Abstract
Although antiretroviral therapy (ART) suppresses viral replication to clinically undetectable levels, HIV-1 persists in CD4+ T cells in a latent form not targeted by the immune system or ART1–5. This latent reservoir is a major barrier to cure. Many individuals initiate ART during chronic infection, and in this setting, most proviruses are defective6. However, the dynamics of the accumulation and persistence of defective proviruses during acute HIV-1 infection are largely unknown. Here we show that defective proviruses accumulate rapidly within the first few weeks of infection to make up over 93% of all proviruses, regardless of how early ART is initiated. Using an unbiased method to amplify near full-length proviral genomes from HIV-1 infected adults treated at different stages of infection, we demonstrate that early ART initiation limits the size of the reservoir but does not profoundly impact the proviral landscape. This analysis allows us to revise our understanding of the composition of proviral populations and estimate the true reservoir size in individuals treated early vs. late in infection. Additionally, we demonstrate that common assays for measuring the reservoir do not correlate with reservoir size. These findings reveal hurdles that must be overcome to successfully analyze future HIV-1 cure strategies.
HIV-1 establishes infection in CD4+ T cells1,2,7, generating a latent reservoir with an extremely long half-life (44 months) that necessitates life-long treatment4,8. Efforts to eradicate this reservoir include the “shock-and-kill” approach in which latent proviruses are induced so that infected cells can be eliminated9,10,11. The latent reservoir was initially defined using a quantitative viral outgrowth assay (QVOA)1,8,12,13. PCR can also detect proviral DNA14–16. However, the QVOA and PCR assays correlate poorly17. We recently examined discrepancies between the assays by sequencing gag+ proviruses from QVOA wells negative for viral outgrowth6. Most proviruses were defective, but 12% were intact, and some of these could be reactivated following a second round of T cell activation6. However, these studies were limited to individuals who initiated ART late in the course of infection and only analyzed gag+ proviruses in cells that had been expanded for weeks in culture6. As immediate ART is now recommended for all infected individuals18, and since the size and perhaps other characteristics of the reservoir differ in individuals treated early versus late in infection19–21, we sought to define the composition of proviral populations in individuals treated during acute infection. As cure efforts advance, it is essential to understand how defective proviruses accumulate, persist in individuals and affect reservoir assay measurements. Therefore, we conducted a proviral analysis in unmanipulated samples from HIV-1 infected adults treated at different stages of infection.
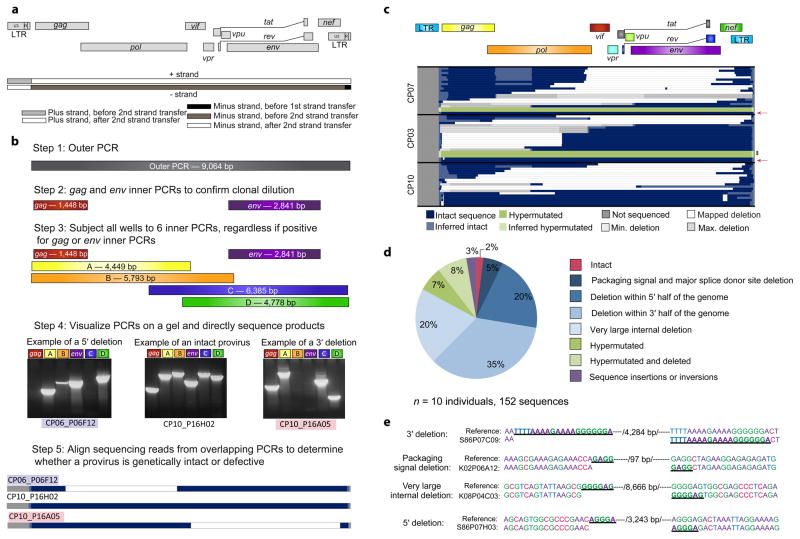
We developed a novel, unbiased single genome amplification method that captures intact and defective proviruses. Since deletions likely occur during minus strand synthesis before the second strand transfer event of reverse transcription, primers were designed to capture deletions arising during this process (Fig. 1a)22–24. PCRs were performed at limiting dilution to prevent in vitro recombination and competition between short and long templates. Following the near full length outer PCR, gag and env inner PCRs were used to confirm clonality, and six overlapping inner PCRs were performed on all wells at limiting dilution (Fig 1b). PCR products were directly sequenced to minimize PCR-induced error.
Figure 1.
