Abstract
tRNA molecules contain numerous chemically altered nucleosides, which are formed by enzymatic modification of the primary transcripts during the complex tRNA maturation process. Some of the modifications are introduced by single reactions, while other require complex series of reactions carried out by several different enzymes. The location and distribution of various types of modifications vary greatly between different tRNA molecules, organisms and organelles.
We have developed a computational method tRNAmodpred, for predicting modifications in tRNA sequences. Briefly, our method takes as an input one or more unmodified tRNA sequences and a set of protein sequences corresponding to a proteome of a cell. Subsequently it identifies homologs of known tRNA modification enzymes in the proteome, predicts tRNA modification activities and maps them onto known pathways of RNA modification from the MODOMICS database. Thereby, theoretically possible modification pathways are identified, and products of these modification reactions are proposed for query tRNAs. This method allows for predicting modification patterns for newly sequenced genomes as well as for checking tentative modification status of tRNAs from one species treated with enzymes from another source, e.g. to predict the possible modifications of eukaryotic tRNAs expressed in bacteria. tRNAmodpred is freely available as web server at http://genesilico.pl/trnamodpred/.
Keywords: tRNA, RNA modification, bioinformatics, sequence similarity, homology, MODOMICS
1. Introduction
tRNA molecules are known to be rich in post-transcriptionally modified nucleosides, which play key roles for their structure and function (reviewed in [1]). This richness is reflected in amount of chemically modified residues in tRNA (up to 25 % of residues can be modified in certain tRNA molecules [2]) and their variety (the majority of more than 100 known types of chemically altered nucleosides can be found in tRNA molecules [3, 4], compared to only 12 different types of modifications found so far in rRNA [5]). Location of modified residues in tRNAs is far from random: it is tightly linked to their properties and functions. A network of modifications located in so-called D- and T -loops is responsible for proper folding and structural dynamics of the core of the tRNA molecule (reviewed in [6, 7]), while a great variety of complex modifications present in the anticodon loop fine-tune mRNA decoding by reducing conformational dynamics of the loop and by ordering the anticodon branch structure (reviewed in [8]). The highly conserved modification landscape of tRNA molecules from all domains of life co-exists with specific modification patterns, characteristic for tRNAs from different phylogenetic groups. Examination of experimentally studied tRNA sequences revealed that different groups of organisms tend to have both specific types of modified residues and characteristic locations of modification [2].
Nucleosides in tRNAs are modified by specific enzymes during tRNA maturation. A chemically modified nucleoside in mature tRNA can be a product of reaction carried out by one enzyme, an enzymatic complex, or results from a whole pathway of modification processes (reviewed in [1]). tRNA modification enzymes are position specific – they introduce the modification(s) in one or more defined places in the tRNA molecule, and their substrate specificity can be defined by sequence or structure determinants, which leads to variability in the spectrum of tRNAs modified by different enzymes. Some of the site-specific enzymes act on essentially all tRNAs containing an appropriate target residue, like TrmA from Escherichia coli, which methylates uridine at position 54 in practically all tRNAs that have this residue [9]. Other enzymes act only on a subset of available substrates that conform to their requirements of sequence, structure and/or presence of other modifications – examples include TrmL [10] and TruA [11] from E. coli.
Nowadays we experience a fast development of high-throughput sequencing-based and mass spectrometry-based methods that allow for detection of chemically modified nucleosides in RNAs at the transcriptome level. Recently developed methods enable quantitative studies of all ribonucleotide modifications in a cell [12] or identification of positions of some modifications, such as N1-methyladenosine, N3-methylcytidine and N1-methylguanosine, in RNA sequence [13]. However, these methods are either limited to a small subset of known types of modifications or they do not allow to specify location of modified nucleoside in the sequence. Hence experimental determination of all modified residues in a given tRNA species or in a tRNA repertoire from a given cell remains laborious and difficult. On the other hand, the success of computational prediction of modification sites in tRNA has been limited by the difficulty in predicting enzymes responsible for modification.
Here we present a computational tool for the prediction of chemically modified nucleosides in tRNA sequences – tRNAmodpred (http://genesilico.pl/trnamodpred/). The prediction is based on detection of homology to known RNA modification enzymes, and hence is applicable for prediction of modifications that have already been discovered in some tRNAs, and for which the enzymatic machinery is known and is expected to be conserved in the evolution. tRNAmodpred allows for predicting modification patterns for newly sequenced genomes as well as for checking tentative modification status of tRNAs expressed in heterologous systems based on input set of unmodified tRNA sequences and a set of protein sequences corresponding to a proteome of a cell.
2. Methods
2.1. Identification of homologous proteins using profile hidden Markov models
2.1.1. Preparation of the enzyme profile-HMM database in tRNAmodpred
Sequences of all known RNA modification enzymes were obtained from the MODOMICS database in August 2014. For each enzyme, similar sequences were collected with PSI-BLAST [14] (3 iterations, E-value threshold = 0.001) by querying the non-redundant sequence database obtained from the NCBI (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/) on 15th January 2014. Before the PSI-BLAST searches the database was clustered at 90 % sequence identity using CD-HIT [15] to reduce bias due to the presence of nearly identical sequences. Homologs of each enzyme from MODOMICS were separately clustered using CD-HIT with sequence identity threshold set to 0.4. Clusters that contained the query enzymes from MODOMICS were chosen and compared to each other. Sequences present in multiple clusters corresponding to different queries were removed. For each chosen cluster, sequences retained were aligned using MAFFT with the FFT-NS-i algorithm [16]. Profile hidden Markov models (HMMs) were built using the hhmake program [17, 18] based on the alignments. As a result, we obtained a set of HMMs, each corresponding to a family of homologous proteins containing a single MODOMICS RNA modification enzyme that catalyzes a known reaction (or a set of reactions) in a defined position (or positions) in tRNA sequence. The obtained library of HMMs was merged with a library of HMMs representing all protein families from the Pfam database [19] downloaded from ftp://toolkit.genzentrum.lmu.de/pub/HH-suite/databases/ (version 27.0). A mapping between Pfam and RNA modification enzymes from MODOMICS was made to identify Pfam families representing known RNA modification enzymes. To this end the Pfam database was searched with HHsearch using the above-mentioned HMMs built for MODOMICS enzymes. Matches with E-value below 1e-3 were stored (Supplementary File 1).
