled aptamers and at 12 hpt, the aptamers were washed out and the cells were infected with A/Victoria/3/75 strain at 2 plaque forming units/cell. At different hpi, immunofluorescence assays were performed using antibodies against HA protein: Green, Aptamers-Alexa-488; Red, HA protein. The experiment was performed twice and a representative image is shown. To show the scale bars present in all the images, magnification of a part of cells infected and treated with the ApControl at 9 hpi is shown in the magnification panel.
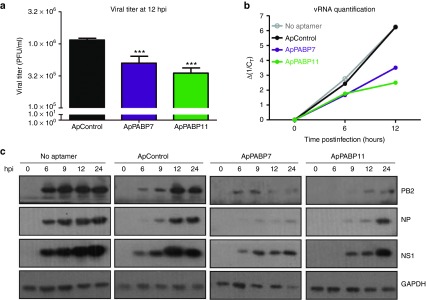
Other parameters related to the effect of PABP1 aptamers on influenza virus infection at high MOI were also analyzed. For that, the cells were transfected with the corresponding aptamers and at 12 hpt, they were infected with the PR8 strain and the viral titers analyzed at 12 hpi. The titers obtained in the infected cells transfected with the ApPABP7 and ApPABP11 aptamers were around three to four times lower than in untransfected cells or cells that were transfected with the control aptamer (Figure 5a). Accordingly, the amount of genomic viral RNA (Figure 5b) and the accumulation of viral proteins (Figure 5c) were reduced concomitantly in cells transfected with the aptamers targeting PABP1. Some reduction in glyceraldehyde 3-phosphate dehydrogenase accumulation in ApPABP7 and ApPABP11 transfected cells was also found. Together these results indicate that aptamers specifically targeting PABP1 produce an important impairment of viral multiplication and influenza virus RNA replication.
Figure 5.
The effect of the PABP1 aptamers on influenza virus replication in single step curves. A549 cells were transfected with the corresponding aptamers at 140 nmol/l (controls; (no aptamer), were mock transfected) and at 12 hpt, the aptamers were removed and the cells were infected with A/PR8/8/34 strain at 2 plaque forming units/cell. (a) At 12 hpi the viral titer was analyzed by the plaque assay method. (b) 12 hpt A549 cells were infected and at different hpi, total RNA of the samples were obtained and used to quantify the genomic viral RNA of nucleoprotein (NP) segment by quantitative real time-PCR (qRT-PCR). Ordinate axis represents the inverse of Ct increase of NP vRNA detected by qRT-PCR. The value obtained in the uninfected cells is taken as 0 time postinfection. (c) At 12 hpt and the specified hpi, the proteins indicated in the total cell extracts were detected in western blots. The bars represent the mean ± standard error of the mean of two different experiments performed in triplicate and the statistical significance is relative to the ApControl. ***P < 0.001.
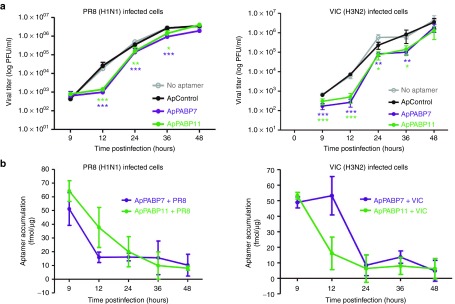
Since natural influenza virus infection occurs at a low MOI, we want to evaluate the effect of the aptamers under these conditions. It should be taken into account that several cycles of cell division are required for these kinetic experiments and the new born cells do not receive the corresponding aptamers since they do not replicate and only the parental cells receive the compounds. A549 cells were transfected with control or specific PABP1 aptamers and at 4 hpt the cells were infected with influenza virus at 10−2 PFU/cell and viral titers were determined at different hpi. Two influenza virus strains of H1N1 (PR8) and H3N2 (VIC) subtypes were used, since natural infections in humans are mainly due to viruses of these subtypes. As can be seen reduction of 1–1.5 logs at early times of infection was observed in cells transfected with ApPABP7 and ApPABP11 compared with untransfected or control-transfected cells infected with the two viruses (Figure 6a). No major differences on viral titers were found at 48 hpi, both in VIC and PR8-infected cells. Our previous observation of the effect of specific PABP1 aptamers on influenza virus multiplication at high moi, (Figures 4 and 5), suggested a higher reduction of virus multiplication in multiple steps kinetics. A reduction of 50–90% of PABP1 aptamers intracellular accumulation (fmol of aptamers/μg of protein) was observed at 24 hpi, in agreement with only a moderate reduction of viral titer at this hpi compared with that obtained at 12 hpi when intracellular accumulation of aptamers is high (Figure 6b). On the other hand, it should be emphasized that influenza virus infection does not trigger aptamer degradation, since similar results are obtained in uninfected cells (Supplementary Figure S2). Together, the lack of replication of aptamers and their intracellular decreased concentration seem to prevent higher reduction of influenza virus particles production in multiple step kinetics.
Figure 6.
