Abstract
Recent advances in fluorescence super-resolution microscopy have allowed sub-cellular features and synthetic nanostructures down to ~15 nm in size to be imaged. However, direct optical observation of individual molecular targets (~5 nm) in a densely packed biomolecular cluster remains a challenge. Here, we show that such discrete molecular imaging is possible using DNA-PAINT (points accumulation for imaging in nanoscale topography) - a super-resolution fluorescence microscopy technique that exploits programmable transient oligonucleotide hybridisation - on synthetic DNA nanostructures. We examined the effects of high photon count, high blinking statistics, and appropriate blinking duty cycle on imaging quality, and developed a software-based drift correction method that achieves <1 nm residual drift (r.m.s.) over hours. This allowed us to image a densely packed triangular lattice pattern with ~5 nm point-to-point distance, and analyse DNA origami structural offset with angstrom-level precision (2 Å) from single-molecule studies. By combining the approach with multiplexed Exchange-PAINT imaging, we further demonstrated an optical nano-display with 5×5 nm pixel size and three distinct colours, and with <1 nm cross-channel registration accuracy. This method opens up possibilities for direct and quantitative optical observation of individual biomolecular features in crowded environments.
Introduction
Biological and synthetic biomolecular systems exhibit complex structures at the nanoscale1-10. Understanding the spatial arrangement of their individual components is critical for unraveling the molecular mechanism underlying complex molecular behaviour (Fig. 1a). Super-resolution fluorescence techniques have bypassed the traditional diffraction limit and demonstrated imaging resolution down to 10-20 nm11-13. In particular, single-molecule localisation microscopy (SMLM) builds up super-resolution images from single-emitter localisations and typically achieves photon-limited localisation precision (down to ~1 nm) for single-emitter blinking events13-15. Previous single-molecule and SMLM studies separately demonstrated single-target visualisation in isolation or sparse arrangements16, 17, and high localisation precision compatible with molecular-scale resolution18-24. However, discrete visualisation and precise localisation of each individual molecular target (~5 nm) in a densely packed biomolecular cluster remains challenging, which we refer to as discrete molecular imaging (DMI).
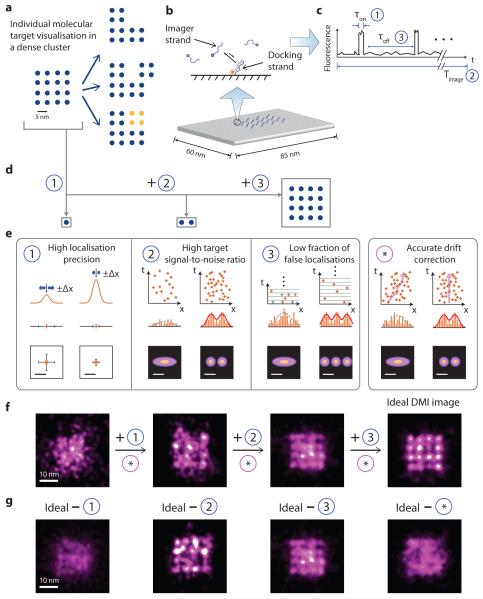
Figure 1. Principle and requirements of discrete molecular imaging (DMI).
(a) Concept of super-resolution discrete molecular imaging, illustrated with point array representation (blue points represent individual molecular targets, yellow points represent chemical modifications). Left, a regular 16-component biomolecular complex; right, its various forms of structural and chemical variations.
(b) Illustration of DNA-PAINT principle: transient binding between a docking strand and dye-conjugated imager strands (top), illustrated on a synthetic DNA origami nanostructure, where each cylinder represents a DNA double helix (bottom).
(c) Schematic DNA-PAINT blinking time trace of a single imaging target. Three blinking characteristics measure (1) blinking on-time, τon, (2) total imaging time, Timage and (3) blinking off-time, τoff, and can be tuned to meet the three blinking requirements in (e).
(d) Schematics of different substructures from the complex in (a): a single target, a pair of close-by targets, and a dense lattice, which need different blinking requirements in (e) to be clearly visualised.
(e) Technical requirements for achieving discrete molecular imaging. Each panel outlines one technical requirement, and depicts schematically the effect on imaging quality before (left column) and after (right column) the requirement is satisfied. For each condition, intensity profile in 1D (top), fitted Gaussian centres in 1D (middle) and 2D (bottom) are shown for requirement (1); localisation time trace in 1D (top), localisation histogram in 1D (middle) and 2D (bottom) are shown for requirements (2), (3) and (*). Orange lines and crosses indicate localisations. Orange bars depict localisation histograms. Solid red lines and dotted grey lines indicate successful and failed Gaussian fittings on localisation histograms, respectively. In panel 3, grey crosses indicate true localisations eclipsed by false double-blinking localisations. The same numbering for technical requirements (1)-(3) is also used in Fig. 2 and 4, and Supplementary Figure S2.