2.1.2. Searches of the enzyme profile-HMM database with user-defined sequences
Input protein sequences provided by the user are used as queries in sequence-profile search of the HMMs library, performed using the HHsearch program from the HH-suite package. The matches with probability below 70 % or E-value above 3.16e-18 are discarded (the 3.16e-18 E-value threshold gave best results in the preliminary benchmark of the method). The E-values of the remaining matches (in ascending order) are analyzed to identify the first biggest drop of E-value between consecutive matches. The number of matches reported before the E-value drop is compared to the number of matches reported with the highest probability. The bigger of these two sets of matches is chosen for further analysis. This approach was applied to prevent rejection of matches to several homologous modification enzymes, which may exhibit slightly different enzymatic activities. If the matches are to profiles representing RNA modification enzymes, these enzymes are considered in further steps of the prediction procedure. Matches to Pfam are checked for the identified similarity to the tRNA modification enzymes, with E-value below 3.16e-28; this particular threshold gave best results in our preliminary benchmark of the method (data not shown).
2.2. Prediction of modifications
The set of predicted enzymes is mapped onto MODOMICS pathways that are composed of reactions leading from completely unmodified or partially modified bases to fully modified (or hypermodified) products. Each specific position (residue number) in tRNA sequence has a list of possible modification pathways that have been previously reported and are stored in MODOMICS. If a reaction depends on a protein complex, then a complex is considered to be present only if more than 50 % of the complex components are identified (i.e., in the case of two-protein complexes like Trm6/Trm61 from S. cerevisiae both components must be identified). The pathways are then analyzed in order to collect all potential modifications, for which a complete reaction path from the unmodified base exists, and intermediate products leading to the formation of hypermodified nucleosides are also reported. The predicted modifications are represented by their type and position in tRNA molecule.
2.3. Mapping of predicted modifications onto input tRNA molecules
The predicted modifications are subsequently mapped onto tRNA sequences provided by the user. The mapping is done based on the identity of the base present in the input tRNA sequence at a given position. Modification is assigned to all sequences that contain an appropriate target base type (i.e., adenosine modifications specific for a given position can be introduced only in sequences that actually possess an adenosine residue at the position considered).
2.4. Filtering based on phylogenetic patterns
Predicted tRNA modifications can be additionally filtered based on query sequences origin (Gram negative bacteria; Gram positive bacteria; Archaea) and localization (cytosol of Viridiplantae; plastids; cytosol of eukaryotic single cell organisms, Fungi and Metazoa; mitochondria). If the query is assigned to one of the predefined phylogenetic/compartment groups then modifications, which were not observed in a given group(s) are excluded. Information about modifications present in each group was obtained from [2] and updated manually (Supplementary File 2).
2.5. tRNA alignment building
The tRNA alignment utility in the tRNAmodpred webserver was built based on the HMMER software v3.1 (http://hmmer.org/). The alignment of the input sequence(s) to the reference tRNA sequence is calculated using a hidden Markov model built for the alignment of tRNA sequences obtained from MODOMICS (version from September 2015).
2.6. Implementation
tRNAmodpred was implemented in Python 3.4. The web interface was developed in the Django framework (http://djangoproject.com). The pathway graphs analysis is performed using the NetworkX Python library (https://networkx.github.io/index.html).
2.7. Benchmarking tRNAmodpred
The performance of the tRNAmodpred server was assessed by performing predictions for six species: Escherichia coli K12 substr DH10B, Bacillus subtilis 168, Lactococcus lactis subsp. cremoris MG1363, Mycoplasma capricolum ATCC 27343, Haloferax volcanii DS2, and Saccharomyces cerevisiae. Fasta-formatted files with complete proteomes were downloaded from ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/ (Please note that NCBI has changed the FTP recently. Currently these data are available at: ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank/Bacteria/). tRNA sequences from E. coli, M. capricolum, H. volcanii, B. subtilis and S. cerevisiae were obtained from the MODOMICS database (version from September 2015). tRNA sequences from L. lactis were obtained from [20]. For each species except L. lactis tRNAmodpred was started either with the complete library of HMMs (referred to as “sanity-check”) or with a library in which HMMs derived from species for which the prediction was being done were excluded (referred to as “cross validation”). Predictions for L. lactis were performed with the complete library only, since the library does not contain any enzymes from this species. They are however shown together with the cross validation predictions results. Both sanity-check and cross validation predictions were carried with and without applying the phylogeny filter.
Predicted modifications reported by tRNAmodpred were compared to data collected in MODOMICS and to data from [20] (for L. lactis) and precision, recall and F-measure were calculated using the following formulas:
where tp is number of true positives, fp is number of false positives, fn is number of false negatives, P is precision, R is recall and F is F-measure. While counting tp, fp and fn not only final modifications were considered but also all intermediate products of the pathway. For example, if the experimental data supported the presence of ms2t6A modification in the position of interest, the tp number was increased by two if both t6A (the intermediate) and ms2t6A (the final modification) were predicted by tRNAmodpred. If only t6A was predicted, then the tp number was increased by one and at the same time the fn number was increased by one. If none of these were reported – then fn number was increased by two. The fp number was increased by one for each predicted modification, which in MODOMICS pathways is not an intermediate of the experimentally detected final modification. Since in some cases the final modification could be a result of different alternative pathways, the tp number was increased by one for each predicted modification, which belongs to any of these alternative pathways. In these cases the fn number was calculated as the minimal number of intermediate modifications missing in any of the alternative pathways. For further explanation and examples see Figure S 9 in Supplementary File 3.