Proviral sequences in chronically-treated subjects are highly defective. (a) Schematic of strand transfer events during reverse transcription relative to the HIV-1 genome. Deletions occur during minus strand synthesis before the second strand transfer event by the same copy choice mechanism that leads to recombination25. (b) Unbiased full genome sequencing strategy based on mechanistic considerations in (a) to capture the majority of defective proviruses. Bands from representative proviruses with a 5′ deletion, an intact sequence, and a 3′ deletion are shown. (c) Maps of proviral clones from three representative subjects treated during the chronic phase of infection. Each horizontal bar represents an individual clone identified through full genome sequencing. In cases where a deletion could not be precisely mapped due to a deletion encompassing multiple forward or reverse primer binding sites, the possible maximum and minimum deletions sizes are plotted (in grey). If sequencing data shows a mapped deletion that removes primer-binding sites for other amplicons, the resulting missing sequence was inferred to be present (light blue or green). A pink arrow denotes full-length, genetically intact sequences. Black asterisks indicate likely full-length hypermutated proviruses that were not fully sequenced due to extreme hypermutation. See Supplementary Fig. 1 for maps of proviruses from seven additional subjects. (d) Summary of 152 proviral sequences. (e) Short repeats (underlined) identified on both ends of the deletion junctions are consistent with a copy choice mechanism of recombination resulting in deletion of the intervening sequence and one homology region (/number of basepairs deleted/).
We examined proviral DNA in purified resting CD4+ T cells from ten subjects initiating ART during the chronic phase of infection (CP; ART started >180 days after infection) (Supplementary Table 1). Analysis was performed on freshly isolated cells to prevent bias from in vitro expansion. Strikingly, 98% of proviruses were defective (Fig. 1c,d and Supplementary Fig. 1). The most common defects were internal deletions (80%), which varied in size (15 bp to ~8 kb) and in location in the genome. Some proviruses had 3′ deletions affecting the env, tat, rev and nef genes. We also identified proviruses with 5′ deletions affecting gag and pol and others with very large deletions (>6 kb) encompassing most HIV-1 genes (Fig. 1c,d). The 5′ deletions and very large deletions, representing 40% of all sequences, were not identified in a previous screen because they contain deletions in the gag gene6. Proviruses with small (15-97bp) deletions at the packaging signal and major splice donor site represented 5% of sequences (Fig. 1d). These proviruses are likely replication-defective due to failure to correctly make spliced HIV-1 RNAs or to package genomes into virions6. Some clones contained multiple deletions suggesting multiple template switching events during reverse transcription (Fig. 1c) and others contained sequence inversions or insertions (Fig. 1d). For each type of deletion, proviruses with sequence homology at the deletion junctions were found (Fig. 1e), consistent with template switching during reverse transcription25. In control experiments, plasmids containing reference (NL4-3) or patient-derived proviruses were diluted into human genomic DNA and amplified by the same procedure with no defects observed (data not shown). Additionally, deletions showed nonrandom distribution within the HIV-1 genome with distinct hotspots (Supplementary Fig. 2) and we observed identical deletions in independent amplifications from the same subject, reflecting clonal expansion of infected cells (see below). Taken together, these results indicate that the deletions we observe occurred in vivo and do not result from PCR recombination. We also identified 7% of proviruses with APOBEC3F/G-induced hypermutation and an additional 8% with both deletions and hypermutation, indicating that these processes can occur together during reverse transcription (Fig. 1d). Almost all hypermutated proviruses contained mutated start codons for the gag, gag-pol and nef ORFs due to the presence of an obligatory position 2 glycine codon which creates the APOBEC3G consensus site, ATGGGT26,27. Additionally, all hypermutated sequences had multiple internal stop codons in the larger ORFs (gag, gag-pol, env, nef) and only a small fraction could make functional gene products (Supplementary Fig. 3). These defects and common deletions affecting key viral ORFs (Fig. 1c,d) likely prevent many defective proviruses from being eliminated by eradication strategies that depend on viral protein expression. All subjects had undetectable viral loads (<50copies/mL) for >8 months prior to sampling and all but one had low frequencies of 2LTR circles in cells (< 25 copies/106 resting CD4+ T cells), which represented less than 8% of the total number of infected cells (Supplementary Fig. 4a,b). The labile nature of linear unintegrated HIV-1 DNA and the low level of 2LTR circles suggest the majority of the sequences examined were integrated proviruses28. Of 152 near full-length sequences examined, only 3 sequences were intact (2%). These data indicate that defective proviruses are much more common than previously shown (Fig. 1d).
