Abstract
Cell-penetrating peptides (CPPs) have shown promise in non-permeable therapeutic drug delivery, owing to their ability to transport a variety of cargo molecules across the cell membranes and their non-cytotoxicity. Drosophila antennapedia homeodomain-derived CPP-Penetratin (RQIKIWFQNRRMKWKK), being rich in positively charged residues, has been increasingly used as a potential drug carrier for the treatment of various purposes. Penetratin can breach the tight endothelial network, blood-brain-barrier (BBB), enabling treatment of several neurodegenerative maladies including Alzheimer's disease, Parkinson's disease, and Huntington's disease. However, detailed structural knowledge and the mechanism of Penetratin action remains unknown. This study defines structural features of the Penetratin derived peptide – DK17 (DRQIKIWFQNRRMKWKK) in several model membranes and describes a membrane-induced conformational transition of the DK17 peptide in these environments. A series of biophysical experiments, including high-resolution NMR spectroscopy, provides the three-dimensional structure of DK17 in different membranes mimicking the BBB or total brain lipid extract. MD simulations support the experimental results showing preferential binding of DK17 to particular lipids at atomic resolution. The peptide conserves the structure of the subdomain spanning residues Ile6-Arg11, despite considerable conformational variation to different membrane models. In vivo data suggest that the wild-type, not a mutated sequence, enters the central nervous system. Together, these data highlight important structural and functional attributes of DK17 that could be utilized in drug delivery for neurodegenerative disorders.
INTRODUCTION
Since their discovery over two decades ago, Cell Penetrating Peptides (CPPs) have been of interest for medicinal biology research. Due to their ability of translocating hydrophobic and hydrophilic molecules across the semi permeable plasma membrane, CPPs have emerged as drug loading agents1-3. Few examples of CPPs include HIV-1 (Human Immunodeficiency Virus type 1)-derived TAT protein4, 5, Drosophila antennapedia homeodomain-derived Penetratin peptide6, 7, Herpes Simplex Virus derived VP22 peptide8, Pep-1 CPP from Simian Virus 40 large T antigen and reverse transcriptase of HIV-19 and simple amphipathic model peptide, MAP10, which deliberately accept the job of translocating the impermeable molecules across the plasma membrane. Features like positively charged amino acid residues, in addition to the hydrophobic counterpart, offers the intrinsic property of non-specific and non-toxic delivery of various bioactive cargos crosswise the cell11, 12. One major challenge in medicinal chemistry is to deliver therapeutic drugs to the central nervous system, the CNS, via the tight endothelial barrier- the Blood-Brain-Barrier (BBB)13, 14. A recent example concerns the mosquito-borne disease, Zika virus, which preferentially affects the spinal cord, bypassing therapeutic approaches that are impermeable to the CNS15, 16.
Recently CPPs have been improved either by conjugating a variety of impermeable drugs to them or by developing more potent synthetic CPPs to advance the targeted systemic cellular uptake17-24. Among these, Penetratin, a 16 residue highly cationic and most importantly non-toxic CPP, efficiently assists in the intracellular delivery of a wide range of cargos via a direct penetration process21, 22. The penetratin peptide is part of the third α-helix homeodomain of Drosophila Antennapedia transcription factor protein (HDAntp), responsible for translocating this protein through the membrane23, 24. The BBB permeability of penetratin makes it a potential drug-delivery agent when conjugated to hydrophobic drugs for treating severe neurological disorders25-28. The study by Alves et al. showed that cell surface binding and cellular internalization can be tuned by mutating penetratin, introducing additional arginine residues29, 30. The driving force behind penetratin's activity appears to involve electrostatic interactions, 30, 31 mediating a conformational transition from random coil to higher order secondary structure in the presence of membranes as has been proposed for antimicrobial peptides, or AMPs32-34. Additionally, lipid structural arrangements may be involved in the difference in activity between eukaryotic and prokaryotic cell membranes35.
The N-terminal region of penetratin preferentially adopts a helical conformation in DPPC bilayers while the C-terminus remains unstructured36. Solution NMR studies of the penetratin peptide in different membrane mimicking micelles, such as DHPC, DMPC, DMPG, and SDS, also reveal conserved helical segments in all cases37. Reports by Mäler et al. propose different translocation mechanisms of penetratin in the presence of bicelles including DPC, CHAPS, and DHPC37, 38. Our current study focuses on the structural characterization of the cationic peptide DK17 (DRQIKIWFQNRRMKWKK-NH2), in the presence of several model membranes to correlate its structure with its function. In particular, three lipid membranes composed of GM1, a BBB model membrane mimic (POPC:POPG:Cholesterol:GM1) and a total brain lipid extract (TLBE) were chosen for detailed structural evaluation. We studied vesicles formed form GM1 ganglioside and total brain lipid extract (excluding GM1 ganglioside) as models of BBB membrane. POPC:POPG:Cholesterol:GM1 model mainly mimics the BBB membrane system in which POPC along with cholesterol balances the structural integrity and POPG preserves the charge of the membrane39.
Herein, we apply several biophysical approaches including fluorescence, CD and NMR spectroscopy to DK17 in different model membrane systems to investigate the conformational manifold of DK17 peptide. We propose to address the following questions. 1) How does the binding propensity of the peptide vary with lipid membrane composition? 2) How can the structural integrity of the Ile6-Arg11 subdomain be conserved despite significant variation in its conformation with different lipids? Molecular dynamics studies provide additional details validating changes in binding affinity of the peptide with lipid membrane composition, as well as membrane-induced conformational changes. This is the first detailed correlation of three-dimensional structure of DK17 and its function of cell permeabilization using in vivo study.
MATERIALS AND METHODS
Materials
Wild-type (DRQIKIWFQNRRMKWKK-NH2) and mutated (DRQIKGGGQNRRMKWKK-NH2) DK17 peptides (purity of > 97%) were acquired from either GL Biochem (Shanghai, China) or GenScript USA. All lipid molecules such as 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC),1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (POPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), GM1 ganglioside (Porcine), Cholesterol and total brain lipid extract (TLBE) (ovine) were purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA). The Alexa Fluor 680 infrared dye was procured from Thermo Fisher Scientific and all other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Vesicle preparation
The large unilamellar vesicles (LUVs) of ganglioside GM1 or POPC:POPG:Cholesterol:GM1 with a molar ratio of 7.65: 0.85: 1: 0.5 or total brain lipid extract (TLBE) (TLBE is mainly composed of PC/PE/PI/PS/PA with more than 55% of lipid constitute is unknown) were prepared by forming a nitrogen-dried lipid film from their chloroform stock solution followed by lyophilization overnight under high vacuum to get rid of the residual solvent. The lipid film was rehydrated in 10mM phosphate buffer, pH 6.0 and then vortexed well until it dissolved in the buffer. Finally the hydrated lipid solutions were extruded at least 23 times through a 100 nm polycarbonate nucleopore membrane filter (Whatman) mounted on a mini-extruder (Avanti Polar Lipids Inc. Alabaster, AL, USA) to obtain homogenous solutions of 100 nm sized LUVs39, 40. The homogeneity of the LUVs in solution was further confirmed through dynamic light scattering experiments.
Fluorescence spectroscopy studies
The intrinsic Tryptophan fluorescence property of DK17 peptide was used to analyze the biophysical properties including binding affinity, solvent accessibility of the peptide to all the model membranes41-43. Tryptophan fluorescence emission spectra of 10 [.proportional]M free peptide (in 10mM phosphate buffer, pH 6.0) were acquired over a spectral range of 300–400 nm in a Hitachi F-7000 FL spectrometer at 25 °C with excitation wavelength of 280 nm. A quartz cuvette of 0.1 cm path length was used to perform all fluorescence experiments. The change in Tryptophan fluorescence emission maximum (F) towards lower wavelength (blue shift) was recorded after successive addition of each LUV and analyzed using the following equation (1)
| (1) |
where λmax nm and λ(o)max nm represent the Tryptophan fluorescence emission maxima of the peptide in the presence and absence of model vesicles respectively.
Using Acrylamide as a neutral quencher, the solvent accessibility of the peptide in proximity of all the model lipid membranes was determined42, 44. Both free and membrane-bound peptide was titrated with increasing concentration of quencher and the resultant quenched fluorescence intensity for both free and bound peptide were related using the Stern-Volmer equation (2).
| (2) |
where the Tryptophan fluorescence intensity of peptide without the addition of any quencher and intensity at each quencher concentration [Q] in units of molarity are denoted by F0 and F, respectively. The Stern–Volmer constant (KSV) values for the free and bound peptides provide a measure of the solvent accessibility of the peptide to acrylamide.
Fluorescence anisotropy
The binding constant (equilibrium dissociation constant, KD) of 10 μM peptide (in 10 mM phosphate buffer, pH 6.0) to model lipid membranes was further measured by steady-state fluorescence anisotropy experiments. A fluorescence spectrophotometer (Hitachi F-7000FL) equipped with a polarizer was used for these experiments, and the fluorescence anisotropy values were calculated using the equation (3)
| (3) |
where IVV and IVH correspond to the Tryptophan fluorescence emission intensities obtained when the excitation polarizer is fixed in the vertical orientation while the emission polarizer's orientation varies from vertical VV to horizontal VH. The sensitivity parameter (G) for the instrument can be represented as IHV/IHH. Equilibrium dissociation constants (KD) were determined for peptide bound to each lipid preparation by plotting anisotropy values against the concentration of lipid membranes and fitting the data to a Hill equation42, 44, 45.
