Abstract
Increasing demand for large-scale synthesis of in vitro transcribed (IVT) mRNA is being driven by the increasing use of mRNA for transient gene expression in cell engineering and therapeutic applications. An important determinant of IVT mRNA potency is the 3′ polyadenosine (poly(A)) tail, the length of which correlates with translational efficiency. However, present methods for generation of IVT mRNA rely on templates derived from circular plasmids or PCR products, in which homopolymeric tracts are unstable, thus limiting encoded poly(A) tail lengths to ~120 base pairs (bp). Here, we have developed a novel method for generation of extended poly(A) tracts using a previously described linear plasmid system, pJazz. We find that linear plasmids can successfully propagate poly(A) tracts up to ~500 bp in length for IVT mRNA production. We then modified pJazz by removing extraneous restriction sites, adding a T7 promoter sequence upstream from an extended multiple cloning site, and adding a unique type-IIS restriction site downstream from the encoded poly(A) tract to facilitate generation of IVT mRNA with precisely defined encoded poly(A) tracts and 3′ termini. The resulting plasmid, designated pEVL, can be used to generate IVT mRNA with consistent defined lengths and terminal residue(s).
Keywords: adenylation; genome engineering; in vitro transcription; mRNA, 3′ untranslated region (UTR)
Introduction
In vitro transcribed (IVT) mRNA has emerged as an important modality of transient heterologous protein expression for therapeutic applications.1,2,3,4,5 Electroporation of IVT mRNA is presently being applied in in vivo and ex vivo translational contexts. For example, IVT mRNA is used for in vivo expression of antigenic proteins in dendritic cells for cancer and infectious disease vaccine purposes,5,6,7,8,9,10,11 as well as for ex vivo expression of nucleases for gene editing in primary hematopoietic stem cells and T cells.12,13,14 It is also used for expression of chimeric antigen receptors in primary human T cells for cancer immunotherapy.15,16,17 Furthermore, nanoparticle-based, IVT-mRNA-mediated transient gene expression is also under development for in vivo administration as a novel modality for protein replacement therapies.18,19
A fundamental goal in any IVT mRNA application is to obtain maximally efficient protein expression. High expression efficiency reduces costs by directly reducing the quantity of mRNA necessary for a given level of expression and may also reduce potential mRNA antigenicity due to the reduced exposure of a patient's cells to a foreign nucleic acid. Numerous determinants of mRNA expression efficiency have been identified, including the nature of the 5′ cap, the efficiency of capping, nature of 5′ and 3′ untranslated regions, codon optimization of protein coding sequence, presence of miRNA target sequences in the protein coding sequence and untranslated regions, and the length of the polyadenosine (poly(A)) tail.20 Of these determinants, the length of the poly(A) tail is most problematic to optimize. Although poly(A) tail length is known to be one of the most important physiological determinants of translational efficiency and mRNA stability,21,22,23 and therefore of total protein synthesis, existing technologies do not allow for production of IVT mRNA with defined length poly(A) tails in the physiological range of a newly synthesized tail (~300 nucleotides).
Here, we demonstrate a novel method for generation of homopolymeric tracts of arbitrary length and content and demonstrate that a previously described linear plasmid system, pJazz, is capable of incorporating poly(A) tracts up to ~500 base pairs (bp) in length. Further, we modify pJazz by removing extraneous BsaI sites while incorporating a unique BsaI site downstream from the poly(A) tract to create p(Extended Variable Length) (pEVL), a linear plasmid that allows for facile generation of IVT mRNAs with defined, extended poly(A) tails using standard T7 phage RNA polymerase chemistry.
Results
Homopolymeric poly(A) tracts 175 bp or longer are extremely unstable in circular plasmids
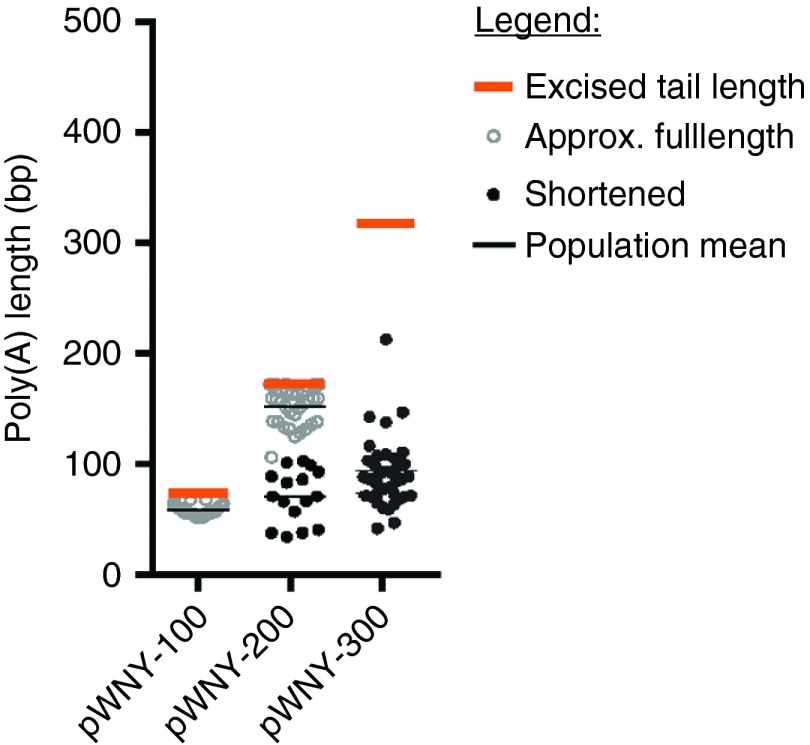
Although there are frequent testaments in the life science literature to the instability of repetitive DNA and extended homopolymeric tracts in circular plasmids (e.g., 5,20), we were unable to find any published data that describe the robustness of the phenomena. To illustrate the difficulties in cloning and propagating extended homopolymeric tracts, we attempted to ligate inserts containing 70, 172, and 325-bp homopolymeric poly(A) tracts, respectively, into a standard circular cloning vector (designated pWNY)—Figure 1. To assess the stability of the inserts, we transformed the plasmids into Escherichia coli and grew them overnight at 25 °C on selective media, and performed PCR across the inserts using DNA obtained from individual bacterial colonies. After analysis by gel electrophoresis, the resulting PCR products resolved primarily into bands of two different sizes—Figure 1, those products reflecting expected insert sizes are represented by open circles, and products representing substantially shortened sizes are represented by closed circles. Although small poly(A) tracts of 70 bp are stable through the standard ligation/transformation process (all maintained their correct length), poly(A) tracts of 172 bp shortened one-third of the time, and longer tracts of 325 bp appear to be extremely unstable, with substantial shortening of the tract occurring without exception.
