Abstract
Influenza virus (IV) is a continuously evolving virus that widely spreads in humans and contributes to substantial morbidity and mortality. Re-emergence of human infection with avian influenza virus H5N1 poses extra challenge to IV control. Artificial microRNA (amiRNA)-mediated RNA interference has become a powerful antiviral approach due to its high specificity and rapid effect. Here, we designed several amiRNAs targeting the hemagglutinin gene of H5N1, a major determinant of pathogenicity. Expression and delivery efficiency were enhanced by presenting functional amiRNA with chimpanzee adenovirus serotype 68 (AdC68). One amiRNA, HA-1405, significantly limited H5N1 replication in vitro and inhibited 96.7% of clade 2.3.2 replication. AdC68-conjugated HA-1405 treatment remarkably decreased different clades of H5N1 plaque formation in Madin–Darby canine kidney cells. Moreover, prophylactic administration with rAd(HA-1405) markedly alleviated clinical symptoms and reduced ~3- to 40-folds of lung viral RNA copies against four clades of H5N1 in Institute of Cancer Research (ICR) mice. Our results further showed that rAd(HA-1405) conferred 70 and 40% immediate protection against lethal clade 2.3.2 and clade 2.3.4 H5N1 challenge, respectively. In conclusion, these data provided information that HA-targeting amiRNA delivered by AdC68 could be pursued as a potential agent for highly pathogenic avian influenza viruses prevention.
Keywords: adenovirus, artificial microRNA, H5N1, influenza virus, RNAi
Introduction
Influenza viruses (IV) widely spread among diverse hosts and lead to seasonal outbreaks annually, which cost millions of human lives and significant economic losses.1 In 1997, highly pathogenic avian influenza (HPAI) H5N1 was firstly reported to transmit from poultry to human in Hong Kong.2 Ever since then, this subtype continuously circulated in domestic and wild birds and sporadically caused human infections. Up to now, 844 cases from 16 countries have been documented in the World Health Organization with the fatality rate of ~53% (2003–2015), among which 52 cases were confirmed in China (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/). Exposure to sick poultry was considered a major risk factor of H5N1 infection.3 However, several episodes of potential human-to-human transmission raised more concerns about H5N1 pandemic.4
Strain-matched vaccines are viewed as the best preventative strategy for IVs, whereas the development of H5N1 vaccines encounters extra obstacles than seasonal flu ones.5 On one hand, H5N1 virus is lethal to chicken embryos, thus the yield of egg-dependent inactivated vaccines is largely restricted. On the other hand, operations with HPAI H5N1 have to be performed in biosafety level 3 facilities. Moreover, neutralizing antibody-based vaccines usually take several days before onset. Thus, novel egg-free avian IV vaccines that could take immediate effect could be an appropriate supplementary.
RNA interference (RNAi) is a conserved sequence-specific silencing mechanism in eukaryotes mediated by short double-stranded RNAs.6 These small RNAs, siRNA and miRNA included, suppress target gene expression posttranscriptionally by mRNA cleavage or translational inhibition.7 Especially, artificial microRNAs (amiRNA) with the backbone of known cellular pre-miRNAs reduce the dependence on host factors and enhance safety. This third-generation gene silencing technology has been widely utilized in treatment of cancers8,9 and infectious diseases.10,11,12,13,14,15 Here, we described the application of amiRNA in the control of H5N1 as a complement of traditional vaccines.
In this study, we exploited the adenovirus delivered, hemagglutinin-specific artificial microRNA as a novel antiviral agent. We tested its prophylaxis efficacy in ICR mice against four clades of H5N1, including clade 2.3.2, clade 2.3.4, clade 7, and clade 9, which were circulating in China from 2004 to 2009 (ref. 16). We proved that influenza-targeting recombinant adenovirus-delivered artificial miRNA (rAd-amiRNA) could combat lethal H5N1 infection and ameliorate severity of symptoms in respect of lung viral yields, inflammation, and survival.
