Abstract
Peptides have been in the limelight, as therapeutic agents for cancer treatment through various applications due to their high target selectivity and exceptional ability to penetrate the cell membrane. Recent studies have revealed that synthesized peptides bind to hairpin structures of RNA that affect their activities such as changing the efficacy of microRNA maturation. MicroRNA-mediated p53 activation by the microRNA-29 (miR29) family is one of the most important regulatory pathways in cancer therapeutics. By targeting the suppressors of p53, a tumor suppressor protein, miR29 induces apoptosis of cancer cells through p53 stabilization. Here, we identify a novel synthesized amphiphilic peptide, LK-L1C/K6W/L8C, which enhances expression of miR29b and promotes p53 activity. In the presence of LK-L1C/K6W/L8C, pre-miR29b preferentially forms a complex with the Dicer protein through interaction of LK-L1C/K6W/L8C with the terminal loop region of pre-miR29b, leading to an increase in Dicer processing. Furthermore, LK-L1C/K6W/L8C stimulates apoptosis by improving p53 stability in miR29-inducible HeLa and MCF7 cells. Collectively, our study shows that a peptide can directly influence the miR29b-mediated p53 activation pathway in cancer cells. Therefore, our findings provide the basis for a new, potentially promising peptide-based drug for cancer therapy.
Keywords: apoptosis, cancer therapy, Dicer processing, miR29b-p53 pathway, synthesized amphiphilic peptide
Introduction
The p53 is an essential tumor suppressor protein that kills cancer cells through apoptosis and senescence. Although many cancer cells possess mutated forms of p53, resulting in severe malignancy, a tumor with the wild type p53 is still affected by stress-induced p53 activation.1 It is well-known that p53 is quickly accumulated under stressful conditions such as DNA damage or oxidative stress, leading to cell death.2 Therefore, p53 is tightly regulated by multiple pathways. MicroRNAs (miRNAs) are one of the p53 regulators in cellular regulatory mechanisms.3
miRNAs are endogenous small noncoding RNAs of approximately 22 nucleotides that can regulate the expression of target genes by binding to target messenger RNA (mRNA) for cleavage or translational repression.4 In recent studies, the role of miRNAs has been highlighted in many types of cancer.5,6,7 Several miRNAs can regulate tumor development by targeting oncogenes and/or genes that control cell differentiation or apoptosis in diverse human cancers.8,9
Many studies have shown that p53 can regulate miRNAs directly and indirectly.10,11,12,13 p53 can transcriptionally activate several miRNAs which target cell cycle related genes that inhibit cell cycle progression, resulting in the induction of apoptosis in tumor cells.3,14 On the other hand, p53 can be regulated by miRNAs which directly bind to the 3′ untranslated region of p53 mRNA resulting in reduced levels of p53.14 In contrast, the miR29 family was the first group of miRNAs identified as p53 positive regulators achieving their regulatory effects through the inhibition of p85a and CDC42.15 Recently, miR542-3p was found to activate p53 by inhibiting proteasomal degradation of p53.16
In the viewpoint of cancer therapy, the modulation of miRNA levels is considered an attractive approach since miRNAs act as key regulators in cancer.17,18 Thus, there have been efforts to find the effective biomolecules (e.g., small molecules and peptide nucleic acids) which can regulate specific miRNAs and their biological functions such as induction of apoptosis and inhibition of cell proliferation.19,20 In this study, we focused on the miR29-p53 regulatory pathway because the modulation of p53 activity leads to apoptosis in cancer cells. Therefore, we attempted to identify novel modulators for miR29-p53 pathway as potent anticancer drugs.
Peptides are feasible therapeutic agents due to their small size, ease of modification, and cell-penetrating ability.21,22 Among various types of peptides, amphiphilic peptides which are a class of cell-penetrating peptides, can be used for powerful applications such as drug delivery because they facilitate cellular uptake of various molecules leading to therapeutic effects in target cells.23,24 Furthermore, previous studies have shown that amphiphilic α-helical peptides can selectively and strongly bind to hairpin RNAs.25,26,27 In accordance with this idea, we hypothesized the existence of peptide(s) that can affect miR29 expression by up-regulating p53 activity in cancer cells.
For many years, there have been extensive efforts to find effective screening methods for the identification of peptide-based therapeutic agents. For example, synthetic peptide libraries have been developed for the selection of peptides, which are effective at disrupting disease pathways.28,29 Here, we performed cell-based screening in order to identify novel peptides, which induce apoptosis via the miR29-mediated p53 pathway in cancer cells. We further investigated a peptide (named LK-L1C/K6W/L8C) that binds to the terminal loop region of pre-miR29b, which improves the complex formation between pre-miR29b and Dicer, promoting Dicer processing. We also showed that LK-L1C/K6W/L8C improves p53 stability and increases apoptosis induction in cancer cells. In addition to identifying novel therapeutic peptides, our study may be able to provide insight into the mechanisms through which peptides achieve anticancer effects.
Results
Anticancer activity of LK-L1C/K6W/L8C through miR29 and p53
To identify potential anticancer peptides, we screened 60 amphiphilic peptides, which were composed of 16 amino acids in length as shown in Supplementary Table S1. Previous studies reported that amphiphilic peptides which are comprised of Leu/Lys (LK) or Leu/Arg (LR) can bind to hairpin RNAs.25,30 Therefore, we synthesized new amphiphilic peptides that were positionally mutated from LK (peptide number 1) and LR (peptide number 39) as the starting peptides (see Supplementary Table S1).
First, to screen for the candidate peptides that could enhance p53-activated apoptosis, we evaluated the cell viability for peptide-treated cells using the MTS (3-[4,5,dimethylthiazol-2-yl]-5-[3-carboxymethoxy-phenyl]-2-[4-sulfophenyl]-2H-tetrazolium, inner salt) assay (see Supplementary Figure S1a). In this screen, we used the stable cell line expressing miR29-inducible system in HeLa to select the peptides, which could promote miR29-mediated p53 activation at physiologically relevant conditions. This was because miR29 levels were very low in HeLa cells (see Supplementary Figure S2a).15 Cells were treated with 60 peptides for 72 hours in the presence or absence of doxycycline (dox). Primary screening showed that the 60 peptides affected cell viability differently. Since we were interested only in the peptides that could up-regulate p53 activity, seven candidates exhibiting the lowest cell viabilities were selected for further experimentation: peptide 5, 8, 23, 28, 35, 44, and 57. In all the experiments, peptide number 60 (named LR-R13Aad/L14W/A15R) was included as a control because its effect on cell viability was similar to that of the mock control.
To narrow down the candidates from the screen, we examined miR29 expression levels in cells after peptide treatment, as well as tested p53 activity. Excluding peptide 5, the other six peptides and the control peptide (LR-R13Aad/L14W/A15R) were examined in the following experiments. Peptide 5 was excluded from further study due to its irreproducibility.