Effect of PABP1 aptamers on influenza virus replication in multiple steps curves. (a) A549 cells were untransfected or transfected with the corresponding aptamers at 140 nmol/l. At 4 hpt, aptamers were removed by washing and cells were infected with influenza virus A/PR8/8/34 (left) or A/Victoria/3/75 (right) strains, at 10−2 plaque forming units/cell and viral titers were determined at different hpi by plaque assays. (b) Aliquots of the samples described in part (a) were obtained and used for intracellular aptamer accumulation detected by polymerase chain reaction analysis. The bars represent the mean ± SEM of three different experiments performed in triplicate and the statistical significance is relative to the ApControl. *P < 0.05, **P < 0.01, ***P < 0.001.
The effects of PABP1 aptamers on the association of viral proteins with translation initiation complexes
Influenza virus efficiently shuts off host cell protein synthesis32 and upon infection the cells translation machinery is fundamentally recruited to the selective translation of viral mRNAs.32,33 A network of viral-host interactions seems to drive this shift, such as the association of viral polymerase complex with translation initiation complexes through the interaction of the PB2 subunit of the viral polymerase with eIF4GI.24,34 Other interactions involve the NS1 protein, which associates with translation initiation factors like PABP123 and eIF4GI.22 Together, these interactions permit selective translation of viral mRNAs in the infected cells, while highlighting the dependence of efficient viral mRNA translation on cellular translation initiation complexes.
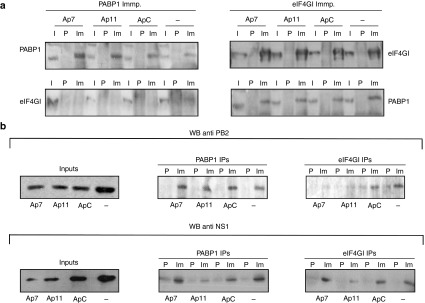
Since PABP1 aptamers efficiently inhibit influenza virus translation and viral multiplication, we assessed whether the aptamers disturb the association of viral proteins with translation initiation complexes. A549 cells transfected with control or specific PABP1 aptamers were infected with the PR8 viral strain at a high MOI and 8 hpi, antibodies against PABP1 and eIF4GI were used to immunoprecipitate protein complexes. None of the aptamers affected the immunoprecipitation of PABP1, and all the complexes recovered with an antibody against eIF4GI from untransfected cells and cells transfected with control or PABP1 aptamers contained eIF4GI as well as PABP1. By contrast, eIF4GI was almost absent from the immunocomplexes recovered by the anti-PABP1 antibody (Figure 7a, left). These results indicate that the aptamers do not interfere with the interaction of eIF4GI with PABP1, and suggest that there is a population of PABP1 that is not associated with eIF4GI since this protein is poorly coimmunoprecipitated by antibodies against PABP1.
Figure 7.
The effect of aptamers on the association of viral protein with translation initiation complexes. A549 cells were transfected with the corresponding aptamers (140 nmol/l) and at 12 hpt, the aptamers were washed out and the cells were infected with A/PR8/8/34 strain at 2 plaque forming units/cell. Cell extracts were obtained at 7 hpi and PABP1 or eIF4GI was immunoprecipitated, and the complexes recovered were analyzed by western blots. (a) anti-PABP1 (left) and anti-eIF4GI (right) immunoprecipitation. (b) PB2 and NS1 immunoprecipitated with the PABP1 (PABP1 IPs) or eIF4GI (eIF4GI IPs) antibody: -, noninfected cells; I, input; P, preimmune serum; Im, immune serum. The experiment was performed three times and a representative image is shown.
Finally, we examined how PABP1 aptamers affected the association of influenza virus PB2 polymerase subunit and NS1 with the translation initiation complexes (Figure 7b). None of the aptamers altered the association of PB2 with PABP1 (PABP1 IPs), whereas the two PABP1-specific aptamers impaired its association with eIF4GI (eIF4GI IPs). In terms of NS1, the PABP11 aptamer dampened its association with both PABP1 and eIF4GI, whereas the PABP7 aptamer did not disturb these interactions. In summary, ApPABP11 disturbs the association of the viral PB2 and NS1 proteins with translation initiation complexes, whereas ApPABP7 impedes the association of the PB2 polymerase subunit with these complexes.
Discussion
The genetic diversity of influenza viruses and their capacity for reassortment are crucial features related to their persistence in humans. The search by private companies and public health services for agents to combat influenza continues, yet the annual hospitalizations or deaths caused by these viruses remain high. Hence, there is still an urgent need for innovative approaches to recognize and prevent this disease in its early stages, especially in high-risk groups.