(f-g) Simulations of imaging effects of the technical requirements for the complex in (a), under increasingly better imaging conditions without stage drift (f), or under non-ideal imaging conditions with one of the four requirements unsatisfied (g).
See online methods and Supplementary Methods S2 for simulation details, and Supplementary Notes S7 for discussions. Scale bars: schematic length scale 5 nm in (a) and (e), 10 nm in (f) and (g).
Several factors limit the performance of current super-resolution techniques, such as finite fluorophore photon budget, unsatisfactory dye imaging efficiency, or limited control over target blinking kinetics9, 22, 25-30. These restrictions respectively translate to limited photon count per localisation, limited number of blinking events per target, and high fraction of false localisations, and ultimately restrict the final imaging resolution, signal-to-noise ratio, and visualisation of individual targets within dense clusters. Moreover nanometre-level accuracy stage noise and drift compensation is critical for high imaging resolution and quality.
We addressed the above challenge and demonstrated DMI using DNA-PAINT super-resolution method10, 23, 31-33 (Supplementary Fig. 1). DNA-PAINT, a variation of PAINT9, exploits the transient binding of fluorophore-labeled imager strands to target-bound docking strands to achieve necessary blinking for super-resolution reconstruction10 (Fig. 1b; Supplementary Fig. 2). The continuous replenishment of imager strands renders DNA-PAINT immune to photobleaching, allowing high localisation precision from extracting a large number of photons per single-molecule localisation and high target separability from collecting a large number of blinking events from each target. Additionally, due to its independent and programmable control of blinking ON/OFF rates, DNA-PAINT permits low imaging background in dense clusters from appropriately adjusted blinking duty cycle based on target density (Fig. 1c).
We developed a framework for achieving DMI for localisation microscopy, including technical requirements and quality assay methods (Supplementary Fig. 1). Specifically, we introduced an image-based assay for measuring localisation precision and maximally achievable resolution, a target signal-to-noise ratio assay for measuring single-target separation, and a method for estimating the fraction of false double-blinking localisations. We systematically studied their effects, and demonstrated stringent control for each of them with DNA-PAINT. In particular, we achieved high localisation precision (<1 nm single-molecule fitting precision, from up to 50,000 photons per single-molecule localisation), high target separability (from ~80 blinking events per target), low imaging background from appropriately tuned blinking duty cycles, and high-accuracy (<1 nm r.m.s. over hours) microscope stage drift correction with a novel method based on synthetic nanostructure drift markers with designed geometric patterns. Finally, we used DMI to visualise individual targets in a compactly labelled molecular grid of targets (with point-to-point spacing ~5 nm), and demonstrated multiplexed DMI on a three-colour nano-display board with ~5 nm pixels.
Technical requirements for DMI
The technical requirements for DMI depend on the spatial distribution of the targets, especially the closest spacing between targets, and their local density26,34 (within a diffraction-limited region, see Supplementary Note 7.1). Even with only two targets, a high localisation precision that allows a full width at half maximum (FWHM) resolution equal to or smaller than the spacing between them, is necessary but not sufficient for their clear separation23,35. Targets arranged in dense clusters impose even more stringent requirements. Although requirements and quality guidelines for super-resolution imaging has been discussed26,27,34,36, a systematic formulation of technical requirements for DMI and quality control methods are still lacking.
We examined the technical requirements for DMI with an example square lattice pattern (Fig. 1d). It is increasingly more difficult to discretely identify and precisely position (1) a single isolated target, (2) a pair of close-by targets, and finally (3) a dense lattice of targets, which require increasingly more stringent imaging conditions as described below (Fig. 1e). (1) High localisation precision. This can be obtained from collecting a high photon count per single-molecule localisation, and allows precise localisation of an isolated target. (2) High target signal-to-noise ratio (target SNR) in the super-resolved image. This can be achieved from collecting a large number of blinking events per molecular target, and allows for clear separation between two close-by targets. (3) Low fraction of false localisations from double-blinking events. This can be achieved by using a low blinking on-off duty cycle, which minimises falsely-localised background noise, and hence allows discrete visualisation of each target within a densely packed complex. (4) Accurate microscope stage drift compensation. This is important for accurate identification and localisation of any molecular structures of interest, especially over extended imaging time. Computer-simulated super-resolution images reveals increasingly better image quality when more of these requirements are satisfied (Fig. 1f), and that each of the four requirements is indispensable for DMI (Fig. 1g). See online methods, Supplementary Method 2, and Note 7.2 for details.
We termed requirements (1)-(3) collectively as the three blinking requirements, as they can all be met by appropriate single-molecule blinking properties (see Fig. 1c). Since DNA-PAINT allows flexible tuning of target blinking kinetics, it provides a promising route for implementing DMI.
