Abstract
Helicobacter pylori infects more than 50% of the worldwide population. It is mostly found deep in the gastric mucus lining of the stomach, being a major cause of peptic ulcers and gastric adenocarcinoma. To face the increasing resistance of H. pylori to antibiotics, antimicrobial nucleic acid mimics are a promising alternative. In particular, locked nucleic acids (LNA)/2'-OMethyl RNA (2'OMe) have shown to specifically target H. pylori, as evidenced by in situ hybridization. The success of in vivo hybridization depends on the ability of these nucleic acids to penetrate the major physical barriers—the highly viscoelastic gastric mucus and the bacterial cell envelope. We found that LNA/2'OMe is capable of diffusing rapidly through native, undiluted, gastric mucus isolated from porcine stomachs, without degradation. Moreover, although LNA/2'OMe hybridization was still successful without permeabilization and fixation of the bacteria, which is normally part of in vitro studies, the ability of LNA/2'OMe to efficiently hybridize with H. pylori was hampered by the presence of mucus. Future research should focus on developing nanocarriers that shield LNA/2'OMe from components in the gastric mucus, while remaining capable of diffusing through the mucus and delivering these nucleic acid mimics directly into the bacteria.
Keywords: fluorescence in situ hybridization, Helicobacter pylori, locked nucleic acid, native gastric mucus, 2'-OMethyl RNA
Introduction
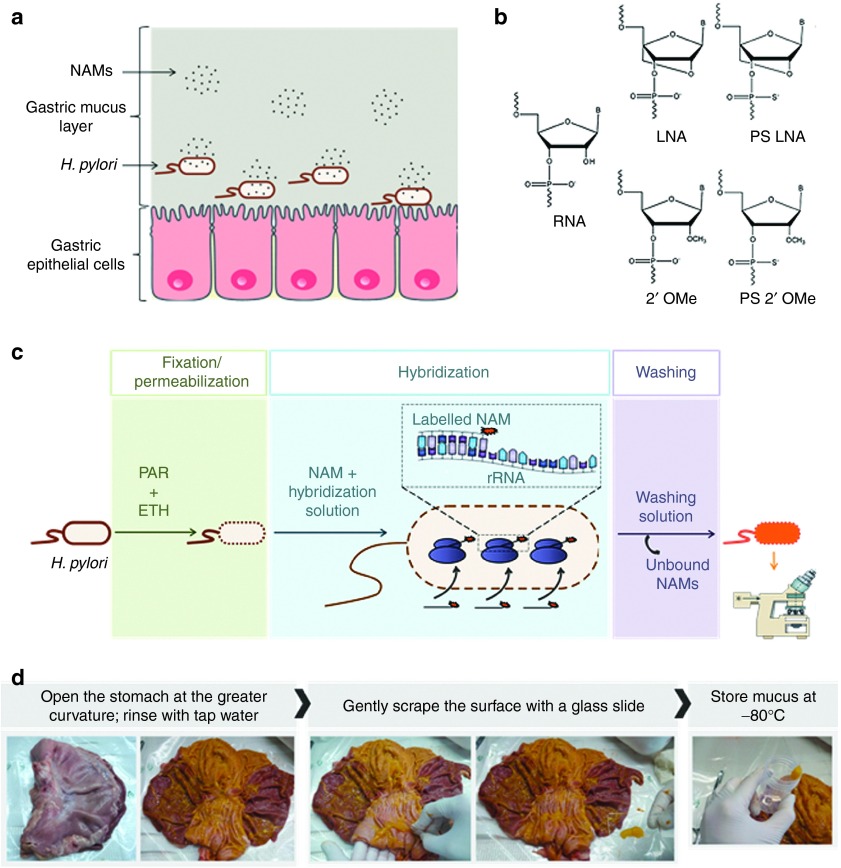
Helicobacter pylori is a stomach-colonizing Gram negative bacterium that infects more than half of the world's population.1,2 Because it increases the susceptibility to gastric cancer, associated with one of the highest mortality rates,3 it is classified as a Group 1 carcinogen by the World Health Organization.4 Moreover, it is associated with 95% of duodenal ulcers and 80% of gastric ulcers.5 H. pylori colonizes almost exclusively the stomach, lying deep in the mucus layer that covers the gastric epithelium,6 and also adhering to the surface of gastric epithelial cells and in deeper regions of the gastric glands (Figure 1a).7,8,9
Figure 1.
Illustration of nucleic acid mimics (NAMs) hybridization to H. pylori and the different components of the implemented model. (a) Schematic representation of NAMs in gastric mucus, on their way to target H. pylori. (b) Monomers of the NAMs used in fluorescence in situ hybridization (FISH), compared to RNA (adapted from32). (c) Illustration of the standard FISH procedure. PAR and ETH being paraformaldehyde and ethanol, respectively (adapted from ref. 30). (d) Procedure followed for the collection of native mucus from the stomach of pigs, obtained from the slaughterhouse. ETH, ethanol; PAR, paraformaldehyde.
The prevailing treatment for H. pylori eradication relies on a standard triple therapy, involving two antibiotics (clarithromycin plus amoxicillin or metronidazole) combined with a proton pump inhibitor.10,11 Even with this triple therapy, full eradication fails in nearly 25% of the patients.12,13 This is caused by (i) the highly acidic gastric pH the drugs need to endure, (ii) the very viscous mucus the drugs have to permeate,14 and (iii) the rising resistance of H. pylori to antibiotics.15 Therefore, alternative treatments are of utmost importance. To fulfil this need, antisense antibacterial therapy, through the use of oligonucleotides to inhibit the expression of essential bacterial genes, is a promising strategy.16,17,18 An attractive aspect of this approach is its flexibility. In case microbial resistance emerges, commonly by point mutations,19 the oligonucleotides can be easily redesigned to target a new mutation, becoming an effective antibacterial again.
Conventional DNA oligonucleotides have relatively low affinity toward RNA and DNA complementary sequences and are susceptible to degradation by endonucleases.20,21 To overcome these drawbacks, nucleic acid mimics (NAMs), have emerged as alternatives.20,22 Among them, locked nucleic acids (LNA), 2'-Omethyl RNA (2'OMe) and phosphorothioate modified oligonucleotides (Figure 1b) have shown the most promising antisense in vivo.23,24,25,26,27 While nucleic acid therapy of bacterial infections is a relatively unexplored field,18 delivery of oligonucleotides in bacteria is well studied to detect microorganisms based on fluorescence in situ hybridization (FISH). FISH (Figure 1c) is based on specific base pairing, at high temperature, of an oligonucleotide with rRNA or mRNA target sequences, obeying to Watson-Crick hydrogen-bonding.28,29,30,31 Recently, Fontenete et al.32 have proven that oligonucleotides composed of both LNA and 2'OMe (which we term LNA/2'OMe), with either phosphodiester (PO) or phosphorothioate (PS) as backbone linkages, allow specific detection of H. pylori. In this work, well-reported agents to permeabilize the bacterial cell envelope (like paraformaldehyde and ethanol) were used; H. pylori both grown in culture as well as in gastric biopsies could be detected in this way at 37 °C. In addition, very recently, H. pylori detection by LNA/2'OMe (PS) oligonucleotides was successful even at a gastric pH and without the use of paraformaldehyde.33 However, the presence of gastric mucus was not addressed in these hybridization experiments.