Results
Design and identification of functional HA-targeting artificial microRNA
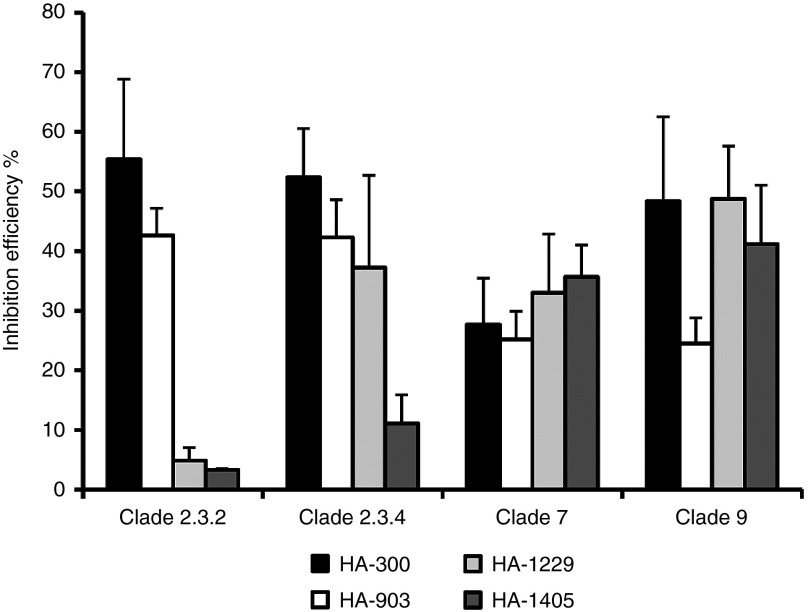
The hemagglutinin gene is one of the major determinants of H5N1 pathogenicity. It has been proved that H5N1 HA was closely related to systemic infection and impaired immune response.17,18 Therefore, we selected H5N1 HA gene as the RNAi targets. Four amiRNAs which scored highest according to online RNAi designer (http://rnaidesigner.invitrogen.com/rnaiexpress/) were generated as described in the Materials and Methods, including HA-300, HA-903, HA-1229, and HA-1405. The pre-amiRNA sequences were displayed in Table 1. To evaluate the inhibitory effect of the amiRNA constructs, we transfected the aforementioned amiRNA plasmids (1 μg) separately into HEK 293T cells 24 hours before influenza challenge. Transduced cells were infected with 0.0005 MOI H5N1 and lysed with Trizol for RNA isolation 24 hours later. Neg-miRNA control plasmid-treated cells were used as mock controls, and its flu gene expression level was set at 100%. As depicted in Figure 1, all amiRNA treatments significantly lowered H5N1 viral production compared to corresponding controls (P < 0.01). Among four amiRNAs, HA-1405 exhibited strongest inhibition on clade 2.3.2 (mean viral load 3.30%) and clade 2.3.4 H5N1 (mean viral load 11.10%). Its mean suppression on clade 9 was 41.17%, only next to HA-903 (mean viral load 24.53%), while the latter one was less efficient against other three H5N1 clades. As to clade 7, all four amiRNAs showed similar effect (HA-300, 27.63%; HA-903, 25.20%; HA-1229, 33.00%; and HA-1405, 35.57%). Based on in vitro screen results, we chose HA-1405 to perform further experiments.
Table 1. Preartificial miRNA sequence designed by BLOCK-iT RNAi designer.

Figure 1.
Screen for effective H5N1 hemagglutinin-targeting artificial microRNAs. HEK 293T cells were transfected with influenza-specific amiRNA constructs (1 μg) and then infected with one clade of H5N1 at the dose of 0.0005 MOI 24 hours later (clade 2.3.2, clade 2.3.4, clade 7, or clade 9). Neg-miRNA plasmid transfectants were used as the positive controls. Influenza matrix gene expression was measured by real-time PCR 24 hours post-infection in cell lysate and normalized to β-actin. Matrix expression level in control groups was designated to be 100%, and the relative Matrix gene level in treated groups was normalized to be a percentage of the control value. Data were shown as mean relative expression to control ± SD from three independent experiments. ANOVA was applied to compare difference among groups. P < 0.01 in all IV-specific amiRNA transfectants when compared to positive controls. ANOVA, analysis of variance.
Adenovirus-expressed HA-1405 inhibited H5N1 replication in vitro
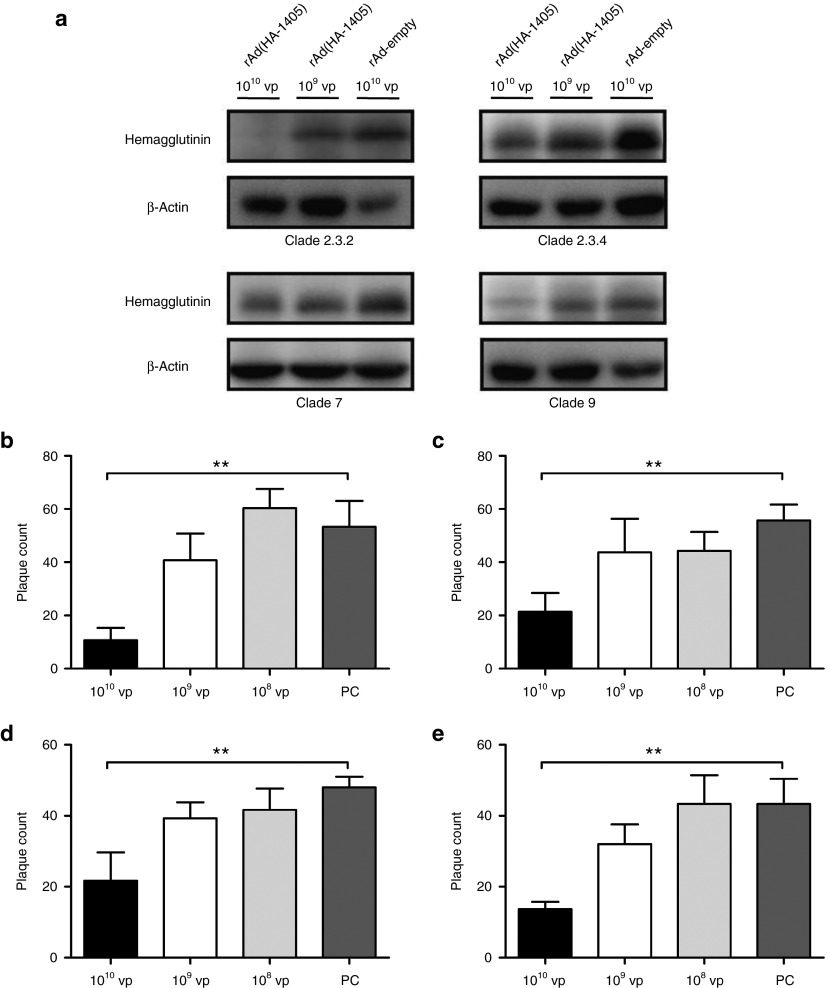
One difficulty in RNAi clinical application is to transduce amiRNA into target sites with high efficiency and minimal toxicity. Our previous data proved that adenovirus vector was one of the ideal candidates considering its tissue tropism and safety.8,10 Here, we inserted the whole amiRNA (HA-1405) expression cassette into the E1 region of AdC68 and rescued the recombinant adenovirus, termed as rAd(HA-1405). To examine whether rAd(HA-1405) could suppress H5N1 growth in vitro, we performed HA-targeting western blot in Vero cells and plaque reduction assay in Madin–Darby canine kidney (MDCK) cells (Figure 2). The expression of HA was significantly reduced in rAd(HA-1405) transduced Vero cells when infected with either clade of H5N1 virus. The inhibition efficiency exhibited a dose-dependent manner (Figure 2a). It is noteworthy that HA expression in clade 2.3.2-infecting cells was nearly abolished in