Random sets of predicted modifications were obtained by randomly selecting (with replacement) tRNA modification enzymatic activities from MODOMICS and taking the products of these activities. These products (modifications) were then randomly assigned to tRNA sequences from the species under consideration, assuring that a given tRNA sequence contains appropriate base type in the position related to the activity associated with the modification. The number of selected activities was equal to the number of predicted activities in the prediction to which the random dataset was compared. For each prediction five independently generated sets of random modifications were prepared and scored. Mapping of tRNA modifications from experimentally studied species (“source”) onto tRNAs of different species (“target”) was done using MODOMICS data on modification pathways and modified tRNA sequences. For tRNA modifications from source species all possible pathway intermediate products were collected and assigned to tRNA sequences in the target species if source and target tRNA anticodon and decoded amino acid type agreed.
3. Results
3.1. tRNAmodpred – a server for computational prediction of posttranscriptional modifications in tRNAs
3.1.1. Method overview
tRNAmodpred is a program for the prediction of position and type of modified residues in tRNA molecules. The prediction is performed based on input protein and tRNA sequences, using information about tRNA modification pathways and location of modifications in experimentally studied tRNA sequences deposited in the MODOMICS database [3, 4]. It is assumed that the presence of a modification in tRNA depends on the presence of appropriate tRNA modification enzymes.
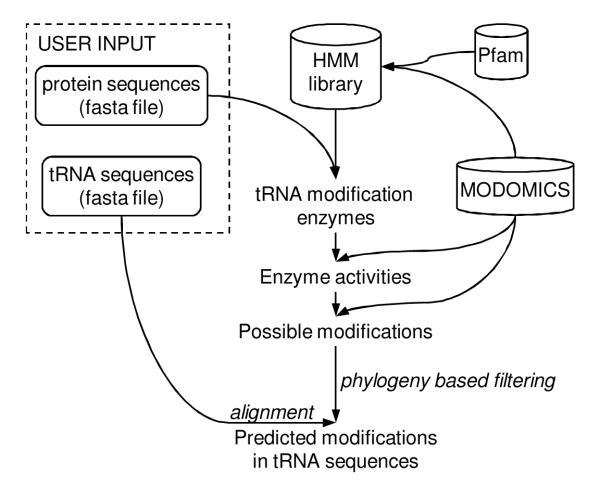
The workflow of the predictions is presented in Figure 1. In the first step, the presence of possible homologs of known tRNA modification enzymes among the input protein sequences is detected. This step is performed using remote protein homology detection method HHsearch [17] and a library of HMMs that contains profiles representing known tRNA modification enzymes. In the second step, the set of identified tRNA modification enzymes homologs is mapped onto MODOMICS pathways. The analysis of the pathways generates a set of modifications, which can be potentially introduced in the presence of identified enzymes. Since in the case of hypermodifications the final product of enzymatic pathway can depend on environmental conditions and partial modifications may occur [21-26], all sub-products of a pathway are reported as possible modifications. Next, the predicted possible modifications are assigned to input tRNA sequences based on the unmodified base type at the position of modification.
Figure 1.
The workflow of the tRNAmodpred program. Optional steps are in italics.
3.1.2. tRNAmodpred webserver
The tRNAmodpred webserver interface allows for submission of user queries and presentation of prediction results. On the submission page the user has to provide input protein and tRNA sequences as files in the Fasta format. Since the prediction of modifications localization in tRNA sequences requires that these sequences are aligned according to the rules described in [27], tRNAmodpred provides an optional utility which aligns input tRNA sequences. The automatically generated alignment can be directly sent for further analyses or downloaded for manual inspection. The submission page also provides a possibility to select whether the optional phylogeny-based filtering step is to be performed. If the user can assign the query to one of the predefined phylogenetic groups, then it is highly recommended to use this filtering option, since it should limit the number of false positive results. Detailed descriptions of the input files format and available options, together with example input files, are available on the Help page.
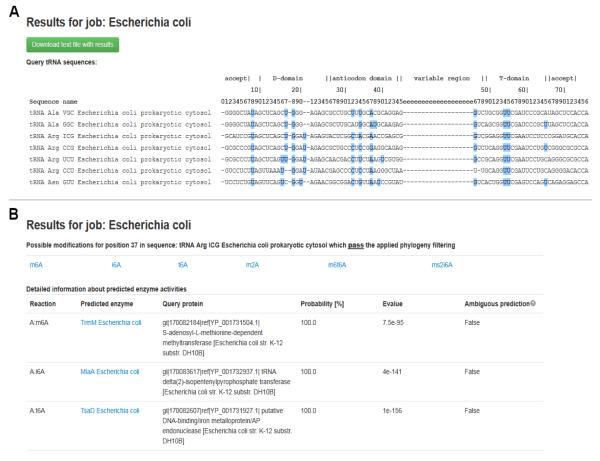
The results of predictions are presented in the results overview page and a collection of detailed results pages (Fig. 2). The results overview page contains an alignment of the input tRNA sequences with residues predicted to be modified marked in blue and clickable. Clicking on the chosen residue allows viewing the detailed prediction for this residue. Details of predictions are presented in a tabular form and include information about modification types predicted for the chosen residue and identified tRNA modification enzymes homologs that can participate in the synthesis of modifications predicted. Since it is possible that one query protein returns significant matches to more than one tRNA modification enzyme, the prediction is labeled as “ambiguous” if these enzymes exhibit different activities. The enzymes are shown together with their appropriate enzymatic activities (represented as reactions). The results can be downloaded as a text file that contains the tabular data for all residues predicted to be modified.
Figure 2.