Clearly, the success of in vivo hybridization (IVH) for treating H. pylori infection depends on the ability of the NAMs to overcome two main barriers: the gastric mucus and the bacterial cell envelope (Figure 1a). Gastric mucus is a highly complex and viscoelastic mixture that forms a major protective barrier for the penetration of particles.34 The rheological properties of the mucus are mostly determined by mucins, the main component of dry mucus, but also affected by the lipids, DNA, cells and cellular debris, proteins and ions present.34,35,36,37 Having crossed the mucus layer, the next barrier encountered by the oligonucleotides will be the cell envelope of H. pylori. Unlike eukaryotic cells, which possess only a cell membrane, bacteria are protected from the outer environment by a multilayered cell envelope, composed of an inner membrane, periplasm, peptidoglycan layer and, in case of Gram negative bacteria, an outer membrane.38 Traditional FISH employs permeabilization agents to allow oligonucleotides to reach the cytoplasm.30 As these agents are typically toxic or noxious,39,40 this step should be circumvented in future IVH therapies.
Considering the potential of IVH for treating H. pylori infection, we studied to which extent gastric mucus may be a barrier for the delivery of oligonucleotides. More specifically, we studied (i) the (chemical) stability of LNA/2'OMe oligonucleotides in native gastric mucus scrapped off from porcine stomachs, (ii) the diffusion of LNA/2'OMe oligonucleotides, PO and PS, through the mucus, and (iii) the effect of gastric mucus on the efficiency of the fluorescence in situ hybridization in H. pylori. On top, we revealed to which extent the bacterial cell envelope is a barrier for the uptake of LNA/2'OMe oligonucleotides. Our experiments disclose important insights which will be of interest for the further development of gastric IVH therapies.
Results
NAMs uptake by H. pylori without prior permeabilization
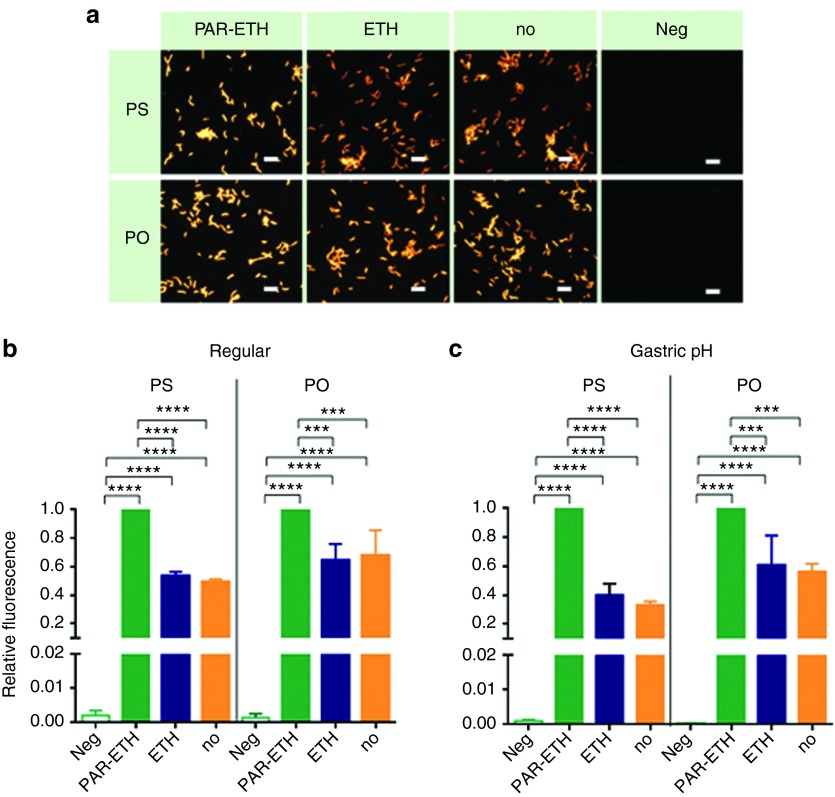
For future H. pylori IVH therapy, fixation and permeabilization of bacteria should be avoided as the reagents used are toxic or noxious. Therefore, we investigated the hybridization efficiency of LNA/2'OMe oligonucleotides without fixation/permeabilization of the bacteria (Figure 2). The hybridization was rather efficient, as nearly 50% (PS) and 70% (PO) of the fluorescence of the fully fixed/permeabilized bacteria (treatment with both paraformaldehyde and ethanol) was achieved (Figure 2b). The treatment with ethanol alone did not have a real benefit.
Figure 2.
Effect of fixation/permeabilization of H. pylori on the hybridization efficiency by PS and PO. The combined use of paraformaldehyde (PAR) and ethanol (ETH) was compared to respectively the use of only ETH and no pretreatment of bacteria (no). Relative fluorescence being the fluorescence of the bacteria normalized to their fluorescence as measured in respective positive control (green bar). Results are presented as mean ± SEM, n = 3. Negative controls (i.e., without the use of oligonucleotides) (Neg) were included as well. (a) Representative epifluorescence microscopy images obtained with regular hybridization and washing at pH 7 and acquired at equal light exposure conditions. Scale bars represent 10 µm. (b) Regular hybridization and washing at pH 7. (c) Hybridization at the average pH found in the collected porcine gastric mucus (pH 5.8), followed by washing with simulated gastric juice without pepsin (pH 1.8). PO, phosphodiester; PS, phosphorothioate.
In Figure 2b, the hybridization and washing steps were performed at pH 7. In an in vivo setting, however, the hybridization will have to happen within the gastric mucus; note that the mucus samples we isolated from pigs showed an average pH of 5.8. In addition, highly acidic gastric juice in the gastric lumen (upwards the gastric mucus) might contribute to wash away free NAMs. Therefore, we performed hybridization experiments at pH 5.8, followed by washing using simulated gastric juice (pH 1.8), instead of water (Figure 2c). It was verified that an efficient uptake of the NAMs occurred without fixation/permeabilization at neutral pH 7 (Figure 2b), as well as in conditions at pH closer to the in vivo (Figure 2c).