presence of 1010 virus particle (vp) of rAd(HA-1405). Furthermore, the inhibition in influenza plaque formation was assessed in MDCK cells. As shown in Figure 2b–e, the numbers of plaques were all significantly lowered in the presence of 1010 vp of rAd(HA-1405) against four clades of H5N1 challenge compared to corresponding controls (clade 2.3.2, P = 0.0024; clade 2.3.4, P = 0.0031; clade 7, P = 0.0060; and clade 9, P = 0.0023). Pretreatment of 109 vp rAd(HA-1405) slightly decreased the average plaque formation of four H5N1 strains, but no significant difference was detected comparing to corresponding controls (clade 2.3.2, P = 0.1928; clade 2.3.4, P = 0.2124; clade 7, P = 0.0502; and clade 9, P = 0.0951). Among four clades, rAd(HA-1405) displayed strongest inhibitory activity against clade 2.3.2 by around fivefold (mean plaque count, 1010 vp of rAd(HA-1405) = 10.7, 1010 vp of rAd-empty = 53.3; Figure 2b), while suppression of other clades ranged from twofold to threefold (Figure 2c–e). The inhibition efficiency of adenovirus-expressed amiRNA was not totally correlated with plasmid screen results (Figure 1). We assumed that difference in cell types, infection doses, and detection sensitivity might lead to this divergence.
Figure 2.
Suppression of H5N1 in vitro replication in the presence of rAd(HA-1405). (a) Expression of HA in adenovirus-treated Vero cells was determined by western blot 24 hours post H5N1 challenge in comparison to β-actin. Cells pretreated with rAd-empty served as controls. (b–e) Plaque reduction assay was conducted in MDCK cells. Cells were preincubated with 10-fold serially diluted rAd(HA-1405) (ranging from 1010 to 108 vp) or rAd-empty (1010 vp). Twenty-four hours after treatment, 40–60 PFU of clade (b) 2.3.2, (c) clade 2.3.4, (d) clade 7, or (e) clade 9 H5N1 viruses were added to each well. The yields of viral progeny were determined by plaque assay. Data were shown as mean plaque counts ± SD from three independent experiments. ANOVA followed by Dunnett's test was used to compare the differences among different doses of rAs(HA-1405) groups and control group (PC). ** indicates P value was less than 0.01 in comparison to PC.
rAd(HA-1405) conferred partial protection to ICR mice against H5N1 challenge
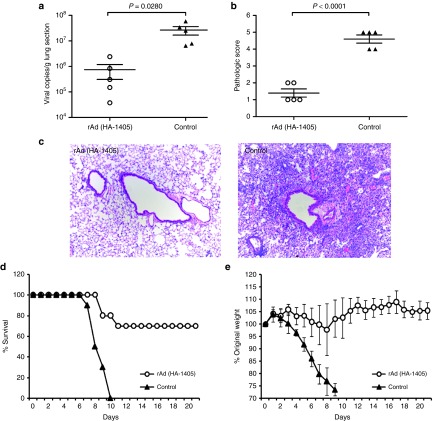
To assess the effect of replication inhibition on clinical outcome, we inoculated ICR mice intranasally with 1011 vp of rAd(HA-1405) or rAd-empty (n = 15) 24 hours prior to 5LD50 H5N1 challenge. Five mice from each group were euthanized for lung viral RNA titration and histological scoring 5 days post infection (dpi). Clinical symptoms of rest 10 mice were monitored on a daily basis for 21 days. Figure 3 described the protection efficiency against clade 2.3.2 H5N1. The mean lung viral copies of rAd(HA-1405)-inoculated mice was 35-fold lower in comparison with rAd-empty group (P = 0.0280; Figure 3a). Hematoxylin and eosin staining proved that lung sections from control mice displayed severe perivascular and interstitial infiltrates (mean histological score = 4.6), while no pathology or small-area perivascular infiltrates was observed in rAd(HA-1405) group (mean histological score 1.4; P < 0.0001; Figure 3b,c). Furthermore, consistent with reduced viral titer and less pronounced inflammation responses, rAd(HA-1405) protected 70% ICR mice from lethal infection of clade 2.3.2 (P = 0.0031; Figure 3d). The survived mice in rAd(HA-1405) group lost at most 5% of initial weight around day4 to day 8 and gradually recovered (Figure 3e). In contrast, control mice suffered constant weight loss and all died or required euthanasia by day 9. Thus, rAd(HA-1405) conferred significant protection against clade 2.3.2 H5N1 challenge.