Circular Dichroism (CD) spectroscopy
In order to define the global conformational transitions of the peptide, CD spectroscopy measurements were carried out in the presence of the model membranes42. The spectral changes in a 25 μM solution of DK17 peptide (in 10mM phosphate buffer, pH 6.0) were recorded on a Jasco J-815 CD spectrometer upon titrating increasing concentrations of different LUVs over a range of 195-260 nm. A constant temperature of 298 K was maintained throughout the experiments. The raw data were acquired in the form of milli-degrees averaging four consecutive scans, at rate of 100 nm/min, subtracting the buffer baseline. A quartz cuvette of 1 mm path length was used. These values were converted to molar ellipticity (θ) (deg cm2 dmol−1) using the following equation (4):
| (4) |
Where m0 is the raw value in milli-degrees, M is molecular weight (g mol−1), L the path length of the cuvette (cm) and C the concentration (molarity). CDNN software (version 2.1) (http://bioinformatik.biochemtech.uni-halle.de/cdnn/) was used to analyze the CD spectrum after deconvolution46.
Two-dimensional (2D) NMR spectroscopy
All NMR experiments were performed using a Bruker Avance III 500 MHz (equipped with 5 mm smart probe) or a Bruker Avance III 700 MHz spectrometer (equipped with 5 mm cryo-probe). The 2D 1H-1H TOCSY and NOESY spectra of DK17 (0.5 mM) were performed using 80 ms and 150 ms mixing times, respectively with excitation sculpting for water suppression47. A spectral width of 12 ppm was used in both dimensions with 2K (t2) × 512 (t1) data points and States TPPI for quadrature detection in the t1 dimension. The experiment was performed in 10 mM phosphate buffer, pH 4.5 and 4,4-dimethyl-4-silapentane-5-sulfonate sodium salt (DSS) was used as an internal reference.
Binding of DK17 to membranes was monitored using a series of one-dimensional 1H NMR spectra of DK17 upon addition of increasing concentration of LUVs delivered from their individual stock solutions. Upon subsequent line broadening, without changing the chemical shift, of the DK17 spectrum in the presence of LUVs the transferred NOESY (trNOESY) experiment was performed with three different mixing times viz. 100, 150 and 200 ms for 24 scans, keeping all the parameters identical to those of the NOESY experiment on DK17 alone in aqueous solution. TOCSY, NOESY and trNOESY spectra were processed using 4K (t2) ×1K (t1) data matrices after zero filling48. The spectra were processed using Topspin 3.1 (Bruker, Switzerland) program suite and peak assignments were performed using SPARKY 3.1 software (https://www.cgl.ucsf.edu/home/sparky)49, 50.
NMR-derived Structure calculations
The three-dimensional NMR structure of the peptide in the presence of following model membranes was determined based on the NOE cross peaks of DK17 in the presence of respective LUVs from a trNOESY spectrum of 150 ms mixing time. The upper bound distance constraints were calculated from the NOE build-up rate to avoid spin diffusion, and were categorized as strong (2.5 Å), medium (2.6 - 3.5 Å) and weak (3.6 – 5.0 Å) for further structure calculations using CYANA program (version 2.1)51. The lower bound distance constraint was fixed to 2 Å to avoid van-der-Waals interaction. As the folding of the peptide structure is sensitive to the medium as well as long-range NOEs, we calculated the structure with CYANA using distance constraints obtained from trNOESY spectra only with no hydrogen bonds or torsional angle constraints. Indeed, the backbone dihedral angles (Phi) were constrained between −30° and −180° in order to conserve an accurate stereochemistry of the calculated structures along with conformational sampling52, 53. The ensemble structure consisted of twenty lowest energy conformations chosen from the 100 calculated structures using Procheck software for the quality check. PyMOL and MOLMOL software were used for visualization of structures54. The three-dimensional high-resolution NMR structures of the peptide in the presence of model lipid membranes including GM1 ganglioside LUVs, POPC:POPG:Cholesterol:GM1 LUVs and TLBE LUVs have been deposited in the PDB (Protein Data Bank) with their respective PDB accession code as 2ND6, 2ND7, and 2ND8, respectively.
Molecular modeling
Molecular dynamic simulations of the DK17 peptide in the membrane models were performed in Desmond module v.2.3, using OPLS_2005 force field55. The NMR resolved coordinates of DK17 in different membrane models were adopted for atomistic simulation studies. The initial assumption of the peptide positioning within the membrane models was obtained using the results of OPM server (Supporting Information S1)56. The model peptide with dummy atoms representing lipid moieties displays the DK17 position to be aligned in a horizontal direction, parallel to the model surface. Similar assumptions were adopted for the generation of starting structure of DK17 positioning within membrane bilayer (POPC:POPG:Cholesterol:GM1) and GM1 system. The equilibrated models of membrane bilayer and GM1 system were prepared using the similar protocol of our earlier work57. Briefly, the equilibration for the systems POPC:POPG:Cholesterol:GM1 and GM1 were performed for 10 ns prior to executing the MD simulation. It is noteworthy to mention that the exact composition of TLBE is not reported in the literature, which renders the difficulty to prepare the model membrane. Owing to such limitation, the MD simulation of DK17 in the presence of TLBE is excluded in the present study.
The model of lipid system with DK17 was further solvated using TIP3P water models at a maximum edge distance of 10Å from solute atoms. The positional equilibration to the system was performed with minimization step with convergence threshold limit of 1.0 Kcal/mol/Å. In the successive equilibration steps, the system was regulated for temperature scaling (Berendsen thermostat 10 K) with and without positional restraint to solute, each for 12 ps time scale. The system was further processed for gradual temperature increment up to 300K with Berendsen NPT ensemble with and without restraint to the heavy solute atoms, each for 24 ps time scale58. The system was also processed with PME method for the consideration of columbic influence with a threshold value of 1e−9 and cutoff value of 10Å, with pressure and temperature maintenance of 1.013 atm and 300 K respectively. M-SHAKE algorithm was used for the repositioning of hydrogen bonds using integration time step of 2fs59. Trajectory frames were collected at the interval of 10 ps for the analysis purpose.
In vivo studies of DK17 permeability in blood-brain barrier (BBB) and blood-spinal cord barrier (BSB) in mice
Wild type (WT) and mutated (mut) DK17 peptides were labeled with Alexa Fluor 680 (AF680) near-infrared fluorescent dye (ThermoFisher Scientific) following the manufacturer's protocol as described earlier60. Briefly, AF680 succinyl ester was allowed to react with the amine group of DK17 in the presence of dimethylformamide. Female SJL/J mice (5-6 week old) were obtained from Harlan (Indianapolis, IN, USA). Mice received AF680-conjugated WT and mut DK17 peptides (200 ng/mouse in a volume of 200μl) via tail-vein injection61. The mutant DK17 is only different from wild type due to its replacement of Ile6-Phe8 motif (Figure 7B) with Gly residues. After 3 hr of injection, spinal cord and various parts of brain were scanned in an Odyssey (ODY-0854; Licor) infrared scanner at two different channels, 700- and 800-nm followed by perfusion of mice with 4% paraformaldehyde. The red background signifies the signal from 800-nm filter while the signal from AF680 at the 700-nm channel was symbolized as green color. The density of the AF680 signal in various segments of CNS was quantified by Quantity One version 4.6.2 software using the volume contour tool analysis module.
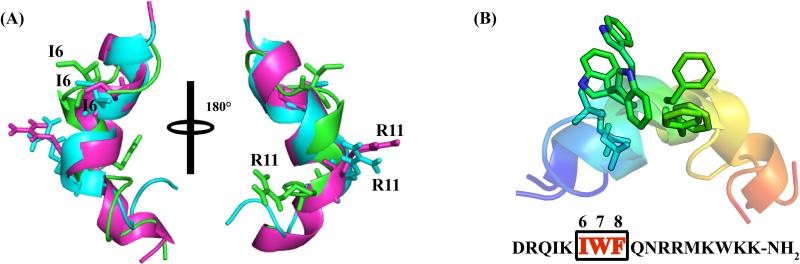
Figure 7.
(A) Overlaid structures of DK17 bound to GM1 (Purple), POPC:POPG:Cholesterol:GM1 (Cyan), and TLBE (Green) LUVs. The central helical segment (Ile6-Arg11) seems to be superimposed nicely with a conserved helical propensity. (B) The Ile6-Phe8 motif is highlighted in the overlaid structures of DK17 in different LUVs.
RESULTS & DISCUSSION
Interaction of DK17 with the model membranes
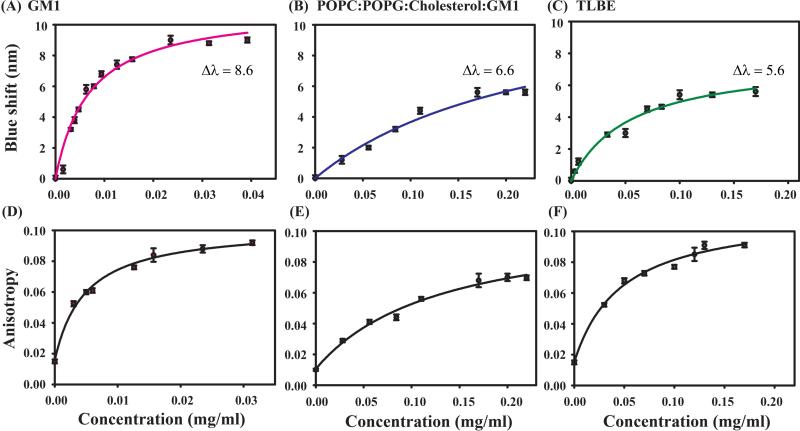
The conformational transition of the peptide and its dynamics in the solvent can readily be studied with the help of naturally abundant intrinsic Trp residues. Owing to such advantage of natural profusion in any protein, its highly sensitive fluorescence emission maxima towards the polarity of the surrounding environment makes these Trp residues more potent as a fluorophore. Interestingly, DK17 possesses two Trp residues, at 7th and 15th positions; hence, the subsequent shift in their fluorescence emission maxima towards lower wavelengths upon successive addition of model membranes helps to explore the interaction between them. The fluorescence emission maxima (λmax) of DK17 in aqueous solution was observed at around 350 nm whereas in the presence of different LUVs the λmax fell at the shorter wavelength (Figure 1, upper panel), indicating that the dynamic property of Trp is restricted, and it is buried into the hydrophobic environment of the LUVs. The peptide underwent a maximum change of around 8.6 nm in GM1 LUVs (Figure 1 A), while the other two membranes, POPC:POPG:Cholesterol:GM1 and TLBE showed the λmax of ~ 6.6 and 5.6 nm, respectively (Figure 1 B,C).
