Abstract
Gene therapy holds considerable promise for the functional cure of HIV-1 infection and, in this context, RNA interference (RNAi)-based approaches represent powerful strategies. Stable expression of small interfering RNAs (siRNAs) targeting HIV genes or cellular cofactors has the potential to render HIV-1 susceptible cells resistant to infection. To inhibit different steps of virus life cycle, self-inactivating lentiviral vectors expressing multiple siRNAs targeting the CCR5 cellular gene as well as vif and tat/rev viral transcripts, under the control of different RNA polymerase III promoters (U6, 7SK, H1) were developed. The use of a single RNA polymerase III promoter driving the expression of a sequence giving rise to three siRNAs directed against the selected targets (e-shRNA) was also investigated. Luciferase assay and inhibition of HIV-1 replication in human Jurkat T-cell line were adopted to select the best combination of promoter/siRNA. The efficacy of selected developed combinatorial vectors in interfering with viral replication was evaluated in human primary CD4+ T lymphocytes. We identified two effective anti-HIV combinatorial vectors that conferred protection against R5- and X4- tropic viruses. Overall, our results showed that the antiviral effect is influenced by different factors, including the promoter used to express the RNAi molecules and the selected cassette combination. These findings contribute to gain further insights in the design of RNAi-based gene therapy approaches against HIV-1 for clinical application.
Introduction
Despite its success in controlling HIV-1 infection and disease progression, antiretroviral drug therapy requires a life-long commitment and it is still associated with considerable comorbidities.1,2,3,4 Thus, the development of strategies to completely eradicate or to control HIV infection without daily drug intake is a priority.1,2,5,6,7,8,9,10 The report of the “Berlin patient”, cured of HIV following hematopoietic stem cell transplant from an individual homozygous for the ▵32 CCR5 deletion, has raised hope in the field.11,12 However, due to the limited chance of finding matching ▵32 CCR5 donors and the high risk associated with allogeneic stem cell transplantation, recapitulating this clinical success on a large scale appears to be difficult.13,14 In this context, gene therapy (GT) represents a viable option, offering the possibility to artificially generate ▵CCR5 cells. Different GT strategies to edit the CCR5 gene or transcript have been tested,15,16,17 including the use of the CRISPR/Cas9 system18,19,20,21 and the intracellular delivery of transcription activator-like effectors nuclease,22,23,24 or zinc finger nucleases.25,26,27,28,29,30 The latter approach is currently being clinically tested (NIH clinical trial NCT01543152). Blocking viral entry has the advantage of leading to the accumulation of uninfected gene-protected cells, thus preventing the continued replenishment of viral reservoirs.13,16,31 However, to increase the potency of gene therapy approaches and to accomplish long-term control of HIV-1 replication, multiple genetic inhibitors interfering with different steps of viral replication should be simultaneously delivered into target cells.31,32,33,34 The multiple targeting GT strategy mimics the antiretroviral drug therapy approach, which combines different drugs in order to decrease the chance of viral escape. Several genetic HIV inhibitors have been developed and tested over the years.31,32,35,36,37,38 Among these, small interfering RNAs (siRNAs) are less immunogenic than protein-based agents and represent the most potent inhibitory effectors, according to preclinical studies.39 Indeed, siRNAs, that trigger homology-dependent, post-transcriptional gene silencing of their targets, have been used to silence not only CCR5,40,41,42,43,44 but also virtually all the HIV-encoded RNAs.45,46,47,48,49,50,51,52,53 Moreover, the expression of multiple anti-HIV siRNAs by means of self-inactivating (SIN) lentiviral vectors has been proven to be effective and safe,53,54,55,56,57 as shown also in humanized mouse models35,58,59 and in patients.60,61 Despite these achievements, further optimization of these GT approaches is required, starting from the selection of new therapeutic targets and the design of innovative genetic platforms.61,62 Furthermore, the need of expressing the therapeutic genes not only efficiently but also as long as it is required for a life-long effect are crucial aspects that have not been entirely addressed so far.
In this study, we investigated the possibility to combine into the same SIN lentiviral vector shRNAs simultaneously targeting CCR5 along with three different viral factors (Tat, Rev, and Vif) with essential roles in different phases of HIV-1 replication/pathogenesis. We analyzed: (i) the effect of different pol-III promoters (H1, U6, and 7SK) on the shRNAs silencing activity, in order to identify the potentially more active combinations; (ii) the possibility to combine two shRNAs with a long harpin RNA (lhRNA) within the same vector, in order to obtain a platform simultaneously encoding four therapeutic molecules; (iii) the efficacy of the extended (e)-shRNA strategy to target the selected cellular/viral transcripts. The e-shRNA strategy is based on the design of an hairpin, optimized in length and sequence, that produces three different and active siRNAs under the transcriptional control of a single pol-III promoter, without inducing the interferon response;63 (iv) the silencing/antiviral activity of the developed vectors in different experimental settings. Our results brought to light different aspects relevant for the design of lentiviral vectors expressing multiple anti-HIV-1 siRNAs. Importantly, we also developed new effective combinatorial strategies that provided protection against HIV-1 in human primary CD4+ T lymphocytes and that deserve further investigations/improvements.
Results
U6 and H1 represent the best promoters driving the expression of different anti-HIV-1 shRNAs within a single transcriptional unit
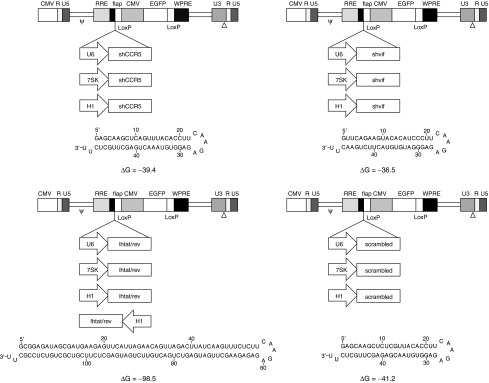
With the aim to combine into the same SIN lentiviral vector shRNAs affecting multiple essential steps of HIV-1 replication/pathogenesis, we selected the cellular CCR5 along with the viral tat, rev, and vif as target transcripts. We took advantage of shRNA sequences already tested for their silencing/antiviral efficacy in different experimental settings. Specifically, in the case of the tat/rev transcripts, in order to increase the efficacy of the strategy, we selected a lhRNA which gives rise, once introduced into the cells, to two different siRNAs targeting the region common to the two viral transcripts.64 In this way, four siRNAs targeting one cellular and three viral genes should be produced. The predicted folding and nucleotide sequence of the selected shRNAs are reported in Figure 1 and in Supplementary Table S1, respectively.
Figure 1.
Schematic representation of vectors expressing single anti-HIV-1 hairpin RNAs. Third-generation self-inactivating lentiviral vector backbone pLL3.7 that was used to derive the antiviral constructs. The selected shRNA was inserted upstream of the CMV-EGFP cassette under the control of either the U6, the 7SK, or the H1 human pol-III promoter. Arrows indicate the transcriptional orientation of the RNAi trigger cassette. Each hairpin molecule encompasses complementary passenger and guide strands separated by a 9 nt loop, followed by the pol-III termination signal (poly-T).82 The predicted folding of each shRNA is shown, as calculated by the mfold web server (version 3.5)83 with default parameters at 37 °C. The thermodynamic stability (▵G) is also reported in Kcal/mol. The shCCR5 and the shvif molecules give rise to a single siRNA against the cellular CCR5 and the HIV-1 vif transcript, respectively. The lhtat/rev leads to the generation of two siRNA both targeting the first overlapping exon of the tat and rev viral genes. The shRNA control encodes for a scrambled sequence.