Diffusion of NAMs in native gastric mucus
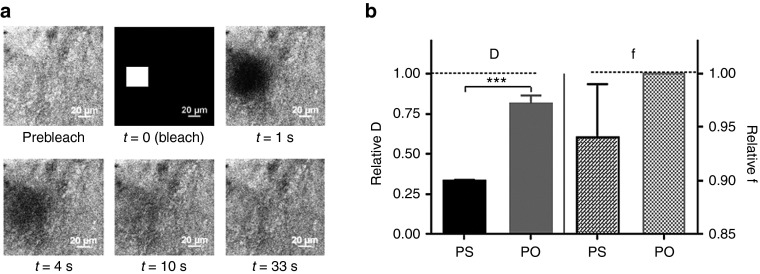
Fluorescence recovery after photobleaching (FRAP) was used to measure the diffusion of both LNA/2'OMe oligonucleotides in native porcine gastric mucus (Figure 3). As Figure 3b shows, when compared to 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) buffer, the diffusion in gastric mucus was slowed down, especially for the PS oligonucleotides. As the f values reveal in Figure 3b, all PO oligonucleotides are mobile in mucus, while a small fraction of the PS oligonucleotides becomes immobilized by the gastric mucus. Visual inspection of the microscopy images also showed a more heterogeneous distribution in mucus of PS, when compared to PO (Supplementary Figure S1 and Supplementary Movies in Supplementary Material).
Figure 3.
Diffusion of NAMs in native gastric mucus. (a) A representative fluorescence recovery after photobleaching measurement on PO in porcine gastric mucus. The first frame shows the prebleach image. Next, a square region (30 × 30 µm) is bleached (at t = 0), followed by a time-lapse recording of the subsequent fluorescence recovery. (b) Average diffusion coefficient (D) and mobile fraction (f) of both NAMs in native gastric mucus normalized to their values in HEPES buffer. Results are presented as mean ± SEM, n = 3. NAM, nucleic acid mimic; PO, phosphodiester.
Stability of NAMs in native gastric mucus
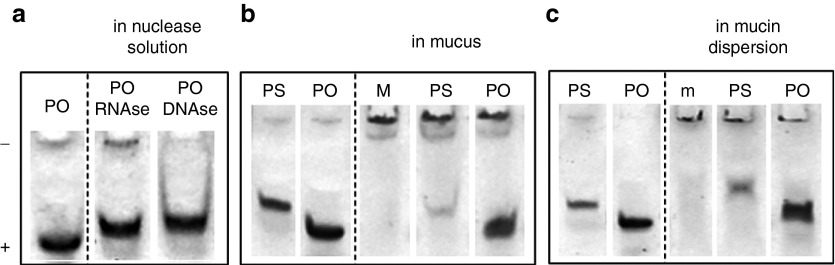
Polyacrylamide gel electrophoresis (PAGE) experiments reveal that both PO and PS oligonucleotides are stable in gastric mucus (Figure 4b). The faster diffusion of the PO in mucus, compared to the PS oligonucleotides (as seen in Figure 3b), cannot be attributed to its enzymatic degradation, since it is stable and DNase and RNase resistant (Figure 4a).
Figure 4.
Stability of NAMs in native gastric mucus. (a) Polyacrylamide gel electrophoresis on PO incubated with RNase and DNase; PO in water was taken as a control. (b) PS and PO incubated in porcine gastric mucus; mucus alone (M) was taken as a control. (c) PS and PO incubated in a commercial mucin dispersion; mucin dispersion alone (m) was run as a control. The lanes at the left of the dotted lines show PS and PO in water. NAM, nucleic acid mimic; PO, phosphodiester; PS, phosphorothioate.
Comparing each oligonucleotide in water and in mucus, we observed that gastric mucus “smeared” the oligonucleotides over the gel, thereby reducing the intensity of the oligonucleotide band (Figure 4b); this smearing was more pronounced in case of the PS oligonucleotides. Similar results were obtained using mucus from two other pigs. When incubating the oligonucleotides in a (commercial) mucin dispersion, the same trend was noticed (Figure 4c), although the smearing of the oligonucleotides was less pronounced. The PAGE results suggest that the NAMs interact with mucus components, likely mucins. In agreement with the outcome of the FRAP experiments (f values in Figure 3b), the PS oligonucleotides seem to adhere more strongly to gastric mucus than the PO oligonucleotides.
H. pylori hybridization in native gastric mucus
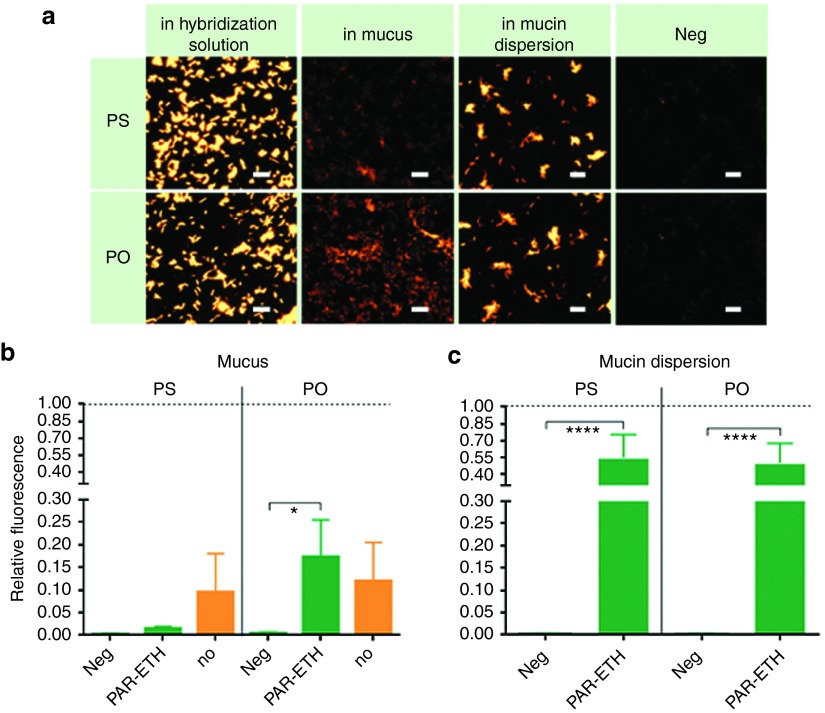
To find out whether the interactions between NAMs and mucus, as observed above, could affect the ability of NAMs to hybridize in H. pylori, FISH was performed with and without mucus in the hybridization step. Additionally, for each of these conditions, we tested both PAR-ETH treated and untreated bacteria (ETH treated was not considered at this stage, since it did not previously show a real effect on NAMs uptake). As shown in Figure 5, the presence of native gastric mucus significantly decreased the hybridization efficiency of both NAMs, in fixed/permeabilized and untreated H. pylori. When using fixed/permeabilized H. pylori, a lower efficiency was noticed for PS than PO, in line with the previously observed stronger interaction of PS to mucus. This was not, however, reflected in untreated H. pylori (Figure 5b, orange bars). Also the mucin dispersion reduced the hybridization efficiency, but much less than native mucus (PS presented 55% and PO 50% of the positive control's fluorescence), suggesting that mucin molecules alone do not fully explain the decreased hybridization in gastric mucus.