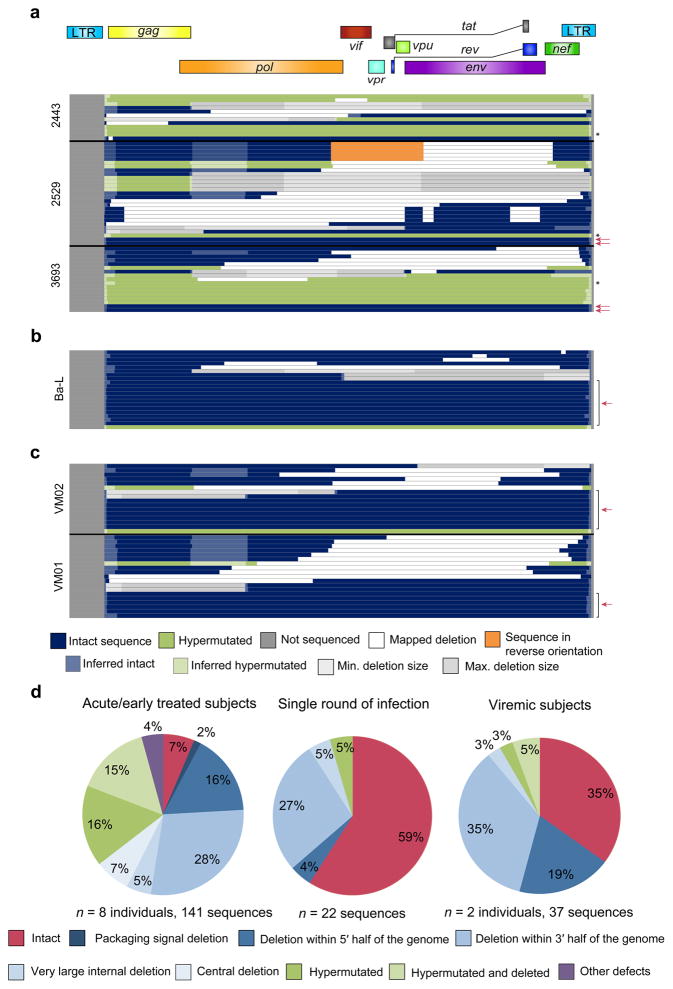
Since the dynamics of proviral accumulation remain unclear, we sought to determine how rapidly defective proviruses accumulate. We examined the proviral populations in subjects treated during the acute/early phase of infection (AP; ART started within 100 days in all subjects and within 60 days in 8 of 9 subjects) (Supplementary Table 1). Surprisingly, only 7% of proviruses were intact, even in subjects 2453, 2454, 3693, and 2443 who initiated ART within the first few weeks of infection (Fig. 2a,d and Supplementary Fig. 5). Although, the number of individuals subjected to this extensive analysis was small, the results were very consistent between subjects. The remaining proviral sequences contained defects similar to those in CP-treated subjects (Fig. 2a,d) and there was no significant difference in the fraction of intact proviruses compared to CP-treated subjects (Supplementary Fig. 6a). AP-treated subjects had a higher fraction of hypermutated sequences (35% vs. 14%, P = 0.0036, Supplementary Fig. 6b) and a significantly lower fraction of deleted sequences (57% vs. 82%, P = 0.0028, Supplementary Fig. 6c) suggesting that hypermutation is particularly important during acute infection, perhaps due to upregulation of APOBEC3G/F by type I interferons29–31. Interestingly, defects are even detectable in a single round of in vitro infection (41% of sequences, Fig. 2b,d). Consistent with this result, we readily detected defective proviruses in unfractionated CD4+ T cells from viremic subjects in the chronic phase of infection, representing 65% of sequences (Fig. 2c,d). In these subjects, the provirus populations include both proviruses in newly infected cells and archived proviruses in the reservoir. These results indicate that defective proviruses are likely generated during the initial rounds of replication after transmission, as defective proviruses are generated at a high frequency from the process of reverse transcription (Fig. 2b). Since many of the proviruses identified have defects that would preclude high-level expression of viral genes, cells carrying these proviruses would likely be less susceptible to elimination through viral cytopathic effects or lysis by cytotoxic T lymphocytes. Thus even individuals treated very early have large numbers of defective proviruses. Together, these results demonstrate that defective proviruses arise commonly, accumulate rapidly within two to three weeks after infection, and persist in vivo.
Figure 2.
Defective proviruses accumulate rapidly during the course of HIV-1 infection. (a—c) Maps of independent proviral clones. Each horizontal bar represents an individual clone identified through full genome sequencing. In cases where a deletion could not be precisely mapped, likely due to a deletion encompassing multiple forward or reverse primer binding sites, the maximum and minimum deletions sizes are plotted (in grey). If sequencing data shows a mapped deletion that removes primer-binding sites for other amplicons, the resulting missing sequence was inferred to be present (light blue or green). A pink arrow denotes full-length, genetically intact sequences. Black asterisks indicate likely full-length hypermutated proviruses that were not fully sequenced due to extreme hypermutation. (a) Sequences from resting CD4+ T cells of three representative subjects treated during acute/early infection (9 subjects total studied). For additional acute/early subject sequences, see Supplementary Fig. 5. (b) Sequences identified following a single round of in vitro infection of healthy donor CD4+ T lymphoblasts with HIV-1 (Ba-L isolate). (c) Sequences from CD4+ T cells from viremic subjects in the chronic phase of infection. (d) Summary of all proviral sequences identified from each study population.