Although vaccination is the most powerful means of mitigating the effects of influenza epidemics, antiviral drugs can also be very useful, particularly in delaying the spread of new pandemic viruses. Neuraminidase inhibitors like oseltamivir, laninamivir, zanamivir, and peramivir are commonly used as antiviral agents to treat influenza infection, especially in Japan.35 Although each of these antiviral agents are sialic acid analogues, they have subtle differences in chemical structure and binding properties, and consequently, the patterns of resistance to them vary. Viruses resistant to each neuraminidase inhibitor have been described, and those resistant to oseltamivir and peramivir are the most abundant.35 Rapid increases in drug-resistant influenza virus isolates provide compelling reasons for the development of novel antiviral drugs. As such, nucleic acid-based drugs represent a promising class of novel antiviral agents and in this category, antisense oligonucleotides, small interfering RNAs (siRNA), nanoRNAs and aptamers represent sequence specific gene-silencing approaches that could be deployed to suppress or inhibit viral protein gene expression.36
Aptamers against influenza virus
Several aptamers against influenza virus have been described, mainly targeting hemagglutinin,37,38,39 and an aptamer was capable of mediating a reduction in viral pathogenicity in mice models.39 Other aptamers targeting NS140 or the PA polymerase subunit41 have also been studied, although aptamers directed against cellular factors that establish essential interactions with influenza virus proteins have not yet been reported. A limited number of aptamers that target the host cell factors that control viral activity have been described. Of these, the use of RIG-I as a target for aptamers to control viral infection should be emphasized.42 RIG-I is a cytosolic receptor for non-self RNA that mediates immune responses against viral infections through IFNα/β production.43 The use of a specific RIG-I aptamer that activates RIG-I, efficiently blocks the replication of the Newcastle disease virus, vesicular stomatitis virus and influenza virus in infected cells,42 evidencing that aptamers targeting cellular factors can act as efficient antiviral agents.
Mechanism of action of PABP1 aptamers in controlling influenza virus infection
Given that cellular translation complexes are required for the translation of influenza virus mRNAs, we explored whether aptamers targeting a component of translational complexes, the PABP1 protein, might serve as a possible antiviral agent against influenza virus. PABP1 plays a key role in mRNA translation as it interacts with eIF4GI, circularizing mRNAs and facilitating the initiation of polyadenylated mRNA translation.18,44,45,46
Mapping studies have shown that the domains within PABP1 that drive its interaction with the eIF4GI and viral NS1 protein reside at positions 1–175 and 365–535, respectively. The specific aptamers used in this study interact with PABP1 between amino acids 307 and 364 of PABP1, a region in the fourth RRM domain close to the NS1 interacting domain. Accordingly, ApPABP11 inhibits the NS1-PABP1 interaction and that of PABP1 with poly(A). Since ApPABP7 does not inhibit these interactions, the specific interaction regions for these two aptamers are distinct, despite their proximity. It is interesting to point out that the two aptamers, mainly ApPABP11, show a lower binding to the PABP1 mutants than to the entire protein. These results clearly indicate that the presence of all the domains is necessary to preserve the correct three-dimensional structure of PABP, which is necessary for the aptamer binding. Further experiments will be necessary to better define these domains.
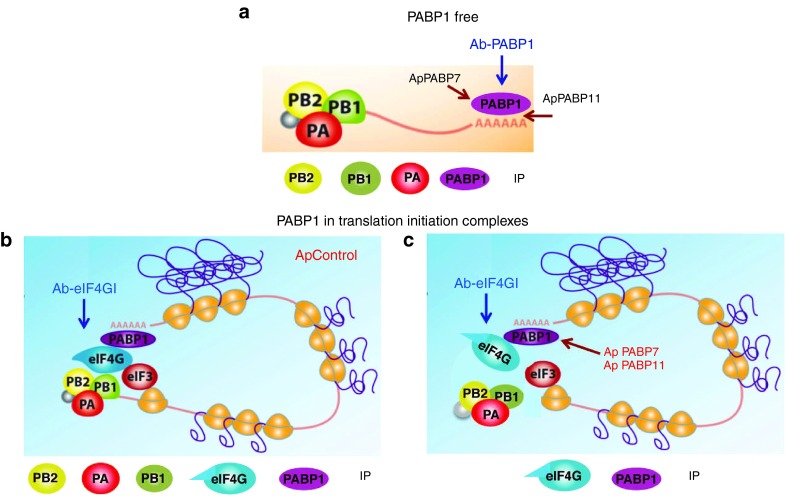
The influenza virus polymerase is a cap-binding complex47 that associates with the translation initiation complex through a direct PB2-eIF4GI interaction. This interaction is consistent with the observed independence of viral mRNA translation for the cellular cap-binding eIF4E, suggesting that the viral polymerase complex that binds to the conserved 5′UTR sequences of viral mRNAs may function as a specific cap-binding factor to drive the translation of viral mRNAs.24,26 Antibodies against PABP1 immunoprecipitate PB2 from cells transfected with the different aptamers, in immunocomplexes virtually free of eIF4GI, although there is slightly less PB2 coimmunoprecipitated from cells transfected with ApPABP11. Hence, it would appear that PB2 would coimmunoprecipitate with PABP1 as a component of the viral polymerase associated with the polyadenylated viral mRNAs. Thus, PABP1 and PB2 would exist in the same complexes, PABP1 at the poly(A) tail and PB2 at the 5′UTR of viral mRNAs, further indicating that viral polymerase accompanies viral cytosolic mRNAs to drive their selective translation (Figure 8a). The mild loss of PB2 in the presence of ApPABP11 is consistent with it inhibiting the PABP1-poly (A) interaction. This aptamer inhibits the NS1-PABP1 interaction, which may be the result of directly impeding the association between NS1 and PABP1 or the association of NS1 with viral mRNA.