We firstly analyzed the silencing activity of each hairpin molecule when expressed under the control of different human pol-III promoters by luciferase assay in 293T cells. The therapeutic cassette was cloned into the vector upstream and in the same orientation of the CMV-EGFP reporter gene cassette (Figure 1). Each vector was transfected into 293T cells along with a reporter plasmid encoding a luciferase gene fused to the respective RNAi target sequence (Supplementary Figure S1). Measurement of luciferase activity provided a readout of vector silencing efficacy, with the most effective vectors resulting in a higher suppression of luciferase activity. The H1 promoter was overall the best performing promoter (Figure 2a–c) and the only one leading to a noteworthy suppression in the case of the vif-specific shRNA (40.8 ± 1.7% of inhibition) (Figure 2b). To assess the impact of orientation on shRNA efficacy, the H1lhtat/rev cassette was also inserted in the opposite orientation with no significant impact on the silencing activity (Figure 2c). To further confirm these data, we evaluated the activity of the shCCR5 by investigating the knockdown of endogenous CCR5 in human primary macrophages. CCR5 downregulation was observed when cells were transduced with the shCCR5 vectors, being the U6 and the H1 the best performing promoters, in agreement with the luciferase assay (Donor 1, Figure 2d and Supplementary Figure S2). These data were confirmed with Donor 2-derived macrophages.
Figure 2.
Silencing activity of the different single shRNA-expressing vectors. (a–c) The shCCR5-, the shvif-, or the lhtat/rev-expressing vector containing the indicated promoter was cotransfected along with its respective reporter plasmid into 293T cells and luciferase activities were measured 48 hours later. To correct for transfection efficiency, relative luciferase activities were calculated from the ratio between Renilla and background firefly luciferase activities. The indicated percentages represent the relative luciferase activity calculated by setting at 100% the value obtained from cells transfected with the corresponding scrambled vector. Black bars refer to vectors harboring the RNAi trigger cassette in the same orientation as the CMV-EGFP reporter cassette, while the gray bar in panel (c) refers to the vector containing the H1lhtat/rev cassette in the opposite orientation. The mean and standard deviation from three replicated experiments are presented (*P < 0.05; **P < 0.01; ***P < 0.001; t-test, relative to the corresponding scrambled). (d) Monocyte-derived macrophages were transduced twice with vectors expressing the shCCR5 or the scrambled sequence under the control of the U6, the 7SK, or the H1 promoter. The cells were harvested 3 days after the second transduction and analyzed by FACS with anti-human CCR5 antibody staining. The indicated percentages represent the reduction of CCR5-positive cells calculated with respect to the values obtained for the scrambled vector-transduced cells. Results from two representative Donors (1 and 2) are shown.
Design and evaluation of different combinatorial anti-HIV-1 vectors
As a next step, to simultaneously express the selected shRNAs, each therapeutic molecule driven by the most efficient promoter, as resulted with the luciferase knockdown assay, was combined generating the U6shCCR5-H1lhtat/rev-H1shvif vector. Moreover, additional combinatorial vectors were designed. In the U6shCCR5-H1lhtat/rev-7SKshvif vector, the shCCR5 and the lhtat/rev were maintained under the control of the U6 and the H1 promoters, respectively, while the 7SK promoter was adopted to express the vif-specific shRNA. In the U6shCCR5-7SKshvif-H1lhtat/rev vector, the H1lhtat/rev and the 7SKshvif cassettes were swapped, as compared to the above-described vector, in order to assess the impact of cassette position on vector antiviral activity. Finally, the 7SKshCCR5-U6lhtat/rev-H1shvif vector was also generated (Figure 3a).
Figure 3.
Inhibition of reporter gene expression by combination of multiple anti-HIV-1 shRNAs. (a) The triple cassette vectors express the shCCR5, the shvif and the lhtat/rev as independent transcriptional units, according to different combinations of promoter-RNAi trigger. Each vector encoding the triple combination of antiviral cassettes driven by the U6, the 7SK, or the H1 promoter was cotransfected along with one (b) (CCR5, vif, or tat/rev) or all (c) (CCR5 + vif + tat/rev) luciferase reporter plasmids into 293T cells and luciferase activities were measured 48 hours later. The indicated percentages refer to the relative luciferase activity calculated as reported above. The average values from three independent experiments, with standard deviations, are given (*P < 0.05; **P < 0.01; ***P < 0.001; t-test, relative to the corresponding scrambled control depending on the promoter driving the shRNAs).
The efficacy of the developed vectors was evaluated by luciferase assay. The results show that the CCR5-, the vif- and the tat/rev-specific shRNAs, when expressed in the context of a triple cassette vector, maintained the expected silencing activity in all the tested conditions, irrespectively of either the employed promoter or the cassette position (Figure 3b,c).
To further exploit combinatorial anti-HIV-1 vectors, we developed an e-shRNA that allows the expression of three siRNAs directed against the selected transcripts under the control of a single pol-III promoter. The hairpin stem length is a critical parameter for proper processing and optimal activity of the siRNAs produced by the e-shRNA. Moreover, G:U wobble pairings were inserted at regular intervals in the sense strand of the e-shRNA, to attenuate the innate immune response to long dsRNAs63 (Figure 4a). In the luciferase assay, under all the tested conditions, silencing activity was achieved only when the e-shRNA was expressed by the H1 promoter (Figure 4b,c).
Figure 4.
Inhibition of reporter gene expression by anti-HIV-1 e-shRNAs. (a) Schematic representation of lentiviral vectors expressing the e-shRNA is displayed. The e-shRNA produces three siRNAs against the CCR5, the tat/rev and the vif transcripts, from the stem base to the loop region of the hairpin, respectively, under the control of a single promoter (U6, 7SK, H1). The predicted folding of e-shRNA is shown, as calculated by the mfold web server (version 3.5)83 with default parameters at 37 °C. The thermodynamic stability (▵G) is also reported in Kcal/mol. G:U pairings are indicated with a black arrowhead. Each vector encoding the e-shRNAs driven by the U6, the 7SK, or the H1 promoter was cotransfected along with one (b) (CCR5, vif or tat/rev#) or all (c) (CCR5 + vif + tat/rev#) luciferase reporter plasmids into 293T cells and luciferase activities were measured 48 hours later. The indicated percentages refer to the relative luciferase activity calculated as reported above. The average values from three independent experiments, with standard deviations, are given (*P < 0.05; **P < 0.01; ***P < 0.001; t-test, relative to the corresponding scrambled control depending on the promoter driving the shRNAs); tat/rev#: luciferase reporter plasmid containing the target sequence of the siRNA against the tat/rev gene produced by the e-shRNA.