Figure 1.
Shortening of poly(A) tracts upon cloning into standard circular plasmid cloning vector at 25 °C. BFP-poly(A) tract inserts of 70, 172, and 325 base pairs bounded by restriction enzyme sites DraIII and SwaI were generated via restriction enzyme digest from the linear plasmid cloning vectors BFP-pEVL-100, BFP-pEVL-200, and BFP-pEVL-300 (described in subsequent portions of the manuscript). The inserts were ligated into a circular cloning vector, designated pWNY, and the resulting plasmids were transformed into Escherichia coli using standard methods and grown at 25 °C. Individual colonies were amplified by PCR using primers flanking the poly(A) tract, and the length of the poly(A) tract was estimated based on the resulting band size. Typically, a band was obtained either near the expected size, or at a smaller size, reflecting shortening of the poly(A) tract during transformation. Colonies were scored for whether the poly(A) tract fragment was approximately of the expected size (open circle), or was substantially shortened (closed circle). BFP, blue fluorescent protein; pEVL, p(Extended Variable Length); poly(A), polyadenosine.
Homopolymeric poly(A) tracts up to ~500 bp can be incorporated into pJazz
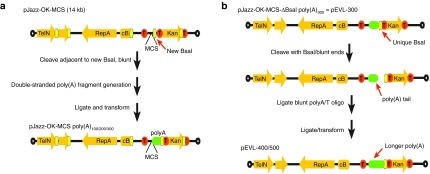
pJazz is a bacteriophage N15-based linear plasmid system that is reportedly superior in the propagation of genomic DNA from organisms with highly repetitive genomes,24 but has not been previously reported to support the cloning or propagation of homopolymeric polynucleotide tracts. To determine whether pJazz was capable of cloning or propagating poly(A) tracts, we performed an in vitro ligation of chemically synthesized DNA oligonucleotides to generate a population of annealed double stranded poly(A/T) polynucleotides of varied lengths, then gel purified those polynucleotides with lengths between ~100 and 850 bp, and attempted to ligate this population of oligonucleotide into a derivative of pJazz-OK with a custom multiple cloning site (MCS) containing a BsaI site (pJazz-OK-MCS, Figure 2a, top). This ligation reaction led to the isolation of pJazz-OK MCS derivatives containing ~70, 172, and 325 bp poly(A) tracts (Figure 2a, bottom) as assessed by gel electrophoresis. To determine the potential for pJazz to stably propagate further extended poly(A) tracts, we developed a method for the generation of extended poly(A) tracts of arbitrary length (Figure 2b). This method required the removal of four BsaI sites in pJazz-OK-MCS to render the BsaI site at the end of the poly(A) tract unique, allowing BsaI, a type-IIS restriction enzyme, to cleave only within the poly(A) tract. This plasmid was designated pJazz-OK-MCS-ΔBsaI (Figure 2b, top). Cloning of previously generated 70, 172, and 325-bp tracts into pJazz-OK-MCS-ΔBsaI generated pEVL-100, pEVL-200, and pEVL-300, respectively, and subsequent cloning of a similarly generated blunt-ended poly(A) polynucleotide population into BsaI-digested/blunted pEVL-300 resulted in incorporation of additional 100- and 200-bp tracts of poly(A) at the end of the pEVL-300 poly(A) tract, thus allowing the isolation of pEVL-400 and pEVL-500 with poly(A) tracts of 425 and 525 bp, respectively (Figure 3a).
Figure 2.
Generation of pEVL: a linear plasmid vector for generation of mRNA with extended encoded poly(A) tracts. (a) Schematic of pJazz and conversion to pEVL. The plasmids are shown with orange arrows denoting genes, red circles with T's denoting transcriptional terminators, open circles denoting terminal hairpin loops, yellow blocks denoting BsaI sites, and green blocks denoting the poly(A) tail. (b) Schematic of pEVL and method used for generation of extended poly(A) tracts in pEVL.
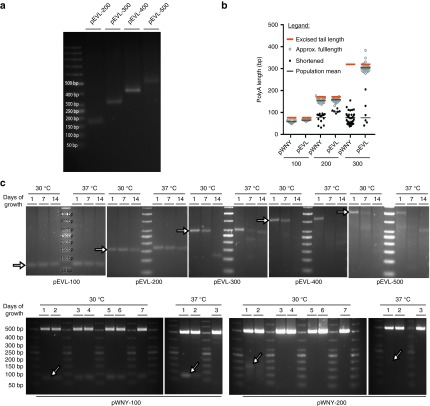
Figure 3.
Characterization of poly(A) tract stability in pEVL. (a) Stability of the encoded poly(A) tracts following overnight induction of pEVL for template preparation. BFP-pEVL-200 through 500 were grown overnight with induction at 30 °C and then maxiprepped. Each maxiprepped sample was digested with BsiWI and BsaI to release the poly(A) tail fragment from the rest of the plasmid. The tail length was determined by gel electrophoresis with comparison to a known molecular weight standard. (b) Shortening of poly(A) tracts upon cloning into standard circular or linear plasmid cloning vectors at 30 °C. BFP followed by poly(A) tract inserts of 70, 172, and 325 base pairs bounded by restriction enzyme sites DraIII and SwaI were generated via restriction enzyme digest from the linear plasmid cloning vectors pEVL-100, pEVL-200, and pEVL-300. The inserts were ligated into the circular cloning vector pWNY or subcloned into pEVL and transformed via electroporation. Transformed bacteria were grown with ampicillin (pWNY) or kanamycin (pEVL) selection at 30 °C. Individual colonies were amplified by PCR using primers flanking the poly(A) tract, and the length of the poly(A) tract was determined based on the resulting band size as in Figure 1. Typically, a band was obtained at the expected size, or a smaller size, reflecting shortening of the poly(A) tract during transformation. Colonies were scored for whether the poly(A) tract fragment was approximately of the expected size (open circle), or was substantially shortened (closed circle). (c) Stability of encoded poly(A) tracts under extended propagation conditions. To test the stability of the poly(A) tail under stringent propagation conditions, pEVL 100 through 500 were grown for 2 weeks at 30 °C and 37 °C with reseeding into fresh media at a 1:1000 dilution every 24 hours. At days 0, 6, and 13, each sample was similarly reseeded into induction media and grown overnight before being miniprepped. Parallel analysis was performed with the circular vectors described in (b), in which the poly(A) tract fragment was sub-cloned into a circular vector (pWNY). As these are already high-copy plasmids, no inducing agent was added to the cultures. For the circular vectors, samples were miniprepped daily for 7 days. For both pEVL and the circular vectors, the tail length of the induced minipreps was determined by gel elctrophoresis as described above. The expected tail band size for each construct is indicated with an arrow.