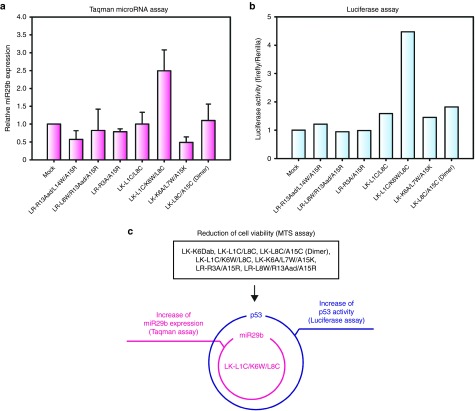
We examined miR29 expression in the selected peptide-treated cells using quantitative real-time polymerase chain reaction. There are three members of the miR29 family: miR29a, miR29b, and miR29c. However, we only observed miR29a and miR29b expression levels because miR29c is not expressed in the miR29-inducible HeLa cell line. Initially, we tested miR29a expression levels in peptide-treated cells. The results showed that all of the candidates had no effect on miR29a levels (see Supplementary Figure S1b). Later, we analyzed miR29b expression levels in peptide-treated cells. Interestingly, mature miR29b expression was ~2.5-fold higher in cells treated with LK-L1C/K6W/L8C (peptide 28) than in cells from the mock control (Figure 1a), indicating that LK-L1C/K6W/L8C can increase the levels of mature miR29b.
Figure 1.
LK-L1C/K6W/L8C increases microRNA-29b (miR29b) expression and enhances p53 activity in miR29-inducible HeLa cells. (a) Relative expression of mature miR29b was examined in the six candidate peptide-treated miR29-inducible HeLa cells by the Taqman miRNA assay. At 24 hours after treatment, mature miR29b expression was estimated. Mock indicates H2O-treated cells as a control. Relative miR29b expression was normalized to the expression levels of U6 snRNA used as endogenous control. To estimate independent effects of miR29, the relative expression of miR29b in miR29-inducible HeLa cells treated with dox was first normalized to that of cells treated without dox in each peptide sample. Values for the relative miR29b expression of peptide-treated cells were further normalized to that of mock control (n = 3, mean ± SD). (b) Luciferase activity for p53 expression was analyzed by the pG13-luciferase assay. miR29-inducible HeLa cells were treated with the six candidate peptides. Luciferase activity was measured after 24 hours incubation and normalized to mock control, as demonstrated in Figure 1a. Data represent mean of the two independent experiments. (c) Summary of screening analysis for candidate peptides that enhance miR29b-induced p53 activity.
Since miR29b activates p53 as described,15 we tested p53 activity with the pG13-luciferase assay. In this assay, the activity of p53 can be quantified by the pG13-luc reporter plasmid contains tandem (x13) p53 binding sites upstream of the luciferase gene.15 Luciferase activity was more than threefold higher in LK-L1C/K6W/L8C-treated cells when compared with the mock control (Figure 1b), suggesting that LK-L1C/K6W/L8C can increase levels of mature miR29b and p53 (Figure 1a, b).
To confirm that both LK-L1C/K6W/L8C and LR-R13Aad/L14W/A15R peptides can readily penetrate into cells, we monitored the intracellular distribution of the peptides by fluorescence microscopy after labeling them with rhodamine (see Supplementary Figure S2b). Cellular uptake of LK-L1C/K6W/L8C and LR-R13Aad/L14W/A15R was observed in low concentration (0.25 μmol/l) compared with the uptake of the hepta-arginine (r7) peptide which is a well-known cell-penetrating peptide.31
Figure 1c summarizes the results of the screening for the peptide candidates that enhance miR29b-induced p53 activity. From these results, LK-L1C/K6W/L8C was determined to be the only candidate that increases both miR29b and p53 activities.
LK-L1C/K6W/L8C promotes Dicer processing of pre-miR29b in vitro
To study how LK-L1C/K6W/L8C increases miR29b levels, we examined whether the process of miR29b biogenesis is affected by LK-L1C/K6W/L8C. Since miRNA is generated consecutively by Drosha and Dicer processing, we performed in vitro processing assays for Drosha and Dicer activities. Primary miR29b (pri-miR29b) substrates were labeled with [α-32P] UTP by in vitro transcription and human Drosha was immunoprecipitated in order to measure Drosha activity (see Supplementary Figure S3d). The in vitro enzymatic activity assay of Drosha with pre-miR29b was carried out with three peptides: LK-L1C/K6W/L8C, LR-R13Aad/L14W/A15R, and LK (see Supplementary Figure S3c). LK peptide is the prototype of LK-L1C/K6W/L8C and was included as a control in this in vitro Drosha processing assay. An analysis of the data shows that LK-L1C/K6W/L8C does not affect Drosha activity during the processing of pri-miR29b.
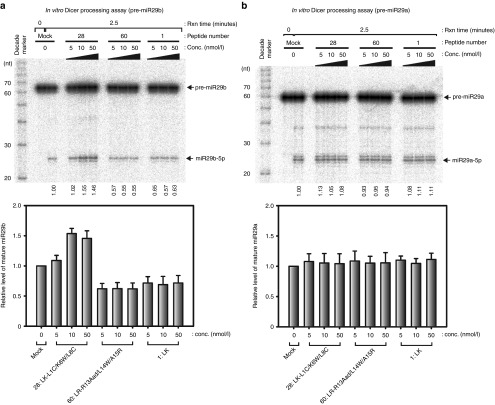
Next, we examined in vitro Dicer processing of pre-miR29b to investigate the effect of LK-L1C/K6W/L8C on Dicer activity. The synthetic pre-miR29 family and pre-let7a substrates were labeled with [γ-32P] ATP at the 5′end, and immunopurified human Dicer was prepared (see Supplementary Figure S3e). The in vitro Dicer processing assay of pre-miR29b was conducted with the same three peptides from the Drosha assay. In the presence of LK-L1C/K6W/L8C, the efficiency of pre-miR29b processing into mature miR29b increased ~1.5-fold when compared with the pre-miR29b processing efficiency of the mock control (Figure 2a). On the other hand, none of the three peptides showed any significant effect on Dicer processing of pre-miR29a (Figure 2b). Furthermore, these peptides had no influence on Dicer processing on both pre-let7a and pre-miR29c (see Supplementary Figure S3a, b). The data suggest that LK-L1C/K6W/L8C can promote in vitro Dicer processing of pre-miR29b, but not of pre-miR29a, pre-miR29c, or pre-let7a.
Figure 2.