Inhibition of HIV-1 replication in CD4+ T lymphoblastoid cells and in human primary CD4+ T cells transduced with the combinatorial antiviral vectors
To address the antiviral activity of the combinatorial vectors, CD4+ T lymphoblastoid Jurkat cells were transduced with recombinant lentiviral particles. Vector titers typically ranged from 106 to 107 transducing units (TU)/ml (Supplementary Figure S3a) and a multiplicity of infection (MOI) of 1 TU/cell was used, obtaining the % of enhanced green fluorescent protein-positive (EGFP+) Jurkat cells reported in Supplementary Figure S3b,c. Once assessed that cells could be efficiently transduced without effect on cell viability (data not shown), transduced Jurkat cells were infected with the HIV-1 HXBc2 Vpr+/Vpu+/Nef+ strain (MOI = 0.1 TCID50/cell). Viral inhibition was assessed by measuring reverse transcriptase (RT) activity in the culture supernatants at different time points postinfection. Effective reduction of HIV-1 replication was achieved with both the U6shCCR5-7SKshvif-H1lhtat/rev and the U6shCCR5-H1lhtat/rev-7SKshvif triple cassette vectors (Figure 5a). The former vector almost completely suppressed viral replication up to 10 days after infection, while the latter determined a 10-fold decrease in virus production. On the contrary, both the U6shCCR5-H1lhtat/rev-H1shvif and the 7SKshCCR5-U6lhtat/rev-H1shvif vectors did not confer protection against HIV-1 infection. Among the e-shRNA-encoding vectors, the H1-driven hairpin provided strong inhibition of viral replication up to 10 days postinfection. According to the results obtained with the luciferase assay, neither the U6- nor the 7SK-driven e-shRNA displayed antiviral activity (Figure 5b). Thus, U6shCCR5-7SKshvif-H1lhtat/rev and the H1e-shRNA were identified as the most effective anti-HIV-1 combinatorial vectors among the ones developed. Therefore, the antiviral activity of these vectors was investigated in a more physiologically relevant setting. To this end, human primary CD4+ T lymphocytes were purified from buffy coats of three healthy donors and transduced with the selected vectors. The transduction efficiency measured as percentage of EGFP-positive cells ranged from 15 to 30% (Supplementary Figure S4). To obtain an almost pure population of transduced cells, 4 days after transduction, lymphocytes were fluorescence-activated cell sorting (FACS) sorted on the basis of EGFP expression. Twenty-four hours later, once assessed that the expression of multiple siRNAs did not affect cell viability as compared to control cells (data not shown), 2.5 × 106 (Donor 3) and 1 × 106 (Donors 4 and 5) cells were challenged with equivalent RT units (10,000 cpm) of either the CXCR4 coreceptor-using HIV-1 HXBc2 Vpr+/Vpu+/Nef+ or the CCR5 coreceptor-using HIV-1 NL4-3-ADA. Infected cells were maintained in 10% fetal bovine serum (FBS) Roswell Park Memorial Institute (RPMI) containing IL-2. The RT activity in culture supernatants was measured at different time points after infection.
Figure 5.
Inhibition of HIV-1 replication in Jurkat T cells expressing multiple siRNAs against the CCR5, the vif and the tat/rev targets. (a,b) Jurkat T cells transduced with the triple cassette vectors, the e-shRNA expressing vectors or the control empty and scrambled vectors were infected with the HIV-1 HXBc2 Vpr+/Vpu+/Nef+ R4-tropic molecular clone (multiplicity of infection = 0.1 TCID50/cell). Culture supernatants were collected at the indicated time points and assayed for RT activity. The reported results have been replicated in at least three independent experiments. The scrambled control plotted in the graphs represents the U6-scrambled vector. Comparable results were obtained with the 7SK- and the H1-scrambled vectors (data not shown).
In the case of Donor 3, both the U6shCCR5-7SKshvif-H1lhtat/rev and the H1e-shRNA vector induced a striking inhibition of viral replication up to 12 days postinfection, when challenged with both HIV-1 strains (Figure 6). Inhibition was also detected in the case of Donors 4 and 5, although to a less extent, likely reflecting the effect of the different cellular density employed compared to the one adopted for Donor 3. Moreover, in the case of the R5 virus, especially for Donor 4, the triple cassette vector displayed, overall, the most potent antiviral effect (Figure 6).
Figure 6.
Inhibition of HIV-1 replication in human primary CD4+ T lymphocytes transduced with the combinatorial vectors. CD4+ T cells were transduced with either the empty vector, the H1e-shRNA or the U6shCCR5-7SKshvif-H1lhtat/rev vector. Four days post-transduction, enhanced green fluorescent protein (EGFP)-expressing cells were sorted by FACS and infected with the CXCR4 coreceptor-using HIV-1 HXBc2 Vpr+/Vpu+/Nef+ or the CCR5 coreceptor-using HIV-1 NL4-3-ADA. Culture supernatants were harvested at the indicated time points postinfection and tested for reverse transcriptase (RT) activity. The indicated percentages represent the reduction of the relative RT activity calculated with respect to the value obtained for the empty vector-EGFP-positive selected-transduced cells.
Discussion
Supported by the success of the “Berlin patient”, over the last years, several gene therapy approaches have been developed to treat HIV-1 infection. As for chemotherapy regimens used in the current clinical practice, a combination of multiple antiviral reagents should be adopted. The strategies adopted so far, despite encouraging results, suffer of different problems that should be further addressed.
Here, we generated different combinatorial vectors based on a self-inactivating HIV-1 platform expressing multiple shRNAs targeting both cellular and viral transcripts. Combinatorial RNAi (co-RNAi) can be achieved by inserting multiple pol-III promoter/shRNA cassettes within the same vector45 or by expressing a single sequence that, once introduced into the cells, gives rise to more than one siRNA (e-shRNA).63 In this study, we directly compared both these strategies. Specifically, we combined in a single construct shRNAs targeting different steps of HIV-1 life cycle (entry, transcription, nuclear export of viral RNAs, and production of infectious particles). In particular, CCR5, tat, rev, and vif were selected as target transcripts. Of note, taking into account that the selected tat/rev target sequence is also present in additional viral transcripts (unspliced and single-spliced), additional silencing activity cannot be excluded. Furthermore, vectors were designed to minimize viral escape not only by targeting multiple genes at the same time, but also by (i) employing an shRNA active against a cellular gene, CCR5, which is characterized by a low mutation rate and (ii) selecting viral target sequences which have been already shown to be highly conserved among different clades.64,65 Indeed, since the aim of our study was to compare the antiviral efficacy of different co-RNAi strategies, we selected shRNAs that were already described for their antiviral activity along with well characterized pol-III promoters (i.e., U6, H1, and 7SK). Moreover, we also combined for the first time two shRNAs (CCR5- and vif- specific) with an lhRNA (tat/rev) obtaining lentiviral vectors simultaneously expressing four siRNAs.
Firstly, we analyzed the impact of the pol-III promoter on the shRNA silencing activity. We show that, when the therapeutic RNA is targeting a viral transcript, the choice of the promoter might be relevant, while its orientation does not seem to have a major impact. Of note, we cannot exclude, at the moment, that the cassette orientation might have an impact on the titer of the recombinant particles, an aspect that is worth to be further investigated. In particular, the H1 is the best promoter in the case of the tat/rev-specific lhRNA and the only one leading to a noteworthy silencing of the vif transcript. It is worth mentioning that a 40.8 ± 1.7% of inhibition in the luciferase assay is not expected to necessarily correlate with a lack of biological effect of the selected vif-specific shRNA especially in vivo where Vif plays a crucial role.66,67,68,69,70 Finally, the CCR5-specific shRNA showed a potent silencing activity independently from the pol-III promoter employed.