Example results pages from tRNAmodpred. A. Results overview page with aligned input tRNA sequences, in which residues predicted to be modified are marked in blue. B. Detailed results page for a chosen residue.
3.1.3. Benchmark
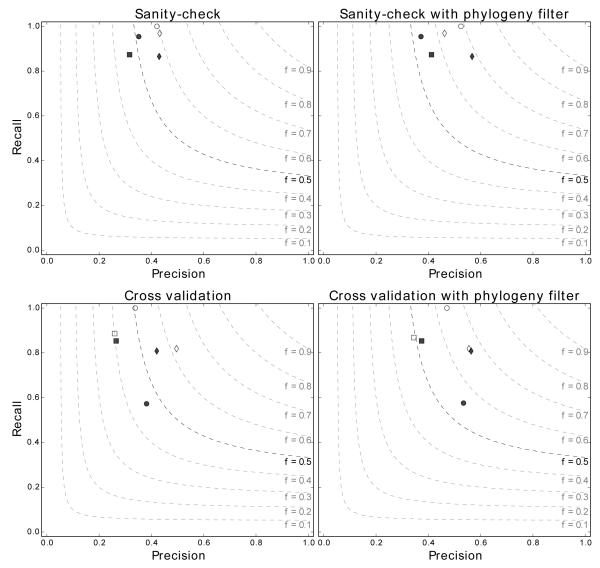
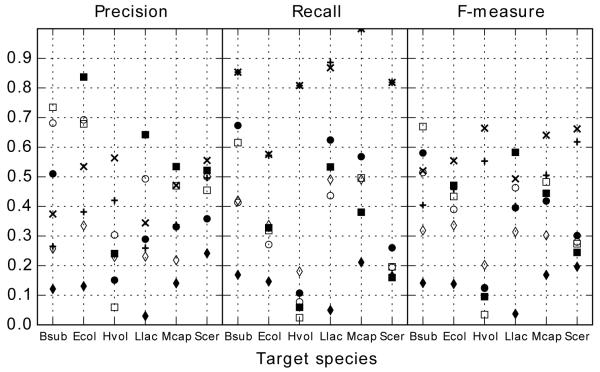
The performance of tRNAmodpred was assessed by carrying out predictions for six species: Escherichia coli, Bacillus subtilis, Haloferax volcanii, Lactococcus lactis, Mycoplasma capricolum, and Saccharomyces cerevisiae. For each species tRNAmodpred was run in four setups: sanity-check and cross validation, each with and without phylogeny filter (see Methods for details). For prediction of complete modifications set for each species the values of precision, recall and F-measure were calculated (Fig. 3, Table S 1-4 in Supplementary File 3).
Figure 3.
Precision, recall and F-measure obtained for predictions for the six tested species in four prediction setups. Species are indicated by symbols: ■ – Bacillus subtilis, ● – Escherichia coli, ◆ Haloferax volcanii, □ – Lactococcus lactis, ○ Mycoplasma capricolum, ◇ Saccharomyces cerevisiae.
Predictions from tRNAmodpred are characterized by high recall (above 0.7 for all cases except cross validation predictions from E. coli) and moderate precision (around 0.4 for predictions without phylogeny filter and 0.5 for predictions with phylogeny filter). In the sanity-check setups all predictions except for B. subtilis without phylogeny filter have F-measure above 0.5. In cross validation the F-measure drops down below 0.5 also for E. coli in the setup without the phylogeny filter. For L. lactis, for which no tRNA modification enzymes are present in the tRNAmodpred profiles library, the F-measure is 0.401 and 0.493 for predictions without and with phylogeny filter, respectively.
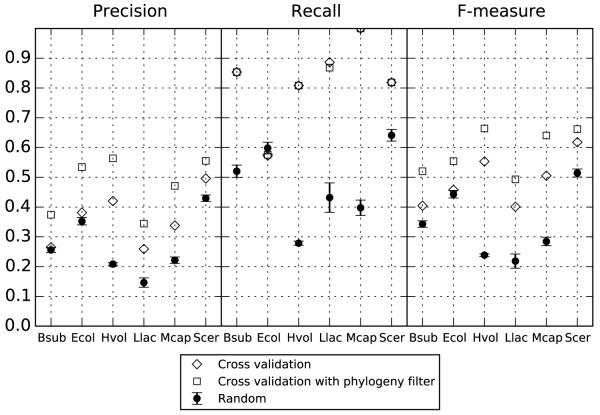
tRNAmodpred performance was compared to results obtained by random assignment of modified residues to tRNA sequences of a species under consideration (see Methods). In the cross validation setup tRNAmodpred is better than random assignment with respect to both precision and recall for all species except E. coli and B. subtilis (Figure 4). F-measures for all predictions done by tRNAmodpred are higher than for random studies. In the sanity-check setup tRNAmodpred predictions obtain higher precision and recall for all tested species except for E. coli in the setup without the phylogeny filter (Figure S 1 in Supplementary File 3).
Figure 4.
Comparison of the tRNAmodpred performance to results obtained by random assignment of modified residues to tRNA sequences from the target species. Scores for tRNAmodpred predictions done in the cross validation setup are shown (for the comparison to sanity-check setup see Figure S 1 in Supplementary File 3). Error bars for random probes depict standard deviation of scores between five random modifications sets. Species names acronyms: Scer – Saccharomyces cerevisiae, Hvol – Haloferax volcanii, Ecol – Escherichia coli, Bsub – Bacillus subtilis, Mcap – Mycoplasma capricolum, Llac – Lactococcus lactis.