Figure 3.
rAd(HA-1405) provided partial protection from lethal challenge of clade 2.3.2 H5N1 in ICR mice. ICR mice (n = 15) pre-inoculated with 1011 vp of rAd(HA-1405) or rAd-empty were infected 1 day later with 5LD50 of clade 2.3.2 H5N1. Randomly five mice from both groups were euthanized 5 dpi, and lung tissues were taken for viral RNA copies titration and histological scoring. (a) Viral yields were measured by real-time PCR in right upper lung lobes (P = 0.0280). (b and c) All data were repeated three times. Hematoxylin and eosin staining was performed with left upper lobes for pathological evaluation (histological score, P < 0.0001). Remaining 10 mice were observed 21 days for (d) survival rates and (e) weight loss (survival rates, P = 0.0031). Student's t-test was performed to compare viral RNA copies and pathological change. The survival rates were compared with chi-square test. dpi, days post infection.
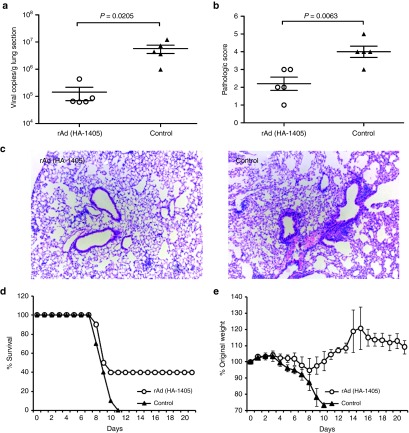
We further tested whether rAd(HA-1405) could provide protection from clade 2.3.4 H5N1. ICR mice were vaccinated and challenged as above. As depicted in Figure 4a–c, rAd(HA-1405) inoculation remarkably decreased ~40 folds of lung viral RNA copies (P = 0.0205) and ~1.8-folds of mean histological scores (P = 0.0063). Four out of 10 rAd(HA-1405)-treated mice survived clade 2.3.4 infection (P = 0.0867) with maximal 5% weight loss peaking at day 8 (Figure 4d,e). All rAd-empty-treated mice showed similar weight change pattern at 4 dpi and then continued to lose weight till death.
Figure 4.
rAd(HA-1405) alleviated flu symptoms in ICR mice against lethal challenge of clade 2.3.4 H5N1. ICR mice (n = 15) immunized with 1011 vp of rAd(HA-1405) or rAd-empty were infected 1 day later with 5LD50 of clade 2.3.4 H5N1. Randomly five mice from both groups were euthanized 5 dpi, and lung tissues were taken for viral RNA copies titration and histological scoring. (a) Viral yields were measured by real-time PCR in right upper lung lobes (P = 0.0205). (b and c) All data were repeated three times. Hematoxylin and eosin staining was performed with left upper lobes for pathological evaluation (histological score, P = 0.0063). Remaining 10 mice were observed 21 days for (d) survival rates and (e) weight loss (survival rates, P = 0.0867). Student's t-test was performed to compare viral RNA copies and pathological change. The survival rates were compared with chi-square test. dpi, days post infection.
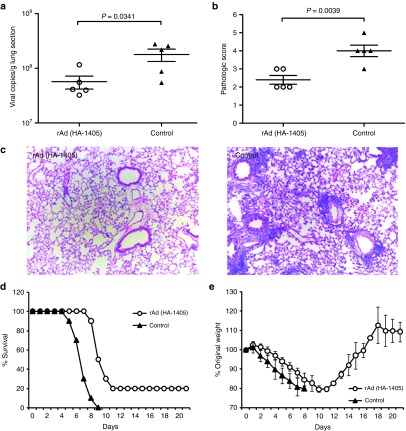
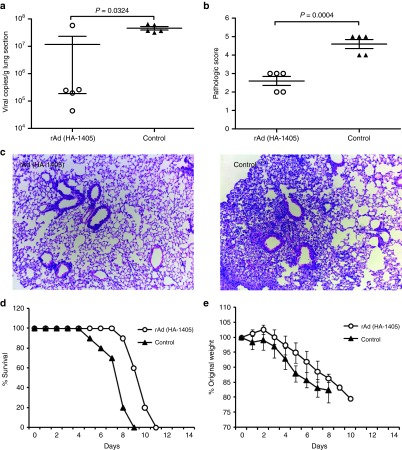
In terms of clade 7, reduction in lung viral copies was also significant (P = 0.0341), but the decline was less obvious than above two clades (~3-folds in decrease; Figure 5a). And average pathologic scores were 2.4 and 4 for rAd(HA-1405) and rAd-empty group (P = 0.0039), respectively (Figure 5b,c). IV-specific amiRNA rescued 20% ICR mice from lethal challenge of clade 7 (P = 0.4737) and alleviated associated symptoms (Figure 5d,e). Figure 6 showed the protection efficiency toward clade 9 H5N1. Although rAd(HA-1405) notably decreased lung viral loads (P = 0.0324) and inflammation (P = 0.0004), all treated mice succumbed to subsequent lethal infection. Therefore, we proved that rAd(HA-1405) could confer substantial protection against clade 2.3.2 H5N1 and ease symptoms caused by other H5N1 clades.