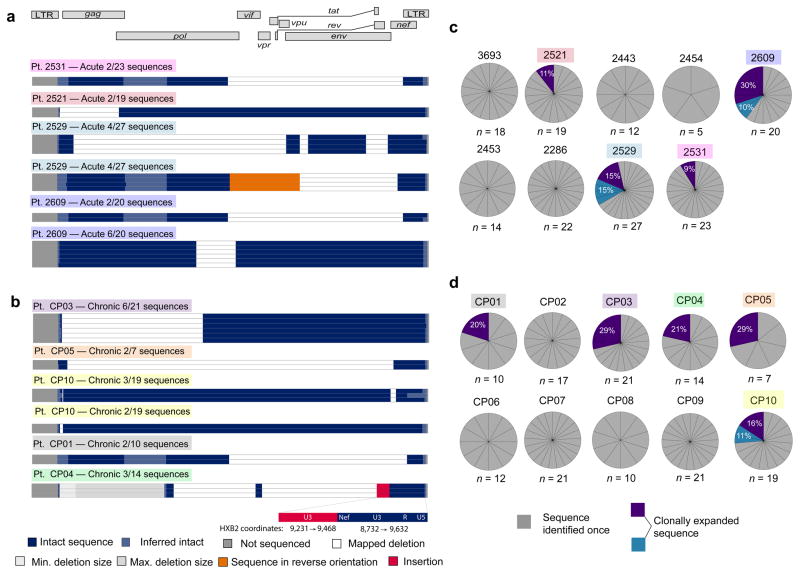
Proliferation of cells carrying replication-competent proviruses could be a major barrier to eradication32–34. Integration-site analysis, pioneered by Schröder et al.35, has provided important evidence for proliferation of infected cells in vivo but does not reveal whether the proviruses are replication-competent33,34,36. Our unbiased analysis allowed us to detect expanded cellular clones in vivo and evaluate the presence of defects. Among 312 sequences from 9 AP- (Fig. 3a) and 10 CP-treated subjects (Fig 3b), 38 were from expanded cellular clones identified as identical sequences arising from independent amplifications. Importantly, all contained gross defects that preclude replication. In subjects 2529, 2609, CP03, CP05, and CP10, expanded clones represented over 25% of all the sequences (Fig. 3c,d). Subject 2521 contained an expanded clone representing 11% of all proviral sequences (Fig. 3a,c) after just 17 months of infection and 8 months on suppressive ART, indicating expanded clones do not require years to accumulate to large proportions. Our results indicate the majority of expanded clones are defective. However, considering that <7% of proviruses are intact, rare expanded clones may carry replication-competent virus and contribute to HIV-1 persistence, as was seen in a recent clinical case study37,38. Although cells harboring defective proviruses can undergo clonal expansion with minimal consequences to the host cells, cells harboring replication-competent viruses could likely expand through either activation of the cell but not the provirus39 or possibly proliferation and survival of the cells despite viral gene expression and virion production.
Figure 3.
Expanded clones identified in chronically and acutely treated subjects are grossly defective. (a,b) Maps of expanded HIV-1 clones identified in resting CD4+ T cells from subjects treated in acute (a) or chronic (b) infection. Expanded clones are defined as clones amplified in completely independent PCR reactions from a single subject that are identical at every nucleotide. The frequency of each clone is shown relative to the total number of clones identified in that subject. Colored boxes denote the subject in which each expanded clone was identified (see c,d) (c,d) Proportion of sequences from subjects treated during acute (c) or chronic (d) infection that are expanded clones. The number of sequences examined for each subject (n) is noted. Expanded clones (purple/blue) are shown as a percentage of total sequences from the relevant subject.