Performance of tRNAmodpred was also compared to results obtained by considering experimentally confirmed tRNA modifications of one species (source) as predictions in another (target) (Fig. 5 and Figures S 2-6 in Supplementary File 3). The assignment of modifications between source and target species was done based on tRNAs anticodons. Since different species have different isoacceptors sets, not all tRNAs in the target species have their counterparts in the source species. Figure 5 shows scores calculated for complete sets of tRNAs of the target species, compared to predictions in the cross validation setup (with and without phylogeny filter). The scores vary considerably depending on the source and target species and can be both higher and lower than scores obtained by tRNAmodpred predictions. For B. subtilis F-measures for tRNAmodpred predictions are higher than F-measures calculated for modifications mapped from H. volcanii and S. cerevisiae, but lower than F-measures calculated for modifications mapped from E. coli and L. lactis, and comparable to M. capricolum. For E. coli tRNAmodpred performance is comparable to mapping modifications from B. subtilis and better than mapping from any other species. For L. lactis mapping from B. subtilis scores better than predictions from tRNAmodpred. Finally, for S. cerevisiae, M. capricolum and H. volcanii tRNAmodpred always outperforms the mapping. In the sanity-check setup the F-measure calculated for the tRNAmodpred predictions is always higher than F-measure calculated for modifications mapping, except for B. subtilis for which mapping from E. coli, M. capricolum and L. lactis obtain comparable scores (Figure S 2 in Supplementary File 3). The performance of tRNAmodpred was also compared to predictions which could be done based on experimental data from closely related species in detail for five randomly chosen L. lactis tRNAs (Table S 9 in Supplementary File 3).
Figure 5.
Comparison of precision, recall and F-measure calculated for cross validation predictions and results of mapping modifications from different species, for all tRNAs from the target species. Different point shapes and colors represent different species used as source of modifications: – Bacillus subtilis, [uni25CF] – Escherichia coli, – Haloferax volcanii, [uni25A1] – Lactococcus lactis, – Mycoplasma capricolum, – Saccharomyces cerevisiae; “x” and “+” markers – predictions done by tRNAmodpred with and without the phylogeny filter, respectively. Species names acronyms – same as in Fig. 4.
3.2. Prediction of pseudouridine modifications in L. lactis
The experimental study of L. lactis tRNAome did not include identification of pseudouridines in tRNA sequences from this species [20]. In Table 1 we present the predictions of pseudouridine locations done by tRNAmodpred with the phylogeny filter.
Table 1.
Predicted location of pseudouridine residues in tRNA sequences from L. lactis. The prediction was performed by tRNAmodpred with phylogeny filtering using L. lactis tRNA sequences reported in [20]. The alignment was built with the tRNA sequence alignment utility implemented in tRNAmodpred and corrected manually. Locations of pseudouridines in sequences are marked with white letter “P” on black background. The underscore in the Leu1 and Lys2 sequences denote the “CACU” insertion.
| 10| | 20| | 30| | 40| | 50| | 60| | 70| | Pseudouridine residue number * |
||
|---|---|---|---|---|---|---|---|---|---|
| tRNA | Anticodon | 012345678901234567-890--1234567890123456789012345eeeeeeeeeeeeeeeeeee6789012345678901234567890123456 | |||||||
| Ala | UGC | -GGGGCCUUAGCUCAGCU-GGG--AGAGCGCCUGCUUUGCACGCAGGAG-------------------GUCAGCGGUPCGAUCCCGCPAGGCUCCACCA | 55, 65 | ||||||
| Arg1 | ACG | -GCAGGCGUGGCUCAACU-GGAU-AGAGUACCUGACUACGAAPCAGGC--------------------GUUGUAGGUPCGAAUCCUACCGCUUGCACCA | 39, 55 | ||||||
| Arg2 | CCG | -GCGUCCGUAGUGUAAU--GGAU-AUCACGUAAGAUUCCGGPPCUUGAA-------------------U--GGGGGUPCGAUUCCCUCCGGACGCACCA | 38, 39, 55 | ||||||
| Arg3 | CCU | -GGUCUCUUAGUUAAAUU-GGAU-AUAACUACUGCCUCCUAAGCAGUGA-------------------U-UGCCAGUPCGAUUCUGGCAGGGGCCACCA | 55 | ||||||
| Arg4 | UCU | -GGUCCGAUAGCUCAGCU-GGAU-AGAGCAUUCGCCUUCUAAGCGAACG-------------------GUCGAGGGUPCGAAUCCCCCUCGGAUCACCA | 55 | ||||||
| Asn | GUU | -UGCGGAUUAGCUCAGUU-GGUA-GUAGCGCAUGACUGUUAAPCAUGAU-------------------GUCGUCAGUPCGAGUCUGACAUCCGCAGCCA | 39, 55 | ||||||
| Asp | GUC | -GGUUCUGUAGUGUAGC--GGUU-AUCACGUCGCCCUGUCACGGCGAAG-------------------AUCGCGGGUPCGAUUCCCGPCAGAACCGCCA | 55, 65 | ||||||
| Cys | GCA | -GGCGGCGUAGUGAAGU--GGU--AACACAUGGCUCUGCAAAAGCUUAA-------------------U-CGUCGGUPCAAAUCCGACCGUCGCCUCCA | 55 | ||||||