Figure 5.
rAd(HA-1405) alleviated flu symptoms in ICR mice against lethal challenge of clade 7 H5N1. ICR mice (n = 15) immunized with 1011 vp of rAd(HA-1405) or rAd-empty were infected 1 day later with 5LD50 of clade 7 H5N1. Randomly five mice from both groups were euthanized 5 dpi, and lung tissues were taken for viral RNA copies titration and histological scoring. (a) Viral yields were measured by real-time PCR in right upper lung lobes (P = 0.0341). (b and c) All data were repeated three times. Hematoxylin and eosin staining was performed with left upper lobes for pathological evaluation (histological score, P = 0.0039). Remaining 10 mice were observed 21 days for (d) survival rates and (e) weight loss (survival rates, P = 0.4737). Student's t-test was performed to compare viral RNA copies and pathological change. The survival rates were compared with chi-square test. dpi, days post infection.
Figure 6.
rAd(HA-1405) alleviated flu symptoms in ICR mice against lethal challenge of clade 9 H5N1. ICR mice (n = 15) immunized with 1011 vp of rAd(HA-1405) or rAd-empty were infected 1 day later with 5LD50 of clade 9 H5N1. Randomly five mice from both groups were euthanized 5 dpi, and lung tissues were taken for viral RNA copies titration and histological scoring. (a) Viral yields were measured by real-time PCR in right upper lung lobes (P = 0.0324). All data were repeated three times. (b and c) Hematoxylin and eosin staining was performed with left upper lobes for pathological evaluation (histological score, P = 0.0004). Remaining 10 mice were observed 21 days for (d) survival rates and (e) weight loss. Student's t-test was performed to compare viral RNA copies and pathological change. The survival rates were compared with chi-square test. dpi, days post infection.
Discussion
Control of HPAI H5N1 human infection has been a severe challenge since the 21st century. This virus has evolved into phylogenetically distinct clades and subclades, among which, clades 0, 1, 2.1, 2.2, 2.3, and 7 have reported to cause human disease.19 In China, clades 2.3.2, 2.3.4, and 7 co-circulated in poultry and threat public health.20 Although vaccination is the most cost-effective IV intervention strategy, production of H5N1 vaccines faces more technical obstacles than season flu vaccines. Moreover, reappearance of drug-resistant H5N1 strains limits the use of currently available antiviral drugs and underscores the urgent demand for novel flu-specific agents.21,22 To this end, we developed the sequence-based H5N1 antiviral compound with the help of RNAi.
RNAi has been exploited as a promising strategy for antiviral purposes. Small RNAs targeting the genome of Ebola virus,15 SARS-coronavirus,14 or hepatitis C virus23 have all been proved to be effective in preclinical studies. Recently, an antisense oligonucleotides, Fomivirsen, has been approved by Food and Drug Administration to treat cytomegalovirus.24 However, to find the way into clinical application, the small RNA compounds have to expand their half-life, lower toxicity, and reach target sites precisely. In the previous study, we demonstrated that chimpanzee adenovirus serotype 68 (AdC68) was a suitable delivery vector for artificial microRNAs in influenza treatment.10 Adenovirus-delivered amiRNAs targeting conserved internal gene, nucleoprotein, or matrix, protected ICR mice completely against homologous A/PR8, whereas only two rAd-amiRNAs conferred 60 to 70% protection from one clade of H5N1 virus. Therefore, we sought to enhance the efficiency toward H5N1 by silencing other flu gene.
In this study, we chose the hemagglutinin gene of H5N1 as the target site for the following reasons: (i) HA was a main flu pathogenicity determinant and the multi-basic cleavage site of highly pathogenic H5N1 HA was closely related with its systemic infection.17 (ii) H5N1 HA, different from H1N1 and H3N2, could reduce perforin expression in CD8+ T cells, which might contribute to impaired virus clearance activity and subsequent increase in proinflammatory cytokines.18 Hence, we designed four amiRNAs based on HA gene from distinct H5N1 clades. Through in vitro screen, one amiRNA, HA-1405, was incorporated into AdC68 genome and extended to in vivo protection experiments. As shown in Figure 3, rAd(HA-1405) remarkably enhanced mice survival by 70% in face of clade 2.3.2 H5N1 challenge. Its efficiency against clade 2.3.4, clade 7, and clade 9 H5N1 viruses was 40, 20, and 0%, respectively (Figure 4–6). Although the survival rates were not significantly different from controls in the latter three clades of infection, rAd(HA-1405) indeed prolonged mean survival time, delayed weight loss, and decreased lung viral yields. The effectiveness of rAd(HA-1405) was comparable but not further enhanced compared to previously reported NP- or M-specific amiRNAs.