Practically, we have found that inserts possessing up to around 300-bp poly(A) sequences are easily subcloned into pEVL (Figure 3a,b). In contrast to pWNY, where 172 bp poly(A) tracts remained full length two-thirds of the time and 325-bp tracts were not able to be subcloned at all without substantial shortening, in pEVL, 172 and 325-bp tracts remained full length 84% of the time (Figure 3b). Moreover, these tracts are also highly stable during extended propagation through at least a week of serial liquid culture at 30 °C (Figure 3c). Consequently, these tracts are maintained during expansion, banking as a glycerol stock, and re-expansion as demonstrated by DNA sequencing in both directions across the pEVL300 poly(A) tail (see Supplementary Figure S1). In contrast, we have noted a tendency for the pEVL-500 poly(A) tract to shorten during cloning, although we have always been able to identify clones possessing inserts with a poly(A) tract of the original ~500 bp size. Similarly, extended propagation of pEVL-500 also shows a tendency for the poly(A) tract to shorten over time (Figure 3c), however, it is sufficiently stable for growth and generation of mRNA with templates with the expected poly(A) length.
mRNA incorporating extended polyA tails generated from pEVL templates exhibit superior translation properties
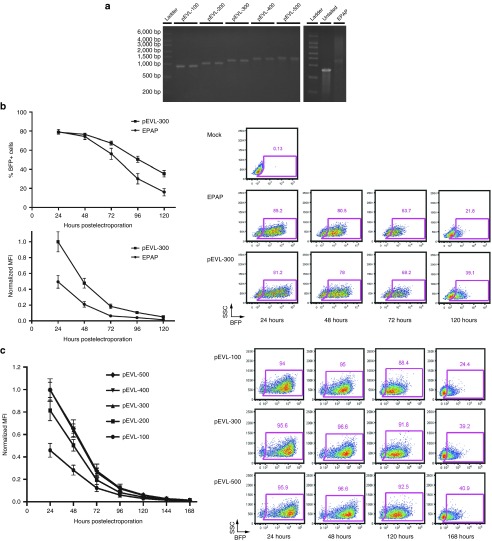
As our goal for developing a means of propagating extended poly(A) tracts was for the purpose of generating IVT mRNA with extended poly(A) tails, we validated pEVL's capacity to function as a template for generating IVT mRNA by cloning a blue fluorescent protein (mTagBFP2) IVT cassette upstream from the poly(A) tracts in pEVL-100/200/300/400/500. We generated IVT mRNA using the BsaI-cleaved pEVL derivatives BFP-pEVL-100 through BFP-pEVL-500 as the IVT mRNA templates and antireverse cap analog capping chemistry. The resulting mRNAs were of predicted size for each pEVL derivative and highly uniform (Figure 4a, left panel) compared with enzymatically polyadenylated RNAs (Figure 4a, right panel, see also5). Enzymatically polyadenylated mRNAs have poly(A) tails varying in length from ~60 to 500 bases, depending on the amount of EPAP used, the amount of time given for the polyadenylation reaction, and the construct size and composition. Importantly, it is difficult to create a consistent tail length across multiple production batches of mRNA, even with standardized protocols.5 In addition, enzymatically added poly(A) tails are not of one homogenous length; the population always contains substantially shortened and lengthened tails, producing a smeared appearance on an RNA gel (Figures 4 and 5). We validated the potency of the mRNA produced from pEVL by comparing BFP expression following electroporation of mRNAs generated through enzymatic polyadenylation and from pEVL-300 into primary human T cells (Figure 4b). We defined potency as mean fluorescence intensity of BFP-positive cells following electroporation with equal molar amounts of mRNA. We observed a striking enhancement of potency, as assessed by differences in BFP mean fluorescence intensity at 24 hours to 5 days postelectroporation, for mRNA generated from pEVL-300 versus enzymatically polyadenylated mRNA. Furthermore, analyses comparing BFP expression driven by mRNA generated from pEVL-100 through pEVL-500 over a period of 7 days (Figure 4c) confirmed the above observations—pEVL-100 and pEVL-200-derived mRNA possessing poly(A) tail lengths of 70 and 172 bp, respectively, exhibited reduced potency versus mRNAs derived from pEVL-300, pEVL-400, or pEVL-500, possessing poly(A) tail lengths of ~325, 425, and 525 bp, respectively. However, we were not able to detect significant potency differences among pEVL-derived mRNAs possessing encoded poly(A) tracts of 300 bp or greater. We did not detect any differences in toxicity between pEVL and enzymatically polyadenylated mRNA, as determined by cell viability in the flow cytometric analysis 24 hours after electroporation.
Figure 4.
Generation and characterization of mRNA from pEVL-encoded templates. (a) IVT mRNA encoding blue fluorescent protein (mTagBFP2) generated from pWNY with enzymatic tailing and pEVL-100 through pEVL-500. BFP-pEVL-100 to 500 were digested with XbaI and BsaI, and pWNY with ScaI and BsiWI, to generate template for IVT. IVT was carried out with antireverse cap analog capping, and for pWNY, enzymatic tailing with EPAP. After purification, 200 ng of each transcript was imaged via gel electrophoresis on the FlashGel system. Typically, pEVL produces a single band of defined length, whereas pWNY with enzymatic tailing produces transcripts of a more heterogenous length. (b) Relative potency of mRNA encoding BFP generated from a circular plasmid vector with enzymatic polyadenylation or from pEVL-300 and representative flow plots. 1 μg of IVT mRNA from the indicated template was electroporated into prestimulated primary human T cells. After a 24-hour cold shock at 30 °C, the cells were analyzed each day for 5 days by flow cytometry for the percentage of cells expressing BFP as well as the mean fluorescence intensity (MFI) of the BFP in BFP+ cells. Flow plots are shown as side scatter (SSC) versus BFP. (c) Relative potency of mRNA encoding BFP generated from pEVL-100 through pEVL-500 and representative flow plots. Equimolar amounts of IVT mRNA from BFP-pEVL-100 to 500 were electroporated into prestimulated primary human T cells. After an initial 24-hour cold shock at 30 °C, the cells were grown at 37 °C for 6 more days. Every 24 hours after electroporation, the percentage of cells expressing BFP and the BFI MFI of the BFP+ cells was analyzed by flow cytometry. Flow plots are shown as side scatter (SSC) versus BFP. BFP, blue fluorescent protein; IVT, in vitro transcribed; pEVL, p(Extended Variable Length).