Figure 5.
Effect of native gastric mucus on the hybridization efficiency by PS and PO. This effect was compared to the one of a mucin dispersion. H. pylori was fixed/permeabilized with paraformaldehyde and ethanol (images and green bars; PAR-ETH) or hybridized without any pretreatment (orange bars; no). Negative controls (in which no oligonucleotides were used; Neg) were included. (a) Representative epifluorescence microscopy images, acquired at equal light exposure conditions. Scale bars represent 10 µm. (b) and (c) Relative fluorescence being the fluorescence of the bacteria normalized to their fluorescence as measured when using the standard hybridization solution, without mucus or mucin dispersion. Results are presented as mean ± SEM, n = 3. PAR-ETH, paraformaldehyde–ethanol; PO, phosphodiester; PS, phosphorothioate.
Discussion
LNA and 2'OMe oligonucleotides are promising nucleic acid mimics for novel gene therapies, due to their biological stability and improved target affinity, compared to unmodified variants.25 Following the discovery of two LNA/2'OMe oligonucleotides, one further modified with PS internucleotide linkages and another possessing normal PO linkages, able to specifically target H. pylori at human body temperature,32 the present work aimed to reveal whether the LNA/2'OMe oligonucleotides hold promise to be used in vivo as antisense strategy against H. pylori infections. Therefore, we investigated if LNA/2'OMe oligonucleotides can hybridize in H. pylori in the presence of native gastric mucus, without prior permeabilization and fixation of the bacteria.
First, we investigated if LNA/2'OMe can penetrate the bacterial cell envelope without prior permeabilization and fixation, employing FISH without mucus. Paraformaldehyde and ethanol are among the most common permeabilization and fixation agents.1,21,30,31,41,42,43 While paraformaldehyde is a fixative, ethanol acts both as a fixative and permeabilizer of the bacterial membrane lipids.44,45 Fixation and permeabilization agents cannot, however, be used for in vivo applications due to their toxicity.44 Our results showed successful hybridization of LNA/2'OMe oligonucleotides in untreated H. pylori, not only when hybridization and washing were performed at pH 7 (Figure 2b), but also under pH conditions representative for the in vivo gastric environment (Figure 2c). These findings are in line with a previous report, where it was shown that FISH targeting of bacterial rRNA can be performed without fixation for identification of bacteria in enhanced biological phosphorus removal sludge samples.46 Since the presence of ethanol by itself did not show a clear advantage in our hands (Figure 2), it can be concluded that permeabilization of the H. pylori envelope is not needed for the penetration of the LNA/2'OMe tested. The relatively small size of the oligonucleotides (10 mers) is certainly an advantage. One has to note, however, that the combined use of paraformaldehyde and ethanol did result in a stronger fluorescence of the bacteria (Figure 2b,c). This may be due to a stronger fixative effect of paraformaldehyde compared to ethanol, providing better preservation of ultrastructures, proteins and nucleic acids and a longer maintenance of the fluorescence, especially for gram negative bacteria.44,45,47
Next, we investigated the mobility and stability of LNA/2'OMe in native porcine gastric mucus. So far the presence of mucus has been neglected in relation to the delivery of macromolecules and antibiotics to H. pylori.12,48,49,50 On the other hand, several studies considering macromolecules/particles diffusion in mucus use mimic models based on commercial mucins or mixtures thereof,51,52,53,54 which are still far from the real native gastric mucus. By PAGE, we could confirm the integrity of both oligonucleotides in native gastric mucus. In addition, by FRAP we found that gastric mucus is not a major diffusion barrier for the oligonucleotides, although the diffusion of PS in mucus was three times slower than in HEPES buffer, while for PO it was 1.2 times slower (DPS = 58.1 ± 8.1 µm2/s and DPO = 161.6 ± 8.0 µm2/s). This difference should not be attributed to the oligos' size (since they differ only in 0.15 kDa), but to the presence of the thioate group (PS). This might lead to more or stronger interactions of oligo PS with mucus components, likely mucins, since a similar interaction trend was observed in a suspension of commercial mucins, by PAGE. In any case, assuming an average thickness of the gastric mucus layer of 180 µm,34 according to Equation 1 this means that crossing this layer would take only 2 minutes for PO and less than 5 minutes for PS.
 |
t being the time, L the thickness of mucus layer, and D the diffusion coefficient.
Therefore, we conclude that the oligonucleotides are sufficiently mobile in gastric mucus and can easily reach the bacteria which are present in the deeper layers of the mucus, adhered to the gastric epithelium.
In the presence of gastric mucus, the hybridization in H. pylori was significantly hampered for both types of oligonucleotides (Figure 5a,b), even at longer hybridization time (90 minutes instead of 30 minutes; results not shown), ruling out the possibility of diffusion being the limiting factor—even though that is not expected from the FRAP diffusion measurements. It is clear that the mucus effect on hybridization was not masked by the dilution upon mixing with the oligonucleotides solution; actually, a relevant dilution may also occur in vivo, especially if a common administration by oral gavage is used. Hybridization within mucus must be hindered by interactions of the NAMs with mucus. Mucus is a complex mixture, composed mainly of water and mucins, but also cellular debris, lipids, DNA, proteins, ions.34,37 Mechanisms of NAMs mucoadhesion may include electrostatic interactions and multiple low-affinity intermolecular bonds, as hydrophobic interactions and hydrogen bonds, given the increased amount of proton donors and acceptors in mucus.55 Adhesion of oligo PS was more evident than PO, likely due to the increased hydrophobicity of PS56 and possible establishment of disulfide bonds with mucins (mucins are formed by glycoprotein monomers cross-linked by disulfide bonds34,36,37), thereby explaining the slower diffusion of PS in mucus compared to PO. Interestingly, replacing mucus by a simple mucin suspension did partially restore hybridization (Figure 5c). This shows that hybridization in mucus is not simply hindered due to interaction of the oligonucleotides with mucins, but rather with other mucus constituents, as well. Although the particular role/identity of mucus components in the interaction with NAMs remains unclear at this time, soluble factors must account for it, as the FRAP experiments did not show substantial immobilization (i.e., f > 0.9). A corollary of the mucin experiment is that clearly a simple mucin dispersion is not a sufficiently accurate model for gastric mucus and may lead to misleading conclusions when used alone. Another factor that might contribute to decreased hybridization in the presence of mucus is that binding of H. pylori to mucins, which has been reported in the colonization of the stomach,57,58 may decrease its membrane surface area available for the NAMs penetration.