In conclusion, our data proved the feasibility of rAd(HA-1405) as an potential antiviral reagent for H5N1. Additional studies, including combination with other amiRNAs or compounds, are required to further optimize prophylactic efficiency.
Materials and methods
Cell culture and transfection. Human embryonic kidney 293 (HEK 293), human embryonic kidney 293T (HEK 293T), Vero and MDCK cell lines were purchased from Shanghai Cell Bank, Chinese Academy of Sciences, and maintained in Dulbecco's Modified Eagle's Medium (HyClone, Beijing, China) supplemented with 10% fetal bovine serum (Gemini Biological Products, Calabasas, CA), 100 U/ml penicillin and 100 μg/ml streptomycin (HyClone).
Transfection was performed with X-treme GENE HP DNA Transfection Reagent (Roche, Indianapolis, IN) in accordance with manufacturer's guidance. In brief, indicated dose of plasmid was mixed with X-treme reagent at the ratio of 1:2 in Opti-MEM (Gbico, Grand Island, NY). The transfection complex was incubated at room temperature for 15 minutes and transferred directly into cells. The plate was cultured at 37 °C in 5% CO2 incubator for 24 hours and processed to further treatment.
Influenza virus. H5N1 strains A/duck/Hunan/3/2007 (clade 2.3.2), A/environment/Hunan/6–69/2008 (clade 2.3.4), A/environment/Hunan/1–35/2007 (clade 7), and A/chicken/Henan/12/2004 (clade 9) were propagated by inoculating the allantoic cavity of 9-day-old specific-pathogen-free (SPF) chicken embryos and incubated for 48 hours at 35 °C. Allantoic fluid was collected and preserved at −80 °C. Experiments involving H5N1 were conducted in the biosafety level 3 containment facilities at Fudan University (Shanghai, China).
Design and generation of artificial microRNA. H5N1 hemagglutinin targeting artificial miRNAs were designed by Invitrogen's BLOCK-iT Pol II miR RNAi Designer (http://rnaidesigner. invitrogen.com/rnaiexpress/). Four best-suited single-stranded oligonucleotides were synthesized, annealed for double-stranded oligos, and further cloned into the miRNA expression vector pcDNA 6.2-GW/miR (Invitrogen, Carlsbad, CA). Pre-miRNA sequences were shown in Table 1. Neg-miRNA control plasmid provided by Block-iT-Pol II miR RNAi Expression Vector Kit served as the negative control.
Generation of recombinant adenoviruses. Recombinant adenoviruses were constructed as previously described.10,25 In brief, the complete artificial amiRNA expression cassettes from the pcDNA6.2-GW/miR vectors were amplified by PCR and ligated with the pShuttle vector (Clonetech, Mountain View, CA). After being digested with I-CeuI and PI-SceI, the fragment was subcloned into the E1 region of the chimpanzee-origin adenovirus molecular clone (AdC68). The constructs were verified by enzyme digestion and sequencing. Recombinant adenoviruses were generated by transfecting PacI linearized plasmids into HEK 293 cells. Rescued Ads were further expended in HEK 293 cells and purified by cesium chloride density-gradient centrifugation. Virus concentration was measured by spectrophotometry at absorbance of 260 nm. rAd-empty was constructed by the same strategy, which was applied as the mock control virus in following studies.
Mice.Six- to eight-week-old female ICR mice were obtained from Shanghai Laboratory Animal Center, China, and housed in biosafety level 3 laboratory (Fudan University). Animal study was conducted in accordance with guidelines of Institutional Animal Care and Use Committee of Institute Pasteur of Shanghai.
Western blot analysis. To prove that rAd(HA-1405) reduced hemagglutinin expression, western blot was conducted in adenovirus (Ad)-treated Vero cells which were subsequently infected with different clades of H5N1. In brief, Vero cells (plated at 5 × 105 per well in 12-well plate on the previous day) were inoculated with indicated dose of rAd(HA-1405). rAd-empty pretreated cells served as controls. Twenty-four hours later, cells were infected individually with four different clades of H5N1 viruses at a dose of 0.001 multiplicity of infection (MOI) and harvested after 24 hours. Protein expression levels were detected with rabbit polyclonal antibody for HA (Sino Biological, Beijing, China), followed by horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich, St Louis, MO). The expression of β-actin (Sigma-Aldrich) was measured as a normalization control for protein loading. Signals were detected using a chemiluminescence detection system (GE Health Life Sciences, Pittsburgh, PA).
Plaque reduction assay. Inhibition of plaque formation was performed with a modified plaque assay protocol.26 MDCK cells were seeded in six-well plate at the density of 1 × 106 cells per well 24 hours prior to Ad incubation. The confluent monolayer was treated with 10-fold serially diluted rAd(HA-1405) (from 1010 vp to 108 vp) or rAd-empty (1010 vp) and cultured at 37 °C for 24 hours. Forty to 60 plaque-forming units of H5N1 virus diluted in 100 μl serum-free Dulbecco's Modified Eagle's Medium (1% bovine serum albumin) were added to each well and absorbed at 37 °C for 1 hour. After incubation, the supernatant was removed. MDCK cells were washed once with phosphate-buffered saline (PBS, Hyclone) and overlaid with minimum essential media (MEM, Gibco) containing 0.8% Agar and 1% BSA. Plates were maintained at 37 °C with 5% CO2. Seventy-two hours later, cells were fixed with 10% formaldehyde and stained with crystal violet. The assay was repeated in triplicate.