Figure 1.
Intrinsic Tryptophan fluorescence emission spectra showing binding property of DK17 peptide towards various membrane models in a buffer solution of 10 mM PO4, pH 6.0. Upper panel represents the change in blue shift in the emission spectra of DK17 peptide in presence of increasing concentrations of GM1 (A), POPC:POPG:Cholesterol:GM1 (B), and TLBE (C) LUVs (from a stock solution of 1mM), whereas the change in fluorescence anisotropy of DK17 as a function of increasing GM1 (D), POPC:POPG:Cholesterol:GM1 (E), and TLBE (F) LUVs concentrations was represented through the mounting trend in curves (lower panel).
On the other hand, as evident from Figure 1 lower panel, the change in fluorescence anisotropy values for peptide continued to increase with increasing concentration of LUVs. The initial low anisotropy values of free peptide represent its rapidly tumbling motion in a solvent. However, the trend of the upward rise of anisotropy values with successive addition of corresponding LUVs (Figure 1 D-F) depicts the restricted motional dynamics of the peptides in the vicinity of membrane models with increased rotational correction time. Overall, these findings clearly support substantial binding affinity of the peptide to various lipid membrane models.
Solvent accessibility of DK17 in the presence of membrane models
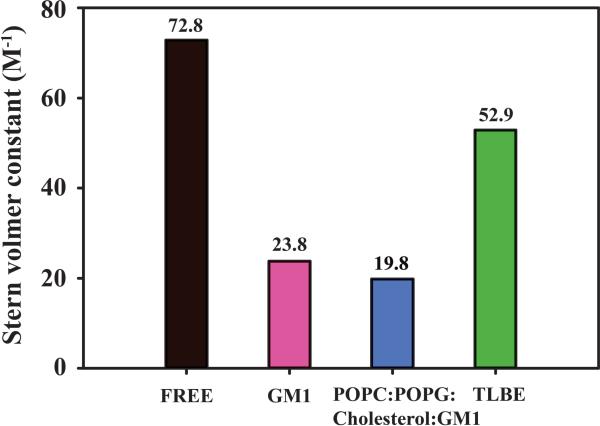
With the help of static quencher- acrylamide, the extent of solvent exposure of DK17 peptide in an aqueous solvent as well as in the presence of various LUVs were analyzed in terms of Stern–Volmer quenching constant (KSV). The higher extent of intrinsic fluorescence quenching of Trp residues of DK17 corresponded to the maximum KSV values and vice versa (Figure 2). DK17 either in free form or in aqueous solution (Black) exhibited the maximum Stern–Volmer quenching constant of around 73 M−1, depicting free accessibility of quencher towards Trp residues in solvent. In contrast, lower extent of Stern–Volmer quenching constant (KSV) values were noticed for peptide in GM1 LUVs (KSV = 23.8 M−1) and POPC:POPG:Cholesterol:GM1 LUVs (KSV = 19.8 M−1) (Figure 2), respectively. These results suggest that the Trp residues are well buried inside the corresponding LUVs, opposite effect to the orientation of Tryptophan residues in aqueous solution. According to Planque et al., Trp residues are often found at the hydrocarbon-water interface of membrane system, which in turn have the potential to dominate the hydrophobic matching effect in peptide/lipid interactions62. Thus, the orientation of Trp residue within LUVs can be assumed to display similar interaction phenomenon. Surprisingly, DK17 in TLBE LUVs (Figure 2, Green color) showed a considerable higher KSV value of around 52.9 M−1, attributing higher extent of solvent accessibility. Therefore, quenching data supports the previous fluorescence experimental results (Figure 1) that the DK17 peptide preferentially interacts either with GM1 or POPC:POPG:Cholesterol:GM1 LUVs in comparison to TLBE LUVs.
Figure 2.
Bar diagram showing Stern–Volmer constant (M−1) of the studied peptide in the presence of GM1 (Pink), POPC:POPG:Cholesterol:GM1 (Blue) and TLBE (Green) LUVs as well as in free state (Black) in 10 mM PO4, pH 6.0.
CD spectroscopic analysis for the structural transition of DK17 in the presence of model membranes
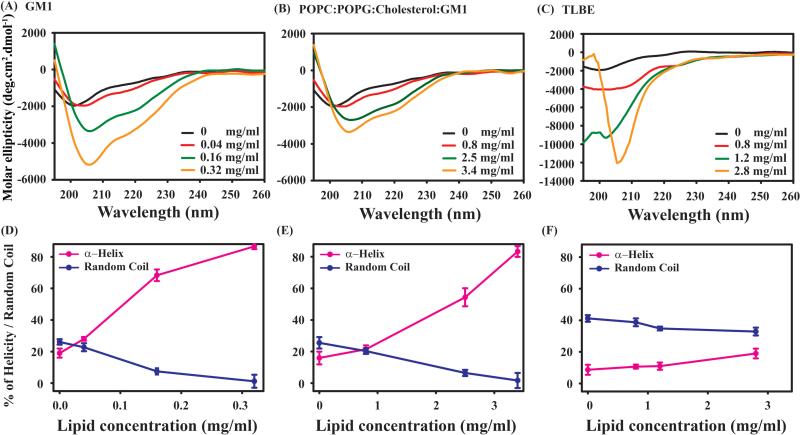
In order to probe the change in the secondary structure of DK17 upon addition of different lipid systems, circular dichroism (CD) spectroscopy was used. The far UV-spectra of free DK17 peptide exhibited strong negative maxima at around 200 nm, featuring mostly its unstructured state along with other minor sub-population conformations (Figure 3). Interestingly, when the peptide was titrated with 0.32 mg/ml GM1 LUVs, a significant change in the signature spectra of peptide majorly towards α-helix was noticed with a shift of negative maxima to ~208 nm along with another comparatively smaller negative hump at ~222 nm (Figure 3 A). The far CD studies of DK17 peptide in another LUVs composed of POPC:POPG:Cholesterol:GM1 revealed the similar helical propensity but at a concentration of 3.4 mg/ml (Figure 3 B). On the other hand, CD spectra of DK17 peptide in TLBE LUVs did not show much conformational change up to a lipid concentration of 2.8 mg/ml (Figure 3 C). Furthermore, CD deconvolution analysis through CDNN software estimated the percentage of secondary structure information of DK17 in terms of α-helix, β-sheet, turn and random coil in the absence as well as in the presence of LUVs. We have found the gradual increase of the percentage of helicity with the consecutive decrease of random coil propensity in a solvent for peptides both in the increasing concentrations of GM1 and POPC:POPG:Cholesterol:GM1 LUVs (Figure 3 D, E). However, no significant changes were observed in terms of secondary structure conformations in the presence of TLBE LUVs (Figure 3F). Overall, the CD results are in good agreement with the previous findings, confirming preferential binding and conformational transitions of DK17 to GM1 and POPC:POPG:Cholesterol:GM1 LUVs, in comparison to its almost negligible interaction with TLBE LUVs. Additionally, the CD results corroborate the fluorescence data, indicating greater binding with GM1 present. It can be hypothesized that the electrostatic interaction between the positively charged Lys and Arg residues of DK17 with the negatively charged phospholipid membranes facilitates the conformational switching of the peptide from random coil state to higher ordered helical structure.
Figure 3.
Comparative variations in secondary structure of DK17 upon titration with GM1 (A), POPC:POPG:Cholesterol:GM1 (B) and TLBE (C) LUVs by far UV-CD spectroscopy (Upper panel). The CD spectra in black color represent the free form the peptide in aqueous solution. Both peptide and LUVs were maintained in 10 mM Na2HPO4 buffer, pH 6.0. The content of secondary structure of DK17 from the CD spectra after deconvolution were plotted with increasing lipid concentrations (Lower panel).
NMR Studies of peptides in the absence and presence of model membrane
One-dimensional 1H NMR spectrum of DK17 was highly dispersed (Figure S2), which motivated us to determine the three-dimensional structure of the DK17 peptide in the absence as well as in the presence of model membrane to correlate the biological function of cell-penetrating activity. The NOESY spectrum of the free peptide in solution (Figure 4A) shows only intra-residue (CαHi/NHi) and sequential (CαHi/NHi+1) NOEs and neither any medium (i to i+2 /i+3 / i+4) nor long range (i to ≥ i+5) NOEs, specifying highly dynamicity of peptide in aqueous solution. Moreover, these results are in harmony with the previous findings of circular dichroism (CD) spectra of free peptide displaying its random coil conformation in aqueous solution (Figure 3).
Figure 4.