Taken together, our study points out that situ hybridization of LNA/2'OMe does not require permeabilization of the cell envelope, which is beneficial towards potential future IVH therapies. On the other hand, while neither NAMs stability is a concern nor their diffusion through native gastric mucus is a major obstacle, binding interactions with mucus components other than mucins reduced the hybridization efficiency. This work reveals the importance of considering the effect of a representative, native, undiluted, gastric mucus on NAMs delivery to mucosal sites. Future work should focus on developing strategies that protect LNA/2'OMe from interacting with mucus components while traveling through the mucus layer. One interesting approach could be the incorporation of LNA/2'OMe into suitable nanocarriers, such as fusogenic liposomes, that shield the oligonucleotides from the environment, while being able to deliver them efficiently into the cells.
Materials and Methods
Isolation of gastric mucus. Native gastric mucus from the stomach of pigs was used in this study, since porcine gastric mucus is considered a suitable model for human gastric mucus.37,51 Moreover, pigs have a gastric physiology similar to humans, making them representative animal models for studies of Helicobacter infection.59
Mucus from three different pigs was included in each assay. Stomachs were collected from a slaughterhouse and immediately opened at the major curvature and rinsed with tap water.60 The mucus from the antrum region was then gently scrapped off using a glass slide and placed in a sterile tube (Figure 1d). Each tube was then aliquoted, placing 200 µl into sterile Eppendorf vials with a sterile plastic syringe, and stored at −80 °C.
Synthesis of NAMs. Two oligonucleotides, named HP_LNA/2OMe_PS and HP_LNA/2OMe_PO,32 complementary to a sequence of the H. pylori 16S rRNA gene, were used in this study. These are composed of LNA and 2'OMe, possess the same sequence and differ only in the internucleotide bonds. One possesses normal phosphodiester oligonucleotides (PO), while the other has one of the two nonbridging oxygen atoms replaced by a sulfur atom at each internucleotide linkage (PS) (Figure 1b). To simplify, these oligonucleotides will be herein designated as PO and PS, respectively. The sequence of PO is 5'-lGmeAmeClTmeAmeAlGmeCmeClC-3', while the sequence of PS is 5'-lG*meA*meC*lT*meA*meA*lG*meC*meC*lC*-3', where “l” represents the LNA monomers, “me” the 2'OMe monomers and * the phosphorothioate linkages. Oligonucleotides labeled at 5' with fluorescein phosphoramidite (FAM, Glen Research, VA) were synthetized and purified according to ref. 32. The same oligonucleotides labeled at the 5'-terminal with Cy3 were acquired from Eurogentec (Ougrée, Belgium).
Measuring the diffusion of NAMs in native gastric mucus by FRAP. Approximately 30 µl of native (undiluted) gastric mucus was taken with a plastic syringe and mixed with 4 µl of 100 µmol/l FAM-labeled PO or PS. The mixture was then placed on a microscopy glass slide sealed with an adhesive spacer (S24735, Secure-Seal, Life Technologies, Paisley, UK) and a cover slip. The sample was placed in a stage top incubation chamber (Tokai Hit, Shizuoka, Japan), to perform measurements at 37 °C. Mucus from three different pigs was tested and for each mucus-NAM sample the diffusion measurements were done in triplicate. Measurements in HEPES buffer were performed as control.
FRAP was used to measure the diffusion of the fluorescently labeled oligonucleotides.61,62 For a detailed description of the FRAP experiments, we refer to a previous publication from our group.61 The measurements were performed using a Nikon C1si confocal laser scanning microscope (CLSM), equipped with a Plan Apo 10x NA 1.4 objective lens. The 488 nm argon ion laser (CVI Melles Griot, CA) was used for photobleaching and imaging. In brief, a FRAP experiment occurs as follows. Using a strong laser beam the fluorescence in a rectangular area (30 × 30 µm) of the mucus-NAM sample is bleached. Next, using an attenuated laser beam, a time-lapse image series is recorded to visualize the fluorescence recovery in the bleached area which is due to diffusion of the fluorescent NAMs from the (nonbleached) surrounding into the bleached zone (see Figure 3a). The local diffusion coefficient (D) and the fraction of mobile (f) and immobile oligonucleotides can be calculated by fitting the fluorescence recovery data to a theoretical model (Supplementary Materials and Methods), as developed in our group by Deschout et al. 62
Measuring the stability of NAMs in native gastric mucus by PAGE. Two microliters of 20 µmol/l PS or PO (labeled with FAM) were gently mixed (using a 1,000 µl pipette tip) with approximately 5 µl of porcine gastric mucus and incubated at 37 °C for 10 minutes (the approximate time needed for a FRAP measurement). Loading buffer (2 µl of Ambion gel loading solution, Life Technologies, Paisley, UK) was then mixed with the mucus-NAM sample. FAM-labeled oligonucleotides alone and mucus alone were run as controls on a PAGE gel. Each empty well on the gel was filled with a blank (a mixture of 7 µl milli-Q water and 2 µl of loading buffer). In addition, the stability of PS and PO was tested in a suspension of commercial mucin from porcine stomach (type II, Sigma-Aldrich, Bornem, Belgium). Therefore, PS and PO were incubated in a 3% (w/v) mucin dispersion (in 0.9% (w/v) NaCl solution).63,64
To investigate the degradation of the oligonucleotides by nucleases, PO was incubated for 2 hours at 37 °C with DNase (TURBO DNase, Life Technologies) and RNase (RNase A, Qiagen Benelux B.V., Venlo, The Netherlands). The reaction mixture contained 4 µl of a 5 µmol/l PO, 1 µl of DNase (2U) or RNase (7U) and 13 µl of the respective supplied reaction buffer.
All the samples were loaded onto 20% nondenaturing polyacrylamide gels prepared in 5× TBE buffer. Electrophoresis was performed at 110 V, during 75 minutes. Thereafter, the fluorescently labeled oligonucleotides on the gels were visualized by UV transillumination and gel photography.