Figure 5.
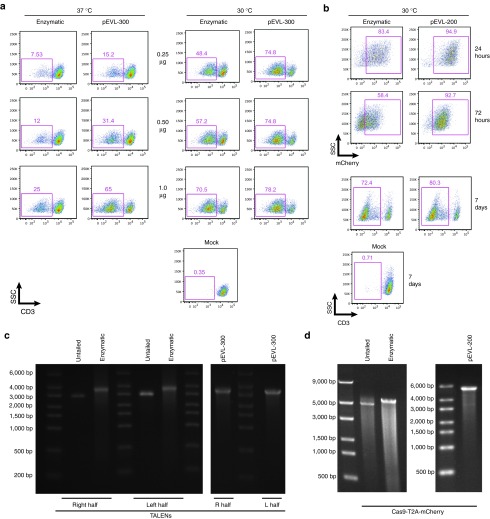
Application of pEVL-encoded mRNA for gene editing in primary human T cells. (a) pEVL-mediated editing of the TCRα locus with TALEN mRNAs in primary human T cells. TALENs targeting the constant region of the TCRα locus were each cloned into pEVL-300. After IVT mRNA production, the indicated amounts of mRNA encoding each TALEN half were electroporated into prestimulated primary human T cells. In the left panel, cells were incubated at 37 °C immediately following electroporation. In the right panel, cells were incubated at 30 °C for 24 hours to increase nuclease protein accumulation after electroporation and then moved to 37 °C. At 72 hours postelectroporation, successful TCR knockout was assayed by flow cytometry for a loss of CD3 expression. (b) pEVL-mediated editing of the TCRα locus with CRISPR/Cas9 in primary human T cells. A Cas9-T2A-mCherry construct was cloned into pEVL-200. After IVT mRNA production, Cas9 was electroporated into prestimulated primary human CD4+ T cells. 3 hours postelectroporation, a CRISPR guide targeting the TCRα constant region was delivered via AAV6 transduction. After electroporation, the cells were incubated at 30 °C for 24 hours before being moved to 37 °C. At 24 and 72 hours postelectroporation, Cas9 electroporation efficiency was determined by FACS analysis of mCherry fluorescence. After 7 days, successful TCR knockout was assayed by flow cytometry for a loss of CD3 expression. (c) Representative RNA FlashGels showing the mRNA transcripts for the left half and right half of the TCRα TALENS produced via enzymatic tailing and from pEVL-300. (d) Representative RNA FlashGels showing the mRNA transcripts for Cas9-T2A-mCherry produced via enzymatic tailing and from pEVL-200. AAV6, adeno-associated vector 6; IVT, in vitro transcribed; pEVL, p(Extended Variable Length).
Previous work has used cellular RNA sequencing to identify correlations between RNA length and the presence of single or double uridine or guanine residues on mRNA 3′ termini.25,26,27 Since the pEVL system allows one to specify the bases present at the 3′ terminus of the IVT template, we chose to validate these correlations by generating mRNAs from templates with termini encoding single or double uridine or guanine bases, and evaluated their capacity to produce fluorescent protein following transfection into primary human T cells (see Supplementary Figure S2). We consistently observed that mRNAs possessing uridine bases at or near the 3′ end (U or UU) exhibited reduced potency (both reduced total magnitude and duration of expression) relative to a standard poly(A) tail of identical length, while those bearing guanine bases at or near the 3′ termini (G or GG) drove BFP expression that was similar to comparable length standard poly(A) tails.
Finally, we evaluated the performance of pEVL-derived IVT mRNA in a gene editing application: disruption of the T-cell receptor α chain (TCRα) locus in primary human T cells (Figure 5). For this purpose, open reading frames encoding transcription activator-like effector nucleases (TALENs) targeting the TCRα constant region locus were cloned into pEVL-300 and used for IVT mRNA production in parallel with production of TALEN mRNA from a standard circular plasmid. Following electroporation of pEVL-derived TALEN mRNA into primary human T cells, we observed that rates of CD3 loss, which occur following disruption of the TCRα gene due to the necessity for TCR α/β holoreceptor/CD3 interaction to support surface expression of CD3, were consistently higher than equal microgram amounts of enzymatically polyadenylated mRNA generated in parallel (Figure 5a). This is consistent with the general observation that increased expression of gene editing nucleases results in higher rates of target site modification (e.g., see28). In comparing TCRα TALEN-mediated knockout using enzymatically polyadenylated or pEVL-300-derived mRNA, we found a dose-dependent response of pEVL-derived mRNA that led to substantially higher gene knockout than enzymatically polyadenylated mRNA (Figure 5a, left panel). When the T cells were incubated at 30 °C for 24 hours after electroporation,29 we again found that we could achieve substantially higher levels of TCR knockout using pEVL-300-derived mRNA and that highest levels of knockout were achieved with a quarter the amount of pEVL-derived mRNA compared with enzymatically tailed mRNA (Figure 5a, right panel). As a second evaluation of the performance of pEVL-derived mRNA for gene editing applications, we used CRISPR-Cas9 as an alternative means of knocking out the TCR. We cloned the spCas9 nuclease linked by a ribosomal skip sequence (T2A)30,31 to mCherry into either a circular plasmid vector or pEVL-200, and used mRNAs derived from these vectors to express Cas9 and mCherry in primary human CD4+ T cells. In our CRISPR system, after mRNA electroporation of Cas9, we deliver the guide RNA (in this case targeting the TCRα constant region) by transducing the T cells with an adeno-associated vector.32 In using this mRNA that coded for a fluorophore-coupled nuclease, we observed that pEVL-derived mRNA (Cas9-T2A-mCherry) produced populations with a tighter distribution of fluorescence and higher average fluorescence compared with enzymatically polyadenylated mRNA (Figure 5b, top panel), and that this correlated with an enhanced efficiency of CRISPR-mediated gene knockout (Figure 5b, bottom panel). Collectively, these data support the concept that efficiency of nuclease expression is an important determinant of the cleavage activity observed for a nuclease at a genomic target site in a living cell,28 and that use of mRNAs with longer, homogenous poly(A) tails are more efficacious for gene editing compared with mRNA with shorter poly(A) tails.
