Abstract
Controlling gene expression in mammalian brain is of utmost importance to causally link the role of gene function to cell circuit dynamics under normal conditions and disease states. We have developed recombinant adeno-associated viruses equipped with tetracycline-controlled genetic switches for inducible and reversible control of gene expression in a cell type specific and brain subregion selective manner. Here, we characterize a two-virus approach to efficiently and reliably switch gene expression on and off, repetitively, both in vitro and in vivo. Our recombinant adeno-associated virus (rAAV)-Tet approach is highly flexible and it has great potential for application in basic and biomedical neuroscience research and gene therapy.
Keywords: brain, inducible and reversible gene expression, Tet systems, recombinant adeno-associated virus
Introduction
The recombinant adeno-associated viruses (rAAVs) are powerful gene delivery vehicles for long-term, cell type-specific gene expression in living animals.1,2 Previously, we deployed the rAAV technology to deliver genetically encoded activity sensors to optically record neuronal activity in selected brain regions.3,4 Further, we have applied the rAAVs to inducibly delete a specific gene in a selective brain region,5,6 thereby linking gene function(s) with learning-dependent synaptic plasticity5 and memory formation.5 With AAVs, after virus infection in cells, maximum transgene expression can be observed in a matter of 2 weeks, both in vitro6 and in vivo.6 The flexibility to engineer designer rAAVs with different capsid proteins7 is an added advantage for selective targeting of diverse cell types. The rAAV DNA exit in the cell nucleus as episomal concatemers,8 and, in some cases, it can integrate at a specific DNA site in the host genome,9 without any observable side effects. Importantly, rAAVs are largely nonpathogenic,10,11 which make them very attractive gene delivery vehicles for human gene therapy. Indeed, promising results have been reported with rAAVs to treat a number of diseases, including Leber's Congenital Amaurosis,12 Hemophilia,13 congestive heart failure,14 and Parkinson's disease.15
Our rAAVs are equipped with tetracycline (tet)-controlled genetic switches for inducible and reversible control of gene expression6 in any brain region(s) and cell type of choice, and gene expression can be controlled by doxycycline (Dox) at any time point chosen by an experimentalist. The use of rAAVs equipped with the tet inducible genetic switches thus paves the way to link gene activity to neuronal circuit function(s) in complex biological processes such as learning and memory,16 emotion,17 addiction,18 stress/anxiety,19 depression,20 pain,21 and aging.22 Here, we have characterized the different parameters for reliable control gene expression in the mammalian brain using a two-virus approach equipped with the tTA (Tet-off) and rtTA (Tet-on) systems. Importantly, to combat various neurological disorders and malfunctions, our two rAAV approach is suitable for gene therapy.
Results
Tet-controlled gene expression in vitro and in vivo
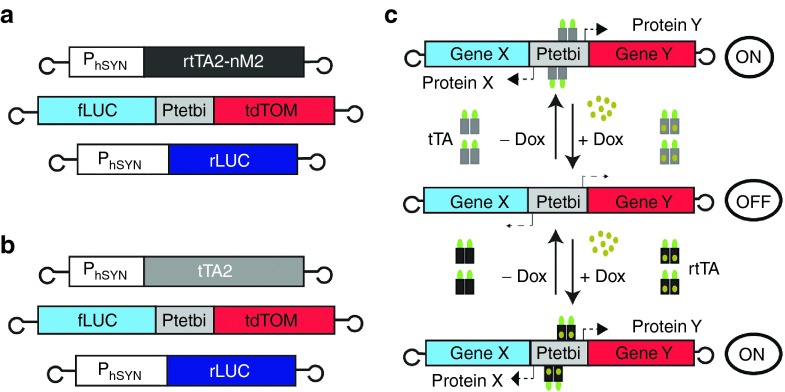
There are two complementary tetracycline (tet) inducible systems,23,24 which are based on three key components (Figure 1a,b): (i) a tet transactivator (tTA)25 or a reverse tTA (rtTA),26,27 which are potent synthetic transcriptional factors, (ii) a unidirectional minimal tet promoter (Ptet)25 or a bidirectional minimal tet promoter (Ptetbi)28 to express multiple gene(s) of interest, when either tTA or rtTA binds to the tet operators (tetO)s located in Ptet/Ptetbi, and (iii) an inducer, doxycycline (Dox), a hydrophobic derivative of tet that controls gene expression and can rapidly distribute in different tissues in vivo, and easily crosses the blood–brain barrier.29 The operating principle of the tet systems (Figure 1c) is as follows: in the absence of Dox, tTA binds to the tetOs located in Ptet/Ptetbi to activate gene transcription, and in the presence of Dox, gene transcription can be switched-off, when the tTA/Dox complex unbinds the tet promoters (Ptet/Ptetbi). The reverse tTA (rtTA) variants binds to Ptet/Ptetbi only in the presence of Dox (rtTA/+Dox) to activate Ptet/Ptetbi mediated gene transcription, and without Dox, rtTA is unable to bind Ptet/Ptetbi, thereby terminating gene transcription.
Figure 1.
The rAAV constructs. (a) Three viral vectors shown here; the reverse tetracycline transactivator (rtTA2-nM2) under control of a human synapsin promoter (hSYN), a bidirectional tetracycline promoter (Ptetbi) driving expression of two genes, firefly luciferase (fLUC) and tdTomato (tdTOM), in opposite orientations and the renilla luciferase (rLUC) under control of the hSYN promoter. (b) Same as (a) except that tetracycline transactivator (tTA) is under control of the hSYN promoter. (c) Turning gene expression ON and OFF using either the tTA system or the rtTA system. With the tTA system, Ptetbi is ‘ON' because tTA binds to Ptetbi and gene expression is switched-OFF in the presence of Dox. The opposite is the case with the rtTA system. Loops at the end of the constructs depict the inverted terminal repeats (ITR).