Next, we tested the impact of the combinatorial vector design on antiviral activity. Since our first aim was to investigate whether a platform expressing multiple siRNAs under the transcriptional control of different pol-III promoters within a SIN vector was stable and efficient in transducing HIV-1 target cells, we selected Jurkat lymphoblastoid T cells as experimental system. This cell line, being target of CXCR4-using HIV-1 strains, offered us the possibility to identify, among the generated vectors, the ones able to efficiently reduce viral replication despite the effect of the potent shCCR5. This aspect was crucial in order to select the best shRNA-based strategy active on viruses regardless their tropism, as, given the essential role of CXCR4 in vivo, only CCR5 can be target of silencing. All the developed combinatorial vectors were able to transduce Jurkat cells, as analyzed in terms of EGFP-positive cells. Specifically, the U6shCCR5-H1lhtat/rev-H1shvif vector was expected to be the most potent with respect to antiviral activity, being characterized by the combination of the different shRNAs cloned under the transcriptional control of the best performing promoter. Even though efficient in silencing all the target transcripts in the luciferase assay, when the U6shCCR5-H1lhtat/rev-H1shvif vector was adopted to transduce Jurkat T cells, HIV-1 replication was not inhibited. This result could be explained by the presence of two H1 promoters within the vector that might lead to recombination events upon transduction, with deletions of one or more therapeutic cassettes, as already reported.57,71 On the other hand, a lentiviral vector expressing multiple shRNAs under the control of repeated pol-III promoters conferred strong resistance to HIV-1 infection in transduced CD34+ progenitor cells65 and it has been adopted in early-phase clinical trial.60 Thus, we cannot exclude that the inserted pol-III/shRNA combination per se, independently from the presence of repeated H1 sequences, might influence vector stability with detrimental effects on the vector antiviral activity. In agreement with this hypothesis, also the 7SKshCCR5-U6lhtat/rev-H1shvif vector, which lacks shRNA expression from repeated promoters, did not protect Jurkat cells from HIV-1 challenge. Of note, the cassette array within this vector, that represents the best combination of pol-III/shRNA in terms of silencing activity without promoter repetition, is the only one with the 7SK positioned as first. Interestingly, both U6shCCR5-H1lhtat/rev-H1shvif and 7SKshCCR5-U6lhtat/rev-h1shvif were consistently characterized by a titer at least 1 log lower than the one obtained for the U6shCCR5-H1lhtat/rev-7SKshvif and U6shCCR5-7SKshvif-H1lhtat/rev vectors. Furthermore, even though target cells were transduced with the same number of TU, the percentages of EGFP-positive cells obtained with U6shCCR5-H1lhtat/rev-H1shvif and 7SKshCCR5-U6lhtat/rev-H1shvif were constantly lower than the ones obtained in the case of the U6shCCR5-H1lhtat/rev-7SKshvif and U6shCCR5-7SKshvif-H1lhtat/rev vectors (Supplementary Figure S3). On the other hand, these two latest vectors efficiently controlled viral replication up to 10 days postinfection in HIV-1 challenged Jurkat cells. Interestingly, in both cases, the lhtat/rev was under the transcriptional control of the H1 promoter. Since in the adopted experimental setting the silencing activity of CCR5 and vif transcripts cannot be appreciated and taking into account that in the luciferase assay the H1lhtat/rev silenced its targets regardless its position with respect to the 7SKshvif cassette, this result also suggests that the U6shCCR5-7SKshvif-H1lhtat/rev and the U6shCCR5-H1lhtat/rev-7SKshvif are the more stable cassette arrays conferring to the vectors the best performance upon transduction. Furthermore, the pol-III/shRNA combination more than the position of the single cassette with respect to each other seems to be the critical aspect. Indeed, the swap between the 7SKshvif and the H1lhtat/rev does not have a major impact on their antiviral activity.
In the case of the e-shRNA-expressing vectors, the sequences generating the siRNAs were positioned across the span of the duplex with the siRNA against CCR5 as first, followed by the siRNAs targeting the tat/rev and the vif transcripts. The results obtained with both the luciferase assay and the viral challenge of transduced Jurkat T cells indicated that H1 was the only effective promoter in driving the production of active siRNAs. Overall, our data clearly indicate that, in the case of the e-shRNA, the silencing activity is strongly influenced by the selected pol-III promoter. To our knowledge, this is the first report directly correlating the impact of different promoters on e-shRNA efficacy. Furthermore, by comparing the results obtained with the two co-RNAi strategies, we can conclude that, in the case of multiple cassette constructs, the design of the vector both in terms of promoter/shRNA cassette selection and its positioning has an impact on its performance upon transduction and thus cannot rely only on luciferase assay. By contrast, in the case of the e-shRNA-expressing vectors, the pol-III promoter appears to be the critical aspect and the luciferase assay might be sufficient to select the best performing platform.
The most promising triple and e-shRNA vectors were further validated in human primary CD4+ T lymphocytes challenged with the CXCR4 coreceptor-using HIV-1 HXBc2 Vpr+/Vpu+/Nef+ and the CCR5 coreceptor-using HIV-1 NL4-3-ADA. Consistent with previous data obtained with the lymphoblastoid T cell line, the triple vector showed on average a higher HIV-1 inhibition than the e-shRNA vector over the 12-day time course of infection. This is not surprising also considering that the multiple-cassette vector encodes four siRNAs, two of which active against tat/rev. Overall, no significant differences in terms of inhibition could be appreciated between the CXCR4 and CCR5 coreceptor-using HIV-1 strains. This result might be simply explained by the fact that also in the primary cells we employed, the major contribution in terms of antiviral activity is likely due to the silencing of viral transcripts targeted by the tat/rev-specific siRNAs. The contribution of CCR5- and vif-specific shRNAs is currently under investigation.
The first lentiviral vector expressing a triple combination of shRNAs targeting viral transcripts is approaching clinical application.58 Our study on the one hand demonstrates that it is feasible to further increase the genetic barrier to viral resistance by including in a single vector sequences expressing four siRNAs targeting one essential cellular and three crucial viral genes. On the other hand, it highlights some important strengths and pitfalls of different platforms/tools used for the design and testing of multiple shRNAs delivery systems, providing valuable insights for the development of an improved reliable combinatorial RNAi-based approach against HIV-1.