Figure 8.
Model of PABP1 aptamers affecting the association of influenza virus proteins with PABP1 and eIF4GI. PABP1would exists in two forms; free (a) or associated in translation initiation complexes (b,c). On the free form (a), PABP1 antibody would coimmunoprecipitate influenza virus polymerase subunits and PABP1; the viral polymerase bound to the cap-5′UTR of viral mRNAs and PABP1 to the 3′polyA-tail of these mRNAs. (b) In the population of PABP1 associated in translation complexes, eIF4GI antibody would coimmunoprecipitate the polymerase subunits, PABP1 and eIF4GI in cells transfected with ApControl. (c) In the population of PABP1 associated in translation complexes, the cells transfected with ApPABP7 and ApPABP11 would disturb eIF4GI-containing complexes abolishing eIF4GI-viral polymerase association. The eIF4GI antibody would coimmunoprecipitate only PABP1 and eIF4GI proteins. IP, immunoprecipitated proteins.
There is less PB2 in the immunocomplexes recovered with antibodies against eIF4GI from cells transfected with ApPABP7 and ApPABP11 than in those from control or untransfected cells (Figure 7b). These immunocomplexes contain PABP1 and probably represent translation initiation complexes. In addition, ApPABP11 also inhibits the NS1-eIF4GI interaction (Figure 7b). The inhibition of the eIF4GI-PB2 interaction by ApPABP7 and ApPABP11 would be mediated by disturbing the complexes containing eIF4GI-PABP1 since these aptamers do not inhibit eIF4GI-PABP1 association. Figure 8 presents a model of PABP1 free (Figure 8a) or associated in translation complexes, and the effect of the control (Figure 8b) or PABP1 aptamers (Figure 8c) in the immunoprecipitation of proteins (IP) using PABP1 or eIF4GI antibodies. Aptamers 7 and 11 target PABP1, and they dampen the production of infective influenza virus particles similarly. Since ApPABP7 affects the PB2-eIF4GI association without altering that between NS1 and eIF4GI, it seems that the interaction of the viral polymerase with eIF4GI plays a major role in controlling influenza virus mRNA translation.
As ApPABP7 efficiently impairs viral mRNA translation while only weakly inhibiting cellular translation, it represents a promising tool to be tested as an anti-flu drug in animal models. In order to use these compounds in vivo, it is necessary to evaluate their selective inhibition (SI) of viral translation against cellular translation, as well as other parameters. To obtain the SI value of the aptamers, we evaluated the dose causing a 50% inhibition of both activities, CTI50 (cellular translation inhibition) and VTI50 (viral translation inhibition): the concentration of aptamers causing a 50% inhibition of cellular translation or viral translation, respectively (Supplementary Figure S3). The SI ratio obtained for aptamer 7 (SI = 7.5) indicates that it could reduce influenza virus yield at nontoxic concentrations and that it could therefore be potentially used in animal models. Additional parameters that should be considered to be able to use the aptamers in vivo include their stability, the periodicity of administration required to achieve adequate concentrations in the lungs and the aptamer delivery.
Intracellular targets are difficult to address in living cells and the problem becomes even harder for the delivery of DNA/RNA molecules in vivo. Although some aptamers can enter the cell in a receptor-mediated way, normally they are transported through some delievery system. Liposomes are also emerging as one of the versatile classes of reagents for the delivery of aptamer-based therapeutics.48 During the last years, aptamer-nanoparticle conjugation forms the basis of a new chemical and biological strategy with wide application starting from assembly to detection. Because of their small size, nanoparticles can interact readily with biomolecules both on surface and within the cells and thus they are considered as a revolutionary approach as a nanodelivery system for therapy of various diseases.49
The search for antiviral compounds against influenza virus is a very active field given that influenza virus constitutes a continuous health challenge for humans. Like other approaches to fight viruses, such as HIV or Hepatitis C, the combined action of several drugs clearly helps to efficiently counteract these pathogens. Aptamers now enlarge the list of the available anti-influenza drugs that can potentially help reduce the spread of this virus, which is particularly important in the case of new emerging viruses.
Materials and methods
Biological materials. DNA aptamers and their derivatives were purchased from IBA GmbH (Göttingen, Germany). Digoxigenin or alexa-fluor 488 labeling is produced during the chemical synthesis by standard procedures. The cell lines used in this study were MDCK (canine), or A549 and HEK293T (human) cells. The influenza virus strains A/Victoria/3/75 (H3N2) and A/PR/8/34 (H1N1) were propagated and titrated in MDCK cells.