H. pylori FISH—NAMs uptake and hybridization in native gastric mucus. H. pylori 26695 (ATCC 700392) was grown in trypticase soy agar (TSA) supplemented with 5% sheep blood (Becton Dickinson GmbH, Germany) for 48 hours, at 37 °C, under microaerobic conditions. The biomass was recovered from the plates using sterile saline (0.9% (w/v) NaCl) and diluted to nearly 1 × 106 colony-forming unit (CFU)/ml. Smears were prepared on glass slides (20 µl per slide well) by drying at 37 °C. FISH on slide was performed according to Fontenete et al.32,33, with slight modifications. The main steps of standard FISH are schematically presented in Figure 1c. For standard fixation/permeabilization, the smears were first immersed in 4% (w/v) paraformaldehyde (Fluka—Sigma-Aldrich) for 10 minutes, followed by 15 minutes in 50% (v/v) ethanol (Fluka—Sigma-Aldrich) and allowed to dry. To test the role of fixation/permeabilization on hybridization efficiency, samples treated only with ethanol, and samples not fixed/permeabilized at all were included. The smears were then covered with 20 µl hybridization solution containing 500 mmol/l urea (Vel—VWR, Haasrode, Belgium), 500 mmol/l Tris-HCl, pH 7 (Sigma-Aldrich), 900 mmol/l NaCl (Sigma-Aldrich), and 400 nmol/l of PS or PO labeled with Cy3. To test the effect of native gastric mucus on hybridization efficiency, approximately 30 µl of gastric mucus was first mixed with 20 µl of hybridization solution, and then applied on the bacteria smear (note that the final concentration of NAMs and hybridization solution applied on the bacteria were kept the same as in standard FISH). Samples were covered with a coverslip, placed in dark moist chambers and allowed to hybridize during 30 minutes, at 37 °C. The coverslips were then removed, together with the mucus mixture (when present). To avoid nonfixed H. pylori to be removed from the slide together with the mucus mixture, slides coated with 0.1% (w/v) Poly-L-lysine (Sigma-Aldrich) were used for better adhesion of H. pylori. The slides were then washed during 15 minutes using warm (37 °C) water at pH 7, allowed to dry and visualized on the microscope. The experiments were performed using gastric mucus from three different pigs, in triplicate. The effect of native gastric mucus was compared with that of a mucin dispersion, by performing hybridization in the presence of a 3% (w/v) suspension of commercial mucin (from porcine stomach), instead of mucus. In addition, the influence of acidic washing with simulated gastric juice without pepsin (sGJ; 0.2% w/v NaCl, 80 mmol/l HCl, pH 1.8, ref. 65) was evaluated, after hybridization at the same average pH found in the collected porcine gastric mucus (pH 5.8). Negative controls using hybridization solution without oligonucleotides, and maintaining the remaining FISH conditions, were performed in all experiments.
Microscopy images were acquired using a Nikon TE2000 microscope equipped with a Nikon DS-Qi 1Mc epifluorescence camera and a Plan Apo VC 100 × 1.4 NA oil immersion objective lens (Nikon). Cy3 fluorescence was visualized using a TRITC filter (excitation: 530–560 nm; emission: 590–650 nm, Nikon). The same exposure time and the same intensity of the fluorescent lamp were used for all the samples. For fluorescence quantification, 10–15 images per well were taken randomly, covering all the areas of the slide well.
Quantification of H. pylori hybridization. The hybridization efficiency in the different conditions was compared through quantification of the fluorescence intensity. This was done using image processing routines available in Matlab (The MathWorks, Natick, MA). First, bacteria were identified in each image based on an automatically determined intensity threshold. After removal of remaining noise (by selecting areas with a minimum number of pixels), a binary mask was generated that corresponds to the location of bacteria in the image. For the regions within the mask, the fluorescence intensity in the original image was quantified as
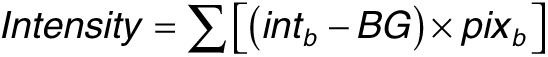 |
where intb corresponds to the fluorescence intensity of each bacterium, BG the background value and pixb the number of pixels of the bacterium.
Per condition, 10 to 15 images were analyzed in this fashion from which the average fluorescence intensity was calculated. These absolute values were normalized to the respective positive control of PS or PO, hybridized in hybridization solution and washed with water or sGJ.
Statistical analysis. Statistical analysis was performed by GraphPad Prism6 software (GraphPad Software, San Diego, CA). FISH results were analyzed using two-way analysis of variance and Tukey's multiple comparison test, comparing the conditions within the same NAM. FRAP results were analyzed by one-way analysis of variance and Sidak's multiple comparisons test. Significance was set at P ≤ 0.05 (****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05).
SUPPLEMENTARY MATERIAL Figure S1. Confocal images of porcine gastric mucus after mixing with respectively PS and PO oligonucleotides. Images from FRAP experiments prior to bleaching. Supplementary Materials and Methods. Supplementary References. Supplementary movie PS. Representative FRAP movie of PS in native gastric mucus. Supplementary movie PO. Representative FRAP movie of PO in native gastric mucus.
Acknowledgments
This work was funded by FEDER funds through the Operational Programme for Competitiveness Factors—COMPETE, ON.2—O Novo Norte—North Portugal Regional Operational Programme and National Funds through FCT—Foundation for Science and Technology under the projects: PEst-C/EQB/UI0511, NORTE-07-0124-FEDER-000025—RL2_ Environment&Health and DNA mimics Research Project PIC/IC/82815/2007, NORTE-07-0124-FEDER-000022, PhD fellowships SFRH/BD/84376/2012 and SFRH/BD/72999/2010, and Post-Doctoral fellowships SFRH/BPD/78846/2011 and SFRH/BPD/33420/2008. The Laboratory of General Biochemistry and Physical Pharmacy, Ghent University is thanked for financial support. This work was performed under the framework of the COST-Action TD1004: Theragnostics for imaging and therapy. We also thank Freddy Haesebrouck, Bram Flahou, Ellen De Bruyne and Iris Bosschem from the Department of Pathology, bacteriology and poultry diseases, Ghent University, for the porcine stomachs, and Tom Coenye from the Laboratory of Pharmaceutical Microbiology, Ghent University, for providing the facilities for bacteria culture.
Supplementary Material
Representative FRAP movie of PS in native gastric mucus.
Representative FRAP movie of PO in native gastric mucus.