Materials and methods
Cell lines and cell cultures. Human embryonic kidney 293T cells were grown in Dulbecco's modified Eagle's medium (Gibco, Life Technologies, Monza, Italy) supplemented with 10% heat-inactivated FBS (Gibco, Life Technologies). Human T lymphoblastoid Jurkat cells (Clone E6-1) were maintained in Roswell Park Memorial Institute's 1640 medium (RPMI) (Gibco, Life Technologies) supplemented with 10% FBS. Human primary monocytes were isolated by plastic adherence from buffy coats of healthy blood donors after Ficoll-Paque PLUS (GE Healthcare, Milan, Italy) purification. Monocytes were cultured in RPMI containing 10% FBS and macrophage colony-stimulating factor (M-CSF) (500 U/ml) (Miltenyi Biotec, Calderara di Reno, Bologna, Italy) for 7 days to differentiate into macrophages. Preparation purity was evaluated by measuring the percentage of CD14+ cells through FACS analysis. The cut-off employed to accept the purity of monocyte-derived macrophage preparation was a CD14+ percentage higher than 90%. Human primary CD4+ T lymphocytes were purified from buffy coats by Rosette Sep (StemCell Technologies, Peschiera Borromeo, Milan, Italy), according to the manufacturer's instructions. The purity of the CD4+ T cell population ranged from 95 to 100%, as estimated by FACS analysis, as follows. 5 × 105 cells were stained with monoclonal antibodies to human CCR5 (APC Mouse Anti-Human CD195, BD Biosciences, Milan, Italy), CD4 (PE-Cy7 mouse anti-human CD4, BD Biosciences), CD14 (CD14-PE, human, Miltenyi Biotec), CD8 (anti-human CD8a APC, eBioscience, Prodotti Gianni, Milan, Italy) or CD19 (CD19-FITC, human, Miltenyi Biotec), according to the manufacturer's instructions. The cells were also stained with isotype controls for each of the specific antibodies, as well as with the 7-Amino-Actinomycin D (7-AAD) viability dye (BD Biosciences). CCR5 expression level from two representative donors is reported in Supplementary Figure S5. When required, cells were incubated with FcR blocking reagent (Miltenyi Biotec) before staining. Samples were acquired on a LSRII (BD Biosciences) instrument and the data analysis was performed with FlowJo (Tree Star, Ashland, OR) software. CD4+ T cells were cultured in RPMI medium supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml) and stimulated with phytohemagglutinin (10 µg/ml) (Sigma-Aldrich, Milan, Italy) for 2 days. Cells were subsequently cultured without phytohemagglutinin.
Construction of multifunctional self-inactivating lentiviral vectors. The human U6 promoter was amplified from 293T genomic DNA with primers 5′-AAGGTCGGGCAGGAAGAGGGCCTA-3′ and 5′-GCACGGTGTTTCGTCCTTTCCACA-3′ (GenBank: X07425.1, nt 1–269). The human 7SK promoter was amplified from HeLa genomic DNA with primers 5′-CTGCAGTATTTAGCATGCCCCACC-3′ and 5′-CCGAGGTACCCAGGCGGCGCACAA-3′ (GenBank: X05490.1, nt 1–246). The EcoRI and MluI sites were included at the 5′ and 3′ end, respectively, of both promoters. The human H1 promoter was derived from the pLVTHM vector,72 after digestion with EcoRI and MluI.
To obtain the shCCR5,42 the shvif47 and the scrambled sequence,42 two complementary DNA oligonucleotides flanked by MluI and ClaI sites were synthesized, annealed, and inserted at the EcoRV site of the pBluescript II KS plasmid (Stratagene, Agilent Technologies, Cernusco sul Naviglio Milan, Italy).
To generate the lhtat/rev64 and the e-shRNA sequences, a two-step polymerase chain reaction (PCR) approach was used, as described elsewhere.73 Briefly, the first PCR reaction was performed using a plasmid containing the human U6 promoter and 153 nt of the downstream snU6 RNA gene as template. The U6 forward primer described above and a reverse primer complementary to the last 14 nt of the snU6 RNA gene, followed by sequences complementary to the sense strand and the 9 nt loop of the hairpin were employed (lhtat/rev reverse primer 1: 5′-TCTCTTGAAGAGAAACTTGATAAGTCTAACTGTTCTAATGAACTCTTCATCGCTATCTCCGCACGCGTAAACAGAAAAACAA-3′; e-shRNA reverse primer 1: 5′-TCTCTTGAAGGGATGTATACTTCTAAACATACTCCACTTCTTCCTACCATGTGGGTATAAACTAAGCTTACTCACGCGTAAACAGAAAAACAA-3′). An MluI site (underlined) was inserted between the end of the snU6 RNA gene and the first nucleotide of the hairpin, to facilitate subsequent cloning. The PCR reaction was carried out as follows: 1 minute at 94 °C, 1 minute at 55 °C, and 1 minute at 72 °C for 30 cycles. A second PCR step was performed employing the same U6 forward primer and a reverse primer harboring sequences complementary to the 9 nt loop, followed by the antisense strand of the hairpin, the pol-III terminator sequence and the ClaI site (lhtat/rev reverse primer 2: 5′-ATCGATAAAAAGCGGAGACAGCGACGAAGAGCTCATCAGAACAGTCAGACTCATCAAGCTTCTCTCTCTTGAA-3′; e-shRNA reverse primer 2: 5′-ATCGATAAAAAGAGCAAGCTCAGTTTACACCCACATGGCAGGAAGAAGCGGAGTATGTTCAGAAGTACACATCCCTCTCTTGAAA-3′). The PCR to obtain the lhtat/rev was carried out as described above, while amplification to obtain the e-shRNA was performed as follows: 40 seconds at 98 °C, 1 minute at 55 °C, and 1 minute at 72 °C for 30 cycles. The resulting PCR products include the U6 promoter and a fragment of the downstream snU6 RNA gene, followed by the MluI site, the sense and the antisense hairpin sequences of either the lhtat/rev64 or the e-shRNA (5′-GAGTAAGCTTAGTTTATACCCACATGGTAGGAAGAAGTGGAGTATGTTTAGAAGTATACATCCCTTCAAGAGAGGGATGTGTACTTCTGAACATACTCCGCTTCTTCCTGCCATGTGGGTGTAAACTGAGCTTGCTCTTTTT-3′) separated by the 9 nt loop, the pol-III terminator signal and the ClaI site. These fragments were ligated into the EcoRV-linearized pBluescript II KS plasmid. Of note, in the e-shRNA, the siRNA against the tat and rev genes has a different target sequence with respect to the ones produced by the lhtat/rev. Indeed, considering that the stem region of the extended hairpin should not exceed 66 bp in length for effective production of multiple and functional siRNAs, the lhtat/rev guide strand sequence was replaced with a shorter one, generating one single siRNA that targets a distinct region of the tat/rev common transcript.63 The U6 promoter was then replaced by either the 7SK or the H1 promoter by enzymatic digestion. To obtain the siRNA-encoding cassettes, the U6, the 7SK, or the H1 promoter was inserted into the pBluescript II KS plasmid immediately upstream of the siRNA, between the EcoRI and the MluI sites.
The third-generation pLentiLox3.7 (pLL3.7) self-inactivating lentiviral vector,74 which contains an EGFP reporter gene and served as the control empty vector, was used to develop the anti-HIV-1 constructs. To obtain vectors expressing one single hairpin molecule, the shRNA, lhtat/rev or e-shRNA transcriptional unit was subcloned into the pLL3.7 plasmid between the XbaI and XhoI sites. Vectors expressing three hairpin molecules were constructed starting from the pBluescript II KS plasmid containing one single hairpin cassette. Initially, the plasmid encoding the first cassette was ClaI digested and protruding ends were filled-in by the Klenow fragment of the DNA polymerase. The linearized plasmid was SalI digested in order to allow the subsequent ligation to the second cassette, encompassed within a SmaI-SalI fragment derived from the relative pBluescript II KS plasmid. The resulting construct, encoding two out of the three hairpin cassettes, was linearized by SalI digestion and cut with XhoI, after treatment with the Klenow enzyme. Next, the third cassette was inserted with the XhoI-SmaI restriction sites. Finally, the fragment containing the triple cassette was excised with XbaI-XhoI and inserted into the pLL3.7 backbone.