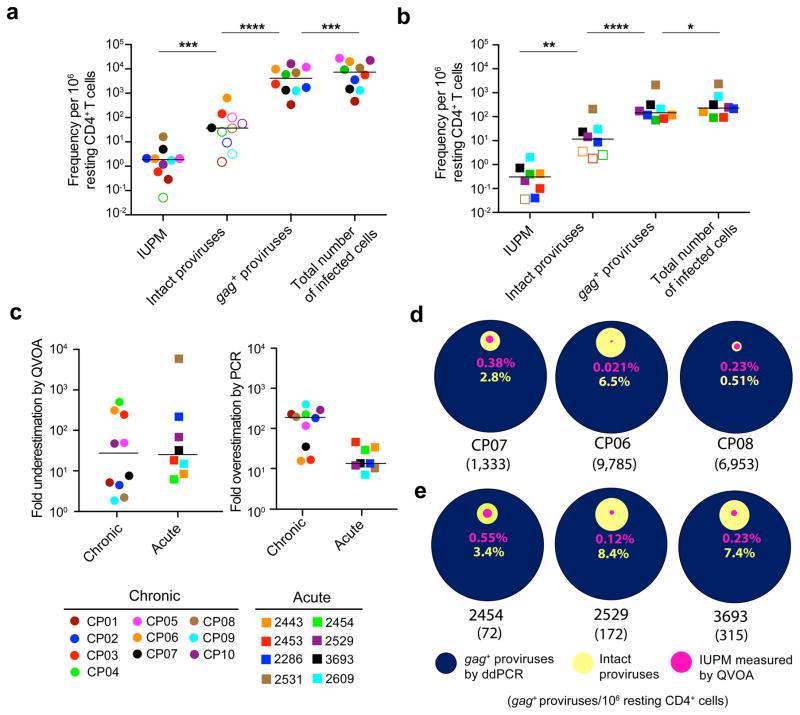
Our analyses show that the fraction of intact proviruses is much lower than originally thought even in individuals treated early. We have previously shown that intact proviruses replicate well in vitro when reconstructed6. Furthermore, when wells that were negative for viral outgrowth in the VOA were reactivated, additional replication competent virus was isolated6. These data taken together suggest that some if not all of these intact proviruses are competent for viral replication and that the number of intact proviruses is likely the closest estimate to the true size of the latent reservoir. Given these results, we sought to more accurately estimate the true reservoir size in AP- and CP- treated individuals, as defined by the number of intact proviruses, and compare that result to current assays such as the QVOA and DNA PCR. We used a validated droplet digital method for proviral DNA with gag primers14,40 (gag+ proviruses) and corrected for proviruses deleted in gag (total number of infected cells). The DNA PCR assay gave infected cell frequencies dramatically higher than the frequency of cells with intact proviruses (P < 0.0001 for CP-treated; P < 0.0001 for AP-treated subjects). The QVOA gave infected cell frequencies that were dramatically lower than the frequency of cells with intact proviruses (P < 0.001 for CP-treated; P < 0.01 for AP-treated subjects) (Fig. 4a,b). We found that the QVOA potentially underestimates the latent reservoir by a median of 27-fold in CP-treated subjects and 25-fold in AP-treated subjects (Fig. 4c). However, DNA PCR overestimated the reservoir size by a median of 188-fold in CP-treated subjects and 13-fold in AP-treated subjects (Fig. 4c). Importantly, there was no correlation between the number of intact proviruses and either the QVOA or DNA PCR results (Supplementary Fig. 7a–c). Additionally, the relationships between the QVOA, intact proviruses, and proviral DNA varied greatly from person to person in both CP-(Fig. 4d and Supplementary Fig. 7d) and AP-treated subjects (Fig. 4e and Supplementary Fig. 7e). This precludes the use of either the QVOA or DNA PCR as a surrogate marker for the number of intact proviruses, which is likely the closest estimate of the true size of the latent reservoir.
Figure 4.
Current assays significantly underestimate or overestimate the size of the latent reservoir. (a,b) Comparison of different reservoir measurements in resting CD4+ T cells from subjects starting ART during chronic (a) or acute (b) infection. The frequency of infected cells was measured by QVOA, which detects cells that release infectious virus after one round of T cell activation and is reported as the number of infectious units per million resting CD4+ T cells (IUPM). For individuals in whom the IUPM was below the limit of detection, the median posterior estimate of infection frequency was plotted instead (open symbols). The predicted total number of infected cells was calculated for each subject by correcting the ddPCR result (gag+ proviruses) for the fraction of proviruses with deletions in gag. The frequency of cells with intact proviruses was calculated as the frequency of infected cells times the fraction of intact proviruses estimated for each subject using an empirical Bayesian model. Open symbols indicate subjects in which no intact proviruses were detected. Horizontal bars indicate median values. Statistical significance was determined using a two-tailed paired t-test and the variance was similar between groups. *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. (c) Underestimation of the latent reservoir size by the QVOA was calculated by dividing the number of intact proviruses by the QVOA result for each subject. Overestimation of the latent reservoir size by DNA PCR was calculated by dividing the gag+ provirus PCR values by the number of intact proviruses. Horizontal bars indicate median values. (d,e) Comparison of infected cell frequencies as determined by QVOA (pink), analysis of intact proviruses (yellow), and ddPCR for gag+ proviral DNA (blue) for three representative CP- (d) and AP- treated (e) subjects. All values are plotted as a percentage of the frequency of cells with gag+ proviral DNA. The number of gag+ proviruses per million resting CD4+ T cells as measured by ddPCR is indicated in parenthesis for each subject. For additional subject comparison plots, see Supplementary Fig. 7d,e.