Systematic characterisation and quality control for DMI
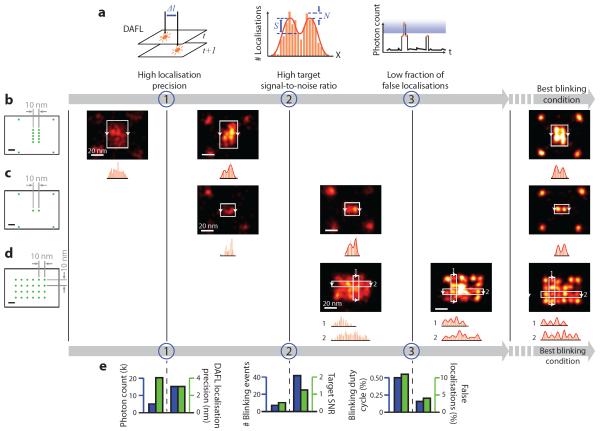
To quantitatively characterise the effects of the above DMI requirements on imaging quality, we proposed a set of assay methods (Fig. 2a): (1) an image-based assay of localisation precision and maximally achievable resolution by comparing the positions of super-localised centres from neighbouring frames (termed distance between adjacent-frame localisations, or DAFL), (2) a target SNR assay based on the analysis of distribution of super-localised centres, which directly measures the separability of neighbouring targets in super-resolved images, and (3) a localisation time trace based assay for estimating false localisation ratio. These assay methods provide a general, sample-agnostic method for stringent quality control of general super-resolution microscopy studies as well as DMI. See online methods, Supplementary Fig. 3, and Method 3 for details.
Figure 2. Systematic characterisation of blinking requirements and optimisation of DNA-PAINT imaging quality.
(a) Methods for systematic characterisation of the three blinking requirements depicted in Fig. 1. (1) Distance between adjacent-frame localisations (DAFL) measures distance between pairs of spatially-close localisations originated from adjacent camera frames. (2) Target signal-to-noise ratio (target SNR) measures separability of peaks in localisation histogram, in the super-resolved image. S, signal; N, noise; red curve indicates two-peak Gaussian fit. (3) Photon count cut-off in blinking trace measures fraction of false localisations. Blue shaded area indicates identified false localisations. Orange markers, bars and curves indicate localisations, histograms and time traces, respectively.
(b-d) Designed origami standards with 10 nm spacing under different blinking conditions. Leftmost column, design schematics of DNA origami standards; green dots indicate DNA-PAINT docking strands; four corners in (b) and (c) are used as alignment markers. Right five columns, DNA-PAINT images under increasingly better blinking conditions (one condition per column). Histograms below images show projection profiles from the areas indicated by white boxes and projected along the directions of arrows.
(e) Quantitative characterisation and pairwise comparisons of imaging conditions used in (b-d), before and after meeting each additional requirement, assayed with methods in (a). For each comparison, left axis (blue) shows the control parameter and right axis (green) shows experimental measurement.
For more details, see Supplementary Figures S3-S5 on origami designs, Supplementary Figures S6-S13 for super-resolution images, online methods and Supplementary Methods S3, S5 for DNA-PAINT imaging conditions and analysis methods. Scale bars, 10 nm in schematics and 20 nm in super-resolution images.
Using these assay methods, we first simulated super-resolution movies with varying photon count, number of blinking events, and blinking duty cycles, and measured the resultant single-molecule localisation precision, target SNR, and fraction of false localisations from double-blinking events (see online methods, Supplementary Fig. 3 and Methods 2, 3 for details). We observed high localisation precision (<1 nm, supporting <2 nm FWHM resolution) with high photon count (>30,000), consistent target separation with high target SNR (>2 under our definition), and low background noise under low fraction of false localisations (down to <5%), allowing DMI imaging.
Applying the same assay methods, the three blinking requirements (localisation precision, target SNR, and false localisation ratio) for DMI were each experimentally verified by subjecting synthetic DNA origami nanostructure34,38 standards with three designed target patterns to different DNA-PAINT imaging conditions (Fig. 2b-d, leftmost columns, designed pattern schematics; five right columns, DNA-PAINT images, see online methods and Supplementary Method S4 for details). Synthetic DNA nanostructures provide a programmable and geometrically precise molecular patterning platform for single-molecule and super-resolution studies. These structures were self-assembled from a long single-stranded DNA scaffold and a collection of short staple strands34. By extending a selected subset of these staple strands, DNA-PAINT docking strands could be arranged into user-prescribed, geometrically precise nano-patterns and used as super-resolution imaging standards or auxiliary markers (see next section, Supplementary Figs. 4-6 and Note 8 for details).