Luciferase assay. Reporter plasmids were obtained, each encoding the shCCR5, the shvif or the lhtat/rev target sequence downstream of the Renilla luciferase open reading frame, followed by the firefly luciferase gene, to control for cell viability and transfection efficiency. Plasmids were constructed by directed insertion of the RNAi target region into the XhoI-NotI sites of the psiCheck2 plasmid (Promega, Milan, Italy), downstream of the Renilla luciferase gene (Supplementary Figure S1). Target fragments were obtained by annealing two complementary DNA oligonucleotides flanked by XhoI and NotI sites. Sequences of the forward oligonucleotide were as follows: shCCR5 target 5′-CAAGAGGCTCCCGAGCGAGCAAGCTCAGTTTACACCCGATCCACTGGGGAGCA-3′ (GenBank:X91492.1, nt 1224–1276); shvif target 5′-CCCTCATCCAAGAATAAGTTCAGAAGTACACATCCCACTAGGGGATGCTAGATTG-3′ (B.FR.83.HXB2, nt 5178–5232); lhtat/rev target 5′-GCGGAGACAGCGACGAAGAGCTCATCAGAACAGTCAGACTCATCAAGCTTCTC-3′ (B.FR.83.HXB2, nt 5983–6035); e-shRNA-derived tat/rev target 5′-CCTTAGGCATCTCCTATGGCAGGAAGAAGCGGAGACAGCGACGAAGAGCT-3′ (B.FR.83.HXB2, nt 5955–6004).
For luciferase assays, 293T cells were plated in 96-well plates at a density of 1 × 104 cells/well in 100 µl of Dulbecco's modified Eagle's medium 10% FBS. The next day, cells were cotransfected with 50 ng of the psiCheck2-derived plasmid(s) and 300 ng of the siRNA-expressing vector, using Lipofectamine 2000 reagent (Invitrogen, Life Technologies, Monza, Italy), as suggested by the manufacturer. Two days post-transfection, firefly and Renilla luciferase activities were assessed using the Dual Glo Luciferase Assay System (Promega), according to the manufacturer's instructions. Relative luciferase activities were calculated from the ratio between Renilla and background firefly luciferase activities. Relative activity for the control vector encoding the scrambled sequence was set to 100% and activities for the corresponding samples calculated accordingly.
Vector production and transduction of target cells. Vesicular stomatitis virus (VSV)-G pseudotyped vector stocks were produced by calcium phosphate transfection of 293T cells. Briefly, 2.5 × 106 cells were seeded on 10 cm Petri dishes and, when subconfluent, cotransfected with 15 µg of the appropriate gene transfer vector, 5 µg of pMDL, 3 µg of pCMV-Rev and 1.5 µg of pCMV-VSV-G (kindly provided by T. Friedman, University of California, San Diego, CA). The culture supernatants were collected on day 2 post-transfection, filtered with a 0.45-µm-pore-size membrane and stored at −80 °C until use. When required, vector particles in the supernatants were concentrated by ultracentrifugation (27,000 rpm, 2 hours, 4 °C). Infectious titer was determined by transducing 293T cells with serial dilutions of the lentiviral stocks, and 72 hours later EGFP expression was assessed by flow cytometry.
Monocyte-derived macrophages (1 × 106) were transduced with lentiviral vectors for 8 hours over 2 consecutive days at a MOI of 1 TU/cell in RPMI containing 10% FBS. After transduction, macrophages were maintained in culture medium supplemented with M-CSF (500 U/ml) for 3 days and analyzed by FACS for CCR5 surface expression. Jurkat cells (1 × 106) were incubated with vectors at an MOI of 1 TU/cell in RPMI 10% FBS. After 3 days of culture, the transduction efficiency was ascertained on the basis of EGFP expression. CD4+ T lymphocytes (1 × 106) were spin-infected with lentiviral vectors (1,200 rpm, 2 hours, 25 °C) at an MOI of 0.5 TU/cell in RPMI supplemented with 10% FBS and polybrene (8 µg/ml). After transduction, fresh culture medium containing penicillin (100 U/ml), streptomycin (100 µg/ml), and IL-2 (100 U/ml) (R&D Systems, Space Import-Export, Milan, Italy) was added to the cells. Four days post-transduction, homogeneous EGFP+ populations were obtained by flow cytometric sorting and used for the next experiments.
HIV-1 stock production and infection. HIV-1 HXBc2 Vpr+/Vpu+/Nef+ was produced by transfection of 5 × 106 Jurkat cells with 10 µg of the pSVC Vpr+/Vpu+/Nef+ construct (kindly provided by Heinrich Göttlinger, University of Massachusetts Medical School, Massachusetts) by the DEAE-dextran method, as previously described.75 This plasmid is a derivative of the pSVC21 construct,76 containing the HIV-1 HXBc2 molecular clone,77 where the vpr, vpu, and nef sequences were substituted with those derived from the pNL4-3 (vpr/vpu)78 and pLAI (nef)79 molecular clones, in order to introduce functional vpr, vpu, and nef genes, respectively. Jurkat cell supernatants were harvested at approximately 48 hours post-transfection, filtered (pore size, 0.45 µm) and stored at −80 °C until use. Viral titer was determined as 50% tissue culture infective doses (TCID50)/ml on C8166 cells by the Reed and Muench end point dilution method, as well as by measuring the RT activity assay, as previously described.80
HIV-1 NL4-3-ADA stocks were produced by calcium phosphate transfection of 2.5 × 106 293T cells with 15 µg of the infectious proviral plasmid, kindly provided by Heinrich Göttlinger. The pNL4-3-ADA plasmid is a derivative of the HIV-1 pNL4-3, where the env sequence was replaced by the CCR5 coreceptor-using HIV-1 ADA envelope.81 The virus was collected from the culture supernatants on day 2 post-transfection, filtered (pore size, 0.45 µm) and stored at −80 °C. The HIV-1 NL4-3-ADA viral titer was determined by the RT assay.
Jurkat cells were seeded in a 12-well plate at the density of 1 × 106/well, and infected with HXBc2 Vpr+/Vpu+/Nef+ at an MOI of 0.1 TCID50/cell 4 days post-transduction. After 1 hour of incubation at 37 °C, the cultures were washed three times and cultured in RPMI 10% FBS medium. Virus replication was monitored by RT activity in cell-free culture supernatants at different days postinfection.
Phytohemagglutinin-activated human CD4+ T lymphocytes were seeded in a 24-well plate at two different densities (2.5 × 106/well and 1 × 106/well) and were challenged with equivalent RT units (10,000 cpm) of either HXBc2 Vpr+/Vpu+/Nef+ or NL4-3-ADA, 24 hours after FACS sorting for EGFP expression. Infection was carried out as described above and cells were maintained in RPMI supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml), and IL-2 (100 U/ml). The RT activity in culture supernatants was measured at different time points after infection.
Statistical analysis. Paired t-test was performed using GraphPad Prism version 5.03 for Windows (GraphPad Software, San Diego, CA). Values of P less than or equal to 0.05, 0.01, and 0.001, were considered statistically significant (*, **, ***) compared to the respective control.