This unbiased screen demonstrates that the fraction of intact proviruses is considerably smaller than previously shown and that defects accumulate as early as two to three weeks after infection. Although surprising, the rapid accumulation is consistent with the observation that defects can be observed in 40% of proviruses generated in a single round in vitro infection (Fig. 2d) and the fact that cells harboring defective proviruses may be less susceptible to immune clearance or cytopathic effects. With these new data, we were able to revise our estimates of the reservoir size in both CP- and AP-treated individuals. If all intact proviruses can be induced in vivo, the true size of the latent reservoir would be on average 12 infectious proviruses (median of AP-treated) in acute/early treated individuals and 37 infectious proviruses (median of CP-treated) per million resting CD4+ T cells in chronically treated individuals, with substantial person-to-person variation. Furthermore, our results change our understanding of the impact of early ART on the proviral landscape. While early ART initiation limits the size of the latent reservoir, it does not have a profound impact on the composition of proviral populations, with the vast majority of proviruses in both CP- and AP-treated adults containing defects. Finally, these results have implications for assessing HIV-1 cure strategies. Since there is no correlation between the QVOA or DNA PCR and the number of intact proviruses, these assays cannot be used to accurately predict the true reservoir size, even in individuals treated early in infection. Indeed any analysis of subgenomic regions of proviruses to evaluate viral evolution or reservoir reduction must be reconsidered in this context, since the majority of sequences studied will be from defective proviruses41. Importantly, since the nature of the defects described here indicates that many defective proviruses may not be eliminated by eradication strategies, defective proviruses could obscure the measurement of real changes in the rarer intact proviruses that must be eliminated to achieve a cure.
Online Methods
Study subjects
The Johns Hopkins Institutional Review Board and the UCSF Committee on Human Research approved this study. All participants provided written consent prior to enrollment. Nineteen HIV-1 infected individuals who met the criteria of suppressive ART and undetectable plasma HIV-1 RNA level (< 50 copies per mL) for a minimum of 8 months were enrolled. Ten of these participants were recruited from the SCOPE and OPTIONS cohorts at the University of California, San Francisco. CP-treated subjects are defined as subjects starting ART >180 days from the estimated date of infection. AP-treated subjects started ART < 100 days after the estimated date of infection. Viremic subjects were either untreated or had viral loads > 4000 copies/ml on ART. Supplementary Table 1 details the characteristics of study participants.
Isolation of resting CD4+ T lymphocytes
Peripheral blood mononuclear cells (PBMCs) were isolated using density centrifugation on a Ficoll-Hypaque gradient. CD4+ T cells were isolated from PBMCs using a negative selection method (CD4+ T cell Isolation Kit II, Miltenyi Biotec). Resting CD4+ lymphocytes (CD4+, CD69−, CD25− and HLA-DR−) were enriched by a second negative depletion (CD25−Biotin; Anti-Biotin MicroBeads; CD69 MicroBead Kit II; Anti–HLA-DR MicroBeads, all from Miltenyi Biotec). Resting CD4+ cell purity was consistently >95% as assessed using flow cytometry.
DNA extraction and limiting dilution PCR
DNA was extracted from 2 x 106 resting CD4+ cells using the Qiagen Gentra Purgene Cell Kit A, which allows for extraction of large fragments of ~200kb in size so as to minimize fragmentation of HIV-1 genomes. DNA was subjected to a nested limiting dilution PCR protocol modified from Ho et al. using Platinum Taq HiFi Polymerase (Life Technologies)6. The outer PCR was nearly full-length from HXB2 coordinates 623 to coordinates 9,686 and employed a touchdown PCR protocol. The outer PCR wells were diluted 1:3 with DI water and 1μL of outer PCR DNA was used for nested amplification of both gag and env to determine clonality. Clonality was determined using Poisson statistics and at dilutions giving a high probability of clonality (P > 0.85), all outer PCR wells, including those that were negative for gag and env, were subjected to six inner PCRs. 1μL of outer PCR DNA was used for each nested PCR reaction. A detailed PCR protocol and primer sequences and locations can be found in Supplementary Table 2. PCR products were visualized on 1% agarose gels. The products were directly sequenced to minimize PCR-induced error using the QIAquick Gel Extraction Kit followed by Sanger sequencing. Sequencing reads from the six overlapping nested PCRs were aligned and compared to the reference genome HXB2 using CodonCode Aligner software to reconstruct near full-length sequences and identify defects. Hypermutation was determined using the full-length sequence for each clone and the Los Alamos hypermut algorithm42. Some hypermutated clones had extensive hypermutation that prevented full sequencing of the entire genome. Since the PCR products were correctly sized on an agarose gel, the sequences were inferred to be full-length hypermutated. We were able to directly map the majority of deletions by direct sequencing of PCR products generated with primers flanking the deletion junction. Some very large internal deletions were identified despite deletion of some inner primer binding sites. Observed bands likely resulted from exponential amplification in the outer PCR and linear amplification in inner PCRs. These deletions were confirmed through sequencing bands from multiple inner PCRs, all of which showed the exact same deletion product.