Expression and purification of recombinant proteins. GST–PABP1 and its mutant derivatives were expressed in E. coli Rosetta cells harboring pGEX-2T-PABP1, pGEX-2T-PABP1 Δ1–307, pGEX-2T-PABP1 319, pGEX-2T-PABP1 Δ1–365, pGEX-2T-PABP1 234, and pGEX-2T-PABP1 Δ1–535 plasmids.15 The proteins were purified with glutathione–Sepharose (GenScript, www.antikoerper-online.de) according to the manufacturer's instructions. Briefly, recombinant protein expression was induced with 1 mmol/l IPTG for 2 hours at 37 °C and the cells were then sonicated in buffer containing 5 mmol/l sodium phosphate (pH 7.4), 150 mmol/l NaCl, 1 mmol/l ethylenediaminetetraacetic acid, 0.1% mercaptoetanol, 0.5% Triton X-100, protease inhibitor cocktail (Roche, Basel, Switzerland). After removal of the cell debris by centrifugation, the supernatant was incubated for 2 hours at 4 °C with glutathione–Sepharose equilibrated in the same buffer with gentle rocking. After extensive washes with the same buffer, the proteins were eluted with 10 mmol/l glutathione in 50 mmol/l Tris-HCl (pH 8.0).
ELONA. ELONA was used to analyze the affinity of the aptamers for their targets, as described previously.50 In order to determine the binding capacity of the individual aptamers for human PABP1, 0.5 µg/well of the purified recombinant protein (GST-PABP1) was diluted in coating solution (KPL) and incubated overnight at 4 °C in a 96-well microtiter plate (NUNC). After washing four times in selection buffer (20 mmol/l Tris-HCl (pH 7.4), 1 mmol/l MgCl2, 150 mmol/l NaCl, 5 mmol/l KCl), the wells were blocked with phosphate-buffered saline (PBS)/bovine serum albumin 5% buffer for 1 hour at room temperature. Subsequently, digoxigenin-labeled aptamers were diluted in selection buffer at the concentrations indicated, denatured for 10 minutes at 95 °C and then cooled for 10 minutes on ice. The aptamer solution (100 µl) was then added to each well and the plate was incubated for 1 hour at 37 °C. After washing four times with selection buffer to remove unbound ssDNA, horseradish peroxidase (POD) conjugated anti-digoxigenin antibody (diluted 1:1,000, Roche) was added to each well and maintained for 1 hour at room temperature on a shaking platform. The plates were then washed four times and antibody binding was visualized with an 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) solution (Boehringer–Mannheim, Mannheim, Germany) according to the manufacturer's instruction. Optical density values at 405 nm were determined on a microplate reader from TECAN (Madrid, Spain).
For mapping studies, 2.5 pmoles of GST-PABP1 or the different mutant derivatives were plated overnight at 4 °C, incubated with 100 µl of the digoxigenin-labeled ApPABP7 or ApPABP11 aptamers diluted in selection buffer (10 nmol/l), and ELONA was performed as indicated above.
Poly A-sepharose binding assay. To test the ability of the individual aptamers to specifically block the interaction between human PABP1 and polyA, binding experiments were carried out with polyA-sepharose microparticles as described elsewhere.21 Lysates of HEK293T cells (25 μg) in 200 μl of binding buffer (20 mmol/l Tris-HCl (pH 7.4), 1 mmol/l dithiothreitol, 1 mmol/l MgCl2, 5 mmol/l ethylenediaminetetraacetic acid, and 1 mmol/l phenylmethylsulfonyl fluoride), were incubated with 50 μl of polyA-Sepharose (50%) for 1 hour at 4 °C in the presence or absence of 400 pmoles of the selected aptamers or the naive RND40. The beads were washed six times with binding buffer and the proteins eluted with loading/sample buffer were analyzed in western blots probed with an anti-human PABP1 antibody (Cell Signaling, Leiden, Holland). The blots were quantified on an analyzer equipped with the ImagequantTL (GE Healthcare, Madrid, Spain) software package.
In vitro translation. In vitro translation assays were performed with the Rabbit Reticulocyte Lysate System, Nuclease Treated (Promega, Madrid, Spain). The aptamers were maintained at 95 °C for 10 minutes in selection buffer and cooled on ice before adding them to the reaction (25 µl) at the concentrations indicated in the legends of the figure, which was performed with 10 μg/ml of luciferase RNA control (provided in the kit) according to the manufacturer's instructions. The reaction was stopped by adding emetine after different incubation times and the luciferase activity in a 2 µl aliquot was measured in a luminometer (Berthold, Zug, Switzerland) with luciferase assay reagent (Promega).
To test the role of the polyA-tail on the effect of the aptamers we used luciferase expressing plasmids (pTNT-LUC-cp) that generate a nonpolyadenylated mRNA once linearized with KpnI or the Luciferase control plasmid (provided by the kit). These plasmids were transcribed and translated using the TNTQuick Coupled Transcription/Translation System. The reaction (25 µl) was performed according to the manufacturer's instructions, using 0.5 µg of plasmid and the aptamers (ApPABP11 or ApControl) at a final concentration of 0.8 µmol/l. The reaction was stopped after 90 minutes and the luciferase activity measured as above.