Discussion
Here, we describe the development and application of a linear plasmid vector system, pEVL, that allows for generation of IVT mRNA with poly(A) tails of up to ~500 nucleotides and possessing defined bases at the 3′ end. Furthermore, we show that IVT mRNAs generated with pEVL have high potency when used in conjunction with electroporation of primary human T cells, and that the observed potency is highest for transcripts possessing 3′ poly(A) tails greater than ~300 nucleotides and either adenine or guanine residues at the 3′ end.
We built pEVL upon the pJazz linear plasmid system, which was developed for cloning highly repetitive DNA from bacterial genomes.24 pJazz is derived from the coliphage N15, which has a linear dsDNA genome, and encodes the phage genes necessary for maintenance of the linear structure within the plasmid. The linear DNA is created by cleavage with a phage enzyme, protelomerase (TelN), which generates closed hairpin DNA ends and maintains them during replication.24 Replication is primed by a phage-derived replication protein (RepA) encoded on the plasmid.24 As schematized in Figure 2b, adaptation of pJazz for facile generation of extended poly(A) tracts required its modification such that it possessed a unique target site for the type-IIS restriction enzyme BsaI. The BsaI site 3′ to the poly(A) tract is central to the method we developed for iterative cloning of blunt end poly(A/T) oligonucleotides to allow extension of the poly(A/T) tract to a length greater than 300 bp. The BsaI site may also be used in a similar fashion for generation of nonadenylate residues at the 3′ end of the mRNA, that, as we show in Supplementary Figure S2, may include one or two terminal guanines following the homo-polymeric poly(A) stretch while still yielding mRNA with potency similar to a homopolymeric poly(A) tract, at least in primary human T cells.
Though traditional cloning and propagation methods can be used successfully with pEVL, several changes are necessary to achieve maximal efficiency. An electrocompetent E. coli host strain that contains the TelN gene (in addition to the TelN gene included in the plasmid), called BigEasy TSA, is commercially available and increases transformation efficiency. Recovery of DNA from plasmid purification is increased by the use of arabinose for copy number induction in liquid culture, the use of terrific broth (TB) as the liquid culture media, and isopropanol precipitation by centrifugation for plasmid recovery after elution from the column.
We anticipate that one of the most important uses of pEVL will be for large-scale generation of mRNA for therapeutic applications. pEVL is easily scaled up for large-scale GMP (good manufacturing practises) plasmid production using kanamycin selection, and allows for generation of uniform sized, capped, polyadenylated mRNAs in a single reaction using standard T7 RNA polymerase chemistry. As validating examples for the utility of pEVL at scale, we have been able to use pEVL-200 and pEVL-300 to generate extremely high-quality and high-potency mRNAs encoding TALENs, BFP, Sleeping Beauty transposase, and Cas9-T2A-mCherry at ~10 mg scale using standard mRNA IVT methods.
pEVL also represents a new tool for investigators interested in mRNA biology. By providing a method for generation of transcripts that possess poly(A) tails of arbitrary length and incorporate defined residues at the 3′ end of the poly(A) tail, pEVL allows detailed investigation of the relationship between mRNA architecture and translational efficiency over a wide range of poly(A) tail lengths in a more simple, rapid, and clinically translatable manner than previously possible.33 Our methods also provide a method for compiling long poly(A) tails with arbitrary internal residues. For example, poly(A) tails may be constructed with single non-A residues at arbitrary positions within the tail.
In summary, we report the development of a linear plasmid vector pEVL that enables generation of templates encoding mRNAs bearing extended encoded poly(A) tracts up to ~500 bp in length. pEVL is a significant advance toward simple, efficient, and consistent large-scale synthesis of highly potent mRNAs for therapeutic application, as well as a new tool for the study of the architecture and function of mRNAs possessing defined poly(A) tracts in the physiologic (~300 nucleotide) range for newly synthesized poly(A) tails.
Materials and methods
Creation of pJazz-OK-MCS-pA-100, -200, and -300. We prepared pJazz-OK blunt (Lucigen, Middleton, WI) for insertion of a polyA tail by inserting an MCS comprising multiple blunt and sticky-ended RE sites as well as a terminal BsaI site [DraIII-FspI-BsiWI-NheI-BsaI] was ligated into pJazz-OK blunt to create pJazz-OK-MCS. pJazz-OK-MCS was digested with NheI (NEB, Ipswich, MA), blunted, and dephosphorylated with calf intestinal alkaline phosphatase (CIP, NEB). We then created a long poly(A) tail by annealing oligos of 200 T and 50 A nucleobases at an equimolar ratio. The annealed oligos were treated with a single cocktail of T4 polymerase (NEB), Klenow fragment (NEB), and T4 PNK (NEB) to trim flaps, fill gaps, and create blunt, phosphorylated ends. The resulting mixture was run on a 3% agarose gel and fragments in the range of ~200–800bp were gel extracted. After purification, these fragments were ligated into pJazz-OK-MCS with T4 DNA ligase (NEB) at room temperature for 2 hours. Resulting colonies were screened for poly(A) insert length by colony PCR as described below. Colonies with inserts of appropriate length were sequenced to screen for correct orientation of the insert. This process resulted in the identification of plasmids with poly(A) lengths of 70, 172, and 325 bp; these constructs were named pJazz-OK-MCS-pA-100, -200, and -300, respectively. The tail lengths were determined by gel electrophoresis as described below.
Tail length screening by colony PCR. To screen large numbers of colonies and identify ones with the desired tail length after cloning and bacterial transformation, bacterial colonies were resuspended in water and used as template in a PCR reaction with primers that bind at the 3′ end of the upstream open reading frame (for mTagBFP2 5′-cgcaaacgctaaaactacc-3′) and in the pEVL vector just 3′ of the tail (5′-tcagttctatgtaccagcaagg-3′). The resulting PCR products were run on a 1% agarose gel, stained with ethidium bromide, and imaged on a GelDoc XR+ imaging system (Bio-Rad, Hercules, CA).