We have developed recombinant rAAVs to inducibly and reversibly control gene expression in the mammalian brain. With our two-virus approach, we placed tTA or rtTA under control of a pan-neuronal specific promoter (human synapsin or hSYN) (Figure 1a,b). For quantifying gene expression, we placed the firefly luciferase (fLUC)30 and the tdTomato (tdTOM)6,31 genes under control of a Ptetbi (Figure 1a–c). With Ptetbi, these two genes were coexpressed for (1) live fluorescence imaging of neurons with tdTOM and (2) quantitative recording of luciferase (LUC) activity (Figure 1a,b). To normalize for the variability in gene expression, which is largely due to variability in virus infection of cells, we included a “tracer” virus to express the renilla luciferase (rLUC) in neurons. The use of specific light-emitting substrates for fLUC and rLUC, D-luciferin and coelenterazine, respectively, enabled us to quantify gene expression in a ratiometric manner (fLUC/rLUC) (Figure 2b), which are displayed here as relative light units (RLU) ± SEM.
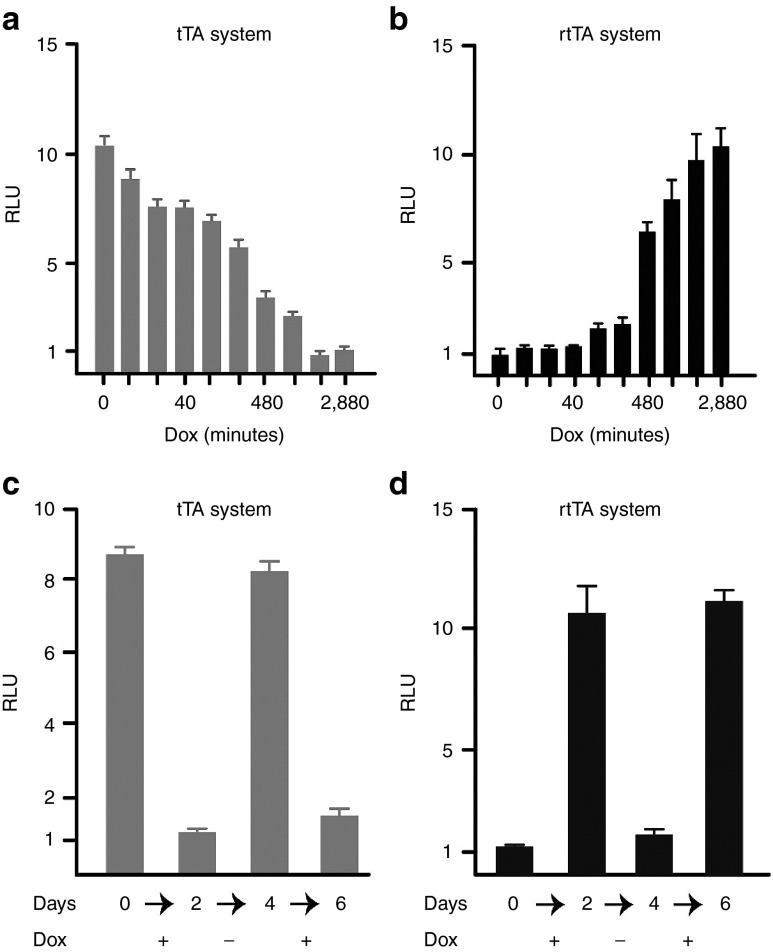
Figure 2.
Functional gene expression in vitro and in vivo. (a) Dox-controlled, rtTA and tTA dependent gene activation in rat organotypic hippocampal slices infected with two viruses as depicted in the images above (scale bar 500 µm). With the rtTA system, tdTOM expression was undetectable in the absence of Dox (left panel, but it was robust with Dox (middle panel). With the tTA system, however, there was very strong tdTOM expression in the absence of Dox (right panel). (b) Firefly luciferase activity measurements of lysates from infected organotypic hippocampal slices treated with or without Dox. The mean of the luciferase activity in the absence of Dox was equated to 1. The data shown are mean of 4 independent infected organotypic slices and expressed as relative light units (RLU; fLUC/rLUC ± SEM). (c) In vivo bioluminescence in the somatosensory cortex in two mice infected with eLUC gene under control of a human hSYN promoter. Bioluminescence signal was detectable only after intraperitoneal injection of D-luciferin.
First, we infected hippocampal organotypic slices with a cocktail of two rAAVs: tTA system (rAAV-hSYN-tTA2 + rAAV-Ptetbi-fLUC/tdTOM) and rtTA system (rAAV-hSYN-rtTA2-nM2 + rAAV-Ptetbi-fLUC/tdTOM). Two weeks after virus infection, we detected robust Dox-induced, rtTA-dependent and tTA-dependent tdTOM expression (Figure 2a, middle and right panels). Expression of tdTOM was undetectable without Dox (Figure 2a, left panel). Because the half-life time of the tdTOM protein is about 24 hours, it takes several days to deplete the tdTOM signal, when using the tTA system after Dox treatment (data not shown).