In Fig. 2b, two 10-nm spaced lines (each consisting of 5 points, spaced 5 nm apart to satisfy Nyquist criterion) only turned from unresolvable speckles (left image) to separable lines (right image) with an increased photon count per single-molecule localisation and hence higher localisation precision. However, the high localisation precision alone under this imaging condition failed to resolve two points spaced by 10 nm (Fig. 2c, left image) due to the reduced number of targets (from 5 to 1 on each side). The two points only became resolvable (Fig. 2c, right image) with a larger number of blinking events per target and hence an increased target SNR. However, this imaging condition with a high localisation precision and target SNR still failed to resolve a 24-target 10-nm grid (Fig. 2d, left image), due to increased target density. The grid points only became individually resolvable (Fig. 2d, right image) with a decreased blinking duty cycle and hence lower false localisation ratio. Quantitative pairwise comparisons of these imaging conditions and imaging quality assay results before and after meeting each of the three blinking requirements are shown in Fig. 2e (see online methods, Supplementary Figs. 7-12 and Methods 5.1, 6.1 for details). Finally, we imaged these structures under the best imaging conditions, and obtained clear images of the designed patterns (Fig. 2b-d, rightmost column, see Supplementary Figs. 13, 14 for details).
Sub-nanometre accuracy software-based drift correction
DMI also imposes stringent requirements on microscope stage drift compensation. For example, clear separation of targets with 5 nm target-to-target spacing requires accurate drift correction of <1 nm (r.m.s. drift), due to the compounded effect from stage drift and finite localisation precision (assuming 1 nm localization precision, Fig. 3a, see online methods and Supplementary Method S2 for details). Previous solutions either used active feedback system to reach <1 nm residual drift, but requires complicated hardware setup and is technically involved to implement21, or used software-based post-processing methods with embedded nanoparticle fiducial markers, but are typically limited by imperfect tracking accuracy5, 7, 22, and do not support DMI resolution (~5 nm).
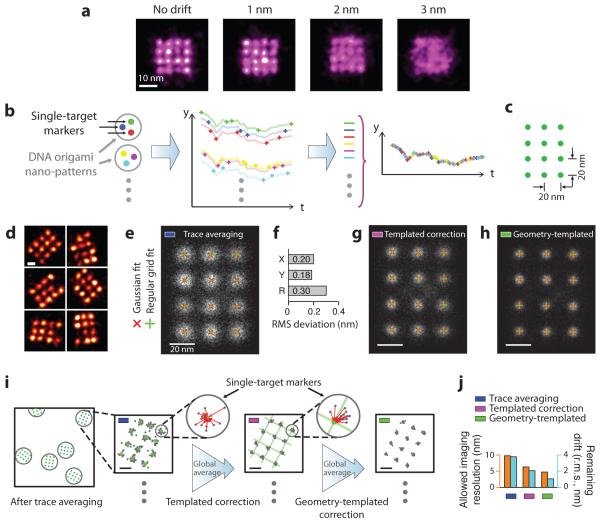
Figure 3. Principle and performance of DNA nanostructure templated drift correction.
(a) Effect of drift on imaging quality, simulated for the biomolecular complex in Fig. 1a with 1 nm localisation precision and different levels of stage drift. Only with 1 nm (r.m.s.) or less drift can the structure be clearly visualised.
(b) Principle of templated drift correction method with pre-designed nanostructure patterns. Illustrated with a three-target marker example, schematics show nano-pattern design with single-target markers (left), localisation time traces from individual single-target markers (middle), and averaged drift correction trace after combining traces from many markers (right). Targets and traces are colour-matched.
(c) Design schematics of a 3 × 4 square grid with 20 nm point-to-point spacing on a DNA origami nanostructure. Each green dot indicates a docking strand.
(d) Representative DNA-PAINT super-resolution images of the 20 nm grid structure in (c), imaged with 300 ms frame time, 30,000 total frames, and 3 nM imager strands. Missing grid points were likely due to synthesis or incorporation defects (see Supplementary Notes S9.1 for more discussions).
(e) Single-particle averages of 20 nm grid images (N = 700) after trace averaging. Overlaid crosses indicate Gaussian fitted centres (red) and regular grid-fitted centres (green) using the red crosses as fitting targets.
(f) Root-mean-square (r.m.s.) deviation between the Gaussian fitted and regular grid-fitted centres in (e).
(g, h) Single-particle averages of 20 nm grid images (N = 700) after templated (g) and geometry-templated (h) drift correction. Overlaid crosses indicate Gaussian fitted (red) and regular grid-fitted centres (green) as in (e). The same colour code for different stages of drift correction in (e), (g), (h) are also used in (i), (j) and Fig. 4.