In vitro infections
HIV-1 negative donor CD4+ T cells were isolated as above and activated with phytohaemagglutinin (PHA) (0.5ug/mL) for 3 days in IL2-containing medium43. Cells were infected with replication-competent HIV-1 Ba-L virus (300 ng p24/106 cells) by spinoculation (2 hrs, 37°C, 1200 x g). Cells were then suspended at 3 x 106/mL in IL2-containing medium and incubated for 6 days at 37°C. Following the incubation, supernatant was collected, and stored at −80°C. 25 x 106 HIV-1 negative donor CD4+ T cells were activated by αCD3/αCD28 stimulation for 72 hours in IL-2-containing medium as previously described43. Activated cells were suspended in 2.5mL of IL-2-containing medium and distributed in 100μL aliquots into a 96-well v-bottom plate. A 500ng inoculum of HIV-1 Ba-L was added to each well and cells were infected by spinoculation (2hrs, 37°C, 1200 x g). Following the spinoculation, cells were suspended in 40mL of IL2-containing medium supplemented with enfuvirtide (10uM) to prevent additional rounds of viral replication. Following a two day incubation at 37°C, genomic DNA was isolated and analyzed as above.
Quantitative viral outgrowth assay
The QVOAs were performed as previously described1,12,13. MOLT-4/CCR5 cells were added on Day 2 of the culture13 and the culture supernatants were examined using the standard protocol, which involves an ELISA for the p24 viral capsid protein (PerkinElmer) after 21 days12. HIV-1 RNA in the culture supernatant was not measured. MOLT-4/CCR5 cells were obtained from NIH AIDS reagent program, were negative for mycoplasma contamination, and were stained to verify expression of CD4, CXCR4, and CCR5. Infectious units per million cells (IUPM) values were calculated using IUPMStats44. If all culture wells were negative for viral outgrowth, the median posterior estimate of infection frequency was used.
Droplet digital PCR
DNA was extracted from 5–10 x 106 resting CD4+ T cells using the QIAamp DNA Blood Mini Kit. DNA was fragmented and a duplex droplet digital PCR was performed with primers in the gag gene as well as primers for 2LTR circles as previously described14,40. For pt. 2609, primers in the pol gene were used instead as the gag PCR primers and probes contained multiple mismatches with the patient sequence and did not amplify40. Measurements of the cellular gene RPP30 in a replicate well by ddPCR were used to calculate the frequency of resting CD4+ T cells. The frequency of gag+ DNA and 2LTR circles were plotted as a frequency per 106 resting CD4+ T cells. In the case of a negative 2LTR circle measurement, the lower limit of detection was determined using the total cellular input for that sample. Since many of the proviruses examined were gag negative, we also estimated the total number of infected cells in study subject resting CD4+ T cells as the copies of gag+ proviral DNA divided by the fraction of proviruses detected that were gag+. For Pt. 2609 the total number of infected cells was estimated as the copies of pol+ proviral DNA divided by the fraction of proviruses detected that were pol+ since the gag PCR primers did not amplify.
Bayesian analysis to calculate number of intact proviruses
The proportion of intact genomes for each subject was estimated using an Empirical Bayesian analysis, with prior distributions generated separately for the acute and chronic subject cohorts. The proportion of clones identified as intact was calculated for each subject, and the mean and variance of this measurement were used to generate beta prior distributions with the same mean and variance. The proviral genome sequences were treated as Bernoulli trials, with intact clones counted as successes. The final estimate for the percent of intact genomes for each subject was calculated as the expected value of the posterior distribution. The number of intact proviruses was calculated as the frequency of cells with total HIV-1 DNA times the frequency of intact proviruses estimated for each subject using an empirical Bayesian model.
Correlation plots
All data sets except the percent of intact proviruses followed a log distribution and were log transformed. The log transformed data met the D’Agostino-Pearson test for normal distribution, and Pearson correlations were performed on log transformed data. For the QVOA data in which one of the samples was below the limit of detection, the median posterior estimate of infection frequency was plotted instead. P < 0.05 was considered statistically significant. GraphPad Prism software was used to perform all statistical tests and correlation plots.
Supplementary Material
Acknowledgments
We thank the study participants who have made this research possible. We also thank G. Laird for critical advice and discussion, L. Alston, H. McHugh, and D. Xu for assistance with study participants, V. Walker-Sperling for providing the Ba-L virus, C. Pohlmeyer for providing a sample from a viremic subject and all of the members of the Siliciano Lab for valuable discussion and advice. This work was supported by the Genomics and Sequencing Core at the UC San Diego Center for AIDS Research (P30AI036214 (D.D.R)), by the Pendleton Charitable Trust and the VA San Diego Healthcare System (D.D.R.), by the Martin Delaney CARE and DARE Collaboratories (US National Institutes of Health grants AI096113 (R.F.S.) and 1U19AI096109 (S.G.D. and R.F.S)), by an ARCHE Collaborative Research Grant from the Foundation for AIDS Research (amFAR 108165-50-RGRL (R.F.S.)), by the Johns Hopkins Center for AIDS Research grant P30AI094189 (R.F.S.), by the US National Institutes of Health grants 43222 (R.F.S.) and R21AI113147-02 (J.D.S.), and by the Howard Hughes Medical Institute (R.F.S.) and the Bill and Melinda Gates Foundation (R.F.S.).