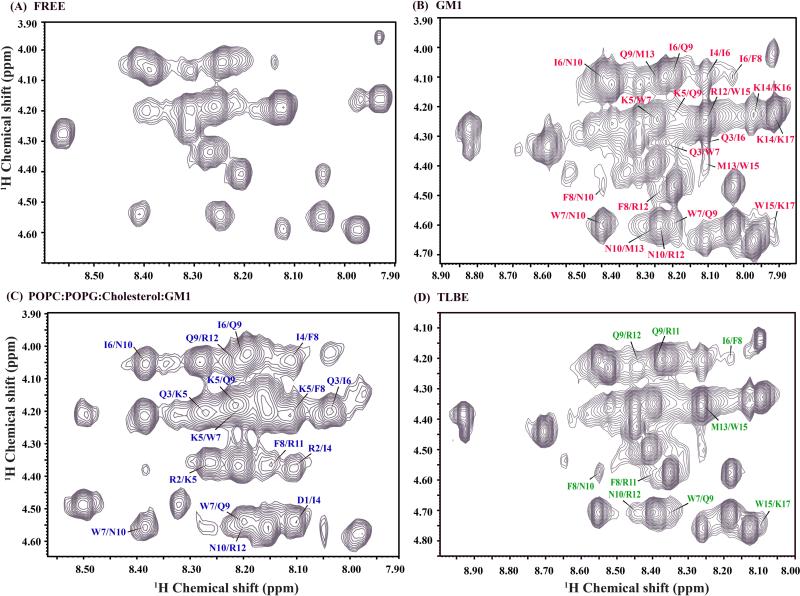
The finger print region of 2D 1H-1H NOESY spectra of DK17 (1mM) in aqueous solution (A) and in the presence of GM1 (B), POPC:POPG:Cholesterol:GM1 (C) and TLBE (D) LUVs in 10 mM Na2HPO4 buffer, pH 4.5 and at 288 K. The stock solution of all the LUVs was 1mM. For clarity, only medium range NOE cross-peaks (i to i+2/i+3/i+4) between CαH/HN resonances are shown here.
In order to understand the structural difference of DK17 in the model membrane at an atomic resolution, we performed a series of one-dimensional 1H NMR spectra of DK17 in the presence of successive addition of lipid membranes (Figure S2). Figure S2 shows the most sensitive part of the one-dimensional spectrum, the amide proton region of the peptide. Upon successive titration, we observed concentration-dependent line shape broadening of these resonances without causing any chemical shift perturbation. The line broadening effect of DK17 in the presence of LUVs clearly demonstrates that the peptide adopts similar T2 relaxation as the large LUVs. In addition, the data also confirms the fast conformational exchange between the free and bound state of the peptide in LUVs at the NMR time scale. This represents an ideal situation for determining the three-dimensional structure of DK17 in LUVs using transferred NOESY (trNOESY) method63-65.
In comparison to NOESY spectrum of DK17 in aqueous solution (Figure 4A), Figure 4 B-D shows the trNOESY spectrum of DK17 in the presence of various membrane models where a sufficient number of medium range CαH/NH (i to i+2/i+3/i+4) NOE cross-peaks along with diagnostic backbone NH/NH (i to i+2) NOEs were well characterized. Interestingly, the peptide showed the comparatively higher number of medium range CαH/NH (i to i+2/i+3/i+4) NOE cross-peaks as well as CαHi/CβHi+3 NOE cross-peaks both in GM1 and POPC:POPG:Cholesterol:GM1 LUVs relative to TLBE LUVs (Figure 5). Few conserved diagnostic backbone NH/NH (i to i+2) NOEs such as Gln3-Lys5 and Asn10-Arg12 were also noted for DK17 peptide either in GM1 or in POPC:POPG:Cholesterol:GM1 LUVs (Figure 5 A-C). These data further support the fact of LUVs-induced conformational transition of the DK17 peptide, from a random coil state to higher-ordered secondary conformations, in the context of GM1 and POPC:POPG:Cholesterol:GM1 LUVs. Surprisingly, the aromatic ring protons of Trp7, Trp15 and Phe8 showed few significant NOEs with the adjacent side chains of neighboring amino acid residues, such as Lys5, Ile6, Gln9, Lys14, implying a rigid side chain orientation of residues in the presence of LUVs. However, intermolecular trNOE peaks were not observed between LUVs and the peptide because of the substantial lower concentration of lipid membrane used in the NMR experiment.
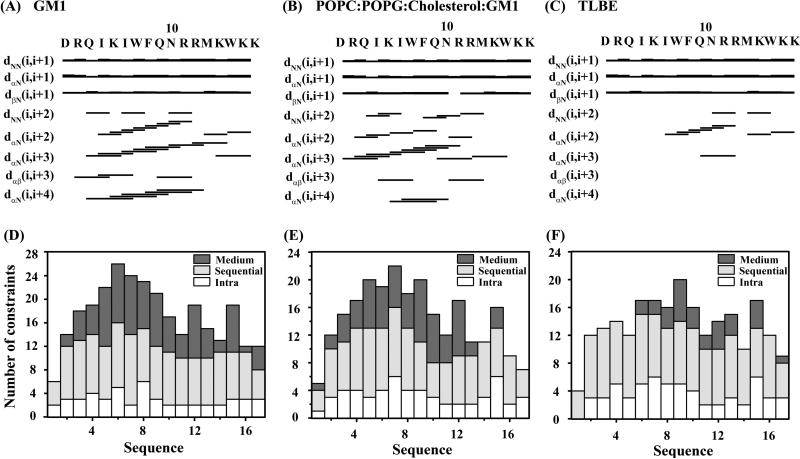
Figure 5.
(Upper panel) NMR structural parameters of DK17 in the context of different LUVs, obtained from trNOESY spectra. Bar diagram representing various NOE (sequential, medium range, and long range) contacts of DK17 in the presence of GM1 (A), POPC:POPG:Cholesterol:GM1 (B) and TLBE (C) LUVs. The thickness of the bars indicates the intensity of the NOESY peaks, which are denoted as strong, medium, and weak. (Lower panel) A histogram representing the number and variety (intra, sequential, medium) of NOEs of DK17 as a function of residue number in complex with GM1 (D), POPC:POPG:Cholesterol:GM1 (E) and TLBE (F) LUVs.
In order to reveal the secondary structural information of DK17 in model membranes, the CαH chemical shift deviation values of the following peptide from standard random coil chemical shift values were calculated (Figure S3). The up-field deviation for the CαH chemical shifts of at least four consecutive amino acid residues is indicative of α-helical structures66. The ΔCαH resonances of most of the residues of DK17 show a remarkable negative trend in GM1 LUVs, indicating a conformational transition to a well-ordered helix form. In the presence of POPC:POPG:Cholesterol:GM1 LUVs, the continuous negative ΔCαH resonances were observed throughout the peptide sequence except for Met13. In contrast, negative ΔCαH was found for the middle segment Phe8-Asn10 of DK17 in the context of TLBE LUVs.
Bound confirmation of DK17 in a model membrane
Based on distance constraints, derived from the trNOESY experiments, the 3D solution structures of DK17 bound to three different membranes were calculated. Among the three membranes studied here, the total number of NOEs including medium range NOEs was found to be maximal in GM1 LUVs (Table 1). Although the total number of NOEs for DK17 is almost similar in the mixed lipid composition (POPC:POPG:Cholesterol:GM1) and TLBE, a drastic difference was observed in the medium range NOEs. The number of medium range NOEs for DK17 in TLBE was 15 while the same peptide showed almost 34 medium range NOEs in the mixed lipid vesicle. These data reveal that the DK17 peptide exhibits higher order stability in mixed lipid vesicles compared to the TLBE LUVs. A superposition of the twenty lowest energy ensemble structures as well as the three-dimensional solution structure of DK17 in all three-model membranes is shown in Figure 6 (A-C). It is worth mentioning that the three-dimensional structure of DK17 peptide in GM1 and POPC:POPG:Cholesterol:GM1 LUVs shows distinct average backbone atoms (N, Cα and C′) of 0.39 ± 0.08 and 0.73 ± 0.30 Å, respectively with a heavy atoms root mean square deviation (RMSD) of 1.28 ± 0.11 and 1.53 ± 0.35 Å, respectively (Table 1). As expected, Table 1 shows a comparatively higher RMSD values for backbone and heavy atoms of the structure of DK17 in TLBE LUVs, around 1.93 ± 0.46 and 3.05 ± 0.54 Å, respectively (Table 1).
Table 1.
Statistical analysis of 20 lowest energy ensemble NMR structures of DK17 in various membrane models.
| Distance restrains | GM1 | POPC:POPG:cholesterol:GM1 | TLBE |
|---|---|---|---|
| Intra-residue (i–j = 0) | 65 | 73 | 76 |
| Sequential (|i–j| = 1) | 74 | 61 | 70 |
| Medium-range (2≤|i–j|≤4) | 49 | 34 | 15 |
| Long-range (|i–j| ≥5) | 0 | 0 | 0 |
| Total | 188 | 168 | 161 |
| Angular restraints | 40 | 40 | 40 |
| Φ | 20 | 20 | 20 |
| Ψ | 20 | 20 | 20 |
| Distance restraints from violation (≥ 0.3Å) | 0 | 0 | 0 |
| Deviation from mean structure (Å) | |||
| Average back bone to mean structure | 0.39±0.08 | 0.73±0.30 | 1.93±0.46 |
| Average heavy atom to mean structure | 1.28± 0.11 | 1.53± 0.35 | 3.05± 0.54 |
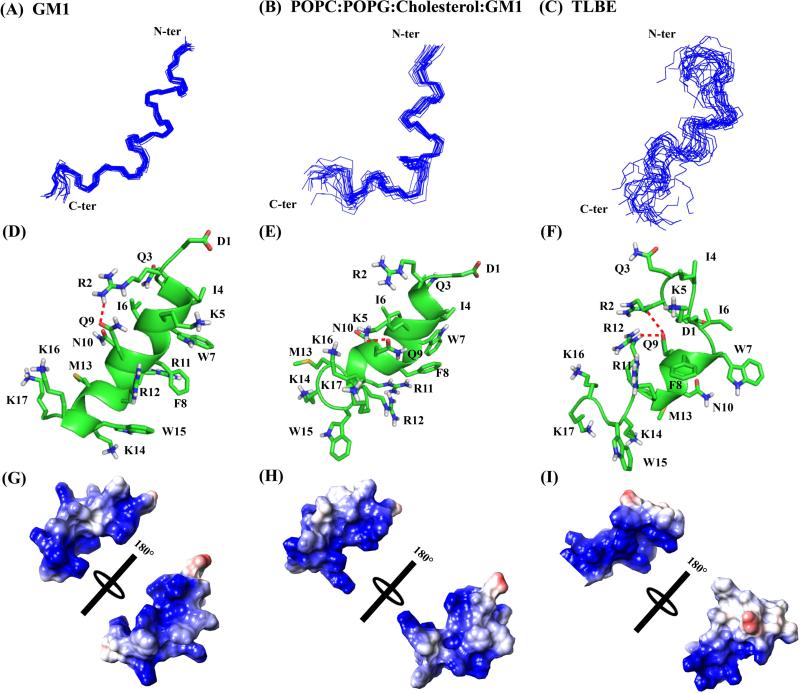
Figure 6.