| Gln | UUG | -UGGGGUAUAGCCAAGC--GGU--AAGGCAAGGGACUUUGACPCCCUCA-------------------UGCGUUGGUPCGAAUCCAGCUACCCCAGCCA | 39, 55 | ||||||
| Glu | UUC | -GGUCCGUUGGUCAAGG--GGUU-AAGACACCGCCUUUUCACGGCGGUA-------------------A-CACGGGUPCGAAUCCCGPACGGACUACCA | 55, 65 | ||||||
| Gly1 | GCC | -GCGAACGUAGUUCAGG--GGU--AGAACACAACCUUGCCAPGGUUGGG-------------------GUCGCGAGUPCGAAUCUCGPCGUUCGCUCCA | 38, 55, 65 | ||||||
| Gly2 | UCC | -GCGGAUGUAGUUUAAU--GGU--AGAACCCCAGCCUUCCAAGCUGGCU-------------------A-CGCGAGUPCGAUUCUCGPCAUCCGCUCCA | 55, 65 | ||||||
| His | GUG | -GCGGAUGUGGUGAAGU--GGUU-AACACACCGGCUUGUGGCGCCGGCA-------------------CUCGCGAGUPCGAUCCUCGPCAUCCG--CCA | 55, 65 | ||||||
| Ile | GAU | -GGGAGUUUAGCUCAGUU-GGUU-AGAGCACUGUGUUGAUAACGCAGGG-------------------GUCCCAGGUPCGAAUCCUGGAAUUCCCACCA | 55 | ||||||
| Leu1 | AAG | -GCGGUGGUGGCGGAAUU-GGCA-GACGCGCAGGAUUAAGGAPCCUGUCGGGAAU-------UUUCUUGGUGCGGGUPCAAGUCCCGCCCACCG_ACCA | 39, 55 | ||||||
| Leu2 | CAG | -GGGGAAGUGGUGGAAU--GGCA-GACACGCAUGCUUCAGGPGCAUGUGCGAGA--------AAUCGCG-UGAGGGUPCAAAUCCCUPCUUCCCCACCA | 38, 55, 65 | ||||||
| Leu3 | UAG | -GCGGAUGUGGCGGAAUU-GGCA-GACGCACUAGAUUUAGGAPCUAGCGCUUA----------ACGGCG-UGGGGGUPCAAGUCCCUPCAUCCGCACCA | 39, 55, 65 | ||||||
| Leu4 | CAA | -GCCGAAGUGGCGGAAUA-GGCA-GACGCGCCAGACUCAAAAPCUGGUUCCUUC--------ACGGGAG-UGCCGGUPCGACCCCGGCCUUCGGCACCA | 39, 55 | ||||||
| Leu5 | UAA | -CCCGGGGUGGCGGAACU-GGCA-GACGCACAGGACUUAAAAPCCUGCGAGGGGU------AACCUUCG-UACCGGUPCGAAACCGGPCCUCGGGACCA | 39, 55, 65 | ||||||
| Leu6 | UAA | -GCUCGAAUGGCGGAAUU-GGCA-GACGCUGCGGACUUAAAAPCCGUUGGUUAU--------UAAACCG-UGAGGGUPCAAGUCCCUCUUUGAGCACCA | 39, 55 | ||||||
| Lys1 | CUU | -GGCCCGGUAGCUCAGUU-GGU--AGAGCAGUAGACUCUUAAPCUAUGG-------------------GUCCAGGGUPCGAGCCCCUGCCGAGCCACCA | 39, 55 | ||||||
| Lys2 | UUU | -GACUCGUUAGCUCAGUU-GGU--AGAGCAUUUGACUUUUAAPCAAAGG-------------------GUCGCUGGUPCGAGCCCAGCACGGGU_ACCA | 39, 55 | ||||||
| Lys3 | UUU | -GACUCGUUAGCUCAGUU-GGU--AGAGCAUUUGACUUUUAAPCAAAGG-------------------GUCGCUGGUPCGAGCCCAGCACGGGUCACCA | 39, 55 | ||||||
| Met1 | CAU | -GGAUCUUUAGCUCAGUU-GGUU-AGAGCUAUCGGCUCAUAACCGAUCG-------------------GUCGCUGGUPCGAAUCCAGCAAGAUCCACCA | 55 | ||||||
| Met2 | CAU | -GGCGGUGUAGCUCAGCU-GGCU-AGAGCGUUCGGUUCAUACCCGAGAG-------------------GUCGGGGGUPCGAUCCCCUPCGCCGCUACCA | 55, 65 | ||||||
| Met3 | CAU | -CGCGGGAUGGAGCAGCUAGGU--AGCUCGUCGGGCUCAUAACCCGAAG-------------------GUCAUAGGUPCAAAUCCUAPUCCCGCAACCA | 55, 65 | ||||||
| Met4 | CAU | -AGUUCUUUAGCUUAGUU-GGUU-AAAGUCCCCCGCUCAUAACGGGGUA-------------------AGCGCUGGUPUGAGUCCAGC---AAGAACCA | 55 | ||||||
| Phe | GAA | -GGCUCGGUAGCUCAGUU-GGU--AGAGCAAUGGAUUGAAGCPCCAUGU-------------------GUCGGCGGUPCGAUUCCGUCUCGCGCCACCA | 39, 55 | ||||||
| Pro | UGG | -CGGGAAGUAGCUCAGCUUGGU--AGAGUACUUGGUUUGGGACCAAGGU-------------------GUCGCAGGUPCGAAUCCUGPCUUCCCGACCA | 55, 65 | ||||||
| Ser1 | GGA | -GGAGAAGUGUCCGAGU--GGCCGAAGGAGCACGCCUGGAAAGPGUGUAUACGUCA-----CAAGCGUAUCGGGGGUPCGAAUCCCCPCUUCUCCUCCA | 40, 55, 65 | ||||||
| Ser2 | CGA | -GGAGCCAUGGCAGAGU--GGU--AAUGCAGCGGACUCGAAAPCCGUCGAACCGUGU---AAAGCGGCG-CAGGGGUPCAAAUCCCCPUGACUCCUCCA | 39, 55, 65 | ||||||
| Ser3 | UGA | -GGAGGAUUACCCAAGCCUGGCUGAAGGGAACGGUCUUGAAAACCGUCAGGCAUGU----AAAGGCGUG-CGUGGGUPCGAAUCCCACAUCCUCCUCCA | 55 | ||||||
| Ser4 | UGA | -GGAGGAUUACCCAAGUCCGGCUGAAGGGAACGGUCUUGAAAACCGUCAGGCGUGU----AAAAGCGUG-CGUGGGUPCGAAUCCCACAUCCUCCUCCA | 55 | ||||||
| Ser5 | GCU | -GGGGGGUUACUCAAGA--GGCUGAAGAGGACGGUUUGCUAAAPCGUUAGGUCGGG----AAACCGGCG-CGAGGGUPCGAAUCCCUPACCCCCCUCCA | 40, 55, 65 | ||||||
| Thr1 | GGU | -GCCGUUGUAGCUCAGUC-GGU--AGAGCAGCACCAUGGUAAGGUGAAG-------------------GUCGACAGUPCGAUUCUGUPCAAUGGCACCA | 55, 65 | ||||||
| Thr2 | CGU | -GCCGAGUUAGCUCAGUC-GGU--AGAGCACUUCACUCGUAACGAAGGG-------------------GUCACAGGUPCGAUUCCUGCACUCGGCACCA | 55 | ||||||
| Thr3 | UGU | -GCCGACUUAGCUCAGUU-GGU--AGAGCAUCUGAUUUGUAAPCAGAGG-------------------GUCGCGUGUPCGAAUCAUGPAGUCGGCACCA | 39, 55, 65 | ||||||
| Trp | CCA | -ACGGGCAUCGUAUAAA--GGU--AGUACAAAGGUCUCCAAAACCUUUA-------------------G-UGUGGGUPCAAUUCCUGCUGCCCGUGCCA | 55 | ||||||
| Tyr | GUA | -GGAAGGGUAGCGAAGA--GGCUAAACGCGGCGGACUGUAAAPCCGCUCCUU------------CGGGUUCGGGGGUPCGAAUCCCUCCCCUUCCACCA | 39, 55 | ||||||
| Val | UAC | -GGGAGUUUAGCUCAGCU-GGG--AGAGCAUCUGCCUUACAAGCAGAGG-------------------GUCAGCGGUPCGAUCCCGUPAACUCCCACCA | 55, 65 | ||||||
Residue numbers according to the universal tRNA numbering system [27].
4. Discussion