References
- Guimarães, N, Azevedo, NF, Figueiredo, C, Keevil, CW and Vieira, MJ (2007). Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimens. J Clin Microbiol 45: 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, D, Nunes, C, Martins, MC, Sarmento, B and Reis, S (2014). Eradication of Helicobacter pylori: Past, present and future. J Control Release 189: 169–186. [DOI] [PubMed] [Google Scholar]
- Dicken, BJ, Bigam, DL, Cass, C, Mackey, JR, Joy, AA and Hamilton, SM (2005). Gastric adenocarcinoma: review and considerations for future directions. Ann Surg 241: 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (1994). Schistosomes, liver flukes and Helicobacter pylori. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. vol. 61. World Heath Organization: Lyon. pp. 1–241. [PMC free article] [PubMed] [Google Scholar]
- Dunne, C, Dolan, B and Clyne, M (2014). Factors that mediate colonization of the human stomach by Helicobacter pylori. World J Gastroenterol 20: 5610–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, AM, Leite, M, Seruca, R and Figueiredo, C (2013). Adherens junctions as targets of microorganisms: a focus on Helicobacter pylori. FEBS Lett 587: 259–265. [DOI] [PubMed] [Google Scholar]
- Sigal, M, Rothenberg, ME, Logan, CY, Lee, JY, Honaker, RW, Cooper, RL et al. (2015). Helicobacter pylori Activates and Expands Lgr5(+) Stem Cells Through Direct Colonization of the Gastric Glands. Gastroenterology 148: 1392–404.e21. [DOI] [PubMed] [Google Scholar]
- Necchi, V, Candusso, ME, Tava, F, Luinetti, O, Ventura, U, Fiocca, R et al. (2007). Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology 132: 1009–1023. [DOI] [PubMed] [Google Scholar]
- Noach, LA, Bosma, NB, Jansen, J, Hoek, FJ, van Deventer, SJ and Tytgat, GN (1994). Mucosal tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-8 production in patients with Helicobacter pylori infection. Scand J Gastroenterol 29: 425–429. [DOI] [PubMed] [Google Scholar]
- Gisbert, JP and Pajares, JM (2010). Treatment of Helicobacter pylori infection: the past and the future. Eur J Intern Med 21: 357–359. [DOI] [PubMed] [Google Scholar]
- Ayala, G, Escobedo-Hinojosa, WI, de la Cruz-Herrera, CF and Romero, I (2014). Exploring alternative treatments for Helicobacter pylori infection. World J Gastroenterol 20: 1450–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira, F, Gonçalves, IC and Martins, MC (2013). Effect of gastric environment on Helicobacter pylori adhesion to a mucoadhesive polymer. Acta Biomater 9: 5208–5215. [DOI] [PubMed] [Google Scholar]
- Sasaki, M, Ogasawara, N, Utsumi, K, Kawamura, N, Kamiya, T, Kataoka, H et al. (2010). Changes in 12-year first-line eradication rate of Helicobacter pylori based on triple therapy with proton pump inhibitor, amoxicillin and clarithromycin. J Clin Biochem Nutr 47: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakil, N (2006). Helicobacter pylori treatment: a practical approach. Am J Gastroenterol 101: 497–499. [DOI] [PubMed] [Google Scholar]
- Malfertheiner, P, Schultze, V, Rosenkranz, B, Kaufmann, SH, Ulrichs, T, Novicki, D et al. (2008). Safety and immunogenicity of an intramuscular Helicobacter pylori vaccine in noninfected volunteers: a phase I study. Gastroenterology 135: 787–795. [DOI] [PubMed] [Google Scholar]
- Good, L, Awasthi, SK, Dryselius, R, Larsson, O and Nielsen, PE (2001). Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol 19: 360–364. [DOI] [PubMed] [Google Scholar]
- Mondhe, M, Chessher, A, Goh, S, Good, L and Stach, JE (2014). Species-selective killing of bacteria by antimicrobial peptide-PNAs. PLoS One 9: e89082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, H, You, Y, Yan, H, Meng, J, Xue, X, Hou, Z et al. (2012). Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials 33: 659–667. [DOI] [PubMed] [Google Scholar]
- Lopes, AI, Vale, FF and Oleastro, M (2014). Helicobacter pylori infection - recent developments in diagnosis. World J Gastroenterol 20: 9299–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira, L, Azevedo, NF, Almeida, C, Jardim, T, Keevil, CW and Vieira, MJ (2008). DNA mimics for the rapid identification of microorganisms by fluorescence in situ hybridization (FISH). Int J Mol Sci 9: 1944–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, C, Azevedo, NF, Santos, S, Keevil, CW and Vieira, MJ (2011). Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS One 6: e14786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, MA and Wengel, J (2011). Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem Soc Rev 40: 5680–5689. [DOI] [PubMed] [Google Scholar]
- Dias, N and Stein, CA (2002). Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther 1: 347–355. [PubMed] [Google Scholar]
- Järver, P, Coursindel, T, Andaloussi, SE, Godfrey, C, Wood, MJ and Gait, MJ (2012). Peptide-mediated cell and in vivo delivery of antisense oligonucleotides and siRNA. Mol Ther Nucleic Acids 1: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin, KE, Højland, T, Hansen, BR, Persson, R, Bramsen, JB, Kjems, J, et al. (2013). Biological activity and biotechnological aspects of locked nucleic acids. In: Theodore Friedmann JCD, Stephen FG (eds). Advances in Genetics. vol. 82. Academic Press: Waltham, MA, pp. 47–107. [DOI] [PubMed] [Google Scholar]
- Lindholm, MW, Elmén, J, Fisker, N, Hansen, HF, Persson, R, Møller, MR et al. (2012). PCSK9 LNA antisense oligonucleotides induce sustained reduction of LDL cholesterol in nonhuman primates. Mol Ther 20: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén, J, Lindow, M, Schütz, S, Lawrence, M, Petri, A, Obad, S et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899. [DOI] [PubMed] [Google Scholar]
- Robertson, KL and Thach, DC (2009). LNA flow-FISH: a flow cytometry-fluorescence in situ hybridization method to detect messenger RNA using locked nucleic acid probes. Anal Biochem 390: 109–114. [DOI] [PubMed] [Google Scholar]
- Bouvier, T and Del Giorgio, PA (2003). Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): A quantitative review of published reports. FEMS Microbiol Ecol 44: 3–15. [DOI] [PubMed] [Google Scholar]
- Amann, R and Fuchs, BM (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol 6: 339–348. [DOI] [PubMed] [Google Scholar]
- Santos, RS, Guimarães, N, Madureira, P and Azevedo, NF (2014). Optimization of a peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) method for the detection of bacteria and disclosure of a formamide effect. J Biotechnol 187: 16–24. [DOI] [PubMed] [Google Scholar]