Vaccination and challenge. Groups of ICR mice (15 mice per group) were intranasally (i.n.) treated with 1 × 1011 vp of rAd(HA-1405) in a volume of 20 μl 1 day prior to IV challenge. Equivalent dose of rAd-empty inoculated mice served as negative controls. 5LD50 of H5N1 was diluted in 30 μl PBS and given i.n. to each mouse after anesthesia. Five mice from each group were sacrificed 5 dpi, and lung tissues were taken for virology and histology examination. Weight loss and survival rates of remaining mice were observed daily for 21 days. Mice in excess of 30% weight loss would be euthanized considering animal welfare.
Viral load titration. Influenza virus RNA copies were determined by quantitative real-time PCR as previously reported,27 which has been proved in good agreement with traditional titration methods with higher sensitivity.28 Briefly, total RNA from treated cells was isolated by Trizol (Invitrogen) following manufacturer's protocols. Purified RNA was dissolved in 100 μl diethylpyrocarbonate-treated water (Takara, Dalian, China). The concentration and quality was determined by NanoDrop spectrophotometrically (Thermo Scientific, Wilmington, DE). One-hundred nanogram of total RNA was retro-transcribed using M-MLV Reverse Transcriptase Kit (Promega, San Luis Obispo, CA) with a universal primer (Uni 12, 5′-AGCRAAAGCAGG-3′) complementary to the conserved end of influenza genome.29 Flu gene expression was subsequently assayed on 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA) with following primers: matrix forward 5′-AAGACCAATCCTGTCACCTCTGA-3′, Matrix reverse 5′-CAAAGCGTCTACGCTGCAGTCC-3′, β-actin forward 5′-ACCGAGCGCGGCTACAG-3′, β-actin reverse 5′-CTTAATGTCACGCACGATTTCC-3′. The amplification was achieved in a 20 μl reaction, containing 2 μl cDNA, 10 μl 2 × SYBR Premix Ex Taq (TliRNaseH Plus) (Takara), 0.4 µmol/l forward primer, 0.4 µmol/l reverse primer and 0.4 µl ROX reference dye2 (50×). Reaction was started with denaturation at 95 °C for 30 seconds, followed by 40 times cycling at 95 °C for 5 seconds, and 60 °C for 30 seconds. Each sample was amplified in triplicate. Relative flu gene expression was quantified by the comparative CT method and normalized to β-actin expression. All data were analyzed with 7900HT System SDS Version 2.4 (Applied Biosystems, Foster City, CA).
Lung tissues were harvested from mice 5 dpi and stored at −80 °C before RNA isolation. Tissues were mechanically homogenized with Precellys 24 (Bertin Technologies, France) in ice-cold PBS. Homogenate was then centrifuged at 12,000 rpm for 10 minutes, and supernatants were applied for viral load titration as above. Viral copies were determined by absolute quantification and the M gene of A/PR8 expressed in pMD18-T vector was used to provide a standard curve. Viral copy numbers were normalized to the mass of lungs.
Histology. Lung tissues were dissected from mice 5 dpi and fixed in 4% paraformaldehide for 24 hours at 4 °C. Hematoxylin and eosin staining was performed as previously described.30 Pathological sections were evaluated double blinded to guarantee objectivity. Histological changes were scored with following criteria27: 1, no observable pathology; 2, perivascular infiltrates; 3, perivascular and interstitial infiltrates affecting <20% of the lobe section; 4, perivascular and interstitial infiltrates affecting 20–50% of the lobe section; 5, perivascular and interstitial infiltrates affecting >50% of the lobe section.
Statistical analyses. SPSS 16.0 (Chicago, IL) was used for statistical analyses. Viral yield, pathological score, and plaque count were compared by Student's t-test or one-way analysis of variance. Chi-square test was applied to assess difference between survival rates. P < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by NIH grants HL113655 and AI101953 to J.W., and grants from “Knowledge Innovation Program,””100 Talent Program” from Chinese Academy of Sciences, Shanghai Pasteur Foundation, and China 863 program (2014AA021003) to D.Z.