(i) Procedure for templated and geometry-templated drift correction with 20 nm grid structures as templates. Schematics shows a large field-of-view image with many drift markers, after simple trace averaging (leftmost). Each grey circle indicates a 20 nm drift marker. Zoomed-in (square) schematics shows a super-resolved 20 nm grid marker, after simple trace averaging (left), after templated drift correction (middle), and after geometry-templated correction (right). Further zoomed-in schematics (round) shows one single-target marker and calculation of offset vectors. In zoomed-in schematics (square and round), grey dots indicate localisations, green dots and lines indicate Gaussian-fitted centres and regular grid-fitted lattices as guides for templated and geometry-templated drift correction calculation, red line segments with arrowheads represent calculated offset vectors.
(j) Comparison of allowable imaging resolution (measured in FWHM, blue) and estimated remaining drift (green) at different stages of drift correction.
For more details, see online methods and Supplementary Methods S2, S5 for simulation and analysis methods, Supplementary Figures S14 for super-resolution images. Scale bars: 10 nm in (a), 20 nm in (d-f) and zoomed-in images in (i).
We demonstrate a novel, synthetic nanostructure fiducial marker based stage drift correction and noise compensation method, “templated drift correction”, which achieves high-accuracy drift and noise cancellation (<1 nm r.m.s. residual drift) over both short and long time scales (from sub-second to ~5 hours), without using specialised hardware. The key principle is to use super-resolved single-molecule targets (such as a single DNA docking strand) on pre-designed geometrically-precise nano-patterns (such as a DNA nanostructure) as drift markers (Fig. 3b). Unlike conventional fiducial markers, these “single-target drift markers” possess unique advantages, as they can be: (1) stably anchored on the surface, (2) localised with high precision and without bleaching, (3) flexibly rotatable around their anchor points and thus avoid fixed-dipole effect, and (4) free of offset from fluorescence centre to drift marker centre position, such as in micron-sized beads and previous DNA origami drift markers10, 31. We arranged these single-target markers in pre-designed, well-separated nano-patterns (the “templates”), to help pack a number of single-target markers within a diffraction-limited area, and to allow their identification and separation during software processing steps (Fig. 3b). Additionally, the precise geometry of a nano-pattern correlates all the single-target drift markers on this pattern and effectively increases their on-fraction (the fraction of time that the marker is bright), producing more accurate drift correction. We term this method “geometry-templated drift correction”.
We designed a square lattice pattern with 20 nm spacing as our nano-pattern template to implement this strategy (Fig. 3c): a grid consists of 12 targets (docking strands), each of which will be treated as a single-target drift marker. After performing DNA-PAINT imaging and applying a round of simple trace averaging drift correction, all 12 targets are clearly separated from each other in the reconstructed image, allowing them to serve as single-target drift markers (Fig. 3d, also see Supplementary Fig. 15). Note that the missing grid points were likely due to defects in origami self-assembly or DNA synthesis (see Supplementary Note 9.1 for discussion), rather than imaging incompleteness. Single-particle analysis confirmed the completeness of the nano-pattern and showed a high degree of regularity of the grid geometry as designed (Fig. 3e, see online methods, Supplementary Fig. 16 and Method 6.2 for details). Specifically, we performed 2D Gaussian fitting on each grid point, followed by a regular grid fit to these 12 Gaussian-fitted centres, and observed that the average deviation between the Gaussian-fitted and regular grid-fitted centres was well below 1 nm (<0.30 nm r.m.s., Fig. 3f, see online methods, Supplementary Method 6.2 and Note 9.2 for details).
We then tested the performance of templated drift correction on the grid structures, following the analysis workflow in Fig. 3i (left arrow, see online methods and Supplementary Method 5.2 for details). Briefly, we first identified a pool of separable single-target drift markers, extracted their blinking time traces, and determined the expected centre position for each target. Then, for every frame, we collected all localisations originating from these drift markers and calculated a global, photon-weighted average of “offset vectors” from each localisation to its expected centre position, and used it as the drift correction vector. Single-particle analysis after templated drift correction showed a sharper image of the grid (Fig. 3g). Furthermore, the superior regularity of these 20 nm grid structures allowed us to perform another round of geometry-templated drift correction, again using these structures as drift markers (Fig. 3i, second arrow, see online methods and Supplementary Method 5.2 for details), and producing an even sharper single-particle averaged image (Fig. 3h). We also compared the imaging resolution before and after templated and geometry-templated drift corrections, respectively, and estimated <1 nm (r.m.s.) residual drift after templated and geometry-templated corrections (Fig. 3j, see online methods and Supplementary Method 6.2 for details).
Note that, during templated and geometry-templated drift correction processes, the global average of all offset vectors calculated from many origami grids across the entire field-of-view (rather than those from a single origami grid) was used for drift correction. In addition, although here we treat the 20 nm grid as both drift markers and imaging samples, in general applications (such as the 5 nm grid image in the next section) no prior knowledge of the imaging sample is required, only that of the origami grid markers is used.