The fLUC on the other hand has a half-life time of only a few hours, enabling determination of Dox-controlled, tTA/rtTA dependent gene expression time course, both in vitro and in vivo. To quantify gene expression, we performed the LUC assays using a cocktail of three viruses; the tTA system (rAAV-hSYN-tTA2 + rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC) and the rtTA system (rAAV-hSYN-rtTA2-nM2 + rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC). With the tTA system, RLU activity was about 18-fold higher over the baseline (without tTA or with tTA/+Dox). In the case of the rtTA system, the RLU activity without Dox was near the baseline levels, and RLU activity increased by about 20-fold by Dox (Figure 2b). We further demonstrated by NeuN immunostaining that the hSYN promoter allows for neuron-specific gene expression both in vitro and in vivo (Supplementary Figure S1).
The raw firefly luciferase (fLUC) activity values in brain tissue (both in vitro and in vivo), infected with the rAAV-Ptetbi-fLUC/tdTOM alone (without tTA), were roughly 10 times higher than the background values (lysate without virus infection) (Supplementary Figure S2). Similarly, raw fLUC activity values with the rtTA system in the absence of Dox were roughly 10 times higher than the background fLUC activity (without virus infection) (Supplementary Figure S2). The observed basal fLUC activity in the uninduced state is thus most likely due to the intrinsic leakiness by the tet responder virus (rAAV-Ptetbi-fLUC/tdTOM), and leakiness increases with increasing number of rAAV-Ptetbi virus particles in infected cells. Consistent with our previous observations,6 intrinsic leakiness by rAAV-Ptetbi module is likely mediated by the rAAV inverted terminal repeat (ITR) enhancer-like activity onto the Ptetbi.32 To reduce leakiness, we had to lower the rAAV-Ptetbi virus titers
We also performed noninvasive bioluminescence imaging with the tet inducible systems using the enhanced firefly luciferase (eLUC), but the eLUC activity was not detected in the mouse brain through an intact skull (data not shown). However, strong bioluminescence signal was detected through an intact skull, skin and fur with the mouse cortex injected with rAAV-hSYN-eLUC (Figure 2c). We further determined that the maximum eLUC activity from brain samples infected with the rAAV-hSYN-eLUC was quite similar to brain samples infected with the Tet-inducible systems (rAAV-hSYN-tTA2 + rAAV-Ptetbi-eLUC/Venus, -Dox and rAAV-hSYN-rtTA2-nM2 + rAAV-Ptetbi-eLUC/Venus, +Dox) (Supplementary Figure S3). In the case of the inducible systems, the low virus titer (rAAV-Ptetbi-eLUC/Venus) hindered signal detection from the intact skull. However, the eLUC signal was detected, when the skull was removed (data not shown). We propose that in future studies long-term regulated gene expression in mouse brain should be feasible by noninvasive in vivo bioluminescence imaging. We further suggest that noninvasive in vivo bioluminescence imaging using the rAAV-hSYN-eLUC can serve as a reliable tool to control for the variability in virus injection and will help to identify properly rAAV-injected animals.
Repetitive ON/OFF gene expression cycles in vitro
Because access and removal of Dox occurs rapidly in cultured cells, we used dissociated neurons as a model system to investigate the time course of tTA-dependent gene expression inactivation by Dox and re-activation upon Dox removal and rtTA-dependent gene expression activation by Dox and inactivation upon Dox removal. In these experiments, we applied a three virus cocktail: the tTA system (rAAV-hSYN-tTA2 + rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC) and the rtTA system (rAAV-hSYN-rtTA2-nM2 + rAAV-Ptetbi-fLUC/tdTOM + hSYN-rLUC). Two weeks after virus infection, neurons were treated with Dox in the medium and RLU activity was calculated at various time points. After Dox treatment for 24 hours, RLU activity with the tTA and rtTA systems reached a baseline and a maximum, respectively, and it remained nearly unchanged after an additional 24 hours (Figure 3a,b). Further, Dox-controlled on/off gene expression cycles with the tTA and rtTA systems were possible with an interval of 48 hours. These experiments were performed by separating the virus-treated primary dissociated neurons into four groups (n = 4 per group, each group) lasting six days and RLU activity was measured from cell lysates. With the tTA and rtTA systems over a period of 6 days, the magnitude of RLU activity regulation was 10- and 12-fold, respectively (Figure 3c,d).
Figure 3.
Dox-controlled, inactivation (tTA) and activation (rtTA) time courses, and regulated gene expression in vitro. (a,b) Time course of Dox-controlled, tTA-dependent luciferase activity in virus-infected dissociated primary neurons. The cells were infected with either the tTA system (three viruses; AAV-hSYN-tTA2, AAV-Ptetbi-fLUC/tdTOM, and AAV-hSYN-rLUC) or the rtTA system (three viruses; AAV-hSYN-rtTA2-nM2, AAV-Ptetbi-fLUC/tdTOM, and AAV-hSYN-rLUC). Dox-induced (1 µg/ml), time course of gene inactivation (with the tTA system) and gene activation (with the rtTA system). fLUC/rLUC activity ratios were calculated from cell lysates harvested at the different time points after Dox treatment, and fLUC/rLUC activity ratios ± SEM were calculated from four independent slices beginning at time 0 up to 2,880 minutes. With tTA and rtTA, the mean of the luciferase activities at time point 2,880 and 0 (shown in minutes) were used as baseline, respectively. (c,d) Repeated cycles of gene activation and inactivation in dissociated primary neurons with addition of Dox (+ sign) and removal of Dox (− sign). Serial Dox addition and removal allowed for ON/OFF cycles testing if the systems. For the tTA and rtTA systems, fLUC/rLUC activity ratios were measured and data calculated as mean ± SEM and displayed here as relative light units (RLU) at time point (day 2) and time point (day 0) were used as the baseline, respectively.