Three-dimensional solution NMR structures of DK17 peptide in various membrane models in 10 mM Na2HPO4 buffer, pH 4.5. Superposition of backbone atoms (N, Cα, C’) of the 20 lowest energy structures of DK17 bound to GM1 (A), POPC:POPG:Cholesterol:GM1 (B), and TLBE (C) LUVs. Cartoon representations of side chain orientation of 2D-NMR derived structure of DK17 in GM1 (D), POPC:POPG:Cholesterol:GM1 (E) and TLBE (F) LUVs showing different residues. The red dotted lines indicate either electrostatic interaction or hydrogen bonding between the side chain residues. Electrostatic surface potential map for DK17 in GM1 (D), POPC:POPG:Cholesterol:GM1 (E) and TLBE (F) LUVs showing charged residues.
As expected, DK17 mainly adopts a well-defined alpha-helical conformation in GM1 LUVs (Figure 6), whereas in the LUVs composed of POPC:POPG:Cholesterol:GM1, the α-helix spans the residues between Arg2-Arg12. The N-terminal of DK17 structure in GM1 is stabilized by hydrophobic interactions between the side chains of Ile4 and aromatic ring protons of Trp7 as well as Phe8. Interestingly, Arg11 is sandwiched between Phe8 and Trp15 to form cation-pi interaction in the central part of the helix, maintaining the three-dimensional architecture. The aliphatic side chain of Arg12 is strongly up-field shifted due to its close proximity to the aromatic ring protons of Phe8 and Trp15 (Figure S4). An additional side chain/side chain electrostatic interaction between Arg2 and Gln9 reinforces the structural stability (Figure 6D). Close inspection of the three-dimensional structure of DK17 in GM1 LUVs clearly indicates that the positively charged residues are highly dispersed throughout the sequence (Figure 6G), and thus could be responsible for the initial electrostatic interaction with the hydroxyl groups of carbohydrate moieties of the GM1 LUVs, followed by folding due to presence of acyl chains of GM1.
The structural pattern of DK17 in a mixed vesicle, consisting of POPC:POPG:Cholesterol:GM1, is less straightforward. The helical region Arg2-Arg12 of DK17 is stabilized by a strong hydrophobic interaction between Ile4-Trp7-Phe8 (Figure 6E). The structure is further stabilized by cation-pi interaction between Phe8 and Arg12; the side chain/side chain electrostatic interaction or hydrogen bonding between Lys5 and Gln9 also plays a crucial role in structural stabilization (Figure 6E). As mentioned earlier, the interesting sequence composition of DK17 helps to create an electrostatic interaction (Figure 6H) with the phosphate groups of the mixed lipid vesicle. It is worth mentioning here that the POPC is zwitterionic, whereas the POPG and GM1 membranes are negatively charged. Additionally, cholesterol stabilizes the lipid membrane by keeping itself in close proximity to the head groups of the phospholipid bilayer and orienting itself parallel to the lipid acyl chains. Due to the stable complex structure of mixed LUVs, DK17 may not adopt straightforward helical conformation; the highly disordered C-terminal positively charged Lys/Arg residues are in close proximity to the phosphate head groups of the lipid while Trp15 is in the interface of the LUVs.
The three-dimensional structure of DK17 in TLBE adopts alpha-helical conformation only at the central Trp7-Arg11 region while both terminal residues Asp1-Ile6 and Arg12-Lys17 are highly dynamic in solution. The architecture of DK17 is tuned in such a way that all positively charged residues Arg2, Gln3, Lys5, Arg11, Arg12, Lys14, Lys16, and Lys17 face one side of the structure (Figures 6F and 6I) and possibly interact with the negatively charged head groups of the LUVs. It was interesting to observe that the Gln9 is juxtaposed between Arg2 and Arg12, governing the side chain/side chain electrostatic interactions and hydrogen bonding for structural stabilization. Taken together, the central segment Ile6-Arg11 of DK17 adopts a conserved alpha-helical conformation (Figure 7), irrespective of the membrane composition, indicating that these hydrophobic or aliphatic residues are chiefly responsible for structural stabilization and could be significant for cell penetrating activity as well.
Dynamics of the peptide in lipid model systems
Molecular dynamics simulations make it possible to visualize residue specific contributions for interaction with membrane systems. The structural stability of the peptide in the model membranes was first investigated using the DSSP module, at various times in the MD simulations67. The central region of DK17, spanning the residues Ile6 - Arg11, was found to form α-helix in the presence of GM1 (Figure 8). In the case of the POPC:POPG:Cholesterol:GM1 system, DK17 was found to retain the α-helical secondary structure in the Phe8 - Arg12 region. The secondary structure plot also indicates the presence of 310-helix during the simulation, as shown in Figure 8. Turn and coil-like structures of DK17 in the GM1 and POPC:POPG:Cholesterol:GM1 systems are also found in the simulation.
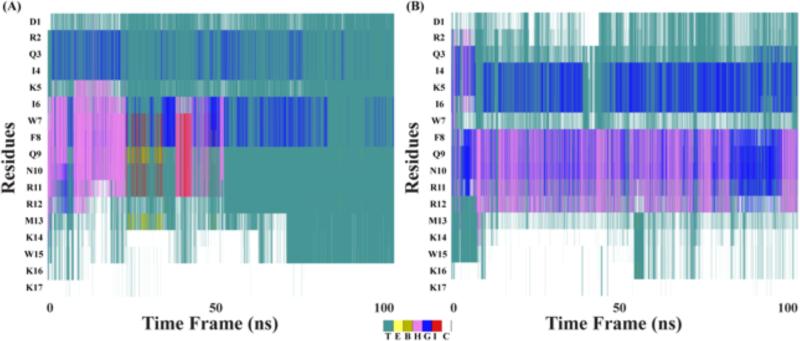
Figure 8.
Account of secondary structure calculation for DK17 in presence of (A) GM1 and (B) POPC:POPG:Cholesterol:GM1.
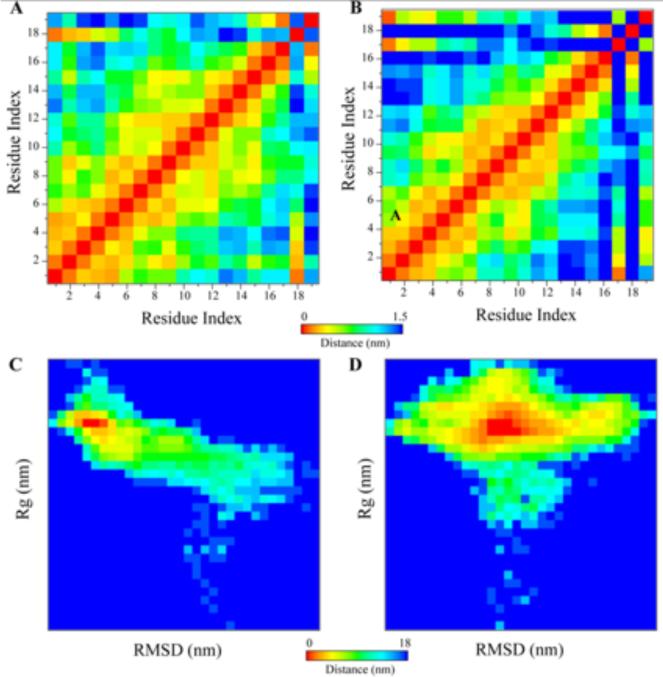
Further evidence of the structural integrity and stability of DK17 in different membrane models was obtained using residue-wise consensus contact maps and analysis of energy basins using RMSD and radius of gyration (Rg) plots (Figure 9). The lower distance region in the 2D plot corresponds to the conservation rate between the residue pairs; while the higher distance region in the 2D plot is indicative of hydrophobic interaction at the inter-residue level of DK17. Importantly, the N-ter and C-ter residues of DK17 are found to be in close proximity to either the GM1 or POPC:POPG:Cholesterol:GM1 LUV's. The C-ter region of DK17 is found to be less dynamic in the presence of GM1 and more dynamic in the POPC:POPG:Cholesterol:GM1 system. This observation is shown by the darker (blue) regions in the 2D contact map for POPC:POPG:Cholesterol:GM1 system compared to GM1 system (Figure 9A and B). Further conformational attributes of DK17 in lipid system were analyzed using energy basin plots correlated with RMSD and Rg plot (Figure 9C and D). The structural rigidity of DK17 in the presence of GM1 is reflective by relatively lesser areas of the scatter plot, compared to POPC:POPG:Cholesterol:GM1 system. Such dynamic flexibility in the later membrane model is attributed to C-ter freedom, as discussed in the above section. The Rg value in the plot is indicative of a constant value throughout the simulation time course in the presence of GM1, whereas an extended value of Rg is reflected in the presence of POPC:POPG:Cholesterol:GM1. Importantly, the free-energy state of DK17 in the presence of GM1 is consistent with a stable conformational ensemble that ranges up to a value of −14.7 Kcal/mol, whereas the same value for POPC:POPG:Cholesterol:GM1 system appears to be −18 Kcal/mol.