5 nm grid DMI and analysis
To finally demonstrate the imaging capability of DMI, we designed a triangular grid structure with ~5 nm point-to-point spacing (Fig. 4a). This is the densest clustering pattern possible on our origami breadboard34, and it also mimics the monomer spacing and arrangement in a microtubule segment (~5 nm × 4 nm monomer size)35.
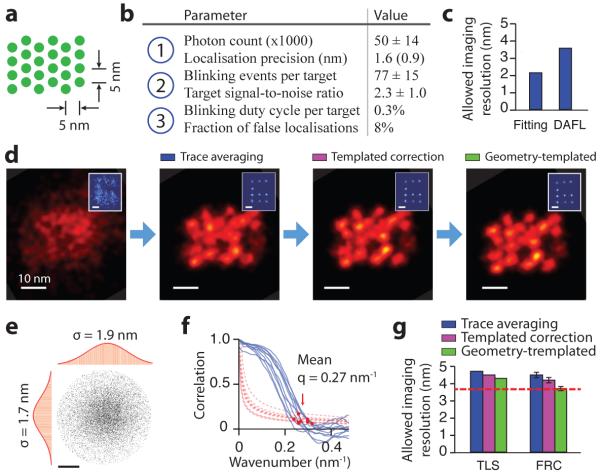
Figure 4. Systematic quality analysis of 5 nm grid super-resolution image.
(a) Design schematics of a 4 × 6 triangular grid structure with ~5 nm point-to-point spacing on a DNA origami nanostructure. Each green dot indicates a docking strand.
(b) Critical imaging quality parameters for the three blinking requirements. Localisation precision value in brackets was measured by single-molecule fitting uncertainty.
(c) Allowable imaging resolution assayed by two methods before drift correction, single-molecule fitting uncertainty (Fitting) and distance between adjacent-frame localisations (DAFL), both estimated in FWHM.
(d) Comparison of DNA-PAINT images of a 5 nm grid structure and a 20 nm grid drift marker (blue, inset) at different stages of drift correction.
(e, f) Measured imaging resolution assayed by two methods after drift correction.
(e) Target localisation spread (TLS). The point cloud shows overlapped localisations from individually separable targets and aligned by centre of mass. Histograms are shown for horizontal (left) and vertical (top) projections. Red curves indicate Gaussian fit.
(f) Fourier ring correlation (FRC). Correlation curves (blue, solid lines) and noise-based cutoff (red, dotted lines) are shown for 10 representative images; red dots indicate crossing points.
(g) Comparison of measured imaging resolution at different stages of drift correction, assayed by TLS and FRC. Red dashed line indicates localisation precision-limited best allowable resolution (as determined by DAFL).
DNA-PAINT imaging conditions used for this experiment: 400 ms frame time, 40,000 total frames, and 1 nM imager strand concentration. See online methods, Supplementary Figures S15-S19 and Methods S3, S5 for more details on assay methods and results. Scale bars, 10 nm in images, 20 nm in insets in (d), 2 nm in (e).
Compared to the 10 nm grid images (Fig. 2b-d), the higher target density in this sample imposes more stringent imaging conditions, namely, even higher photon count, larger number of blinking events per target, and lower blinking duty cycle. We used a short (7 nt) docking strand to accommodate the high target density and avoid potential spatial cross-talk between neighbouring targets, and carefully adjusted the imaging conditions to satisfy all three blinking requirements (Fig. 4b), especially with high single-molecule localisation precision (1.6 nm by DAFL, <1.0 nm from single-molecule fitting, 1.5 nm by theoretical estimate15, Fig. 4c). See Supplementary Figs. 17-21 for details on imaging quality characterisation.
We used the 20 nm grid structures as drift markers. After applying each step of the drift correction procedure (Fig. 3i), both the 20 nm grid drift markers (Fig. 4d, insets) and 5 nm grid samples (Fig. 4d) became increasingly sharper and more regular. We employed two methods for assaying integral imaging quality: target localisation spread (or TLS, by overlaying localisation clouds from well-separated targets and measuring the spread of the overlaid cloud with 2D Gaussian fitting, Fig. 4e), and Fourier ring correlation (FRC, by computing the correlation between 2D Fourier transform spectra of independent half images, Fig. 4f). A comparison of maximal allowable resolution is shown in Fig. 4g, with 4.3 nm (by TLS) and 3.7 nm (by FRC) after geometry-templated drift correction, which allowed us to estimate the residual drift to be <1 nm (r.m.s.). See online methods for details on image quality characterisation.