LK-L1C/K6W/L8C promotes in vitro Dicer processing of precursor-microRNA-29b (pre-miR29b), but not that of pre-miR29a. (a) In vitro processing assay for pre-miR29b was performed by treating the indicated peptides with immunopurified human Dicer. The dicing reaction was conducted for 2.5 minutes. The RNA size markers (decade marker, Ambion) were displayed on the left side of the gel. Values indicate ratios of mature miRNA to mature and precursor miRNA was quantified and normalized with mock sample. Relative levels of mature miR29b were presented below (n = 3, mean ± SD). (b) In vitro Dicer processing of pre-miR29a was carried out with peptides. Proportion of mature miRNA was quantified and normalized, as described in Figure 2a. Relative levels of mature miR29a were indicated below (n = 3, mean ± SD).
LK-L1C/K6W/L8C facilitates complex formation between pre-miR29b and Dicer by binding to the terminal loop region of pre-miR29b
To understand better, how LK-L1C/K6W/L8C enhances the Dicer processing of pre-miR29b, we examined whether LK-L1C/K6W/L8C binds to pre-miR29b. The RNA footprinting assays of pre-miR29a and pre-miR29b were conducted with RNase T1 and RNase A (see Supplementary Figure S4). As LK-L1C/K6W/L8C concentration increased, the cleavage products of pre-miR29b were reduced (see Supplementary Figure S4a), whereas those of pre-miR29a were not (see Supplementary Figure S4b), indicating that pre-miR29b was protected from both RNase T1 and RNase A. These results suggest that LK-L1C/K6W/L8C can bind to pre-miR29b, but not to pre-miR29a.
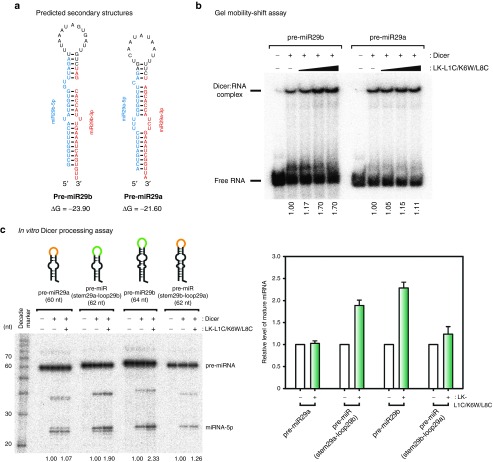
Next, we monitored the interaction between pre-miR29a and Dicer; and the interaction between pre-miR29b and Dicer in the presence of LK-L1C/K6W/L8C. We investigated whether LK-L1C/K6W/L8C can affect complex formation between pre-miR29a or pre-miR29b and Dicer protein by a gel mobility-shift assay. Recombinant human Dicer protein was prepared using the baculovirus expressing system, and both pre-miR29a and pre-miR29b substrates were labeled with [γ-32P] ATP at the 5' end of the RNA. As LK-L1C/K6W/L8C concentration increased, the formation of the Dicer and pre-miR29b complex became more efficient than that of Dicer and pre-miR29a (Figure 3a,b). As can be seen in Figure 2a, the results showed that the efficiency of in vitro Dicer activity increased for pre-miR29b by ~1.7-fold, presumably because Dicer binds to pre-miR29b more easily in the presence of LK-L1C/K6W/L8C (Figure 3b). This indicates that LK-L1C/K6W/L8C can improve the complex formation between pre-miR29b and Dicer, resulting in an increase of processed pre-miR29b by Dicer.
Figure 3.
LK-L1C/K6W/L8C preferentially aids the complex formation between precursor microRNA-29b (pre-miR29b) and Dicer protein via an interaction with the terminal loop region of pre-miR29b. (a) Secondary structures of pre-miR29a and pre-miR29b were indicated. Guide strands of miRNAs are represented in red, and passenger strands of those are in blue. These structures are based on miRbase (http://www.mirbase.org). Value of Gibbs energy was estimated by mfold server (http://mfold.rna.albany.edu/). (b) Gel mobility-shift assay for either pre-miR29a or pre-miR29b and human Dicer was performed. Recombinant human Dicer was incubated at 30 nmol/l with [γ-32P]-labeled pre-miR29a or pre-miR29b. As indicated in Figure 2, LK-L1C/K6W/L8C was treated in a dose-dependent manner (5, 10, and 50 nmol/l). Values indicate ratios of bound-fraction to bound- and unbound-fraction, which were quantified and normalized with each mock sample. (c) In vitro processing assay for pre-miRNAs was performed. Radiolabeled pre-miRNAs was incubated for 1 hour with 30 nmol/l recombinant human Dicer in the absence or presence of 10 nmol/l LK-L1C/K6W/L8C. Values indicate that each ratio of mature miRNA was quantified and normalized, as described in Figure 2a. Relative levels of mature miRNA was presented in right panel (n = 3, mean ± SD).
Recent reports demonstrated that certain peptides affect Dicer activity by binding to the terminal loop regions of pre-miRNA.27,29 Therefore, we tested the location of the region that is crucial for enhanced Dicer processing of pre-miR29b in the presence of LK-L1C/K6W/L8C. Synthetic pre-miRNAs were prepared, similar to pre-miR29a and pre-miR29b but with the terminal loop regions switched (Figure 3a, Supplementary Figure S5). In vitro Dicer processing assays of pre-miRNA were performed with recombinant human Dicer. In the presence of LK-L1C/K6W/L8C, pre-miRNA containing the terminal loop of pre-miR29b was processed ~twofold more efficiently by Dicer when compared with the same pre-miRNA in the absence of LK-L1C/K6W/L8C (Figure 3c). Interestingly, these data suggest that the LK-L1C/K6W/L8C peptide preferentially acts on the terminal loop region of pre-miR29b to promote its Dicer processing.
LK-L1C/K6W/L8C improves p53 protein stability and promotes apoptosis
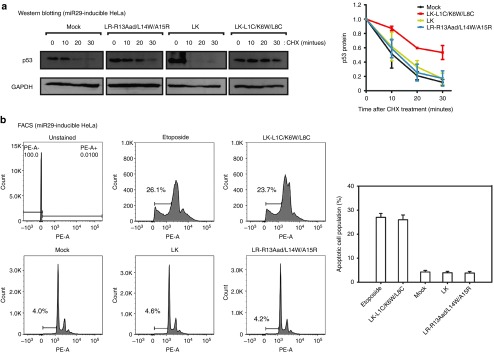
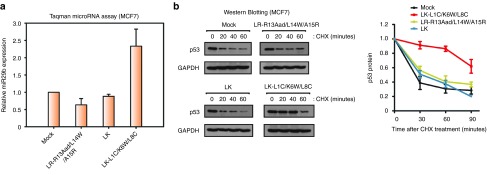
To examine why p53 expression was higher in the LK-L1C/K6W/L8C-treated cells (Figure 1b), we initially measured p53 protein levels by western blot analysis. However, since there was no significant effect of LK-L1C/K6W/L8C on p53 protein levels (data not shown), we instead monitored the stability of the p53 protein by treating it with cycloheximide, an inhibiter of de novo protein synthesis (Figure 4a). After treatment with LK-L1C/K6W/L8C, LR-R13Aad/L14W/A15R, and LK for 24 hours, cycloheximide was added to miR29-inducible HeLa cells. The half-life of the p53 protein is short in most cells but it is remarkably short in the HeLa cells.32 However, we observed a longer p53 half-life in HeLa cells treated with LK-L1C/K6W/L8C. As shown in Figure 4a, the p53 protein was more stable in miR29-inducible HeLa cells treated with LK-L1C/K6W/L8C than in cells treated with other peptides, suggesting that LK-L1C/K6W/L8C activates p53 by stabilizing the protein.