SUPPLEMENTARY MATERIAL Figure S1. Schematic representation of the reporter plasmids used for the luciferase assay. Figure S2. Reduction of CCR5 surface expression on human primary macrophages transduced with the shCCR5 vectors. Figure S3. Titer and transduction efficiency of the triple cassette vectors. Figure S4. Transduction efficiency of human primary CD4+ T lymphocytes transduced with the combinatorial vectors. Figure S5. CCR5 surface expression on human primary CD4+ T lymphocytes. Table S1. Sequences of the shRNAs cloned into the SIN lentiviral vector.
Acknowledgments
This work was supported by grants from ANRS (Agence Nationale de Recherche sur le Sida et les hépatites virales, Agreement n. 13018 to C.P. and n. 13043 to M.C.), Istituto Superiore di Sanità (Rome-AIDS Project 40H98 to C.P.), Regione Veneto (Ricerca Sanitaria Finalizzata n.312/10 to C.P.). We thank Vittoria Raimondi for technical help in some experiments. The authors state that there are no conflicts of interest.
Supplementary Material
References
- Deeks, SG (2011). HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med 62: 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaes, CP and Sáez-Cirión, A (2014). HIV cure research: advances and prospects. Virology 454-455: 340–352. [DOI] [PubMed] [Google Scholar]
- Sabin, CA (2013). Do people with HIV infection have a normal life expectancy in the era of combination antiretroviral therapy? BMC Med 11: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volberding, PA and Deeks, SG (2010). Antiretroviral therapy and management of HIV infection. Lancet 376: 49–62. [DOI] [PubMed] [Google Scholar]
- Barouch, DH and Deeks, SG (2014). Immunologic strategies for HIV-1 remission and eradication. Science 345: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré-Sinoussi, F, Ross, AL and Delfraissy, JF (2013). Past, present and future: 30 years of HIV research. Nat Rev Microbiol 11: 877–883. [DOI] [PubMed] [Google Scholar]
- Durand, CM, Blankson, JN and Siliciano, RF (2012). Developing strategies for HIV-1 eradication. Trends Immunol 33: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis, DM (2014). How Might We Cure HIV? Curr Infect Dis Rep 16: 392. [DOI] [PubMed] [Google Scholar]
- Richman, DD, Margolis, DM, Delaney, M, Greene, WC, Hazuda, D and Pomerantz, RJ (2009). The challenge of finding a cure for HIV infection. Science 323: 1304–1307. [DOI] [PubMed] [Google Scholar]
- Siliciano, JD and Siliciano, RF (2013). HIV-1 eradication strategies: design and assessment. Curr Opin HIV AIDS 8: 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers, K, Hütter, G, Hofmann, J, Loddenkemper, C, Rieger, K, Thiel, E et al. (2011). Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117: 2791–2799. [DOI] [PubMed] [Google Scholar]
- Hütter, G, Nowak, D, Mossner, M, Ganepola, S, Müssig, A, Allers, K et al. (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360: 692–698. [DOI] [PubMed] [Google Scholar]
- Burke, BP, Boyd, MP, Impey, H, Breton, LR, Bartlett, JS, Symonds, GP et al. (2014). CCR5 as a natural and modulated target for inhibition of HIV. Viruses 6: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter, G and Ganepola, S (2011). Eradication of HIV by transplantation of CCR5-deficient hematopoietic stem cells. ScientificWorld Journal 11: 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, P and June, C (2011). CCR5 knockout strategies. Curr Opin HIV AIDS 6: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didigu, C and Doms, R (2014). Gene therapy targeting HIV entry. Viruses 6: 1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter, G, Bodor, J, Ledger, S, Boyd, M, Millington, M, Tsie, M et al. (2015). CCR5 targeted cell therapy for HIV and prevention of viral escape. Viruses 7: 4186–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, SW, Kim, S, Kim, JM and Kim, JS (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Wang, W, Ye, C, Liu, J, Zhang, D, Kimata, JT and Zhou, P (2014). CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One 9: e115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C, Guan, X, Du, T, Jin, W, Wu, B, Liu, Y et al. (2015). Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol 96: 2381–2393. [DOI] [PubMed] [Google Scholar]
- Kang, H, Minder, P, Park, MA, Mesquitta, WT, Torbett, BE and Slukvin, II (2015). CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol Ther Nucleic Acids 4: e268. [DOI] [PubMed] [Google Scholar]
- Mussolino, C, Morbitzer, R, Lütge, F, Dannemann, N, Lahaye, T and Cathomen, T (2011). A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers, M, Maggio, I, Liu, J, Janssen, JM, Miselli, F, Mussolino, C et al. (2013). Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res 41: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock, U, Machowicz, R, Hauber, I, Horn, S, Abramowski, P, Berdien, B et al. (2015). mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res 43: 5560–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badia, R, Riveira-Muñoz, E, Clotet, B, Esté, JA and Ballana, E (2014). Gene editing using a zinc-finger nuclease mimicking the CCR5Δ32 mutation induces resistance to CCR5-using HIV-1. J Antimicrob Chemother 69: 1755–1759. [DOI] [PubMed] [Google Scholar]
- Holt, N, Wang, J, Kim, K, Friedman, G, Wang, X, Taupin, V et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L, Krymskaya, L, Wang, J, Henley, J, Rao, A, Cao, LF et al. (2013). Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther 21: 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, DA, Brennan, AL, Jiang, S, Binder-Scholl, GK, Lee, G, Plesa, G et al. (2013). Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther 24: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath, N, Yi, G, Dang, Y and Shankar, P (2013). Newer gene editing technologies toward HIV gene therapy. Viruses 5: 2748–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas, P, Stein, D, Tang, WW, Frank, I, Wang, SQ, Lee, G et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, A, Garg, H, Ablan, S, Freed, EO, Nagashima, K, Manjunath, N et al. (2011). Targeting the HIV entry, assembly and release pathways for anti-HIV gene therapy. Virology 415: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiem, HP, Jerome, KR, Deeks, SG and McCune, JM (2012). Hematopoietic-stem-cell-based gene therapy for HIV disease. Cell Stem Cell 10: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, CW, Younan, P, Jerome, KR and Kiem, HP (2013). Combinatorial anti-HIV gene therapy: using a multipronged approach to reach beyond HAART. Gene Ther 20: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, PS and Schaffer, DV (2010). Gene therapy takes a cue from HAART: combinatorial antiviral therapeutics reach the clinic. Sci Transl Med 2: 36ps30. [DOI] [PubMed] [Google Scholar]
- Bennett, MS and Akkina, R (2013). Gene therapy strategies for HIV/AIDS: preclinical modeling in humanized mice. Viruses 5: 3119–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, JM (2013). HIV gene therapy research advances. Blood 121: 1483–1484. [DOI] [PubMed] [Google Scholar]
- Rossi, JJ, June, CH and Kohn, DB (2007). Genetic therapies against HIV. Nat Biotechnol 25: 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lunzen, J, Fehse, B and Hauber, J (2011). Gene therapy strategies: can we eradicate HIV? Curr HIV/AIDS Rep 8: 78–84. [DOI] [PubMed] [Google Scholar]
- Zeller, SJ and Kumar, P (2011). RNA-based gene therapy for the treatment and prevention of HIV: from bench to bedside. Yale J Biol Med 84: 301–309. [PMC free article] [PubMed] [Google Scholar]