Virus infection. Cells were transfected with 140 nmol/l of the different aptamers using jetPRIME (Polyplus-transfection)). According to the manufacturer's recommendations, 2 μl of jetPRIME per μg of aptamer was added. Then transfected cells were infected at the MOI indicated. After 1 hour of virus adsorption, the cells were washed with PBS and overlaid with growth medium (Dulbecco's Modified Eagle Medium), and at the times indicated postinfection the supernatants were obtained and used to determine the viral titers in plaque assays.
Immunofluorescence. Cultured A549 cells were transfected with 140 nmol/l of the different aptamers and after 12 hours, they were infected with the VIC strain at a MOI of 2 PFU per cell. At the hpi indicated, the cells were fixed in 3.7% formalin for 20 minutes at room temperature and stored in PBS. For immunofluorescence studies, the cells were permeabilized for 5 minutes in PBS containing 0.5% Triton X-100 and incubated with a mouse monoclonal antibody against HA diluted in PBS/0.1% bovine serum albumin (w/v).
Coimmunoprecipitation. A549 cells were transfected with 140 nmol/l of each aptamer and 12 hpt, the cells were infected with the PR8 strain at 2 PFU/cell. At 7 hpi, the cells were collected and lysed in buffer A (150 mmol/l NaCl, 1.5 mmol/l MgCl2, 10 mmol/l Tris/HCl (pH 8.5), 0.2% Igepal) containing protease (“Complete”) inhibitors. The lysates were centrifuged at 10,000 × g and immunocomplexes were immunoprecipitated from the supernatants with antibodies against PABP1 or eIF4GI.22,24 The immunocomplexes were washed eight times with buffer A and analyzed in western blots.
Western blotting. Western blotting was performed as described previously51 and the membranes were probed with the following primary antibodies: for translation initiation factor eIF4GI a mixture of two rabbit polyclonal antibodies was used (each diluted 1:2,000; ref. 22); a rabbit antiserum raised against GST-PABP1 fusion protein (diluted 1/1,000; ref. 23); monoclonal antibodies 8 and 28 against PB2 (each diluted 1:100, ref. 51 #1183; and a rat polyclonal antiserum against His-NS1 (diluted 1:400; ref. 52).
Metabolic labeling. In vivo effect of aptamers in HEK293T mRNA translation was performed by transfecting 60 nmol/l aptamers using the jetPRIME transfection reagent. Three hours after transfection, 8 μCi/well of 3H-Methionine was added and the incorporation of methionine into protein over 1 hour was determined by precipitation with trichloroacetic acid, as described previously.53 The results are calculated as the cpm of 3H-Methionine incorporated into the protein and expressed relative to the control.
To evaluate the effect of aptamers in cellular and influenza virus mRNA translation in parallel experiments, cultures of A549 cells transfected with 140 nmol/l aptamers were mock-infected or infected with PR8 at 2 PFU/cell at 12 hpt. At 6 hpi, the cells were starved for 1 hour in methionine and cysteine-free Dulbecco's Modified Eagle Medium and labeled for 1 hour with a mixture of 35S-Met/Cys (Promix, Amersham, Henley-on-Thames, United Kingdom) in 100 μl of the same medium. Finally, the cells were washed with PBS buffer and analyzed by polyacrylamide gel electrophoresis and autoradiography.
RNA analysis. For RNA extraction, cell pellets were resuspended in 1 ml of TRIZOL reagent (Invitrogen, Carlsbad, CA)/p35 well and the RNA was treated with Turbo DNA-free kit (2U/µl: Ambion, Foster City, CA), according to the manufacturer's instructions. Specific real-time RT-PCR to detect influenza virus genomic RNA for nucleoprotein segment was performed essentially as described previously.54 This method is based on hot-start reverse transcription using tagged primers to add a “tag” sequence to the 5′-end. Real-time PCR was performed in 96-well PCR plates using a SYBR green PCR master mix (Applied Biosystems, Foster City, CA), and a tagged forward primer and a segment-specific reverse primer to ensure the specific quantification of mRNAs and vRNAs. The reactions were followed in a PRISM 7500 Sequence detection system (Applied Biosystems) and the cycle threshold (Ct) was determined with analytical software (sodium dodecyl sulfate: Applied Biosystems), using serial dilutions of cDNA to ensure amplification.
Aptamer quantification. For aptamer quantification, cells were pelleted at the hpt indicated, washed twice with PBS, lysed in 30 μl of H20 by homogenization, vortexed and boiled at 90 °C for 10 minutes. Afterwards, the lysates were centrifuged at 12,000 g for 10 minutes and the intracellular aptamers in the supernatants quantified by quantitative PCR. The pellet was resuspended in 30 μl of NaOH 0.1 M and the protein concentration analyzed by the BCA Kit (Pierce). Quantification was performed with the QUANTIMIX ASY kit (Biotools, Madrid, Spain) in an iQ5 apparatus, and with the F3 and R3 oligonucleotides according to the manufacturer's instructions (Bio-Rad, Barcelona, Spain). The reaction mixture consisted of a 1× master mix, 0.2 µmol/l oligonucleotide and 1 µl of template (previously diluted 1/50) in a 20 µl/tube final volume. The aptamers were quantified using a standard curve for each aptamer (100 fmol–10 fmol).