Figure 4.
LK-L1C/K6W/L8C induces apoptosis by improving p53 stability in miR29-inducible HeLa cells. (a) Western blotting for p53 protein visualized protein stability by treating cycloheximide (CHX). Peptides and dox were cotreated on miR29-inducible HeLa cells and then incubated for 24 hours. Prior to cell harvesting, CHX (100 μg/ml) was treated for the indicated time. Protein levels of GAPDH served as a loading control (left panel). The p53 protein levels were quantified and normalized (right panel, n = 3, mean ± SD). (b) Fluorescence-activated cell sorting (FACS) was examined for apoptosis. miR29-inducible HeLa cells were treated with peptides, a mock control and etoposide (50 μmol/l) used as a positive control. The cells were stained propidium idodide (PI) and analyzed by FACS (left panel). Apoptotic cell population was evaluated by sub-G1 peak from FACS data (right panel, n = 3, mean ± SD).
To examine whether LK-L1C/K6W/L8C-treated cells facilitated induction of apoptosis, we evaluated apoptotic cell population using fluorescence-activated cell sorting analysis. miR29-inducible HeLa cells were treated with peptides (LK-L1C/K6W/L8C, LR-R13Aad/L14W/A15R, and LK) and incubated for 48 hours. Prior to analysis for apoptosis, the cells were stained with propidium iodide, a fluorogenic compound, to measure DNA content within the cells. The hypodiploid (sub-G1) peak was assessed to estimate apoptotic cell population. As shown in Figure 4b, the population of apoptotic cells increased as much in the sample treated with LK-L1C/K6W/L8C as it did in the etoposide-treated sample used as the positive control group for apoptosis induction. The results indicate that LK-L1C/K6W/L8C induces p53-activated apoptosis.
To confirm whether LK-L1C/K6W/L8C could induce apoptosis due to its cytotoxicity (i.e., nonspecific toxicity), we tested the cytotoxicity of the peptide by a cell viability assay. In this experiment, to exclude p53-induced apoptosis effect of the peptide, we used the p53 null mutant cell lines (HCT116 p53KD) that stably express shRNA against p53 (see Supplementary Figure S6a). After the treatment of peptides (LK-L1C/K6W/L8C and LR-R13Aad/L14W/A15R) in a dose-dependent manner, we evaluated the cell viabilities by the MTS (3-[4,5,dimethylthiazol-2-yl]-5-[3-carboxymethoxy-phenyl]-2-[4-sulfophenyl]-2H-tetrazolium, inner salt) assay (see Supplementary Figure S6b). The data show that the peptides have little cytotoxicity as low as the mock control.
LK-L1C/K6W/L8C elevates miR29b expression and enhances p53 activity in MCF7 cells
Cervical malignant HeLa cells are developed by the human papillomaviruses, containing the E6 protein, which stimulates p53 degradation.32 Therefore, it was possible that LK-L1C/K6W/L8C improved p53 stability by interfering with the interaction of the E6 proteins with p53. In order to rule out this alternative explanation, we investigated whether LK-L1C/K6W/L8C could also extend the turnover rate of p53 protein in other cells, such as MCF7 breast carcinoma cells. Like the HeLa cells, MCF7 cells encoded normal p53 genes and expressed low levels of miR29.15 Moreover, LK-L1C/K6W/L8C can also penetrate into the MCF7 cell membranes at low concentrations as shown in see Supplementary Figure S7a. In accordance with the data described above (Figure 1a), relative expression of miR29b in MCF7 cells treated with LK-L1C/K6W/L8C was elevated ~2.5-fold (Figure 5a). In addition, p53 protein was accumulated in MCF7 cells incubated with LK-L1C/K6W/L8C (Figure 5b), demonstrating that LK-L1C/K6W/L8C could improve the stability of p53 induced by miR29b. Consequently, LK-L1C/K6W/L8C can also induce apoptosis in MCF7 cells (see Supplementary Figure S7b).
Figure 5.
LK-L1C/K6W/L8C enhances expression of microRNA-29b (miR29b) and promotes p53 stability in MCF7 breast carcinoma cells. (a) Relative expression of miR29b in the indicated peptide-treated MCF7 cells was evaluated by quantitative polymerase chain reaction. Mock, LR-R13Aad/L14W/A15R, and LK peptides were used as controls. Values for relative miR29b expression of samples treated with each peptide were normalized to that of mock sample (n = 3, mean ± SD). (b) Western blotting of p53 protein visualized protein stability after treating the indicated peptides. GAPDH served as a loading control. Quantification of the p53 protein levels was presented in graph (n = 3, mean ± SD).
LK-L1C/K6W/L8C promotes p53 protein stability via the increase in miR29b levels
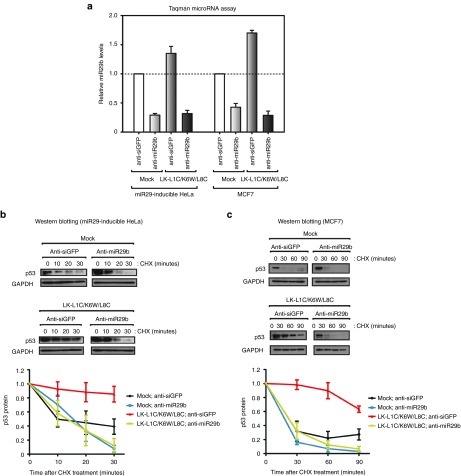
Several studies have reported that the pro-apoptotic peptide (KLAKLA)2, a peptide from one of the typical classes of amphiphilic peptides, disrupts mitochondrial membranes to cause apoptosis.33,34 To confirm that the mechanism of apoptosis induction by LK-L1C/K6W/L8C was through miR29b-mediated p53 activation, endogenous miR29b was inhibited using 2′-O-methylated antisense miR29b (anti-miR29b). This material is a synthetic reverse complement of mature miRNA and generally used to suppress the function of miRNA by binding tightly to the miRNA.35 As shown in Figure 6a, relative miR29b levels were assessed by the Taqman miRNA assay, confirming that endogenous levels of miR29b were successfully down-regulated in both miR29-inducible HeLa and MCF7 cells. It should be noted that the miR29b levels were elevated in the presence of LK-L1C/K6W/L8C in anti-siGFP-treated cells (control).