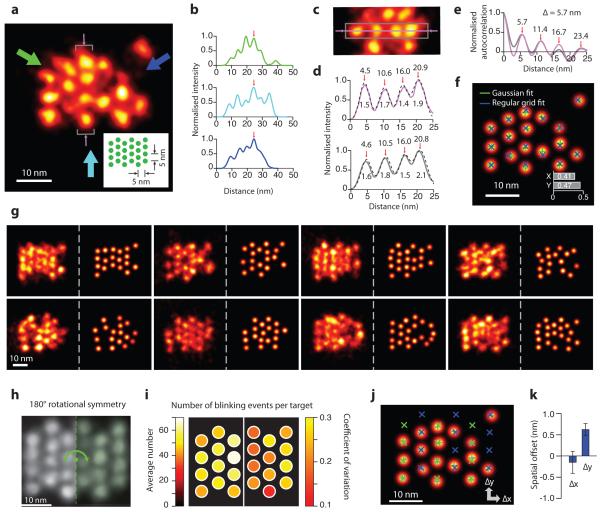
Plotting the projection profiles along the three symmetry axes of the triangular grid structure further confirmed DMI imaging quality and the structural regularity of nanostructures on a representative single-molecule image of a 5 nm grid (Fig. 5a,b). Variations in the peak intensities resulted from missing grid points, which we note again, were likely due to nanostructure synthesis defects (see Supplementary Note 9.1 for more details). A projection histogram of the four targets in a central column and auto-correlation analysis showed an averaged 1.7 nm target standard deviation (equivalent to 4.0 nm resolution in FWHM, Fig. 5c,d) and 5.7 nm inter-target spacing, consistent with the design (Fig. 5e). We next performed automatic target detection and 2D Gaussian fitting to each target, followed by regular grid fitting to the Gaussian-fitted centres (Fig. 5f, see online methods and Supplementary Method 6.2 for details). The average deviation between the Gaussian-fitted and grid-fitted centres was measured to be below 1 nm (single-target localisation precision 0.18 nm, Fig. 5f).
Figure 5. Discrete molecular imaging of 5 nm grid structure.
(a) Representative DMI image of a 5 nm triangular grid structure obtained with DNA-PAINT. Inset shows design schematics, where each green dot indicates a docking strand. Arrows indicate projection directions and areas of study for panels (b-e). Missing grid points were likely due to synthesis or incorporation defects (see Supplementary Notes S9.1 for more discussions).
(b) Intensity projection profiles from the image in (a), along the directions indicated by colour-matched arrows. Profiles are aligned by central peaks indicated by red arrows.
(c) Cropped-out image from (a), showing central region (grey rectangle) and central pixel line (magenta line and arrows) used for analysis in (d, e), also marked by grey brackets and thin magenta arrows in (a).
(d) Intensity profile along the central line (magenta), and projection from the central region (grey), as indicated by colour-matched regions in (c), and four-peak Gaussian fit for both (black, dashed lines). Numbers indicate fitted centre positions and standard deviation values for each peak, with an average of 1.7 nm, supporting a 4.0 nm FWHM resolution.
(e) Auto-correlation analysis from colour-matched profiles in (d), showing consistent periodicity of 5.7 nm.
(f) Automatic multi-target fit of the 5 nm grid image in (a). Overlaid crosses indicate Gaussian-fitted centres (green) and regular grid-fitted centres using the green crosses as targets (blue). Inset shows r.m.s. deviation between the green and blue crosses (<0.5 nm in 1D and <0.7 nm in 2D).
(g) More representative images of the 5 nm grid structures, showing structural regularity and heterogeneity. For each structure, left panel shows super-resolution rendered image, right panel shows automatic fitted image.
(h) Single-particle class average of the 5 nm grid (N = 25). Green dashed line and arrow indicate symmetry axis and operation of the structure.
(i) Uniformity of blinking kinetics, as represented on a 5 nm degenerate grid. colour maps show averages (left) and coefficients of variation (right) of the number of blinking events for each distinguishable target.
(j) Automatic multi-target fit (grey) and two-component grid fit of 5 nm image in (a), allowing an offset between two groups of targets with opposite staple strand orientations, coloured in green and blue respectively.
(k) Offsets between the two groups of staples in (j) measured from single-molecule images, error bars indicate standard deviation (N = 10).
It is important to note that no prior knowledge of the sample structure (the 5 nm grid) was used to produce the above results. DNA-PAINT imaging condition used for this experiment: 400 ms frame time, 40,000 total frames, and 1 nM imager strand concentration. See Supplementary Figures S15, S20-S22 and Methods S5, S6 for super-resolution images and analysis details, and Supplementary Notes S9 for discussions. Scale bars: 10 nm in all panels.
Each imaging session produced ~50-100 single-molecule images of well-resolved 5 nm grids (Fig. 5g). We next performed single-particle averaging on them, and studied single-molecule imaging heterogeneity among them on the degenerate (half) grid (due to the 180° rotational symmetry, see Fig. 5h, Supplementary Fig. 16 and Note 9.2). Specifically, we computed the coefficient of variation in the number of blinking events across different targets in the degenerate grid (0.07), and across different single-molecule images at the same target (0.20). These results demonstrated a relatively uniform imaging efficiency (Fig. 5i, see Supplementary Fig. 22 and Method 6.2 for details), which further supports the hypothesis that the missing grid points likely resulted from strand synthesis or incorporation defects, rather than non-uniform imaging efficiency (see Supplementary Note 9.1 for details).