Determination of tail length by gel electrophoresis. To determine the length of the poly(A) tract without relying on imprecise polymerase-base methods, pEVL was transformed by electroporation into BigEasy TSA E. coli (Lucigen) and grown overnight with induction at 30 °C in TB+Kan+10% arabinose media and then either miniprepped or maxiprepped. Minipreps were done with the QIAPrep Spin Miniprep kit (Qiagen, Valencia, CA) with doubled amounts of buffers P1, P2, and N3. Maxipreps were done with the NucleoBond Xtra Maxi kit (Macherey-Nagel, Bethlehem, PA), using the manufacturer's instructions for low-copy plasmids (differs from the standard protocol namely in using 8–12 ml resuspension, lysis, and neutralization buffers per gram of pellet weight). For each miniprepped or maxiprepped pEVL plasmid, 2 µg was digested with BsiWI and BsaI. These restriction enzymes cut at the 5′ and 3′ ends of the poly(A) tract, respectively. The digested plasmid was run on a 3% agarose gel with 1–2 µl of GeneRuler 50-bp DNA ladder and poststained with GelStar Nucleic Acid Stain. It was then imaged on a GelDoc XR+ imaging system and the tail length was determined by comparison to the ladder using Quantity One 1-D Analysis Software (Bio-Rad).
Creation of pJazz-OK-MCS-▵BsaI. To create longer poly(A) tails, it was necessary to mutate the four BsaI sites native to pJazz-OK to allow for unique digestion at the BsaI site directly 5′ to and that cleaves within the poly(A). One site was removed by Gibson assembly using Gibson Assembly Master Mix (NEB) and designed with the NEBuilder tool (NEB). The remaining three sites were removed by restriction digest of a fragment containing the BsaI site to be mutated as well as a pair of flanking RE sites, blunting and ligation of this fragment into pUC57 digested with EcoRV (NEB), site-directed mutagenesis of the BsaI site, and then subcloning the mutated fragment back into pJazz-OK-MCS using the included flanking RE sites. This plasmid was designated pJazz-OK-MCS-▵BsaI.
Creation of pEVL series plasmids. Once all native BsaI sites were removed, the fragment containing the poly(A) tail from pJazz-OK-MCS-pA-100, -200, and -300 was cloned into pJazz-OK-MCS-▵BsaI, creating pEVL-100, -200, and -300. pEVL-300 was then digested with BsaI, blunted with Klenow fragment, dephosphorylated with calf intestinal alkaline phosphatase, and ligated with additional blunt double stranded poly(A) fragment prepared as above. Screening as above resulted in plasmids with 425 and 525bp p(A) tails; these were designated as pEVL-400 and pEVL-500.
Cloning of CRISPR-associated protein 9 (Cas9) and TCRα guides. The Cas9 expression construct was obtained from Addgene (plasmid #41815). To ensure nuclear expression of Cas9 in primary human T cells, nuclear localization signals (NLS) were added at both 5′ and 3′ ends (2X-NLS). To track transfection efficiency, Cas9 was fused in-frame with T2A-mCherry and cloned into an in-house modified version of pUC57 with a T7 promoter called pWNY to obtain pWNY-2X-NLS-Cas9-T2A-mCherry. Subsequently, 2X-NLS-Cas9-T2A-mCherry was excised from pWNY and cloned into pEVL-200.
Guides targeting the constant region of TCRα were designed using an online CRISPR design tool (http://crispr.mit.edu). Guides were designed with a U6 promoter and ordered as gblocks (IDT, San Jose, CA). Subsequently, the gblock comprising U6 promoter and sgRNA (crRNA+tracrRNA) was cloned into a self-complementary adeno-associated vector backbone, and checked for maintenance of intact ITRs. Four different G-block-guide constructs generated in this way were used for adeno-associated vector 6 synthesis, and following a series of pilot experiments, a best-performing guide was chosen for further comparative analyses. The target sequence of this guide is TCAAGAGCAACAGTGCTG.
Production of IVT mRNA. For BFP and TALEN constructs, inserts containing a T7 promoter, Kozak sequence, and the TALEN or mTagBFP34 open reading frame were cloned into pWNY or pEVL-100 through pEVL-500. The sequence of the T7 promoter/spacer/Kozak sequence is: taatacgactcactatagggagagcggccgctttttcagcaagattaagccgccaccatggcg. The target site for the TCRα constant region TALEN is tTGTCCCACAGATATCCagaaccctgaccctgCCGTGTACCAGCTGAGa.35 BFP, TALENs, and Cas9 pEVL constructs were digested with BsaI (NEB or Thermo Scientific) to create a template with a terminal poly(A) tail. Templates were simultaneously digested with at a site upstream of the T7 (XbaI or SpeI, NEB or Thermo Scientific) to reduce the overall size of the template-containing fragment and enhance purification. pWNY constructs were linearized at the BsiWI at the end of the open reading frame, and with a second enzyme (typically ScaI or DraIII, NEB or Thermo Scientific) to ensure complete linearization. After checking the completeness of the digest on a 1% agarose gel, the digested DNA was purified on a silica column and eluted in water or 10 mmol/l Tris. IVT with antireverse cap analog capping was carried out using the mMessage mMachine T7 Ultra kit (Life Technologies, Carlsbad, CA) per the manufacturer's directions with 200–1,000 ng of the T7 to poly(A) template fragment; IVT reactions were routinely extended to 150 minutes. Additional DNAse and/or incubation time was used to counterbalance any increased amount of template DNA in the reaction (e.g., double DNAse or double incubation time for double template). For pEVL constructs, the reaction was cleaned up after DNAse treatment using the RNeasy Mini kit (Qiagen) following the manufacturer's directions. For pWNY constructs, the enzymatic tailing step of the mMessage mMachine T7 Ultra kit was carried out for 1 hour according to the manufacturer's instructions before RNA clean-up by the same method as above. RNA quality was determined by NanoDrop spectrophotometry (Thermo Scientific) and gel analysis with the FlashGel RNA system (Lonza). All mRNA was aliquoted in single-use aliquots and stored at −80 °C.