Repetitive ON/OFF gene expression cycles in vivo
To determine an optimal Dox concentration required to switch-off and switch-on gene expression in tTA and rtTA-dependent manner, respectively, we injected a three-virus cocktail bilaterally into cortex and hippocampus of 10 wild-type mice. Two weeks after virus injection, mice were treated with different concentrations of Dox (3, 10, 30, and 100 μg/g body weight, n = 2 mice per group). Two days after intraperitoneal (i.p.) Dox injection, RLU activity was measured from virus injected brain lysates (four samples per mouse, i.e., cortices and hippocampi of both brain hemispheres). With the tTA system, 10 and 100 µg/g body weight of Dox reduced gene expression levels by 7- and 10-fold, respectively (Figure 4a). With the rtTA system, however, 30 and 100 µg/g body weight of Dox increased gene expression by 13- and 16-fold, respectively (Figure 4b).
Figure 4.
Dox doses, inactivation (tTA) and activation (rtTA) time courses, and Dox-controlled regulated gene expression in vivo. Wild-type mice were injected with the tTA system (three viruses; AAV-hSYN-tTA2, AAV-Ptetbi-fLUC/tdTOM, and AAV-hSYN-rLUC) or the rtTA system (three viruses; AAV-hSYN-rtTA2-nM2, AAV-Ptetbi-fLUC/tdTOM, and AAV-hSYN-rLUC). For each time point, measurements were made from 12 cortical and hippocampal extracts from three independently injected mice, and fLUC/rLUC activity ratios were measured and data calculated as mean ± SEM and displayed here as relative light units (RLU). (a,b) Mice were treated with different concentrations of Dox per gram body weight (ranging from 0 to 100 µg/g). For the tTA and rtTA systems, Dox-treated (100 µg/g) and Dox-untreated mean luciferase/renilla ratios were used as baselines, respectively. (c,d) Time course (days) for tTA-dependent gene re-activation with a single intraperitoneal Dox injection (10 µg/g, P < 0.0001) and rtTA-dependent gene inactivation after a single intraperitoneal Dox injection (30 µg/g, P < 0.0001). (e,f) With the tTA system, gene inactivation (single Dox i.p.) and reactivation (after single Dox i.p.) was 2 and 6 days, respectively. With the rtTA system, gene activation (single Dox i.p.) and inactivation (after single Dox i.p.) was also 2 and 6 days, respectively.
To determine the number of days required for gene reactivation after a single i.p. Dox injection (with tTA) and gene inactivation (with rtTA), cortex and hippocampus of mice from these two groups (10 mice per group) were injected with a three-virus cocktail. Two weeks after virus injection, a single dose of Dox was delivered by a single intraperitoneal (i.p.) injection into tTA (10 µg/g body weight) and rtTA (30 µg/g body weight) mice, and RLU activity was measured from brain lysates on day 0, 2, 4, 8, and 16 (two mice per time point and four brain samples per mouse). With a single i.p. Dox injection, tTA-dependent gene expression was switched “off” and “on” after 2 and 6 days, respectively (Figure 4c). The rtTA system, gene expression was switched “on” and “off” after 2 and 6 days, respectively (Figure 4d).
For repeated on/off gene expression cycles, 8 tTA and 8 rtTA mice were placed into four separate groups, and treated with Dox by a single i.p. injection (10 µg/g body weight of Dox for tTA mice and 30 µg/g body weight of Dox for rtTA mice). In the case of tTA mice, it took 2 days to switch-off gene expression by Dox, and after Dox clearance from the brain gene expression was fully “on” after 6 days. The rtTA showed the reverse; in 2 days gene expression was fully switched-on by a single i.p. Dox and switched-off after 6 days (Figure 4e,f).
Discussion
We have generated two recombinant rAAVs with tet controlled genetic switches for long-term, inducible and reversible regulation of gene expression in neurons. The fLUC and the tdTOM genes were placed under a bidirectional tet promoter (Ptetbi), allowing for Dox-controlled, tTA- and rtTA-dependent regulated gene expression. The fLUC protein has a short half-life of roughly 2–4 hours and a large dynamic range that is five orders of magnitude. The LUC activity assay is highly sensitive, with minimal background activity, and is ideally suited for quantitative measurement of gene expression.
We characterized our two-virus approach to determine the time course of gene activation and inactivation in the mammalian brain. Our results show that both tTA and rtTA systems provide about 12-fold change in gene regulation. With the tTA system, gene expression (without Dox) is enabled from the very beginning. Although Dox treatment terminates gene transcription within minutes, the preexisting fLUC protein, for example, with its intrinsic half-life time of a few hours, will reach undetectable background levels in about 48 hours. With the tTA system, when it is desired that to keep gene expression switched-off, Dox must be constantly administered. However, upon Dox removal, gene expression can be switched-on in about a week. This slow time course is due to slow Dox clearance from the brain. With the rtTA system, on the other hand, gene expression can be rapidly and fully switch-on within 24 hours, but it takes about a week to switch-off gene expression, again due to slow Dox clearance from the brain. These results clearly show that the tTA and rtTA systems are highly complementary and can be flexibly applied to investigate specific biological processes, and they are also highly suited for gene therapy applications. Previous studies have shown that controlling the amounts of Dox levels allow for graded control of gene expression, demonstrating that the tet systems operate as a rheostat.33 However, the rheostat can be converted to on/off switches, when transactivator is combined with a repressor.34 These features allow for flexible application of the tet systems for biological studies.