SUPPLEMENTARY MATERIAL Figure S1. PABP1 aptamer toxicity. Figure S2. Accumulation of the PABP1 aptamers in uninfected cells. Figure S3. Selective inhibition ratio of PABP1 aptamers.
Acknowledgments
We are indebted to J. Ortin and P. Gastaminza for their criticism of the manuscript. The technical assistance of N. Zamarreño and M. García-Hernández are also grateful acknowledged. P. Rodriguez was a fellow of the Ministry of Science and Competitiveness. M.E. Martin and V.M. Gonzalez are researchers from FIBio-HRC. This work was supported by the Spanish Ministry of Economy and Competitiveness, Plan Nacional de Investigacion Científica, Desarrollo e Innovacion Tecnologica (BFU2011-26175 and BFU2014-57797-R) and the Ciber de Enfermedades Infecciosas (A. Nieto), and by grant RTC-2014-1986-1 from the Plan Estatal de Investigación Científica y Técnica y de Innovación 2013–2016 (V.M. Gonzalez).
Supplementary Material
References
- Parvin, JD, Moscona, A, Pan, WT, Leider, JM and Palese, P (1986). Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol 59: 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama, A, Mizumoto, K, Kawakami, K, Kato, A and Honda, A (1986). Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J Biol Chem 261: 10417–10421. [PubMed] [Google Scholar]
- Horimoto, T and Kawaoka, Y (2005). Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 3: 591–600. [DOI] [PubMed] [Google Scholar]
- Cox, NJ and Subbarao, K (2000). Global epidemiology of influenza: past and present. Annu Rev Med 51: 407–421. [DOI] [PubMed] [Google Scholar]
- Taubenberger, JK, Baltimore, D, Doherty, PC, Markel, H, Morens, DM, Webster, RG et al. (2012). Reconstruction of the 1918 influenza virus: unexpected rewards from the past. MBio 3:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta, M, Gao, P, Halfmann, P and Kawaoka, Y (2001). Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293: 1840–1842. [DOI] [PubMed] [Google Scholar]
- Neumann, G, Noda, T and Kawaoka, Y (2009). Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459: 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman, K and Sasisekharan, R (2015). Influenza surveillance: 2014-2015 H1N1 “swine”-derived influenza viruses from India. Cell Host Microbe 17: 279–282. [DOI] [PubMed] [Google Scholar]
- Watanabe, T, Watanabe, S, Maher, EA, Neumann, G and Kawaoka, Y (2014). Pandemic potential of avian influenza A (H7N9) viruses. Trends Microbiol 22: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, LL, Pritlove, DC, Sharps, J and Brownlee, GG (1998). The RNA polymerase of influenza virus, bound to the 5' end of virion RNA, acts in cis to polyadenylate mRNA. J Virol 72: 8214–8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritlove, DC, Poon, LL, Devenish, LJ, Leahy, MB and Brownlee, GG (1999). A hairpin loop at the 5' end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol 73: 2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, LL, Pritlove, DC, Fodor, E and Brownlee, GG (1999). Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol 73: 3473–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, GX, Luytjes, W, Enami, M and Palese, P (1991). The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol 65: 2861–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze, MG, DeCorato, D and Krug, RM (1986). Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus. J Virol 60: 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, AC, Raught, B and Sonenberg, N (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Burd, CG, Matunis, EL and Dreyfuss, G (1991). The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol 11: 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, U and Pieler, T (1996). Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol 256: 20–30. [DOI] [PubMed] [Google Scholar]
- Imataka, H, Gradi, A and Sonenberg, N (1998). A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17: 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, SZ Jr and Sachs, AB (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Gallie, DR and Tanguay, R (1994). Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem 269: 17166–17173. [PubMed] [Google Scholar]
- Michel, YM, Poncet, D, Piron, M, Kean, KM and Borman, AM (2000). Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem 275: 32268–32276. [DOI] [PubMed] [Google Scholar]
- Aragón, T, de la Luna, S, Novoa, I, Carrasco, L, Ortín, J and Nieto, A (2000). Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol 20: 6259–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgui, I, Aragón, T, Ortín, J and Nieto, A (2003). PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J Gen Virol 84(Pt 12): 3263–3274. [DOI] [PubMed] [Google Scholar]
- Burgui, I, Yángüez, E, Sonenberg, N and Nieto, A (2007). Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J Virol 81: 12427–12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez, E, Castello, A, Welnowska, E, Carrasco, L, Goodfellow, I and Nieto, A (2011). Functional impairment of eIF4A and eIF4G factors correlates with inhibition of influenza virus mRNA translation. Virology 413: 93–102. [DOI] [PubMed] [Google Scholar]