Figure 9.
Visualization of structural integrity and stability in presence of membrane system. 2D contact map of DK17 in presence of (A) GM1 and (B) POPC:POPG:Cholesterol:GM1 system. Energy basin corresponding to bi-plot of RMSD and Rg in presence of (C) GM1 and (D) POPC:POPG:Cholesterol:GM1 system.
What is the driving energy in the interaction of DK17 with membrane models?
Calculation of the energies in reference to solute (TIP3P water models) and the membrane structures can reveal the forces responsible for the interactions. Electrostatic and hydrophobic forces dominate the interaction between DK17 and water or membranes.
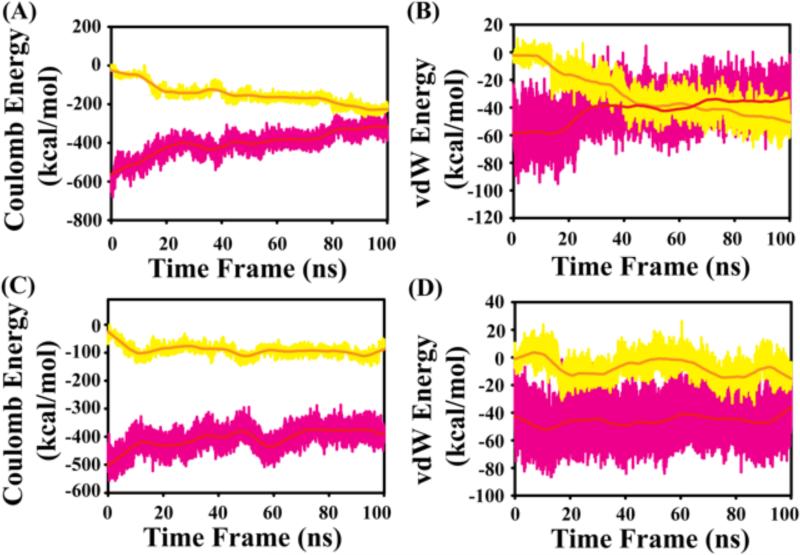
Based on energy contribution plots, the coulombic energy contribution appears to exceed that of the van der Waal energy. The contribution of solvent is found to be greater than that of the polar head groups of lipid molecules in both systems. In the presence of GM1, the electrostatic contribution by both water molecules and polar groups in the head region of GM1 are comparatively similar in the course of simulation time scale. This is reflected in the convergence of the values after 100 ns (Figure 10 A). In contrast, for the POPC:POPG:Cholesterol:GM1 system, the electrostatic interaction between polar solvent is found to contribute more to stability than the polar moieties of GM1, POPG and POPC molecules (Figure 10 C). The average contribution of coulombic energy by solute is −149.3 ± 54.2 Kcal/mol and −91.4 ± 19.5 Kcal/mol in the presence of GM1 and POPC:POPG:Cholesterol:GM1 system, respectively. Likewise, the coulombic energy contribution by solvent is −402.7 ± 65.9 Kcal/mol and −406.1 ± 38.0 Kcal/mol in the presence of GM1 and POPC:POPG:Cholesterol:GM1, respectively, indicating that the electrostatic contribution by the solvent are comparable in both systems. The van der Waals contribution to GM1-peptide interaction is lower at the initial stages of simulation, but dominates after 50 ns (Figure 10 B). Interestingly, the hydrophobic contribution in the POPC:POPG:Cholesterol:GM1 is comparable to that from solvent molecules (Figure 10 D): the contribution by solute is −30.8 ± 16.1 Kcal/mol and −7.0 ± 8.0 Kcal/mol in GM1 and POPC:POPG:Cholesterol:GM1 system, respectively. The van der Waals energy contribution by the solvent molecules for DK17 is −42.3 ± 13.7 Kcal/mol and −45.8 ± 11.5 Kcal/mol, respectively in the presence of GM1 and POPC:POPG:Cholesterol:GM1 system. Overall, structural stability of DK17 is higher in the presence of GM1 compared to POPC:POPG:Cholesterol:GM1.
Figure 10.
Energy contribution from solute (yellow) and solvent (pink) that governs the DK17 interaction with micelle models. Energy contribution corresponding to Coulomb energy in (A) GM1 and (B) POPC:POPG:Cholesterol:GM1 system in Kcal/mol. Energy contribution corresponding to van der Waals energy in (C) GM1 and (D) POPC:POPG:Cholesterol:GM1 system in Kcal/mol.
Monitoring blood-brain barrier (BBB) and blood-spinal cord barrier (BSB) permeability of DK17 in mice
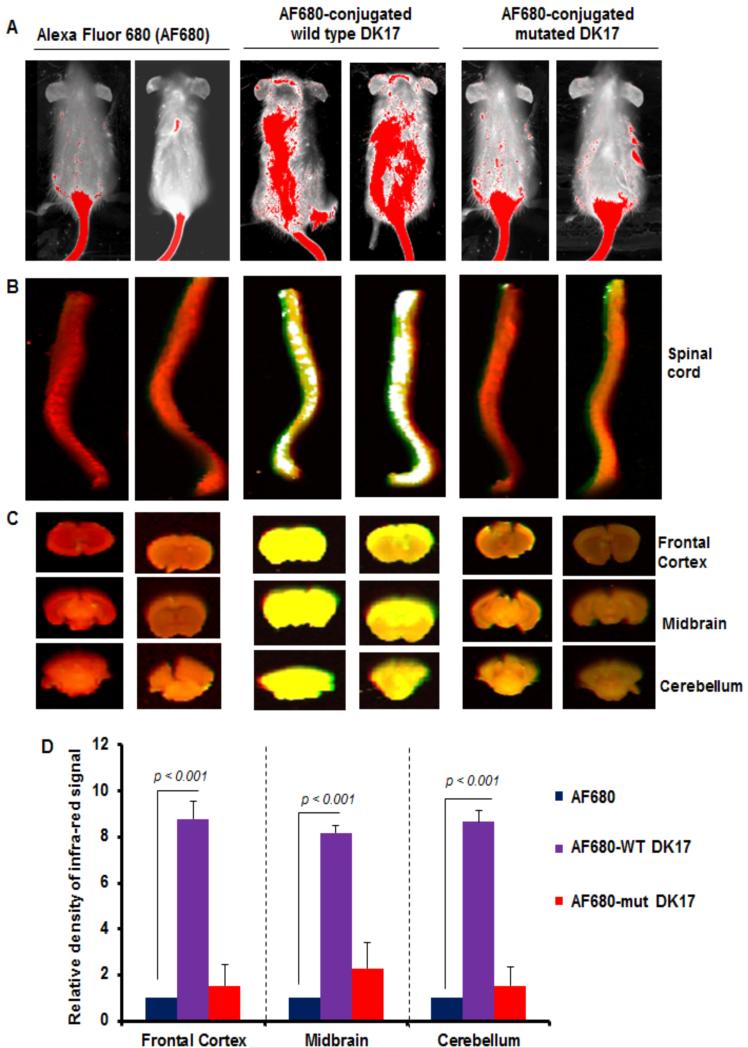
The two membrane structures, blood-brain barrier (BBB) and blood-spinal cord barrier (BSB), are known to be the most compact barrier in the human body that prohibit the passage of most of the chemicals and peptides into the brain and the spinal cord, respectively. So far, it has been significant from the NMR studies that the central segment Ile6-Arg11 of DK17 surprisingly preserves its alpha-helical conformation in a variety of model membranes irrespective of the membrane compositions, and hence provides the signal regardless of its contribution in membrane permeability. Therefore, in order to evaluate the prominence of those residues in cell membrane permeabilization into the brain and spinal cord, we injected AF 680 (an infrared dye)-conjugated DK17 peptide in live mice via tail vein and after 3 h of incubation, the signals were scanned with an Odyssey infrared scanner. As evident from Figure 11A (first two lanes), no such significant infrared signals were visible on areas of the brain and the spinal cord in Alexa Fluor 680-injected mice. However, some infrared signals were detected on areas of the brain and the spinal cord (Figure 11B, 3rd, and 4th lanes), in mice receiving WT DK17-conjugated Alexa Fluor 680, thus signifying the cell membrane permeability of the peptide through BBB and BSB. In contrast, these infrared signals were not found in mice receiving mutated DK17-conjugated Alexa Fluor 680 (Figure 11B, 5th, and 6th lanes) where the hydrophobic residues Ile6-Phe8 were substituted with Gly residues. The density of the AF680 signal was further quantified in different parts of the brain where it seems like the relative density of AF680 infra-red signal is comparatively much higher in all sections of the brain than that of mut-DK17 (Figure 11D). CD experiments confirm that mut-DK17 in aqueous solution as well as in lipid environment remains as random coil conformation (Figure S5). Moreover, the mut-DK17 appears to be trailing its translocation mechanism to spinal cord and brain region due to the absence of hydrophobic contacts, associated with the side chains of Ile6-Phe8 with lipid acyl groups. Overall, these results are able to set resilient implication to the importance of the hydrophobic interaction facilitated by the Ile6-Phe8 motif of the DK17 peptide to lipid membrane which chiefly governs its translocation ability to CNS.