The Ptetbi vectors are intrinsically leaky. Although this leakiness is minimal, it can produce enough gene-product to interfere with cell physiology. The internal terminal repeat (ITR) of rAAVs have potential enhancer-like activity.6,32 Previously, we discovered that two oppositely oriented DNA sequences flanking the Ptetbi, insulate the Ptetbi from the ITR. However, removal of one such DNA sequence (a gene, for example) brings the ITR very close to the Ptetbi, thereby increasing Dox-independent expression or leakiness.6,32 In our current study, we have observed that the minimal leakiness of a rAAV-Ptetbi can be substantially decreased by reducing the responder virus (rAAV-Ptetbi) titer.
We have demonstrated here that our rAAV-based approach is highly suitable for long-term regulated gene expression in living mammals. It should be pointed out that a previous study elegantly demonstrated the use of a single rAAV system, harboring a transactivator (tTA or rtTA) together with a Ptet for regulated gene expression.35 However, due to the limited cloning capacity of the rAAVs, a single rAAV approach can accommodate small DNA sequence lengths, which substantially limits the utility of a single virus approach for many applications. Our two rAAV approach on the other hand provides a flexible solution to simultaneously control the expression of multiple genes.
In summary, we have characterized a two-virus approach equipped with Dox-controlled genetic switches for regulated gene expression in the mammalian brain. The implementation of our flexible rAAV based inducible and reversible gene expression approach has a promising future for neuroscience and biomedical studies, and gene therapy applications.
Materials and methods
Animal welfare. All experiments were performed in accordance with the animal welfare guidelines of the Max Planck Society and approved by the local authorities (Regierungspräsidium Karlsruhe). We housed 6–10-week-old C57BL/6 wild-type and NMRI albino mice under standard conditions in a 12-hour light/dark cycle in Makrolon cages type 2A with food and water.
Plasmid constructs. In the current study, we generated new rAAV plasmids, (i) pAAV-Ptetbi-fLUC/tdTOM and (ii) pAAV-hSYN-eLUC (enhanced luciferase)36 and (iii) pAAV-hSYN-rLUC. The other plasmids (pAAV-hSYN-tTA, pAAV-hSYN-rtTA, and pAAV-hSYN-EGFP) were described previously.6,37 To generate the plasmid pAAV-Ptetbi-fLUC/tdTOM, firefly luciferase gene (1.6 kb fragment) was excised from the plasmid pB1-5 (ref. 28) with Pst1/Xba1, blunted and subsequently ligated into a HindIII/Sph1 blunted plasmid pAAV-Ptetbi-iCre/tdTOM, replacing iCre with fLUC. Similarly, the plasmid pAAV-hSYN-rLUC was generated by blunt-end ligation by replacing the EGFP from pAAV-hSYN-EGFP (BamHI/HindIII) with a 947 bp rLUC fragment from the plasmid pRL-CMV (Nhe/Xba1). To generate the plasmid pAAV-hSYN-eLUC, the enhanced luciferase gene (eLUC) including SV40pA was excised from the plasmid peLUC-test (a gift from Dr. Nakajima) using EcoR1/BamH1 and a 1.9 kb fragment was ligated into the vector pAAV-hSYN-vector (without tTA) using EcoR1/BamH1. To generate the plasmid pAAV-Ptetbi-eLUC/Venus, the 1.6 kb eLUC fragment was amplified by PCR from the plasmid peLUC with the forward primer EcoRV 5′-AATTCGATATCGAATTCCTGCAGCCCAC-3′ and the reverse primer SpeI 5′-TGTCTGACTAGTGGCGGCCGCTCTAGATTA-3′. The PCR generated fragment was cloned into the vector pAAV-Ptetbi-IKK2DN/Venus (unpublished plasmid), by removing IKK2DN with EcoRV and SpeI and replacing it with eLUC to generate the plasmid pAAV-Ptetbi-eLUC/Venus.
Preparation and purification of rAAVs via Heparin column. Recombinant adeno-associated viruses (rAAVs) serotype 1 and 2 were generated by cotransfection of HEK293 cells and purified by modified heparin column purification as described.6 In brief, plasmids corresponding to the various rAAVs used in this study were individually cotransfected with pDp1 and pDp2 (ratio 2:1:1) helper plasmids in HEK293 cells by the calcium-phosphate method.38 Forty-eight hours after transfection, cells were harvested into a 50 ml falcon tube followed by centrifugation at 1,000 rpm for 15 minutes. The cell pellet was resuspended in 45 ml of 150 mmol/l NaCl, 20 mmol/l Tris.HCl (pH 8.0), frozen in liquid nitrogen and stored at −70 °C. Cells were thawed at room temperature (RT) and incubated at 37 °C with 40 U/ml of benzonase and 0.5% NaDOC for 60 minutes with frequent mixing. Lysed cells were centrifuged and frozen at −70 °C for at least a few hours and subsequently thawed and centrifuged at 3,900 rpm for 15 minutes, supernatant collected and frozen at −70 °C overnight. Frozen supernatant was thawed and spin for 15 minutes at 4,000 rpm. The cleared supernatant was run through a pre-equilibrated 1 ml heparin column. The HiTrap Heparin HP column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) was serially washed with 20 ml of 100 mmol/l NaCl, 20 mmol/l Tris.HCl (pH 8), 1 ml of 200 mmol/l NaCl, 20 mmol/l Tris.HCl (pH 8) and 1 ml of 300 mmol/l NaCl, 20 mmol/l Tris.HCl (pH 8). The virus was eluted from the heparin column serially with 1.5 ml of 400 mmol/l NaCl, 20 mmol/l Tris.HCl, 3ml of 450 mmol/l NaCl, 20 mmol/l Tris.HCl (pH 8) and 1.5ml of 500 mmol/l NaCl, 20 mmol/l Tris.HCl (pH 8). The eluate was concentrated in a 15 ml Amicon Ultra (100,000 cut-off) concentrator (Merck Millipore, Darmstadt, Germany). The virus was concentrated further into a final volume of 250 µl and filtered through 0.2 µm Acrodisc filter (Merck Millipore, Darmstadt, Germany).