Figure 6.
Inhibition of microRNA-29b (miR29b) has no effect on p53 stability in both LK-L1C/K6W/L8C-treated miR29-inducible HeLa and MCF7 cells. (a) Relative miR29b levels were estimated in miR29-inducible HeLa and MCF7 cells treated with antisense miR29b (anti-miR29b) in the presence or absence of LK-L1C/K6W/L8C. Anti-siGFP served as control small interfering RNA (siRNA). Values were normalized to controls (anti-siGFP and mock)-treated samples (n = 3, mean ± SD). (b, c) Western blotting of p53 protein visualized protein stability for p53 in both (b) miR29-inducible HeLa and (c) MCF7 cells cotreated with anti-miR29b and LK-L1C/K6W/L8C. Quantification of the p53 protein levels was presented in below (n = 3, mean ± SD).
Subsequently, a p53 stability test was performed in cells treated with anti-miR29b in the presence or absence of LK-L1C/K6W/L8C using western blot analysis (Figure 6b, c). In miR29-inducible HeLa and MCF7 cells, p53 proteins were degraded in samples treated with anti-miR29b in spite of a cotreatment with LK-L1C/K6W/L8C, exhibiting similar stability compared with the p53 proteins in the mock control samples. These data indicate that LK-L1C/K6W/L8C improves p53 protein stability by increasing the level of mature miR29b in cancer cells.
Discussion
There is clear consensus that p53 is a key element in overcoming cancer diseases.36,37 Additionally, miRNAs are critical regulators for controlling cancer-associated genes.5,6,7 Between many miRNAs in the p53 pathway, miR29 family is a good therapeutic target because it can up-regulate p53 activity and induce apoptosis in cancer cells.15 Unfortunately, direct introduction of miR29 into cells is an unfeasible therapeutic method because miRNA and siRNA have poor cell penetration capabilities.38,39 Therefore, we tried to find alternative approaches to increasing miR29 levels in cancer cells.
A great majority of available drug therapies for human diseases rely on functional peptides to achieve therapeutic results.40 Peptides have several benefits that can be easily modified for diverse applications.21,22,40 Cell-penetrating peptides enhance penetration to intracellular regions due to their cationic residues, so they are commonly used as delivery agents of drugs.23,24 In particular, amphiphilic peptides, a class of the cell-penetrating peptides, show efficient uptake into cell membranes even at lower concentrations compared with r7, a common cell-penetrating peptides (see Supplementary Figures S2b and S7a).
Unlike several studies of peptides as delivery vehicles,41,42,43 we attempted to screen for synthesized amphiphilic peptides that have therapeutic activities (e.g., anticancer activity) and could facilitate their own delivery. For example, many reports found different synthetic peptides that could restrict cancer development through independent mechanisms, involving induction of apoptosis, inhibition of angiogenesis and cell proliferation.29,44,45
In this study, we identified the LK-L1C/K6W/L8C peptide as a directed modulator of the miR29b-mediated p53 pathway, achieving its effect through the regulation of Dicer activity. We performed cell-based screening for peptide candidates by evaluating their ability to decrease cell viability, to increase the expression of miR29b, and to promote p53 activity (Figure 1c). Our data revealed that LK-L1C/K6W/L8C could bind to pre-miR29b and promote its processing to generate mature miR29b by Dicer, which facilitates apoptosis induction by improving the stability of p53 protein in cancer cells.
Interestingly, our results showed that the Dicer processing of pre-miR29b was stimulated in the presence of LK-L1C/K6W/L8C, but not pre-miR29a and pre-miR29c, which are the other members of the pre-miR29 family, or pre-let7a (Figure 2a,b and Supplementary Figure S3a, b). These results suggest that LK-L1C/K6W/L8C may selectively affect pre-miR29b processing by Dicer. In addition, our data demonstrate that LK-L1C/K6W/L8C preferentially interacts with the terminal loop region of pre-miR29b, leading to the promotion of cleavage activity by Dicer on pre-miR29b (Figure 3b,c and Supplementary Figure S4). As supporting evidences, independent groups reported that synthetic peptides can bind to the terminal loop regions of pre-miRNAs and affect their processing by Dicer.27,29 Moreover, the synthetic peptides could change the conformation of pre-miRNAs and enhanced the processing activity of Dicer.27 Therefore, we propose that LK-L1C/K6W/L8C functionally binds to the terminal loop region of pre-miR29b, and improves the complex formation between pre-miR29b and the Dicer protein.
Activation and stability of the p53 protein are essential for defending against cancer development.1 We show herein that LK-L1C/K6W/L8C can improve p53 protein stability and induce apoptosis in HeLa and MCF7 cancer cells (Figures 4 and 5b and see Supplementary Figure S7b). In addition, we show further evidence that LK-L1C/K6W/L8C directly promotes p53 stability via increased miR29b levels in cancer cells (Figure 6). These results suggest that LK-L1C/K6W/L8C affects directly on miR29b-p53 pathway and it could be a promising candidate for peptide-based anticancer drugs.
In conclusion, we identified the first directed modulator of miR29b-p53 pathway. Moreover, our study revealed that the synthetic peptide LK-L1C/K6W/L8C can bind to terminal loop region of pre-miR29b and promotes its Dicer processing by fostering the complex formation between pre-miR29b and the Dicer protein. This leads to elevated levels of miR29b, thus resulting in an increase in p53 activity and subsequently a higher rate of apoptosis induction in cancer cells. We strongly suggest that LK-L1C/K6W/L8C can primarily bind to pre-miR29b due to the abundance of pre-miR29b, which is the main target of the peptide, thus having predominant influence on the miR29b-p53 pathway in cancer cells. Therefore, we suggest that LK-L1C/K6W/L8C is a potent anticancer peptide and potentially applicable as therapeutic drugs.
In this study, we selected the effective peptide on the miR29b-p53 pathway from rather small scale of library. Furthermore, to utilize the peptide as a therapeutic agent for cancer treatment, we will develop secondary libraries systematically with the strategy as previously described,25 and improve the peptide more effective and stable in cells. In addition, the effect of LK-L1C/K6W/L8C in vivo system (animal model) should be tested in further investigation. Nevertheless, our study may contribute to the development of novel therapeutic drugs.
Materials and methods
Cell culture. Tetracycline-inducible miR29a/b HeLa cell lines (gifts from V. Narry Kim) were established by transfecting cells with pcDNA-miR29a/b plasmid, as described.15 Doxycycline (dox, 5 mg/ml), a tetracycline analog, was treated to induce miR29 for 24–72 hours. These cells were maintained in Dulbecco's Modified Eagle's Medium supplemented with 10% (v/v) Fetal Bovine Serum. Also, HeLa, MCF7, and HCT116p53KD (gifts from V. Narry Kim) cells were cultured in Dulbecco's Modified Eagle's Medium with 10% Fetal Bovine Serum. HCT116, SW480, and SNU-638 were cultured in Roswell Park Memorial Institute 1640 (RPMI1640) with 10% (v/v) Fetal Bovine Serum. All cells were cultured in a 37°C incubator at 5% CO2.