mRNA electroporation of primary human T cells. Freshly isolated primary human T cells were stimulated with αCD3/αCD28 microbeads (Dynal, Thermo Scientific) in RPMI (Roswell Park Memorial Institute) (Corning Cellgro, Tewksbury, MA) with 10–20% fetal bovine serum (Gibco, Thermo Scientific), 2.5% HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) solution (Corning Cellgro, Tewksbury, MA), and 1% L-glut or Glutamax (Thermo Scientific). For isolated pan T cells, IL-2 (Chiron, Emeryville, CA) and IL-15 (Miltenyi, San Diego, CA) at final concentrations of 5 ng/ml IL-2 and 1 ng/ml IL-15 were added. For isolated CD4+ T cells, IL-2, IL-7 (Peprotech, Rocky Hill, NJ), and IL-15 were added at final concentrations of 50 ng/ml IL-2, 5 ng/ml IL-7, 5 ng/ml IL-15, and 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO) was added at a final concentration of 55 μmol/l. After 48–72 hours, the stimulation beads were removed, and the T cells were rested for 1–16 hours in complete media with cytokines. If the initial stimulation was carried out for more than 48 hours, half of the media was replaced with fresh media with cytokines at 48 hours poststimulation. After resting, the T cells were washed in PBS and resuspended at 3 × 107 cells/ml in Neon Electroporation system Buffer T (Life Technologies) with 0.25–2 µg mRNA per 300,000 cells. Electroporation was carried out in a 10 µl tip on the Neon electroporation system (Life Technologies) at 1400 V, 10 ms, and 3 pulses. Immediately following electroporation, cells were resuspended in complete T-cell media with cytokines (as above). Cells were either placed in a 37 °C incubator or in a 30 °C incubator for 24 hours and then moved to a 37 °C incubator. A cold-shock step of this nature has been observed to increase the amount of protein per cell for the period when the mRNA is present36,37,38,39 as well as gene editing efficiency.40 For CRISPR-Cas9 experiments, cells were rested for 3 hours after electroporation and then transduced with adeno-associated vector containing the CRISPR guide at a constant 10% culture volume, equating to an MOI of ~1.33 × 104.
Flow cytometry. Expression of the encoded protein was assayed by flow cytometry at 24–168 hours postelectroporation on an LSRII flow cytometer (BD Biosciences, San Jose, CA). TCR knockout by TALEN or CRISPR was assayed by loss of TCR expression 3–7 days postelectroporation after staining with anti-CD3 (clone HIT3a, direct Alexa-488 or PerCP-Cy5.5 conjugate, BioLegend, San Diego, CA). Data were analyzed using FlowJo software (Treestar, Ashland, OR). Cells were gated on live singlet cells based on the forward and side scatter.
Evaluation of transformation-associated tail shortening. BFP-pEVL100, 200, and 300 were digested with DraIII (which cuts 5′ of BFP) and SwaI (which cuts ~75bp 3′ of poly(A)/BsaI site). This fragment was gel purified and ligated back into pEVL digested with DraIII and SwaI as well as into pWNY digested with DraIII and SmaI. Following ligation, the ligase was heat-killed, and the pEVL and pWNY products were transformed by electroporation into BigEasyTSA electrocompetent cells (Lucigen) and Top10 electrocompetent cells (Life Technologies), respectively. After recovery at 30 °C for 1 hour in SOC (super optimal broth with catabolite repression), aliquots of each transformation were plated in triplicate on kanamycin (kan) plates and grown at 25 °C (72 hours), 30 °C (24 hours), or 37 °C (18 hours). Colonies were screened for tail length by colony PCR as described above. For ligations of the 325 bp tail into pWNY, we were unable to recover any clones with full-length tail. To confirm that the poly(A) tail lengths determined by colony PCR were not biased by PCR polymerase slippage, tail lengths were also determined by gel electrophoresis, as described above. These digests confirmed that the poly(A) tail lengths determined by colony PCR are accurate.
Evaluation of propagation-associated tail shortening. Colonies grown for 18 hours at 30 °C after transformation of intact pEVL-200 to -500 in BigEasy TSA (Lucigen, Middleton, WI) E. coli were immediately screened for tail length by colony PCR. Colonies with the correct tail lengths were seeded into four aliquots of 1 ml TB+Kan and grown overnight, two at 30 °C and two at 37 °C. At each temperature, one aliquot was grown under arabinose induction and one without. After 24 hours, 1 µl of each culture without arabinose was seeded into another 1 ml of TB+Kan without induction. This was carried out for 2 weeks. At the time of seeding each day, another 1 μl was seeded into a 1 ml culture of TB+Kan+arabinose. Each induced culture was spun down and the pellet was frozen after 24 hours of growth. At the end of 2 weeks, all of the frozen induced pellets were miniprepped (Qiagen) and the tail length was determined by gel electrophoresis as described above.
SUPPLEMENTARY MATERIAL Figure S1. Sequencing electropherograms covering the poly(A) tract in pEVL-300. Figure S2. Potency of mRNA generated from pEVL-300 variants with encoded terminal U, -UU, G-, -GG residues. Supplemental Methods
Acknowledgments
We would like to acknowledge the support of the Program in Cell and Gene Therapy at the Center for Immunity and Immunotherapies (CIIT), Seattle Children's Research Institute. We would also like to acknowledge the expert help of the CIIT Viral Core for generating and providing AAV6 vector. This work was supported by NIH U19 AI096111, the Ben Towne Foundation, and through a grant from the Foundation for the National Institutes of Health through the Vector-Based Control of Transmission: Discovery Research (VCTR) program of the Grand Challenges in Global Health to support the Target Malaria consortium. A.M.S. is a consultant for, sits on the scientific advisory board of, and holds equity in Bluebird Bio. A.M.S., K.J., and A.E.G. are inventors on a patent application for technology related to propagating long poly(A) tracts in bacteria. M.C.J. is a cofounder of, receives research support from, and holds equity in Juno Therapeutics.