Virus infection of hippocampal organotypic slices and dissociated neurons. Organotypic slices were prepared as described previously39 and slices were infected with 500 nl of virus cocktails by local application on individual slices. The two different virus cocktails are as follows: the tTA system (rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC + rAAV-hSYN-tTA) and the rtTA system (rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC + rAAV-hSYN-rtTA). Media were changed 5 days after infection, and subsequently replaced every 3 days. Dissociated neurons on the other hand, were infected by adding 1 μl of the virus cocktail into 1 ml of medium. Two weeks of virus infection, brain slices were lysed for quantitative luciferase activity measurements or fixed for fluorescent imaging of tdTOM.
Quantifying luciferase gene activity in dissociated neurons. Dissociated primary neurons were infected with a tTA virus cocktail or rtTA virus cocktails. Two weeks later, cells infected with the tTA and rtTA systems were incubated for 48 hours either without or 1 µg/ml of Dox, respectively. Subsequently, cells were lysed with 100 µl of passive lysis buffer and 20 µl of the lysates were used for measuring firefly and renilla luciferase activities. For the repeated cycle of gene activation and inactivation, dissociated primary neurons in 24-well plates were infected with tTA and rtTA systems. Two weeks after infection, cells were put into five groups each group consisting of four wells. Each group was incubated with or without Dox over 48 hours. The Dox-treated medium was replaced with a conditioned medium without Dox also for another 48 hours. This was repeated for five cycles of Dox addition or withdrawal. The removal of Dox was done by washing three times with conditioned medium without Dox. At the end of two cycles, cells were harvested for luciferase assay. The firefly luciferase (fLUC) and renilla luciferase (rLUC) assays were performed sequentially in one reaction tube according to the manufacturer's instruction (Promega Corporation, Wisconsin). To normalize gene activity, fLUC/rLUC mean ratios were calculated as relative light units (RLU) ± SEM, and for each time point, measurements were performed from four independent cell lysates. For detailed dual luciferase reporter assay protocol, refer to Dual-Luciferase Reporter Assay System protocol instructions for use of products e1910 and e1960 (Promega Corporation).
Stereotactic virus injection into the mouse brain. Six- to 10-week-old C57BL/6 and NMRI albino mice were deeply anesthetized (ketamine 100 mg/kg, and xylazine 5 mg/kg) and secured in a Kopf stereotaxic setup (Kopf Instruments, California). The surgical procedure was started only after animals no longer responded to tail or paw pinch. Throughout the surgery, animals were kept on a heating pad to prevent hypothermia. Cream (Bepanthen, Bayer, Leverkusen, Germany) was applied to the eyes of mice to prevent dehydration. The foreskin on the skull was cut open to expose the skull. With the help of a drill, small holes (about 50–100 µm) were drilled into the head of the mice at different coordinates. Approximately 300 nl of viral cocktail was injected via glass pipette (tip diameter 10–20 µm) into each brain region. For in vivo bioluminescence imaging, mice were injected in the motor cortex. For in vivo luciferase activity measurement, mice were injected in the somatosensory cortex and in the hippocampus of both brain hemispheres. After injection, the skin was sutured and the wound disinfected. Newly injected mice were kept on heating blocks at 37 °C until they woke up and were fed wet food. The coordinates used for the different injections with reference to the bregma are as follows: for hippocampus (−1.70 mm bregma, 1.5 mm lateral, 1.5 mm deep), for motor cortex (0.5 mm bregma, 1 mm lateral, 500 µm deep) and for somatosensory cortex (−1.70 mm bregma, 1.5 mm lateral, 500 µm deep). To avoid postoperative dehydration, mice were injected with physiological saline solution. To reduce moderate acute pain, buprenorphine (Buprenex, 0.1 µg/g body weight) was injected after the surgical procedure. Mice were placed on a heating-pad while recovering from surgery.
Quantifying luciferase gene activity in brain tissues. To measure luciferase activities from brain lysates, wild-type mice were injected with the tTA system (rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC + rAAV-hSYN-tTA) and the rtTA system (rAAV-Ptetbi-fLUC/tdTOM + rAAV-hSYN-rLUC + rAAV-hSYN-rtTA). Two weeks after virus injection, mice were either injected with Dox (rtTA injected mice) or without (tTA injected mice), and 48 hours later mice were anaesthetized with isoflurane and decapitated or virus infected brain regions (somatosensory cortex and hippocampus) were carefully dissected and frozen in liquid nitrogen. Brain extracts were prepared and later sonicated in 200 µl PBS and 20 μl of the extract was used to measure firefly and renilla luciferase activities as described above.
Bioluminescence imaging in vivo. Bioluminescence imaging was performed with an EM-CCD Digital camera C9100-13 (Hamamatsu Photonics Deutschland GmbH, Herrsching am Ammersee, Germany) in combination with a dark box (AEQUORIA MDS) (Hamamatsu Photonics Deutschland GmbH, Herrsching am Ammersee). At the beginning of the experiment, BALB/c mice were stereotactically injected with rAAV-hSYN-eLUC into the cortex of the right brain hemisphere. After recovery from surgery the animals were anesthetized with a mixture of ketamine (65 µg/g) and xylazine (14 µg/g) by i.p injection. To avoid dehydration, all mice were injected with 500 μl of physiological saline (0.9 % NaCl). Furthermore, the animals were i.p. injected with an aqueous solution of D-luciferin (300 µg/g). Right after the last injection, mice were placed on a stage in the Hamamatsu dark box. At first, a bright field image was taken for later analysis followed by bioluminescence recording using the streaming mode (six images possible with an exposure time of 10 minutes each). All data was recorded with the aid of Hokawo imaging software version 2.1, also provided by Hamamatsu Photonics Deutschland GmbH, Herrsching am Ammersee.