Peptide synthesis. All peptides were synthesized by a standard solid phase peptide synthesis method using Fluoren-9-ylmethoxy carbonyl-protecting group links amide 4-methylbenzhydrylamine resin (0.6 mmol/g) as 25 μmol scale. The amino acids were purchased from NovaBiochem (Darmstadt, Germany). The synthesized peptides were acetylated at the N-terminus, purified by HPLC (Agilent 1,100, Santa Clara, CA, column Zorbax C18) and validated using MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA), as previously described.27 For the rhodamine conjugation, rhodamines were synthesized at the N-terminus of peptides without acetylation.
Cell viability assay. For primary screening, miR29-inducible HeLa cells were seeded in 96-well plates. One day later, cells were treated with peptides (0.25 μmol/l) and dox (Sigma, Darmstadt, Germany). At 72 hours after treatment, 20 μl of MTS (3-[4,5,dimethylthiazol-2-yl]-5-[3-carboxymethoxy-phenyl]-2-[4-sulfophenyl]-2H-tetrazolium, inner salt) solution (Promega, Madison, WI) was added to each well and then the cells were incubated at 37°C for 1 hour. Absorbance at 490 nm was then measured by a microplate reader (Tecan, Männedorf, Switzerland). Viability values were double-normalized to a mock control, as mentioned in Figure 1a.
For the cytotoxicity assay, HCT116 p53KD cells were seeded in 96-well plates. One day later, peptides (LK-L1C/K6W/L8C and LR-R13Aad/L14W/A15R) and the mock control were treated at concentrations of 50, 100, 250, 500, and 1,000 nmol/l. After 24 hours incubation, the MTS (3-[4,5,dimethylthiazol-2-yl]-5-[3-carboxymethoxy-phenyl]-2-[4-sulfophenyl]-2H-tetrazolium, inner salt) solution was added and the samples were measured by a microplate reader. All experiments were conducted in triplicates.
Northern blotting of miRNA. Total RNAs were extracted by using TRI Reagent solution (Ambion, Waltham, MA) according to the manufacturer's protocol. Total RNAs were resolved on 15% UREA-PAGE and transferred onto Hybond-NX membrane (GE healthcare, Chicago, IL), as described previously.46 The oligonucleotide probes were labeled with [γ-32P] ATP using T4 polynucleotide kinase (Takara, Shiga, Japan). These DNA probes were as follows: probe for miR29a, 5′-TAACCGATTTCAGATGGTGCTA-3′; probe for miR29b, 5′-AACACTGATTTCAAATGGTGCTA-3′; probe for U6 snRNA, 5′-TGCGTGTCATCCTTGCGCAGG-3′.
Quantitative real-time PCR (Taqman miRNA assay). miR29-inducible HeLa or MCF7 cells were seeded in 24-well plates. Peptides (1 μmol/l) and dox were treated for 24 hours before total RNA extraction. Taqman probes were used for measuring mature miR29a, miR29b, and U6 snRNA expression. The reaction was performed by TaqMan MicroRNA assay kit (Applied Biosystem, Waltham, MA) in accordance with the manufacturer's instructions and then conducted using Light Cycler II 480 (Roche, Basel, Switzerland). Mature miR29 expression was normalized to U6 snRNA. All experiments were carried out in triplicates.
Luciferase assay. miR29-inducible HeLa cells were plated into 24-well plates. After 1 day, peptides (1 μmol/l) were treated with or without dox. pG13-luciferase plasmid (a gift from V. Narry Kim) and Renilla luciferase control plasmid were cotransfected into the cells using Lipofectamine 2,000 (Invitrogen, Waltham, MA).15 After incubation for 24 hours, luciferase activity was measured by performing dual-luciferase assay using a Glomax96 Microplate Luminometer (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Antisense miRNA transfection. For inhibition of miR29b expression, antisense miR29b and siGFP RNA were synthesized with 2′-O-methyl RNA modification (Bioneer, Daejeon, Republic of Korea): anti-miR29b, 5′-AACACUGAUUUCAAAUGGUGCUA-3′ and anti-siGFP, 5′-AACAUCGCCAUCUAAUUCA-3′. One day before transfection, miR29-inducible HeLa or MCF7 cells were seeded. After treating 1 μmol/l of LK-L1C/K6W/L8C peptide, 200 nmol/l of anti-siGFP or anti-miR29b was transfected into cells using Lipofectamine 2,000 (Invitrogen). Prior to extraction of total RNAs or proteins, the cells were incubated at 37°C for 24 hours.
Immunoprecipitation and in vitro processing assay. For immunoprecipitation of Flag tagged human Dicer or human Drosha protein, transfected HEK293T cells were incubated in lysis buffer, as previously described.47,48 Whole cell extracts were rotated constantly with anti-Flag M2 affinity gel (Sigma) at 4°C for 1 hour. The beads were washed with lysis buffer.
The Dicer processing assay reaction was performed as described previously.47 Precursor miRNA substrates were generated and labeled with [γ-32P] ATP at 5′ end of RNA. The radiolabeled RNAs were incubated at 37°C for 1.5–2.5 minutes with the immunopurified or baculovirus-expressed recombinant Dicer and the peptides. The reaction was stopped by adding Proteinase K solution (Roche). The processed RNAs were resolved on 12% UREA-PAGE and analyzed using BAS-2,500 (Fuji, Tokyo, Japan).
The Drosha processing assay was conducted as described.48 Primary miR29b (pri-miR29b) transcripts were synthesized with [α-32P] UTP using general in vitro transcription method.49 The radiolabeled pri-miR29b transcripts were incubated at 37°C for 15 minutes with immunopurified Drosha and the peptides. The processed RNAs were resolved on 12% UREA-PAGE and analyzed using BAS-2,500 (Fuji).
Gel mobility-shift assay. Gel mobility-shift assay was carried out with the following condition: 30 nmol/l of recombinant human Dicer, 0.2 nmol/l of radiolabeled pre-miR29a or pre-miR29b, 1 μg of bovine serum albumin (Takara), 1 mmol/l Dithiothreitol in binding buffer (20 mmol/l HEPES at pH 7.4, 50 mmol/l KCl, 0.5 mmol/l EDTA, 10% (v/v) Glycerol). The reaction was incubated with LK-L1C/K6W/L8C peptide (5, 10, and 50 nmol/l) at 30°C for 20 minutes. Free pre-miRNAs and Dicer-RNA complexes were resolved on 6% nondenaturing PAGE and analyzed using BAS-2,500 (Fuji).