- Yángüez, E and Nieto, A (2011). So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res 156: 1–12. [DOI] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Tuerk, C and Gold, L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Guerra-Pérez, N, Ramos, E, García-Hernández, M, Pinto, C, Soto, M, Martín, ME et al. (2015). Molecular and functional characterization of ssDNA aptamers that specifically bind Leishmania infantum PABP. PLoS One 10: e0140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra, N, Vega-Sendino, M, Pérez-Morgado, MI, Ramos, E, Soto, M, Gonzalez, VM et al. (2011). Identification and functional characterization of a poly(A)-binding protein from Leishmania infantum (LiPABP). FEBS Lett 585: 193–198. [DOI] [PubMed] [Google Scholar]
- Levitz, SM and Diamond, RD (1985). A rapid colorimetric assay of fungal viability with the tetrazolium salt MTT. J Infect Dis 152: 938–945. [DOI] [PubMed] [Google Scholar]
- Garfinkel, MS and Katze, MG (1993). Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5'-untranslated region. J Biol Chem 268: 22223–22226. [PubMed] [Google Scholar]
- Park, YW and Katze, MG (1995). Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem 270: 28433–28439. [DOI] [PubMed] [Google Scholar]
- Yángüez, E, Rodriguez, P, Goodfellow, I and Nieto, A (2012). Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation. Virology 422: 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt, AC, Hui, DS, Hay, A and Hayden, FG (2015). Overview of the 3rd isirv-Antiviral Group Conference–advances in clinical management. Influenza Other Respir Viruses 9: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, JP, Christopher, ME, Salazar, AM, Sun, LQ, Viswanathan, S, Wang, M et al. (2010). Broad-spectrum and virus-specific nucleic acid-based antivirals against influenza. Front Biosci (Schol Ed) 2: 791–800. [DOI] [PubMed] [Google Scholar]
- Kwon, D, Shin, K, Kim, S, Ha, Y, Choi, JH, Yang, JS et al. (2010). Replication and pathogenesis of the pandemic (H1N1) 2009 influenza virus in mammalian models. J Microbiol 48: 657–662. [DOI] [PubMed] [Google Scholar]
- Jeon, SH, Kayhan, B, Ben-Yedidia, T and Arnon, R (2004). A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J Biol Chem 279: 48410–48419. [DOI] [PubMed] [Google Scholar]
- Musafia, B, Oren-Banaroya, R and Noiman, S (2014). Designing anti-influenza aptamers: novel quantitative structure activity relationship approach gives insights into aptamer-virus interaction. PLoS One 9: e97696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, HM, Kim, KS, Lee, JM, Shim, HS, Cho, SJ, Lee, WK et al. (2013). Single-stranded DNA aptamer that specifically binds to the influenza virus NS1 protein suppresses interferon antagonism. Antiviral Res 100: 337–345. [DOI] [PubMed] [Google Scholar]
- Yuan, S, Zhang, N, Singh, K, Shuai, H, Chu, H, Zhou, J et al. (2015). Cross-protection of influenza A virus infection by a DNA aptamer targeting the PA endonuclease domain. Antimicrob Agents Chemother 59: 4082–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, SY, Sun, HY, Lee, KH, Oh, BH, Cha, YJ, Kim, BH et al. (2012). 5'-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res 40: 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, F (2015). The catcher in the RIG-I. Cytokine 76: 38–41. [DOI] [PubMed] [Google Scholar]
- Gray, NK, Coller, JM, Dickson, KS and Wickens, M (2000). Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J 19: 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, SZ Jr, Wells, SE, Deardorff, JA and Sachs, AB (1997). Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA 94: 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama, M, Imataka, H and Sonenberg, N (2000). Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol 10: 1147–1150. [DOI] [PubMed] [Google Scholar]
- Resa-Infante, P, Jorba, N, Coloma, R and Ortin, J (2011). The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol 8: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattal, E., Dubernet, C. and Coubreur, P. (2001) Liposome based formulations for the delivery of oligonucleotides. STP Pharma Sciences, 11, 31–44. [Google Scholar]
- Kanwar, JR, Roy, K and Kanwar, RK (2011). Chimeric aptamers in cancer cell-targeted drug delivery. Crit Rev Biochem Mol Biol 46: 459–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, E, Piñeiro, D, Soto, M, Abanades, DR, Martín, ME, Salinas, M et al. (2007). A DNA aptamer population specifically detects Leishmania infantum H2A antigen. Lab Invest 87: 409–416. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A, Pérez-González, A and Nieto, A (2007). Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J Virol 81: 5315–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta, U, Falcón, AM and Ortín, J (2005). Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. J Virol 79: 15246–15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz, F, Quevedo, C, Martín, ME, Alcázar, A, Salinas, M and Fando, JL (1998). Increased activity of eukaryotic initiation factor 2B in PC12 cells in response to differentiation by nerve growth factor. J Neurochem 71: 1905–1911. [DOI] [PubMed] [Google Scholar]
- Kawakami, E, Watanabe, T, Fujii, K, Goto, H, Watanabe, S, Noda, T et al. (2011). Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.