Supplementary Material
References
- Morgan, RA and Kakarla, S (2014). Genetic modification of T cells. Cancer J 20: 145–150. [DOI] [PubMed] [Google Scholar]
- Zhao, Y, Zheng, Z, Cohen, CJ, Gattinoni, L, Palmer, DC, Restifo, NP et al. (2006). High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol Ther 13: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, ML, Ljungberg, K, Liljeström, P and Johansson, DX (2014). Intradermal electroporation of RNA. Methods Mol Biol 1121: 147–154. [DOI] [PubMed] [Google Scholar]
- Smits, E, Ponsaerts, P, Lenjou, M, Nijs, G, Van Bockstaele, DR, Berneman, ZN et al. (2004). RNA-based gene transfer for adult stem cells and T cells. Leukemia 18: 1898–1902. [DOI] [PubMed] [Google Scholar]
- Holtkamp, S, Kreiter, S, Selmi, A, Simon, P, Koslowski, M, Huber, C et al. (2006). Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108: 4009–4017. [DOI] [PubMed] [Google Scholar]
- Shindo, Y, Hazama, S, Maeda, Y, Matsui, H, Iida, M, Suzuki, N et al. (2014). Adoptive immunotherapy with MUC1-mRNA transfected dendritic cells and cytotoxic lymphocytes plus gemcitabine for unresectable pancreatic cancer. J Transl Med 12: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benencia, F (2014). Antigen-specific mRNA transfection of autologous dendritic cells. Methods Mol Biol 1139: 77–86. [DOI] [PubMed] [Google Scholar]
- Coosemans, A, Vanderstraeten, A, Tuyaerts, S, Verschuere, T, Moerman, P, Berneman, Z et al. (2013). Immunological response after WT1 mRNA-loaded dendritic cell immunotherapy in ovarian carcinoma and carcinosarcoma. Anticancer Res 33: 3855–3859. [PubMed] [Google Scholar]
- Benteyn, D, Van Nuffel, AM, Wilgenhof, S and Bonehill, A (2014). Single-step antigen loading and maturation of dendritic cells through mRNA electroporation of a tumor-associated antigen and a TriMix of costimulatory molecules. Methods Mol Biol 1139: 3–15. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, IA, Hoyer, S, Gerer, KF, Voll, RE, Knippertz, I, Gückel, E et al. (2014). Triggering of NF-κB in cytokine-matured human DCs generates superior DCs for T-cell priming in cancer immunotherapy. Eur J Immunol 44: 3413–3428. [DOI] [PubMed] [Google Scholar]
- Coosemans, A, Vanderstraeten, A, Tuyaerts, S, Verschuere, T, Moerman, P, Berneman, ZN et al. (2013). Wilms' Tumor Gene 1 (WT1)–loaded dendritic cell immunotherapy in patients with uterine tumors: a phase I/II clinical trial. Anticancer Res 33: 5495–5500. [PubMed] [Google Scholar]
- Berdien, B, Mock, U, Atanackovic, D and Fehse, B (2014). TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther 21: 539–548. [DOI] [PubMed] [Google Scholar]
- Holt, N, Wang, J, Kim, K, Friedman, G, Wang, X, Taupin, V et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L, Krymskaya, L, Wang, J, Henley, J, Rao, A, Cao, LF et al. (2013). Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther 21: 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, S, Shimasaki, N, Suwanarusk, R, Ho, ZZ, Chia, A, Banu, N et al. (2013). A practical approach to immunotherapy of hepatocellular carcinoma using T cells redirected against hepatitis B virus. Mol Ther Nucleic Acids 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörrie, J, Krug, C, Hofmann, C, Müller, I, Wellner, V, Knippertz, I et al. (2014). Human adenovirus-specific γ/δ and CD8+ T cells generated by T-cell receptor transfection to treat adenovirus infection after allogeneic stem cell transplantation. PLoS One 9: e109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, DM, Zhao, Y, Liu, X, Jiang, S, Carpenito, C, Kalos, M et al. (2011). Treatment of advanced leukemia in mice with mRNA engineered T cells. Hum Gene Ther 22: 1575–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y, Eltoukhy, AA, Alabi, CA, Khan, OF, Veiseh, O, Dorkin, JR et al. (2014). Lipid-like nanomaterials for simultaneous gene expression and silencing in vivo. Adv Healthc Mater 3: 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitragotri, S, Burke, PA and Langer, R (2014). Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov 13: 655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin, U, Karikó, K and Türeci, Ö (2014). mRNA-based therapeutics—developing a new class of drugs. Nat Rev Drug Discov 13: 759–780. [DOI] [PubMed] [Google Scholar]
- Gallie, DR (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5: 2108–2116. [DOI] [PubMed] [Google Scholar]
- Munroe, D and Jacobson, A (1990). mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol 10: 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, U and Wahle, E (2004). Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678: 67–84. [DOI] [PubMed] [Google Scholar]
- Godiska, R, Mead, D, Dhodda, V, Wu, C, Hochstein, R, Karsi, A et al. (2010). Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli. Nucleic Acids Res 38: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, D and Tian, B (2014). Sizing up the poly(A) tail: insights from deep sequencing. Trends Biochem Sci 39: 255–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H, Lim, J, Ha, M and Kim, VN (2014). TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Subtelny, AO, Eichhorn, SW, Chen, GR, Sive, H and Bartel, DP (2014). Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo, MT, Ryu, BY, Annis, JE, Garibov, M, Jarjour, J, Rawlings, DJ et al. (2011). Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods 8: 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, Y, Choi, VM, Xia, DF, Vo, TD, Gregory, PD and Holmes, MC (2010). Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 7: 459–460. [DOI] [PubMed] [Google Scholar]
- Ryan, MD, King, AM and Thomas, GP (1991). Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol 72: 2727–2732. [DOI] [PubMed] [Google Scholar]
- Kim, JH, Lee, SR, Li, LH, Park, HJ, Park, JH, Lee, KY et al. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather, BD, Romano Ibarra, GS, Sommer, K, Curinga, G, Hale, M, Khan, IF et al. (2015). Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med 7: 307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, U, Gündel, M, Knoth, A, Kerwitz, Y, Rüdel, S and Wahle, E (2009). Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 284: 22803–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach, OM, Cranfill, PJ, Davidson, MW and Verkhusha, VV (2011). An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6: e28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot, L, Philip, B, Schiffer-Mannioui, C, Le Clerre, D, Chion-Sotinel, I, Derniame, S et al. (2015). Multiplex genome-edited T-cell manufacturing platform for “Off-the-Shelf” adoptive T-cell immunotherapies. Cancer Res 75: 3853–3864. [DOI] [PubMed] [Google Scholar]
- Kumar, N, Gammell, P, Meleady, P, Henry, M and Clynes, M (2008). Differential protein expression following low temperature culture of suspension CHO-K1 cells. BMC Biotechnol 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N, Gammell, P and Clynes, M (2007). Proliferation control strategies to improve productivity and survival during CHO based production culture: a summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology 53: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, H, Mazur, X, Marone, R, Bailey, JE and Fussenegger, M (2001). Comparative analysis of two controlled proliferation strategies regarding product quality, influence on tetracycline-regulated gene expression, and productivity. Biotechnol Bioeng 72: 592–602. [PubMed] [Google Scholar]
- Kaufmann, H, Mazur, X, Fussenegger, M and Bailey, JE (1999). Influence of low temperature on productivity, proteome and protein phosphorylation of CHO cells. Biotechnol Bioeng 63: 573–582. [DOI] [PubMed] [Google Scholar]
- Doyon, Y, Choi, VM, Xia, DF, Vo, TD, Gregory, PD and Holmes, MC (2010). Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 7: 459–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.