In vitro RNA footprinting assay. The [γ-32P] 5′ end labeled RNA (pre-miR29b) was heated in buffer (20 mmol/l HEPES at pH 7.4, 1 mmol/l MgCl2, 5 mmol/l KCl, and 140 mmol/l NaCl) at 80°C for 10 minutes, and then slowly cooled down to room temperature. RNase T1 (0.01 U, Ambion), and RNase A (0.5 U, Ambion) were added with LK-L1C/K6W/L8C peptide (5, 10, and 50 nmol/l) and incubated at room temperature for 15 minutes and 2 minutes, respectively. Alkaline hydrolysis of pre-miR29b was carried out with alkaline hydrolysis buffer (Ambion) and yeast RNA (3 μg, Ambion) and heated at 95°C for 10 minutes. Both RNase T1 and RNase A ladders were generated according to the manufacturer's protocol (Ambion). The processed RNA samples were resolved on 20% UREA-PAGE and analyzed using BAS-2500 (Fuji).
Western blotting. Total cell lysates were prepared by RIPA buffer (25 mmol/l Tris-Cl at pH 7.6, 150 mmol/l NaCl, 1% (v/v) NP-40) with protease inhibitor cocktail (Roche). Protein samples were separated on 8–10% SDS-PAGE and transferred to PVDF membrane (Millipore, Darmstadt, Germany). The membrane was incubated with primary antibodies to anti-Flag rabbit (Sigma), anti-p53 mouse (Santa Cruz Biotechnology, Dallas, TX), anti-GAPDH mouse (Abcam, Cambridge, UK) and anti-α-tubulin rabbit (Abcam). Secondary antibodies were goat anti-rabbit IgG HRP-conjugated (Jackson, West Grove, PA) antibody and donkey anti-mouse (Jackson). Proteins were detected by ECL solution (Thermo Scientific, Waltham, MA) and exposed to X-ray film.
Fluorescence-activated cell sorting for apoptosis analysis. miR29-inducible HeLa cells or MCF7 cells were seeded. One day later, 1 μmol/l peptides and 50 μmol/l etoposide (Sigma) were treated. After 48 hours incubation, total cells were harvested in phosphate-buffered saline and fixed by 70% (v/v) ethanol for 16 hours. Following phosphate-buffered saline washing, the cells were stained in propidium iodide (50 μg/ml) with RNase A (5 μg/ml). Stained cells were then analyzed for DNA content by fluorescence-activated cell sorting AriaII (Becton Dickinson, Franklin Lakes, NJ) and displayed in a graph using FlowJo software, Ashland, OR. All experiments were conducted in triplicates.
Fluorescence microscopy. For observation of the peptide penetration, miR29-inducible HeLa or MCF7 cells were seeded in 24-well plate. One day later, rhodamine-labeled peptides were treated in a dose-dependent manner (0, 0.25, 1, 2.5, 5, and 10 μmol/l). In addition, Hoechst (5 μg/ml, Invitrogen) was cotreated with the labeled peptides. The cells were then incubated at 37°C for 6 hours. Following phosphate-buffered saline washing, 4% (w/v) paraformaldehyde (Sigma) was treated for 10 minutes. Then, the cells were washed twice with phosphate-buffered saline and observed using fluorescence microscopy (Nikon Eclipse Ti, New York, NY ) equipped with 40X objective.
SUPPLEMENTARY MATERIAL Figure S1. Screening for peptides promoting p53-activated apoptosis and increasing miR29a levels. Figure S2. miR29-inducible HeLa cells express both miR29a and miR29b in a dox-dependent manner and facilitate cellular uptake of amphiphilic peptides. Figure S3. LK-L1C/K6W/L8C has no influence on Dicer processing of pre-let7a and pre-miR29c, and Drosha processing of pri-miR29b in vitro. Figure S4. LK-L1C/K6W/L8C binds to pre-miR29b, but not to pre-miR29a. Figure S5. Scheme of pre-miRNA switching the terminal loop region between pre-miR29a and pre-miR29b. Figure S6. The cytotoxicity of the peptides in a dose-dependent manner. Figure S7. LK-L1C/K6W/L8C readily penetrates and induces apoptosis in MCF7 cells. Table S1. Names and sequences of 60 amphiphilic peptides.
Acknowledgments
We are grateful for helpful discussion with members of the Shin laboratory. We also thank V. Narry Kim for providing pG13-luciferase plasmid and miR29-inducible HeLa cells. This work was supported by the Next-Generation BioGreen 21 Program (No. PJ01115601), Rural Development Administration, Republic of Korea and the Seoul National University Research Grant (Brain Fusion Program: 500-20120248). The authors declare that there are no conflicts of interest.
Supplementary Material
References
- Kim, E, Giese, A and Deppert, W (2009). Wild-type p53 in cancer cells: when a guardian turns into a blackguard. Biochem Pharmacol 77: 11–20. [DOI] [PubMed] [Google Scholar]
- Ben-Porath, I and Weinberg, RA (2005). The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 37: 961–976. [DOI] [PubMed] [Google Scholar]
- Hermeking, H (2012). MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 12: 613–626. [DOI] [PubMed] [Google Scholar]
- Bartel, DP (2009). MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, GA and Croce, CM (2006). MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- Garzon, R, Calin, GA and Croce, CM (2009). MicroRNAs in Cancer. Annu Rev Med 60: 167–179. [DOI] [PubMed] [Google Scholar]
- Farazi, TA, Spitzer, JI, Morozov, P and Tuschl, T (2011). miRNAs in human cancer. J Pathol 223: 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B, Pan, X, Cobb, GP and Anderson, TA (2007). microRNAs as oncogenes and tumor suppressors. Dev Biol 302: 1–12. [DOI] [PubMed] [Google Scholar]
- Volinia, S, Calin, GA, Liu, CG, Ambs, S, Cimmino, A, Petrocca, F, et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney, DC, Flesken-Nikitin, A, Godwin, AK, Wang, W and Nikitin, AY (2007). MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res 67: 8433–8438. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira, N, Marciano, E, Meiri, E, Spector, Y, Rosenfeld, N, Moskovits, N et al. (2007). Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743. [DOI] [PubMed] [Google Scholar]
- Suzuki, HI, Yamagata, K, Sugimoto, K, Iwamoto, T, Kato, S and Miyazono, K (2009). Modulation of microRNA processing by p53. Nature 460: 529–533. [DOI] [PubMed] [Google Scholar]
- Su, X, Chakravarti, D, Cho, MS, Liu, L, Gi, YJ, Lin, YL et al. (2010). TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nat 467: 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, K and Ochiya, T (2014). Genetic networks lead and follow tumor development: microRNA regulation of cell cycle and apoptosis in the p53 pathways. Biomed Res Int 2014: 724–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, SY, Lee, JH, Ha, M, Nam, JW and Kim, VN (2009). miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 16: 23–29. [DOI] [PubMed] [Google Scholar]