Figure 11.
In-vivo study of DK17 permeability to blood-brain barrier (BBB) and blood-spinal cord barrier (BSB) in mice models. Wild type (WT) and mutated DK17 peptides were conjugated with Alexa Fluor 680 (AF680) infrared dye followed by treatment of control female SJL/J mice (5-6 week old) with AF680-conjugated WT and mutated DK17 peptides (200 ng/mouse) via tail-vein injection. After 3 h, mice were scanned in an Odyssey (ODY-0854; Licor) infrared scanner at the 700- and 800-nm channels (A). Mice were perfused with 4% paraformaldehyde. Spinal cord (B) and different parts of the brain (C) were scanned in an Odyssey infrared scanner. The red background comes from 800-nm filter, whereas the green signal is from AF680 at the 700-nm channel. Wild type DK17 is able to translocate through BSB and BBB, therefore it displays signal both in different sections of brain and spinal cord. However, the translocation of mut-DK17 seems to be restricted in the tail region only indicating its impermeability to CNS. The density of the AF680 signal in different parts of the brain (D) was quantified. Data (relative to AF680 control) are expressed as the mean ± SEM of five different mice (n=5) per group. ap<0.001 vs AF680.
CONCLUSIONS
In summary, the present report focuses on high-resolution structural analysis of the penetratin peptide DK17 in model membranes, providing new details relevant to the mode of action of CPP's. By investigating the binding of DK17 to membrane systems, we have identified the significance of lipid bilayer composition, in the early stages of peptide-lipid interaction. Fluorescence and CD analysis provide conformational and dynamic information on differential lipid properties. High-resolution NMR studies have defined the role of the central segment Ile6-Arg11 of DK17, which adopts α-helical conformation irrespective of the (brain) model membrane compositions. Moreover, this central segment appears to stabilize the structure of the bound peptide through a hydrophobic interaction between the side chains of Ile6-Arg11 and acyl chains of lipid moieties. Dynamic behavior and secondary structure of DK17 in the presence of model membranes correlated with results from MD simulation studies. Interestingly, the atomic level information (secondary structure and interaction energy), elucidated by the computational techniques, is consistent the spectroscopic studies. Mutational change in the central region of DK17 efficiently loses its membrane permeability activity to BBB and BSB in mice models, while in contrary the wild type DK17 is capable of retaining its intrinsic membrane permeabilization ability. Our biophysical approach revealing atomic details of DK17 interaction in different lipid enviroments may assist rational designing of robust medicinal therapeutics. Input from such enhanced therapeutic designs might offer novel treatments for several neurodegenerative diseases including Alzheimer, Parkinson, Zika virus where effective treatment is still lacking.
Supplementary Material
Acknowledgments
Funding
This study was supported by Bose Institute (Plan Project-II (to AB)), India, and grants (AG050431 and NS83054) from National Institutes of Health and merit awards (I01BX002174 and 1I01BX003033) from Veterans Affairs to KP. AB would like to acknowledge DBT, Government of India, for infrastructure development fund (BT/PR3106/INF/22/138/2011) to Bose Institute for purchasing 700 MHz NMR spectrometer with cryoprobe. SB and RKK are grateful to UGC and CSIR, Government of India, for their junior and senior research fellowships, respectively. Central Instrument Facility (CIF) of Bose Institute is greatly acknowledged. The structures of DK17 bound to GM1, POPC:POPG:Cholesterol:GM1 and TLBE have been deposited to protein data bank (PDB) with accession numbers 2ND6, 2ND7 and 2ND8, respectively.
ABBREVIATIONS
- CPPs
Cell Penetrating Peptides
- BBB
Blood Brain Barrier
- CD
Circular Dichroism
- NMR
Nuclear Magnetic Resonance
- MD
Molecular Dynamics Simulation
- TOCSY
Total Correlation Spectroscopy
- NOESY
Nuclear Overhauser Effect Spectroscopy
Footnotes
AUTHOR CONTRIBUTION
AB and KP designed, analyzed as well as funded the research work for biophysical part and in vivo studies, respectively; SB conducted biophysical experiments, analyzed the results, calculated the three-dimensional structure using NMR spectroscopy, and wrote the manuscript; RKK performed molecular dynamics simulation and wrote the manuscript; SM conducted and analyzed in vivo experiments; KP and AB analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
SUPPLEMENTARY INFORMATION
Supporting Information
This information is available free of charge via the Internet at http://pubs.acs.org.
Prediction of DK17 peptide position in the membrane system using OPM server (Figure S1); 1H NMR spectra of DK17 in the presence of various LUVs (Figure S2); Bar diagram of CαH chemical shift deviation of DK17 in different LUVs (Figure S3); The trNOESY spectra of the aliphatic side chains of DK17 in presence of GM1 LUVs (Figure S4); Structural changes of mut-DK17 in aqueous solution as well as in lipid environment (Figure S5).
REFERENCES
- 1.Lindgren M, Hällbrink M, Prochiantz A, Langel U. Cell-penetrating peptides. Trends Pharmacol Sci. 2000;21(3):99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 2.Lindgren M, Langel U. Classes and prediction of cell-penetrating peptides. Methods Mol Biol. 2011;683:3–19. doi: 10.1007/978-1-60761-919-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Kauffman WB, Fuselier T, He J, Wimley WC. Mechanism Matters: A Taxonomy of Cell Penetrating Peptides. Trends Biochem Sci. 2015;40(12):749–64. doi: 10.1016/j.tibs.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55(6):1189–93. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 5.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55(6):1179–88. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 6.Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc Natl Acad Sci U S A. 1991;88(5):1864–8. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prochiantz A. Homeodomain-derived peptides. In and out of the cells. Ann N Y Acad Sci. 1999;886:172–9. doi: 10.1111/j.1749-6632.1999.tb09410.x. [DOI] [PubMed] [Google Scholar]
- 8.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88(2):223–33. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 9.Chaloin L, Vidal P, Lory P, Méry J, Lautredou N, Divita G, Heitz F. Design of carrier peptide-oligonucleotide conjugates with rapid membrane translocation and nuclear localization properties. Biochem Biophys Res Commun. 1998;243(2):601–8. doi: 10.1006/bbrc.1997.8050. [DOI] [PubMed] [Google Scholar]
- 10.Oehlke J, Scheller A, Wiesner B, Krause E, Beyermann M, Klauschenz E, Melzig M, Bienert M. Cellular uptake of an alpha-helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior non-endocytically. Biochim Biophys Acta. 1998;1414(1-2):127–39. doi: 10.1016/s0005-2736(98)00161-8. [DOI] [PubMed] [Google Scholar]
- 11.Hällbrink M, Florén A, Elmquist A, Pooga M, Bartfai T, Langel U. Cargo delivery kinetics of cell-penetrating peptides. Biochim Biophys Acta. 2001;1515(2):101–9. doi: 10.1016/s0005-2736(01)00398-4. [DOI] [PubMed] [Google Scholar]
- 12.Zorko M, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57(4):529–45. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. 2015;7(1):a020412. doi: 10.1101/cshperspect.a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders NR, Habgood MD, Møllgård K, Dziegielewska KM. The biological significance of brain barrier mechanisms: help or hindrance in drug delivery to the central nervous system? F1000Res. 2016;5:313. doi: 10.12688/f1000research.7378.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajra A, Bandyopadhyay D, Hajra SK. Zika Virus: A Global Threat to Humanity: A Comprehensive Review and Current Developments. N Am J Med Sci. 2016;8(3):123–8. doi: 10.4103/1947-2714.179112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saiz JC, Vázquez-Calvo Á, Blázquez AB, Merino-Ramos T, Escribano-Romero E, Martín-Acebes MA. Zika Virus: the Latest Newcomer. Front Microbiol. 2016;7:496. doi: 10.3389/fmicb.2016.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshayes S, Morris MC, Divita G, Heitz F. Cell-penetrating peptides: tools for intracellular delivery of therapeutics. Cell Mol Life Sci. 2005;62(16):1839–49. doi: 10.1007/s00018-005-5109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157(2):195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramsey JD, Flynn NH. Cell-penetrating peptides transport therapeutics into cells. Pharmacol Ther. 2015;154:78–86. doi: 10.1016/j.pharmthera.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Snyder EL, Dowdy SF. Cell penetrating peptides in drug delivery. Pharm Res. 2004;21(3):389–93. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- 21.Terrone D, Sang SL, Roudaia L, Silvius JR. Penetratin and related cell-penetrating cationic peptides can translocate across lipid bilayers in the presence of a transbilayer potential. Biochemistry. 2003;42(47):13787–99. doi: 10.1021/bi035293y. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Gehring W. Cellular uptake of the Antennapedia homeodomain polypeptide by macropinocytosis. Biochem Biophys Res Commun. 2014;443(4):1136–40. doi: 10.1016/j.bbrc.2013.12.062. [DOI] [PubMed] [Google Scholar]
- 23.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269(14):10444–50. [PubMed] [Google Scholar]