In this work we describe tRNAmodpred – a tool for computational prediction of post-transcriptionally modified residues in tRNAs. The post-transcriptional RNA modifications are synthesized in cells by a variety of enzymes with different activities and specificities. Since the availability of appropriate enzymes is crucial for the synthesis of modifications, tRNAmodpred predicts the presence of modified residues based on identification of homologs of known tRNA modification enzymes. tRNAmodpred allows to use the knowledge about tRNA modification enzymes and pathways that were studied experimentally to infer expected patterns of modifications in tRNAs of any species with fully sequenced genome or in tRNAs expressed in heterologous system.
We tested tRNAmodpred by performing predictions for six model species, in several setups: “sanity-check” (with complete library of sequence profiles) or “cross validation” (after excluding from the library profiles representing enzymes from the target species) and with or without use of an additional filtering step based on the phylogeny of the organism. We observed that in the cross validation setups the recall of the predictions tends to drop, however except for E. coli, this drop is not significant. Our results show that phylogenetic origin of the species and subcellular localization are important factors influencing prediction of modifications and applying the phylogeny-based filtering allows for improving the precision.
The benchmarking of tRNAmodpred was performed on currently available, known tRNA sequences from the chosen six model species. However, these sets of tRNA sequences are not always complete (e.g. not all tRNAs from B. subtilis or from S. cerevisiae mitochondria have been sequenced). Although it is possible that after including the yet unknown tRNA sequences the scores obtained by tRNAmodpred would change slightly, we believe that the general conclusions regarding the applicability of the method would remain valid.
The main limitation of our approach that may lead to false negative predictions is that the predictions are based on homology, i.e. the procedure implemented in tRNAmodpred can predict only modifications that were previously characterized and that are introduced by an evolutionarily conserved process. Thus, tRNAmodpred cannot predict any new modifications or known modifications at new positions or introduced by analogous enzymes. Another assumption that may lead to false positive predictions is that all tRNA modification enzymes can modify any tRNA species that encodes the correct target nucleotide at the position to be modified. In reality, tRNA modification enzymes often have specificity determinants like sequence patterns or presence of other modifications, which narrow down the repertoire of tRNAs that are actually used as substrates for a given enzyme. This results in the relatively low precision of our method, which could be improved in the future by introduction of additional parameters that allow for capturing the enzymes specificities (e.g., addition of filters that consider specificity determinants for particular enzymes or mutual dependencies between different modifications).
It should be kept in mind that the set of input protein sequences should represent the proteins that are expected to be able to interact with the input tRNAs. It is especially important if the aim is to predict tRNA modifications in organelles. Since it is beyond the scope of tRNAmodpred to predict the subcellular localization of proteins or tRNAs the user should provide input data appropriate for the task: e.g., the input should consist of a set of sequences of proteins expected to be present in mitochondria (both encoded by the mitochondrial genome and imported), and a set of mitochondrial tRNA sequences, if the goal is to predict tRNA modifications in mitochondria. We have provided a short description and scores for the prediction of S. cerevisiae mitochondrial tRNAs in Supplementary File 3 (page 9, Table S 10).
We compared tRNAmodpred performance to random predictions and to an approach in which tRNA modifications in one species (target) are predicted by assignment of modifications known to exist in different species (source). We observed that while tRNAmodpred in general performs better than an approach with random assignment of modified residues to tRNAs from the target species, the outcome of comparison between our method and the mapping of tRNA modifications from another species depends highly on the case under consideration. We observe that in cases when experimental data are available for an organism closely related to the target species, the mapping approach gives better results than predictions done by tRNAmodpred (for example as in the case of B. subtilis and L. lactis, Fig. 5). However, when no data from close relatives are available, tRNAmodpred rather than mapping from distant organism should be used.