References
- Thompson, WW, Comanor, L and Shay, DK (2006). Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis 194 (suppl. 2): S82–S91. [DOI] [PubMed] [Google Scholar]
- Subbarao, K, Klimov, A, Katz, J, Regnery, H, Lim, W, Hall, H et al. (1998). Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279: 393–396. [DOI] [PubMed] [Google Scholar]
- Mounts, AW, Kwong, H, Izurieta, HS, Ho, Y, Au, T, Lee, M et al. (1999). Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong, 1997. J Infect Dis 180: 505–508. [DOI] [PubMed] [Google Scholar]
- Ungchusak, K, Auewarakul, P, Dowell, SF, Kitphati, R, Auwanit, W, Puthavathana, P et al. (2005). Probable person-to-person transmission of avian influenza A (H5N1). N Engl J Med 352: 333–340. [DOI] [PubMed] [Google Scholar]
- Treanor, JJ, Wilkinson, BE, Masseoud, F, Hu-Primmer, J, Battaglia, R, O'Brien, D et al. (2001). Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 19: 1732–1737. [DOI] [PubMed] [Google Scholar]
- Fire, A, Xu, S, Montgomery, MK, Kostas, SA, Driver, SE and Mello, CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Hannon, GJ (2002). RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Chi, Y, Wang, X, Yang, Y, Zhang, C, Ertl, HC and Zhou, D (2014). Survivin-targeting artificial microRNAs mediated by adenovirus suppress tumor activity in cancer cells and xenograft models. Mol Ther Nucleic Acids 3: e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z, Wu, H, Reddy, S, Zhu, A, Wang, S, Blevins, D et al. (2007). Blockade of invasion and metastasis of breast cancer cells via targeting CXCR4 with an artificial microRNA. Biochem Biophys Res Commun 363: 542–546. [DOI] [PubMed] [Google Scholar]
- Zhang, H, Tang, X, Zhu, C, Song, Y, Yin, J, Xu, J et al. (2015). Adenovirus-mediated artificial MicroRNAs targeting matrix or nucleoprotein genes protect mice against lethal influenza virus challenge. Gene Ther 22: 653–662. [DOI] [PubMed] [Google Scholar]
- Israsena, N, Supavonwong, P, Ratanasetyuth, N, Khawplod, P and Hemachudha, T (2009). Inhibition of rabies virus replication by multiple artificial microRNAs. Antiviral Res 84: 76–83. [DOI] [PubMed] [Google Scholar]
- Ye, X, Liu, Z, Hemida, MG and Yang, D (2011). Targeted delivery of mutant tolerant anti-coxsackievirus artificial microRNAs using folate conjugated bacteriophage Phi29 pRNA. PLoS One 6: e21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Q, McManus, MT, Nguyen, T, Shen, CH, Sharp, PA, Eisen, HN et al. (2003). RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA 100: 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, BJ, Guan, Y, Tang, Q, Du, C, Xie, FY, He, ML et al. (2004). Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir Ther 9: 365–374. [PubMed] [Google Scholar]
- Geisbert, TW, Hensley, LE, Kagan, E, Yu, EZ, Geisbert, JB, Daddario-DiCaprio, K et al. (2006). Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J Infect Dis 193: 1650–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y, Shi, J, Zhong, G, Deng, G, Tian, G, Ge, J et al. (2010). Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J Virol 84: 8389–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer, DA (1999). Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258: 1–20. [DOI] [PubMed] [Google Scholar]
- Hsieh, SM and Chang, SC (2006). Insufficient perforin expression in CD8+ T cells in response to hemagglutinin from avian influenza (H5N1) virus. J Immunol 176: 4530–4533. [DOI] [PubMed] [Google Scholar]
- Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza AV, Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH et al. Update on avian influenza A (H5N1) virus infection in humans. The New England journal of medicine 2008; 358: 261–273. [DOI] [PubMed] [Google Scholar]
- Jiang, WM, Liu, S, Chen, J, Hou, GY, Li, JP, Cao, YF et al. (2010). Molecular epidemiological surveys of H5 subtype highly pathogenic avian influenza viruses in poultry in China during 2007-2009. J Gen Virol 91: 2491–2496. [DOI] [PubMed] [Google Scholar]
- de Jong, MD, Tran, TT, Truong, HK, Vo, MH, Smith, GJ, Nguyen, VC et al. (2005). Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med 353: 2667–2672. [DOI] [PubMed] [Google Scholar]
- He, G, Qiao, J, Dong, C, He, C, Zhao, L and Tian, Y (2008). Amantadine-resistance among H5N1 avian influenza viruses isolated in Northern China. Antiviral Res 77: 72–76. [DOI] [PubMed] [Google Scholar]
- Kapadia, SB, Brideau-Andersen, A and Chisari, FV (2003). Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA 100: 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, RM (2001). Technology evaluation: fomivirsen, Isis Pharmaceuticals Inc/CIBA vision. Curr Opin Mol Ther 3: 288–294. [PubMed] [Google Scholar]
- Zhou, D, Zhou, X, Bian, A, Li, H, Chen, H, Small, JC et al. (2010). An efficient method of directly cloning chimpanzee adenovirus as a vaccine vector. Nat Protoc 5: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H, Goldsmith, C, Thawatsupha, P, Chittaganpitch, M, Waicharoen, S, Zaki, S et al. (2007). Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J Virol 81: 12439–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D, Wu, TL, Lasaro, MO, Latimer, BP, Parzych, EM, Bian, A et al. (2010). A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol Ther 18: 2182–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, CL, Dempsey, MH, Ring, CJ, Kempson, RE, Zhang, L, Gor, D et al. (2004). Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol 29: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, CH, Lin, KL, Chan, Y, Wang, YL, Chi, YT, Tu, HL et al. (2006). Amplification of the entire genome of influenza A virus H1N1 and H3N2 subtypes by reverse-transcription polymerase chain reaction. J Virol Methods 136: 38–43. [DOI] [PubMed] [Google Scholar]
- McAuley, JL, Hornung, F, Boyd, KL, Smith, AM, McKeon, R, Bennink, J et al. (2007). Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]