- Fontenete, S, Guimarães, N, Leite, M, Figueiredo, C, Wengel, J and Filipe Azevedo, N (2013). Hybridization-based detection of Helicobacter pylori at human body temperature using advanced locked nucleic acid (LNA) probes. PLoS One 8: e81230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenete, S, Leite, M, Guimarães, N, Madureira, P, Ferreira, RM, Figueiredo, C et al. (2015). Towards Fluorescence In Vivo Hybridization (FIVH) Detection of H. pylori in Gastric Mucosa Using Advanced LNA Probes. PLoS One 10: e0125494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, RA (2009). Barrier properties of mucus. Adv Drug Deliv Rev 61: 75–85. [DOI] [PubMed] [Google Scholar]
- Khanvilkar, K, Donovan, MD and Flanagan, DR (2001). Drug transfer through mucus. Adv Drug Deliv Rev 48: 173–193. [DOI] [PubMed] [Google Scholar]
- Bansil, R, and Turner, BS (2006). Mucin structure, aggregation, physiological functions and biomedical applications. Current Opinion in Colloid & Interface Science 11: 164–170. [Google Scholar]
- Lai, SK, Wang, YY, Wirtz, D and Hanes, J (2009). Micro- and macrorheology of mucus. Adv Drug Deliv Rev 61: 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, AP. Microbial glycobiology. Structures, Relevance and Applications. Elsevier, London, 2009. [Google Scholar]
- Acton, A, Harvey, T and Grow, MW (2005). An examination of non-formalin-based fixation methods for Xenopus embryos. Dev Dyn 233: 1464–1469. [DOI] [PubMed] [Google Scholar]
- van Essen, HF, Verdaasdonk, MA, Elshof, SM, de Weger, RA and van Diest, PJ (2010). Alcohol based tissue fixation as an alternative for formaldehyde: influence on immunohistochemistry. J Clin Pathol 63: 1090–1094. [DOI] [PubMed] [Google Scholar]
- Moter, A and Göbel, UB (2000). Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods 41: 85–112. [DOI] [PubMed] [Google Scholar]
- Almeida, C, Azevedo, NF, Fernandes, RM, Keevil, CW and Vieira, MJ (2010). Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples. Appl Environ Microbiol 76: 4476–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, A, Almeida, C, Carvalho, A, Boyen, F, Haesebrouck, F, Rodrigues, L et al. (2013). Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Lactobacillus spp. in milk samples. Int J Food Microbiol 162: 64–70. [DOI] [PubMed] [Google Scholar]
- Chao, Y and Zhang, T (2011). Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl Microbiol Biotechnol 92: 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo, GJ. The basics of in situ hybridization. In: Nuovo GJ (ed.). In Situ Molecular Pathology and Co-expression Analyses. Academic Press, San Diego, 2013. pp. 81–131. [Google Scholar]
- Yilmaz, S, Haroon, MF, Rabkin, BA, Tyson, GW and Hugenholtz, P (2010). Fixation-free fluorescence in situ hybridization for targeted enrichment of microbial populations. ISME J 4: 1352–1356. [DOI] [PubMed] [Google Scholar]
- Amann, RI, Ludwig, W and Schleifer, KH (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umamaheshwari, RB and Jain, NK (2003). Receptor mediated targeting of lectin conjugated gliadin nanoparticles in the treatment of Helicobacter pylori. J Drug Target 11: 415–23; discussion 423. [DOI] [PubMed] [Google Scholar]
- Lin, YH, Chang, CH, Wu, YS, Hsu, YM, Chiou, SF and Chen, YJ (2009). Development of pH-responsive chitosan/heparin nanoparticles for stomach-specific anti-Helicobacter pylori therapy. Biomaterials 30: 3332–3342. [DOI] [PubMed] [Google Scholar]
- Gonçalves, IC, Magalhães, A, Fernandes, M, Rodrigues, IV, Reis, CA and Martins, MC (2013). Bacterial-binding chitosan microspheres for gastric infection treatment and prevention. Acta Biomater 9: 9370–9378. [DOI] [PubMed] [Google Scholar]
- Groo, AC and Lagarce, F (2014). Mucus models to evaluate nanomedicines for diffusion. Drug Discov Today 19: 1097–1108. [DOI] [PubMed] [Google Scholar]
- Grießinger, J, Dünnhaupt, S, Cattoz, B, Griffiths, P, Oh, S, Gómez, SB et al. (2015). Methods to determine the interactions of micro- and nanoparticles with mucus. Eur J Pharm Biopharm 96: 464–476. [DOI] [PubMed] [Google Scholar]
- Adebisi, AO and Conway, BR (2014). Lectin-conjugated microspheres for eradication of Helicobacter pylori infection and interaction with mucus. Int J Pharm 470: 28–40. [DOI] [PubMed] [Google Scholar]
- Iqbal, J, Shahnaz, G, Dünnhaupt, S, Müller, C, Hintzen, F and Bernkop-Schnürch, A (2012). Preactivated thiomers as mucoadhesive polymers for drug delivery. Biomaterials 33: 1528–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X, Forier, K, Steukers, L, Van Vlierberghe, S, Dubruel, P, Braeckmans, K et al. (2012). Immobilization of pseudorabies virus in porcine tracheal respiratory mucus revealed by single particle tracking. PLoS One 7: e51054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox, KA, Owczarzy, R, Thomas, DM, Walder, JA and Behlke, MA (2013). Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Mol Ther Nucleic Acids 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman, TL, McGee, DJ, and Mobley, HLT. Adherence and colonization. In: Mobley HLT, Mendz GL and Hazell SL (eds.). Helicobacter Pylori: Physiology and Genetics. ASM Press, Washington (DC), 2001. [PubMed] [Google Scholar]
- Gold, BD, Dytoc, M, Huesca, M, Philpott, D, Kuksis, A, Czinn, S et al. (1995). Comparison of Helicobacter mustelae and Helicobacter pylori adhesion to eukaryotic cells in vitro. Gastroenterology 109: 692–700. [DOI] [PubMed] [Google Scholar]
- Akira, N, and Toshio, F (2004). Animal models for the study of helicobacter infection. Handbook of laboratory animal science, second edition. CRC Press. pp. 151–167. [Google Scholar]
- Baele, M, Decostere, A, Vandamme, P, Ceelen, L, Hellemans, A, Mast, J et al. (2008). Isolation and characterization of Helicobacter suis sp. nov. from pig stomachs. Int J Syst Evol Microbiol 58(Pt 6): 1350–1358. [DOI] [PubMed] [Google Scholar]
- Xiong, R, Deschout, H, Demeester, J, De Smedt, SC and Braeckmans, K (2014). Rectangle FRAP for measuring diffusion with a laser scanning microscope. Methods Mol Biol 1076: 433–441. [DOI] [PubMed] [Google Scholar]
- Deschout, H, Hagman, J, Fransson, S, Jonasson, J, Rudemo, M, Lorén, N et al. (2010). Straightforward FRAP for quantitative diffusion measurements with a laser scanning microscope. Opt Express 18: 22886–22905. [DOI] [PubMed] [Google Scholar]
- Laohavaleeson, S, Tessier, PR and Nicolau, DP (2008). Pharmacodynamic characterization of ceftobiprole in experimental pneumonia caused by phenotypically diverse Staphylococcus aureus strains. Antimicrob Agents Chemother 52: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodvold, KA, Nicolau, DP, Lodise, TP, Khashab, M, Noel, GJ, Kahn, JB et al. (2009). Identifying exposure targets for treatment of staphylococcal pneumonia with ceftobiprole. Antimicrob Agents Chemother 53: 3294–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, KY and Heo, TR (2000). Survival of Bifidobacterium longum immobilized in calcium alginate beads in simulated gastric juices and bile salt solution. Appl Environ Microbiol 66: 869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative FRAP movie of PS in native gastric mucus.
Representative FRAP movie of PO in native gastric mucus.