Apart from tRNAmodpred there is another computational tool that deals with the problem of the prediction of tRNA modifications. tRNAmod is a machine-learning based program which predicts modifications of uridine based on tRNA sequence [28]. The reported performance of the method is very good – AUC (area under the curve) of about 0.9. However, it must be noted that tRNAmod has limited application: it can only predict localization of three most common uridine modifications (pseudouridine, dihydrouridine and 5-methyluridine) or report that a particular uridine residue is modified or not. We checked whether combining tRNAmod and tRNAmodpred results improves the accuracy of predictions, but unfortunately we have not observed significant improvement compared to predictions done by tRNAmodpred alone (Tables S 5-8 in Supplementary File 3). This is presumably due to the fact that predictions for pseudouridine, dihydrouridine and 5-methyluridine obtained by tRNAmod and tRNAmodpred achieve comparable precision and recall values (Figures S 7-8 in Supplementary File 3).
5. Conclusion and future perspectives
We present here a computational method to predict structure and localization of modified nucleosides in tRNA molecules based on identification of homologs of known tRNA modification enzymes and knowledge about tRNA modification pathways. tRNAmodpred is freely available to the scientific community as a web server. It can provide valuable initial predictions that could be a starting point for experimental analyses. The quality of tRNAmodpred predictions depends on the data about modified tRNA sequences and RNA modification enzymes, hence it is expected to improve with time, with the accumulation of experimental data. The approach used in tRNAmodpred is in principle applicable to other conserved RNA molecules, such as rRNA, and we intend to develop this tool in this direction in the future. Another direction of potential future development, which would be particularly relevant for prediction of rRNA modification is the consideration of guide RNAs that introduce methylation and pseudouridylation [29-31], reviewed in [32].
Supplementary Material
Acknowledgements
We would like to thank Dr Lukasz Kozlowski, Dr Jo Marie Bacusmo and Marcin Magnus for valuable discussions, help in the webserver development and testing tRNAmodpred.
This analysis was initially supported by the Foundation for Polish Science (FNP, grant TEAM/2009–4/2 to J.M.B.) and subsequently by the Polish National Science Center (NCN, grant 2012/04/A/NZ2/00455 to J.M.B.). M.A.M. was additionally supported by funds from the European Union (European Social Fund - stipend for Mazovian PhD students, Human Capital Operational Program 2007–2013). S.D.H. was supported by Polish National Science Centre (NCN) (grant 2011/03/D/NZ8/03011 to S.D.H.) and by a fellowship for outstanding young scientists from the Polish Ministry of Science and Higher Education. This work was also supported by the National Institutes of Health (grant number R01 GM70641 to V.dC-L.).
Footnotes
Appendix A. Supplementary data
Supplementary File 1. Matches between the Pfam database and the library of HMMs built for MODOMICS enzymes.
Supplementary File 2. Information about modifications present in each phylogenetic/subcellular localization group used by the phylogeny filter in tRNAmodpred.
Supplementary File 3. Supplementary Tables and Figures with results of tRNAmodpred and tRNAmod benchmarks.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Björk GR, Hagervall TG. EcoSal Plus. 2014;6 doi: 10.1128/ecosalplus.ESP-0007-2013. [DOI] [PubMed] [Google Scholar]
- [2].Machnicka MA, Olchowik A, Grosjean H, J M. Bujnicki. RNA Biol. 2014;11:1619–1629. doi: 10.4161/15476286.2014.992273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. Nucleic Acids Res. 2006;34:D145–149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. Nucleic Acids Res. 2013;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sharma S, Lafontaine DL. Trends Biochem Sci. 2015;40:560–575. doi: 10.1016/j.tibs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- [6].Helm M, Alfonzo JD. Chem Biol. 2014;21:174–185. doi: 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jackman JE, Alfonzo JD. Wiley Interdiscip Rev RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Agris PF. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gu X, Ivanetich KM, Santi DV. Biochemistry. 1996;35:11652–11659. doi: 10.1021/bi9612125. [DOI] [PubMed] [Google Scholar]
- [10].Zhou M, Long T, Fang ZP, Zhou XL, Liu RJ, Wang ED. RNA Biol. 2015;12:900–911. doi: 10.1080/15476286.2015.1050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H. J Biol Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- [12].Rose RE, Quinn R, Sayre JL, Fabris D. RNA. 2015;21:1361–1374. doi: 10.1261/rna.049429.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li W, Godzik A. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- [16].Katoh K, Misawa K, Kuma K, Miyata T. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Soding J. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- [18].Remmert M, Biegert A, Hauser A, Soding J. Nat Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- [19].Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Nucleic Acids Res. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Puri P, Wetzel C, Saffert P, Gaston KW, Russell SP, Cordero Varela JA, van der Vlies P, Zhang G, Limbach PA, Ignatova Z, Poolman B. Mol Microbiol. 2014;93:944–956. doi: 10.1111/mmi.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tyagi K, Pedrioli PG. Nucleic Acids Res. 2015;43:4701–4712. doi: 10.1093/nar/gkv322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moukadiri I, Garzon MJ, Björk GR, Armengod ME. Nucleic Acids Res. 2014;42:2602–2623. doi: 10.1093/nar/gkt1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roe BA, Stankiewicz AF, Rizi HL, Weisz C, DiLauro MN, Pike D, Chen CY, Chen EY. Nucleic Acids Res. 1979;6:673–688. doi: 10.1093/nar/6.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kuchino Y, Borek E, Grunberger D, Mushinski JF, Nishimura S. Nucleic Acids Res. 1982;10:6421–6432. doi: 10.1093/nar/10.20.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- [26].Buck M, Ames BN. Cell. 1984;36:523–531. doi: 10.1016/0092-8674(84)90245-9. [DOI] [PubMed] [Google Scholar]
- [27].Steinberg S, Misch A, Sprinzl M. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Panwar B, Raghava GP. BMC Bioinformatics. 2014;15:326. doi: 10.1186/1471-2105-15-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- [30].Ni J, Tien AL, Fournier MJ. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- [31].Ganot P, Bortolin ML, Kiss T. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- [32].Watkins NJ, Bohnsack MT. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.