- An, DS, Donahue, RE, Kamata, M, Poon, B, Metzger, M, Mao, SH et al. (2007). Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci USA 104: 13110–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J and Akkina, R (2005). HIV-1 resistance conferred by siRNA cosuppression of CXCR4 and CCR5 coreceptors by a bispecific lentiviral vector. AIDS Res Ther 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, M, Kamata, M, Chen, KN, Pariente, N, An, DS and Chen, IS (2010). Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction. J Gene Med 12: 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, XF, An, DS, Chen, IS and Baltimore, D (2003). Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA 100: 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, S, Kamata, M, Kittipongdaja, P, Chen, KN, Kim, S, Pang, S et al. (2009). Characterization of a potent non-cytotoxic shRNA directed to the HIV-1 co-receptor CCR5. Genet Vaccines Ther 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J, DiGiusto, DL and Rossi, JJ (2013). Combinatorial RNA-based gene therapy for the treatment of HIV/AIDS. Expert Opin Biol Ther 13: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque, JM, Triques, K and Stevenson, M (2002). Modulation of HIV-1 replication by RNA interference. Nature 418: 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, SK, Dykxhoorn, DM, Kumar, P, Ranjbar, S, Song, E, Maliszewski, LE et al. (2005). Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood 106: 818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, YP, Haasnoot, J and Berkhout, B (2007). Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res 35: 5683–5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, YP, Haasnoot, J, ter Brake, O, Berkhout, B and Konstantinova, P (2008). Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res 36: 2811–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, KV, Chung, CH, Witke, W and Looney, DJ (2005). Inhibition of HIV-1 replication by siRNA targeting conserved regions of gag/pol. RNA Biol 2: 17–20. [DOI] [PubMed] [Google Scholar]
- Naito, Y, Nohtomi, K, Onogi, T, Uenishi, R, Ui-Tei, K, Saigo, K et al. (2007). Optimal design and validation of antiviral siRNA for targeting HIV-1. Retrovirology 4: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina, CD, Murray, MF, Dykxhoorn, DM, Beresford, PJ, Riess, J, Lee, SK et al. (2002). siRNA-directed inhibition of HIV-1 infection. Nat Med 8: 681–686. [DOI] [PubMed] [Google Scholar]
- Zhou, J and Rossi, JJ (2011). Current progress in the development of RNAi-based therapeutics for HIV-1. Gene Ther 18: 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, DS, Qin, FX, Auyeung, VC, Mao, SH, Kung, SK, Baltimore, D et al. (2006). Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther 14: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, MJ and Rossi, JJ (2005). Lentiviral vector delivery of recombinant small interfering RNA expression cassettes. Methods Enzymol 392: 218–226. [DOI] [PubMed] [Google Scholar]
- Li, M and Rossi, JJ (2008). Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol Biol 433: 287–299. [DOI] [PubMed] [Google Scholar]
- ter Brake, O, ‘t Hooft, K, Liu, YP, Centlivre, M, von Eije, KJ and Berkhout, B (2008). Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther 16: 557–564. [DOI] [PubMed] [Google Scholar]
- Centlivre, M, Legrand, N, Klamer, S, Liu, YP, Jasmijn von Eije, K, Bohne, M et al. (2013). Preclinical in vivo evaluation of the safety of a multi-shRNA-based gene therapy against HIV-1. Mol Ther Nucleic Acids 2: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brake, O, Legrand, N, von Eije, KJ, Centlivre, M, Spits, H, Weijer, K et al. (2009). Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(-/-)gammac(-/-)) mouse model. Gene Ther 16: 148–153. [DOI] [PubMed] [Google Scholar]
- DiGiusto, DL, Krishnan, A, Li, L, Li, H, Li, S, Rao, A et al. (2010). RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med 2: 36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiusto, DL, Stan, R, Krishnan, A, Li, H, Rossi, JJ and Zaia, JA (2013). Development of hematopoietic stem cell based gene therapy for HIV-1 infection: considerations for proof of concept studies and translation to standard medical practice. Viruses 5: 2898–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuyasu, RT, Zack, JA, Macpherson, JL and Symonds, GP (2011). Phase I/II clinical trials using gene-modified adult hematopoietic stem cells for HIV: lessons learnt. Stem Cells Int 2011: 393698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, YP, von Eije, KJ, Schopman, NC, Westerink, JT, ter Brake, O, Haasnoot, J et al. (2009). Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol Ther 17: 1712–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, M, Li, H, Nakanishi, M and Rossi, JJ (2008). Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: advantages and limitations. Mol Ther 16: 170–177. [DOI] [PubMed] [Google Scholar]
- Li, MJ, Kim, J, Li, S, Zaia, J, Yee, JK, Anderson, J et al. (2005). Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther 12: 900–909. [DOI] [PubMed] [Google Scholar]
- Cruz, NV, Amorim, R, Oliveira, FE, Speranza, FA and Costa, LJ (2013). Mutations in the nef and vif genes associated with progression to AIDS in elite controller and slow-progressor patients. J Med Virol 85: 563–574. [DOI] [PubMed] [Google Scholar]
- Desimmie, BA, Delviks-Frankenberrry, KA, Burdick, RC, Qi, D, Izumi, T and Pathak, VK (2014). Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol 426: 1220–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves, J and Santa-Marta, M (2004). HIV-1 Vif and APOBEC3G: multiple roads to one goal. Retrovirology 1: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani, R, Chen, D, Schröfelbauer, B, Navarro, F, König, R, Bollman, B et al. (2003). Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114: 21–31. [DOI] [PubMed] [Google Scholar]
- Martins, MA, Wilson, NA, Piaskowski, SM, Weisgrau, KL, Furlott, JR, Bonaldo, MC et al. (2014). Vaccination with Gag, Vif, and Nef gene fragments affords partial control of viral replication after mucosal challenge with SIVmac239. J Virol 88: 7493–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, W and Telesnitsky, A (2001). Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 286: 475–482. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz, M and Trono, D (2003). Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 77: 8957–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto, D, Li, H and Rossi, JJ (2002). Functional siRNA expression from transfected PCR products. RNA 8: 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson, DA, Dillon, CP, Kwiatkowski, AV, Sievers, C, Yang, L, Kopinja, J et al. (2003). A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33: 401–406. [DOI] [PubMed] [Google Scholar]
- Smale, ST (2010). DEAE-Dextran transfection of lymphocyte cell lines. Cold Spring Harb Protoc pdb.prot5373. [DOI] [PubMed]
- Sodroski, JG, Rosen, CA and Haseltine, WA (1984). Trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science 225: 381–385. [DOI] [PubMed] [Google Scholar]
- Ratner, L, Haseltine, W, Patarca, R, Livak, KJ, Starcich, B, Josephs, SF et al. (1985). Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313: 277–284. [DOI] [PubMed] [Google Scholar]
- Adachi, A, Gendelman, HE, Koenig, S, Folks, T, Willey, R, Rabson, A et al. (1986). Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59: 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, K, Emerman, M and Montagnier, L (1991). Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185: 661–672. [DOI] [PubMed] [Google Scholar]
- Parolin, C, Taddeo, B, Palú, G and Sodroski, J (1996). Use of cis- and trans-acting viral regulatory sequences to improve expression of human immunodeficiency virus vectors in human lymphocytes. Virology 222: 415–422. [DOI] [PubMed] [Google Scholar]
- Theodore, TS, Englund, G, Buckler-White, A, Buckler, CE, Martin, MA and Peden, KW (1996). Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses 12: 191–194. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, TR, Bernards, R and Agami, R (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Zuker, M (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]