Immunostaining with NeuN antibody. Immunostaining was carried out on wild-type mice or rat organotypic slices infected with a tTA system (rAAV-Ptetbi-fLUC/tdTOM and rAAV-hSYN-tTA). In brief, mice were perfused with warm PBS and fixed in 4% paraformaldehyde in 1× phosphate buffered saline (PBS) prior to decapitation. Brains were removed and postfixed in 4% paraformaldehyde at 4 °C overnight, followed by embedding in 2.5% agarose. Coronal sections with 60–100 μm in thickness were prepared by vibratome slicing and stored in PBS 4 °C. Tissues were blocked in 4% normal goat serum supplemented with 1% bovine serum albumin (BSA), 0.3% Triton X-100, and incubated overnight at room temperature in anti-NeuN (mouse monoclonal, Chemicon, California) primary antibody diluted in 1× PBS, 1% (BSA), and 1% normal goat serum/0.3% Triton X-100. The next day, tissues were washed twice in 1× PBS/0.3% BSA/0.1% Triton X-100 followed by anti-mouse FITC secondary antibody (1:200, Jackson Immuno Research, Pennsylvania) for 1 hour at room temperature. Slices were washed 2× in PBS and mounted with Aqua Poly/Mount on glass slides with cover slips. Immunostained sections were analyzed with an Axiovert 200 M confocal microscope with LSM 5 PASCAL (Zeiss, Jena, Germany) coupled to 543 nm HeNe and 450–530 nm Argon lasers (Lasos Lasertechnik GmbH, Jena, Germany).
Statistical analysis. All data are presented as mean ± standard error of the mean. Figure 2b was analyzed with t-tests P < 0.0001. Multiple group comparisons were performed with one-way analysis of variance with P < 0.05 is considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Neuron-specific gene expression. Figure S2. Firefly luciferase activity in relative light units. Figure S3. Enhanced firefly luciferase expression using different rAAV expression systems.
Acknowledgments
We thank Peter H. Seeburg for support. We thank Andy Migala, Sabine Grünewald and Annette Herold for technical assistance. Max Planck Society, the Fritz Thyssen Stiftung (M.T.H.) and the SFB636/A4 (R.S.) supported this work. M.T.H. designed and supervised the project. G.K.D. performed the experiments and analyzed the data. M.R. performed in vivo bioluminescence imaging and analyzed the data. R.S. provide reagents and scientific input. G.K.D. and M.T.H. wrote the manuscript with input from M.R. and R.S. The authors declare that they have no competing financial interests.
Supplementary Material
References
- Mastakov, MY, Baer, K, Kotin, RM and During, MJ (2002). Recombinant adeno-associated virus serotypes 2- and 5-mediated gene transfer in the mammalian brain: quantitative analysis of heparin co-infusion. Mol Ther 5: 371–380. [DOI] [PubMed] [Google Scholar]
- Shevtsova, Z, Malik, JM, Michel, U, Bähr, M and Kügler, S (2005). Promoters and serotypes: targeting of adeno-associated virus vectors for gene transfer in the rat central nervous system in vitro and in vivo. Exp Physiol 90: 53–59. [DOI] [PubMed] [Google Scholar]
- Lütcke, H, Murayama, M, Hahn, T, Margolis, DJ, Astori, S, Zum Alten Borgloh, SM et al. (2010). Optical recording of neuronal activity with a genetically-encoded calcium indicator in anesthetized and freely moving mice. Front Neural Circuits 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, DJ, Meyer zum Alten Borgloh, S, Astori, S, Yang, Y, Bausen, M, Kügler, S et al. (2008). Single-spike detection in vitro and in vivo with a genetic Ca2+ sensor. Nat Methods 5: 797–804. [DOI] [PubMed] [Google Scholar]
- Hasan, MT, Hernández-González, S, Dogbevia, G, Treviño, M, Bertocchi, I, Gruart, A et al. (2013). Role of motor cortex NMDA receptors in learning-dependent synaptic plasticity of behaving mice. Nat Commun 4: 2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogbevia, GK, Marticorena-Alvarez, R, Bausen, M, Sprengel, R and Hasan, MT (2015). Inducible and combinatorial gene manipulation in mouse brain. Front Cell Neurosci 9: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J, Faust, SM and Rabinowitz, JE (2011). The next step in gene delivery: molecular engineering of adeno-associated virus serotypes. J Mol Cell Cardiol 50: 793–802. [DOI] [PubMed] [Google Scholar]
- Schnepp, BC, Jensen, RL, Chen, CL, Johnson, PR and Clark, KR (2005). Characterization of adeno-associated virus genomes isolated from human tissues. J Virol 79: 14793–14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotin, RM, Siniscalco, M, Samulski, RJ, Zhu, XD, Hunter, L, Laughlin, CA et al. (1990). Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA 87: 2211–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, YJ, Wang, J, Kearns, WG, Loiler, S, Poirier, A and Flotte, TR (1999). Latent adeno-associated virus infection elicits humoral but not cell-mediated immune responses in a nonhuman primate model. J Virol 73: 8549–8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirmule, N, Propert, K, Magosin, S, Qian, Y, Qian, R and Wilson, J (1999). Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther 6: 1574–1583. [DOI] [PubMed] [Google Scholar]
- Bainbridge, JW, Smith, AJ, Barker, SS, Robbie, S, Henderson, R, Balaggan, K et al. (2008). Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med 358: 2231–2239. [DOI] [PubMed] [Google Scholar]