- Wang, Y, Huang, JW, Castella, M, Huntsman, DG and Taniguchi, T (2014). p53 is positively regulated by miR-542-3p. Cancer Res 74: 3218–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, PM, and Pedroso de Lima, MC (2013). MicroRNAs as molecular targets for cancer therapy: On the modulation of MicroRNA expression. Pharmaceuticals 6: 1195–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y, Wang, X, Yang, Y, Zhang, C, Ertl, HC, and Zhou, D (2014). Survivin-targeting artificial MicroRNAs mediated by adenovirus suppress tumor activity in cancer cells and Xenograft models. Mol Ther Nucleic Acids 3: e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z, Li, CH, Chan, SL, Xu, F, Feng, L, Wang, Y et al. (2014). A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res 74: 6236–6247. [DOI] [PubMed] [Google Scholar]
- Brognara, E, Fabbri, E, Aimi, F, Manicardi, A, Bianchi, N, Finotti, A et al. (2012). Peptide nucleic acids targeting miR-221 modulate p27Kip1 expression in breast cancer MDA-MB-231 cells. Int J Oncol 41: 2119–2127. [DOI] [PubMed] [Google Scholar]
- Craik, DJ, Fairlie, DP, Liras, S and Price, D (2013). The future of peptide-based drugs. Chem Biol Drug Des 81: 136–147. [DOI] [PubMed] [Google Scholar]
- Sun, L (2013). Peptide-based drug development. Mod Chem appl 1: e103. [Google Scholar]
- Mohammadi, S, Shahbazi Mojarrad, J, Zakeri-Milani, P, Shirani, A, Mussa Farkhani, S, Samadi, N et al. (2015). Synthesis and in vitro evaluation of amphiphilic peptides and their nanostructured conjugates. Adv Pharm Bull 5: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järver, P, Coursindel, T, Andaloussi, SE, Godfrey, C, Wood, MJ and Gait, MJ (2012). Peptide-mediated cell and in vivo delivery of antisense oligonucleotides and siRNA. Mol Ther Nucleic Acids 1: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, SJ, Hyun, S, Kieft, JS and Yu, J (2009). An approach to the construction of tailor-made amphiphilic peptides that strongly and selectively bind to hairpin RNA targets. J Am Chem Soc 131: 2224–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai, J, Yoon, T, Kim, ND, Lee, IS, Yu, J and Shin, I (2012). High-throughput profiling of peptide-RNA interactions using peptide microarrays. J Am Chem Soc 134: 19287–19296. [DOI] [PubMed] [Google Scholar]
- Hyun, S, Han, A, Jo, MH, Hohng, S and Yu, J (2014). Dicer nuclease-promoted production of Let7a-1 microRNA is enhanced in the presence of tryptophan-containing amphiphilic peptides. Chembiochem 15: 1651–1659. [DOI] [PubMed] [Google Scholar]
- Dietrich, U, Dürr, R and Koch, J (2013). Peptides as drugs: from screening to application. Curr Pharm Biotechnol 14: 501–512. [DOI] [PubMed] [Google Scholar]
- Bose, D, Nahar, S, Rai, MK, Ray, A, Chakraborty, K and Maiti, S (2015). Selective inhibition of miR-21 by phage display screened peptide. Nucleic Acids Res 43: 4342–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, S, Han, A and Yu, J (2009). Photocrosslinking of RNA and photoMet-containing amphiphilic alpha-helical peptides. Chembiochem 10: 987–989. [DOI] [PubMed] [Google Scholar]
- Mitchell, DJ, Kim, DT, Steinman, L, Fathman, CG and Rothbard, JB (2000). Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res 56: 318–325. [DOI] [PubMed] [Google Scholar]
- Scheffner, M, Werness, BA, Huibregtse, JM, Levine, AJ and Howley, PM (1990). The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63: 1129–1136. [DOI] [PubMed] [Google Scholar]
- Ellerby, HM, Arap, W, Ellerby, LM, Kain, R, Andrusiak, R, Rio, GD et al. (1999). Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med 5: 1032–1038. [DOI] [PubMed] [Google Scholar]
- Law, B, Quinti, L, Choi, Y, Weissleder, R and Tung, CH (2006). A mitochondrial targeted fusion peptide exhibits remarkable cytotoxicity. Mol Cancer Ther 5: 1944–1949. [DOI] [PubMed] [Google Scholar]
- Lennox, KA and Behlke, MA (2011). Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther 18: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Fei, P, Bernhard, EJ and El-Deiry, WS (2002). Tissue-specific induction of p53 targets in vivo. Cancer Res 62: 7316–7327. [PubMed] [Google Scholar]
- Selivanova, G and Wiman, KG (2007). Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene 26: 2243–2254. [DOI] [PubMed] [Google Scholar]
- Thomas, M and Deiters, A (2013). MicroRNA miR-122 as a therapeutic target for oligonucleotides and small molecules. Curr Med Chem 20: 3629–3640. [DOI] [PubMed] [Google Scholar]
- Lam, JK, Chow, MY, Zhang, Y and Leung, SW (2015). siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol Ther Nucleic Acids 4: e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, P, Lisowski, V, Martinez, J and Khrestchatisky, M (2010). Synthetic therapeutic peptides: science and market. Drug Discov Today 15: 40–56. [DOI] [PubMed] [Google Scholar]
- Watt, PM (2006). Screening for peptide drugs from the natural repertoire of biodiverse protein folds. Nat Biotechnol 24: 177–183. [DOI] [PubMed] [Google Scholar]
- Lim, KJ, Sung, BH, Shin, JR, Lee, YW, Kim, da J, Yang, KS et al. (2013). A cancer specific cell-penetrating peptide, BR2, for the efficient delivery of an scFv into cancer cells. PLoS One 8: e66084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, JS, Lee, JY, Choi, YS, Chung, CP, Chong, PC and Park, YJ (2013). Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomat 34: 4347–4359. [DOI] [PubMed] [Google Scholar]
- Griffioen, AW, van der Schaft, DW, Barendsz-Janson, AF, Cox, A, Struijker Boudier, HA, Hillen, HF et al. (2001). Anginex, a designed peptide that inhibits angiogenesis. Biochem J 354(Pt 2): 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, HS, Park, CB, Kim, JM, Jang, SA, Park, IY, Kim, MS et al. (2008). Mechanism of anticancer activity of buforin IIb, a histone H2A-derived peptide. Cancer Lett 271: 47–55. [DOI] [PubMed] [Google Scholar]
- Park, JH and Shin, C (2015). Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res 43: 9418–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, JE, Heo, I, Tian, Y, Simanshu, DK, Chang, H, Jee, D et al. (2011). Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nat 475: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y, Ahn, C, Han, J, Choi, H, Kim, J, Yim, J et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nat 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lee, Y, Jeon, K, Lee, JT, Kim, S and Kim, VN (2002). MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.