Footnotes
Accession Codes: Accession codes are pending and will be included in the final version of the manuscript.
Author Contributions
K.M.B., R.A.P., S.G.D. and R.F.S. designed experiments; K.M.B., A.J.M., R.A.P., M.G.S., A.A.C., J.L., M.C.S. and S.M.L. performed experiments; K.M.B., A.J.M., S.B.L., Y.-C.H., D.D.R., J.D.S., R.F.S. analyzed the data; R.H. and A.A.C. managed study participant recruitment; S.G.D. provided patient samples; K.M.B. and R.F.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 5.Ruelas DS, Greene WC. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 8.Crooks AM, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J INFECT DIS. 2015;212:1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archin NM, Margolis DM. Emerging strategies to deplete the HIV reservoir. Curr Opin Infect Dis. 2014;27:29–35. doi: 10.1097/QCO.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks SG. HIV: Shock and kill. Nature. 2012;487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 11.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 13.Laird GM, et al. Rapid Quantification of the Latent Reservoir for HIV-1 Using a Viral Outgrowth Assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strain MC, et al. Highly Precise Measurement of HIV DNA by Droplet Digital PCR. PLoS ONE. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouzioux C, Mélard A, Avéttand-Fénoël V. In: Human Retroviruses. Vicenzi E, Poli G, editors. Humana Press; 2013. pp. 1087pp. 261–270. [Google Scholar]
- 16.Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods. 2012;186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson S, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günthard HF, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312:410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 19.Archin NM, et al. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc Natl Acad Sci USA. 2012;109:9523–9528. doi: 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain V, et al. Antiretroviral therapy initiated within 6 months of HIV infection is associated with lower T-cell activation and smaller HIV reservoir size. J INFECT DIS. 2013;208:1202–1211. doi: 10.1093/infdis/jit311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng K, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delviks-Frankenberry K, et al. Mechanisms and factors that influence high frequency retroviral recombination. Viruses. 2011;3:1650–1680. doi: 10.3390/v3091650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jetzt AE, et al. High rate of recombination throughout the human immunodeficiency virus type 1 genome. Journal of Virology. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Jetzt AE, Ron Y, Preston BD, Dougherty JP. The nature of human immunodeficiency virus type 1 strand transfers. J Biol Chem. 1998;273:28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]
- 25.Hwang CK, Svarovskaia ES, Pathak VK. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc Natl Acad Sci USA. 2001;98:12209–12214. doi: 10.1073/pnas.221289898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 27.Kieffer TL, et al. G->A Hypermutation in Protease and Reverse Transcriptase Regions of Human Immunodeficiency Virus Type 1 Residing in Resting CD4+ T Cells In Vivo. Journal of Virology. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierson TC, et al. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. Journal of Virology. 2002;76:8518–8531. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai SK, et al. Role of retroviral restriction factors in the interferon-α-mediated suppression of HIV-1 in vivo. Proc Natl Acad Sci USA. 2012;109:3035–3040. doi: 10.1073/pnas.1111573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper MS, et al. Interferon-α Subtypes in an Ex Vivo Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms. PLoS Pathog. 2015;11:e1005254. doi: 10.1371/journal.ppat.1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. Journal of Virology. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearney MF, et al. Origin of Rebound Plasma HIV Includes Cells with Identical Proviruses that are Transcriptionally Active Before Stopping Antiretroviral Therapy. Journal of Virology. 2015;90:1369–1376. doi: 10.1128/JVI.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schröder ARW, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 36.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M, Siliciano RF. Reservoir expansion by T-cell proliferation may be another barrier to curing HIV infection. Proc Natl Acad Sci USA. 2016;113:1692–1694. doi: 10.1073/pnas.1600097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 2011;7:e1002288. doi: 10.1371/journal.ppat.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massanella M, Gianella S, Lada SM, Richman DD, Strain M. Quantification of Total and 2-LTR (Long terminal repeat) HIV DNA, HIV RNA and Herpesvirus DNA in PBMCs. Bio-protocol. 2015;5:e1492. doi: 10.21769/bioprotoc.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenzo-Redondo R, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530:51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 43.Kim M, et al. A primary CD4(+) T cell model of HIV-1 latency established after activation through the T cell receptor and subsequent return to quiescence. Nat Protoc. 2014;9:2755–2770. doi: 10.1038/nprot.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenbloom DIS, et al. Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus-1. Open Forum Infect Dis. 2015;2:ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.