- Nathwani, AC, Tuddenham, EG, Rangarajan, S, Rosales, C, McIntosh, J, Linch, DC et al. (2011). Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 365: 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup, M, Greenberg, B, Mancini, D, Cappola, T, Pauly, DF, Jaski, B et al.; Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) Investigators. (2011). Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation 124: 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWitt, PA, Rezai, AR, Leehey, MA, Ojemann, SG, Flaherty, AW, Eskandar, EN et al. (2011). AAV2-GAD gene therapy for advanced Parkinson's disease: a double-blind, sham-surgery controlled, randomised trial. Lancet Neurol 10: 309–319. [DOI] [PubMed] [Google Scholar]
- Tonegawa, S, Nakazawa, K and Wilson, MA (2003). Genetic neuroscience of mammalian learning and memory. Philos Trans R Soc Lond B Biol Sci 358: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, JP, Cain, CK, Ostroff, LE and LeDoux, JE (2011). Molecular mechanisms of fear learning and memory. Cell 147: 509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noori, HR, Spanagel, R and Hansson, AC (2012). Neurocircuitry for modeling drug effects. Addict Biol 17: 827–864. [DOI] [PubMed] [Google Scholar]
- Duval, ER, Javanbakht, A and Liberzon, I (2015). Neural circuits in anxiety and stress disorders: a focused review. Ther Clin Risk Manag 11: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzung, C, Turiault, M and Griebel, G (2014). Optogenetics to study the circuits of fear- and depression-like behaviors: a critical analysis. Pharmacol Biochem Behav 122: 144–157. [DOI] [PubMed] [Google Scholar]
- Kim, SK, Eto, K and Nabekura, J (2012). Synaptic structure and function in the mouse somatosensory cortex during chronic pain: in vivo two-photon imaging. Neural Plast 2012: 640259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, KM, Nadon, NL, Morrison, JH, Thibault, O, Barnes, CA and Blalock, EM (2006). The neurobiology of aging. Epilepsy Res 68 Suppl 1: S5–20. [DOI] [PubMed] [Google Scholar]
- Sprengel, R and Hasan, MT (2007). Tetracycline-controlled genetic switches. Handbook of Experimental Pharmacology 178:49–72. [DOI] [PubMed] [Google Scholar]
- Heindorf, M and Hasan, MT (2015). Fluorescent calcium indicator protein expression in the mouse brain using recombinant adeno-associated viruses. Cold Spring Harb Protoc 2015: 697–709. [DOI] [PubMed]
- Gossen, M and Bujard, H (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89: 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger, S, Baron, U, Thellmann, M, Hasan, MT, Bujard, H and Hillen, W (2000). Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA 97: 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff, S, Köster, M, Wirth, M, Schaper, F, Gossen, M, Bujard, H et al. (1995). Identification of mammalian cell clones exhibiting highly regulated expression from inducible promoters. Trends Genet 11: 219–220. [DOI] [PubMed] [Google Scholar]
- Baron, U, Freundlieb, S, Gossen, M and Bujard, H (1995). Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res 23: 3605–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcker, R, Estler, CJ, Maywald, M and Weber, D (1981). Comparison of distribution of doxycycline in mice after oral and intravenous application measured by a high-performance liquid chromatographic method. Arzneimittelforschung 31: 2116–2117. [PubMed] [Google Scholar]
- Hasan, MT, Schönig, K, Berger, S, Graewe, W and Bujard, H (2001). Long-term, noninvasive imaging of regulated gene expression in living mice. Genesis 29: 116–122. [DOI] [PubMed] [Google Scholar]
- Shaner, NC, Steinbach, PA and Tsien, RY (2005). A guide to choosing fluorescent proteins. Nat Methods 2: 905–909. [DOI] [PubMed] [Google Scholar]
- Hatfield, L and Hearing, P (1991). Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology 184: 265–276. [DOI] [PubMed] [Google Scholar]
- Kistner, A, Gossen, M, Zimmermann, F, Jerecic, J, Ullmer, C, Lübbert, H et al. (1996). Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA 93: 10933–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, FM, Kringstein, AM, Spicher, A, Guicherit, OM and Blau, HM (2000). Transcriptional control: rheostat converted to on/off switch. Mol Cell 6: 723–728. [DOI] [PubMed] [Google Scholar]
- Manfredsson, FP, Burger, C, Rising, AC, Zuobi-Hasona, K, Sullivan, LF, Lewin, AS et al. (2009). Tight long-term dynamic doxycycline responsive nigrostriatal GDNF using a single rAAV vector. Mol Ther 17: 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, Y, Yamazaki, T, Nishii, S, Noguchi, T, Hoshino, H, Niwa, K et al. (2010). Enhanced beetle luciferase for high-resolution bioluminescence imaging. PloS One 5: e10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, P, Aller, MI, Baron, U, Cambridge, S, Bausen, M, Herb, J et al. (2007). Silencing and un-silencing of tetracycline-controlled genes in neurons. PLoS One 2: e533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C and Okayama, H (1987). High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7: 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini, L, Buchs, PA and Muller, D (1991). A simple method for organotypic cultures of nervous tissue. J Neurosci Methods 37: 173–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.