- 24.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J Biol Chem. 1996;271(30):18188–93. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 25.Dinca A, Chien W-M, Chin MT. Intracellular Delivery of Proteins with Cell-Penetrating Peptides for Therapeutic Uses in Human Disease. Int J Mol Sci. 2016;17(2):263. doi: 10.3390/ijms17020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kristensen M, Birch D, Mørck Nielsen H. Applications and Challenges for Use of Cell-Penetrating Peptides as Delivery Vectors for Peptide and Protein Cargos. Int J Mol Sci. 2016;17(2):185. doi: 10.3390/ijms17020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGowan JW, Bidwell GL, Vig PJ. Challenges and new strategies for therapeutic peptide delivery to the CNS. Ther Deliv. 2015;6(7):841–53. doi: 10.4155/tde.15.30. [DOI] [PubMed] [Google Scholar]
- 28.Zhang D, Wang J, Xu D. Cell-penetrating peptides as noninvasive transmembrane vectors for the development of novel multifunctional drug-delivery systems. J Control Release. 2016;229:130–9. doi: 10.1016/j.jconrel.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Alves ID, Correia I, Jiao CY, Sachon E, Sagan S, Lavielle S, Tollin G, Chassaing G. The interaction of cell-penetrating peptides with lipid model systems and subsequent lipid reorganization: thermodynamic and structural characterization. J Pept Sci. 2009;15(3):200–9. doi: 10.1002/psc.1070. [DOI] [PubMed] [Google Scholar]
- 30.Alves ID, Bechara C, Walrant A, Zaltsman Y, Jiao CY, Sagan S. Relationships between membrane binding, affinity and cell internalization efficacy of a cell-penetrating peptide: penetratin as a case study. PLoS One. 2011;6(9):e24096. doi: 10.1371/journal.pone.0024096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jobin ML, Alves ID. On the importance of electrostatic interactions between cell penetrating peptides and membranes: a pathway toward tumor cell selectivity? Biochimie. 2014;107(Pt A):154–9. doi: 10.1016/j.biochi.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 32.Bahnsen JS, Franzyk H, Sandberg-Schaal A, Nielsen HM. Antimicrobial and cell-penetrating properties of penetratin analogs: effect of sequence and secondary structure. Biochim Biophys Acta. 2013;1828(2):223–32. doi: 10.1016/j.bbamem.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Caesar CE, Esbjörner EK, Lincoln P, Nordén B. Membrane interactions of cell-penetrating peptides probed by tryptophan fluorescence and dichroism techniques: correlations of structure to cellular uptake. Biochemistry. 2006;45(24):7682–92. doi: 10.1021/bi052095t. [DOI] [PubMed] [Google Scholar]
- 34.Bahnsen JS, Franzyk H, Sayers EJ, Jones AT, Nielsen HM. Cell-penetrating antimicrobial peptides - prospectives for targeting intracellular infections. Pharm Res. 2015;32(5):1546–56. doi: 10.1007/s11095-014-1550-9. [DOI] [PubMed] [Google Scholar]
- 35.Maniti O, Alves I, Trugnan G, Ayala-Sanmartin J. Distinct behaviour of the homeodomain derived cell penetrating peptide penetratin in interaction with different phospholipids. PLoS One. 2010;5(12):e15819. doi: 10.1371/journal.pone.0015819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pourmousa M, Karttunen M. Early stages of interactions of cell-penetrating peptide penetratin with a DPPC bilayer. Chem Phys Lipids. 2013;169:85–94. doi: 10.1016/j.chemphyslip.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Mäler L, Gräslund A. NMR studies of three-dimensional structure and positioning of CPPs in membrane model systems. Methods Mol Biol. 2011;683:57–67. doi: 10.1007/978-1-60761-919-2_5. [DOI] [PubMed] [Google Scholar]
- 38.Mäler L. Solution NMR studies of cell-penetrating peptides in model membrane systems. Adv Drug Deliv Rev. 2013;65(8):1002–11. doi: 10.1016/j.addr.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Nicastro MC, Spigolon D, Librizzi F, Moran O, Ortore MG, Bulone D, Biagio PL, Carrotta R. Amyloid β-peptide insertion in liposomes containing GM1-cholesterol domains. Biophys Chem. 2016;208:9–16. doi: 10.1016/j.bpc.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Kakio A, Nishimoto SI, Yanagisawa K, Kozutsumi Y, Matsuzaki K. Cholesterol-dependent formation of GM1 ganglioside-bound amyloid beta-protein, an endogenous seed for Alzheimer amyloid. J Biol Chem. 2001;276(27):24985–90. doi: 10.1074/jbc.M100252200. [DOI] [PubMed] [Google Scholar]
- 41.Domadia PN, Bhunia A, Ramamoorthy A, Bhattacharjya S. Structure, interactions, and antibacterial activities of MSI-594 derived mutant peptide MSI-594F5A in lipopolysaccharide micelles: role of the helical hairpin conformation in outer-membrane permeabilization. J Am Chem Soc. 2010;132(51):18417–28. doi: 10.1021/ja1083255. [DOI] [PubMed] [Google Scholar]
- 42.Bera S, Ghosh A, Sharma S, Debnath T, Giri B, Bhunia A. Probing the role of Proline in the antimicrobial activity and lipopolysaccharide binding of indolicidin. J Colloid Interface Sci. 2015;452:148–59. doi: 10.1016/j.jcis.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Bhunia A, Saravanan R, Mohanram H, Mangoni ML, Bhattacharjya S. NMR structures and interactions of temporin-1Tl and temporin-1Tb with lipopolysaccharide micelles: mechanistic insights into outer membrane permeabilization and synergistic activity. J Biol Chem. 2011;286(27):24394–406. doi: 10.1074/jbc.M110.189662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakowicz JR, Joseph R. Principles of Fluorescence Spectroscopy. 3 ed. Springer; US, USA: 2006. [Google Scholar]
- 45.Ghosh S, Pradhan SK, Kar A, Chowdhury S, Dasgupta D. Molecular basis of recognition of quadruplexes human telomere and c-myc promoter by the putative anticancer agent sanguinarine. Biochim Biophys Acta. 2013;1830(8):4189–201. doi: 10.1016/j.bbagen.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Böhm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5(3):191–5. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 47.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J Magn Reson. 1993;102:241–245. [Google Scholar]
- 48.Ikura M, Tschudin R, Bax A. Rapid Recording of 2D NMR Spectra Without Phase Cycling. Application To The Study of Hydrogen Exchange in Proteins. J Magn Reson. 1989;85:393–399. [Google Scholar]
- 49.Kneller DGK, Irwin D. UCSF Sparky An NMR display, annotation and assignment tool. Journal Of Cellular Biochemistry. 1993 [Google Scholar]
- 50.Wüthrich K. NMR of Proteins and Nucleic Acids. 1986:320. [Google Scholar]
- 51.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–98. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 52.Schibli DJ, Hwang PM, Vogel HJ. Structure of the antimicrobial peptide tritrpticin bound to micelles: a distinct membrane-bound peptide fold. Biochemistry. 1999;38(51):16749–55. doi: 10.1021/bi990701c. [DOI] [PubMed] [Google Scholar]
- 53.Eisenmesser EZ, Post CB. Insights into tyrosine phosphorylation control of protein-protein association from the NMR structure of a band 3 peptide inhibitor bound to glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1998;37(3):867–77. doi: 10.1021/bi971445b. [DOI] [PubMed] [Google Scholar]
- 54.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 55.Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J Chem Theory Comput. 2010;6(5):1509–19. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 56.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40(Database issue):D370–6. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bera S, Korshavn KJ, Kar RK, Lim MH, Ramamoorthy A, Bhunia A. Biophysical insights into the membrane interaction of the core amyloid-forming Aβ40 fragment K16-K28 and its role in the pathogenesis of Alzheimer's disease. Phys Chem Chem Phys. 2016 doi: 10.1039/c6cp02023b. [DOI] [PubMed] [Google Scholar]
- 58.Todorova N, Makarucha AJ, Hine ND, Mostofi AA, Yarovsky I. Dimensionality of carbon nanomaterials determines the binding and dynamics of amyloidogenic peptides: multiscale theoretical simulations. PLoS Comput Biol. 2013;9(12):e1003360. doi: 10.1371/journal.pcbi.1003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kräutler V, van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 60.Ghosh A, Roy A, Liu X, Kordower JH, Mufson EJ, Hartley DM, Ghosh S, Mosley RL, Gendelman HE, Pahan K. Selective inhibition of NF-kappaB activation prevents dopaminergic neuronal loss in a mouse model of Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104(47):18754–9. doi: 10.1073/pnas.0704908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma L, Yu P, Veerendra B, Rold TL, Retzloff L, Prasanphanich A, Sieckman G, Hoffman TJ, Volkert WA, Smith CJ. In vitro and in vivo evaluation of Alexa Fluor 680-bombesin[7-14]NH2 peptide conjugate, a high-affinity fluorescent probe with high selectivity for the gastrin-releasing peptide receptor. Mol Imaging. 2007;6(3):171–80. [PubMed] [Google Scholar]
- 62.de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, Separovic F, Watts A, Killian JA. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42(18):5341–8. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 63.Clore GM, Gronenborn AM. Theory and applications of the transferred nuclear overhauser effect to the study of the conformations of small ligands bound to proteins. Journal of Magnetic Resonance. 1969;48(3):402–417. [Google Scholar]
- 64.Wang Z, Jones JD, Rizo J, Gierasch LM. Membrane-bound conformation of a signal peptide: a transferred nuclear Overhauser effect analysis. Biochemistry. 1993;32(50):13991–9. doi: 10.1021/bi00213a032. [DOI] [PubMed] [Google Scholar]
- 65.Bhunia A, Domadia PN, Mohanram H, Bhattacharjya S. NMR structural studies of the Ste11 SAM domain in the dodecyl phosphocholine micelle. Proteins. 2009;74(2):328–43. doi: 10.1002/prot.22166. [DOI] [PubMed] [Google Scholar]
- 66.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5(1):67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 67.Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32(Web Server issue):W500–2. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.