We further analysed the origami staple wiring pattern on single-molecule DMI images with angstrom-level precision (<2 Å). By performing an automatic two-component grid fitting algorithm on single-molecule images (Fig. 5j), we determined the structural offset between two groups of staples with opposing orientations (0.6 ± 0.1 nm, Fig. 5k), consistent with structural analysis prediction (0.6 nm, see Supplementary Fig. 23 for details).
DMI in complex and multiplexed samples
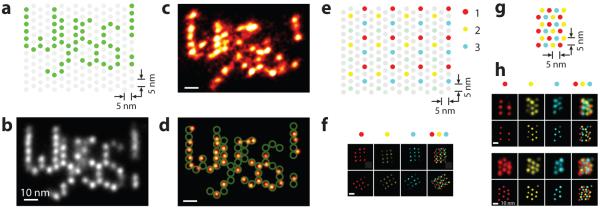
We further demonstrated DMI in a complex arrangement, by constructing and visualising a custom-designed letter pattern (“Wyss!”) on the 60 nm × 85 nm origami nano-display breadboard, with 5 nm display pixel size (Fig. 6a, Supplementary Fig. 24). Single-particle class average showed an average resolution of 4.6 nm in FWHM, allowing visualisation of individual targets (Fig. 6b). A representative single-molecule super-resolution image (Fig. 6c) displays individually distinguishable targets arranged in the designed pattern (Fig. 6d).
Figure 6. Discrete molecular imaging with complex patterns and multiplexed visualisation.
(a-d) DMI of a five-character pattern “Wyss!” on a DNA origami nano-display board with 5 nm pixel size.
(a) Design schematics. Each dot indicates a staple strand. Green dots were extended with DNA-PAINT docking strands.
(b) Single-particle class average of the “Wyss!” pattern (N = 85).
(c) Representative single-molecule image of the “Wyss!” pattern under DMI.
(d) Overlay of the design schematics on top of automatically fitted single-molecule image in (c).
(e-h) Three-colour multiplexed DMI, each colour indicates a separate imaging channel with an orthogonal DNA-PAINT sequence.
(e) Design schematics of a three-colour dual-purpose drift and alignment marker.
(f) Cross-channel alignment. Thee single-channel images (left three columns) and one composite image (rightmost column) are shown for two example alignment markers.
(g) Design schematics of a three-colour 5 nm grid structure.
(h) Representative multiplexed DMI image of three-colour 5 nm grid pattern as in (g). DNA-PAINT super-resolution images (top row) and automatically fitted image (bottom row) are shown for all three single-colour channels (left three columns) and the combine image (rightmost column), for two representative 5 nm grid structures.
DNA-PAINT imaging conditions used in these experiments are as follows. “Wyss!” letter pattern image: 500 ms frame time, 100,000 total frames, and 0.4 nM imager strand concentration. Multi-colour pattern image: 400 ms frame time, 2-3 nM imager strand concentration, 20,000 total frames for each colour channel. See Supplementary Figures S23, S24 for more super-resolution images, online methods and Supplementary Methods S5, S6 for image analysis methods. Scale bars: 10 nm in (b-d), 20 nm in (f), and 10 nm in (h).
Multiplexed DMI further requires accurate registration between multiple imaging channels. For example, to achieve 5 nm imaging resolution, <1 nm registration accuracy is required. Traditional multi-target imaging and co-localisation studies on molecular scale rely on registration between multiple spectral channels, and either (1) require specialised and complicated hardware setup and calibration process, and hence could be practically difficult to extend beyond two-colour registration or to the entire field of view, or (2) achieve suboptimal registration accuracy that does not allow molecular resolution imaging21, 36.
We combined DMI with Exchange-PAINT10 and demonstrated highly-accurate (<1 nm) “three-colour” registration, in addition to highly-accurate drift correction (<1 nm r.m.s.) within each channel. The method uses only one fluorophore and one optical path, thus greatly simplifies cross-channel registration, and naturally extends to the entire field of view. To implement this method, we first designed a three-colour nano-grid dual-purpose drift and alignment marker, where each colour comprises a regular square grid with 20 nm lattice spacing, similar to the 20 nm grid used above (Fig. 6e). We designed three orthogonal imager sequences with optimised binding on-time, and labelled the substructure in each colour with a unique sequence. We performed multiplexed DMI imaging through serial buffer exchange (Exchange-PAINT), and then performed drift correction using these dual-purpose markers for each channel individually, followed by cross-channel alignment between each pair of different colours (Fig. 6f, see online methods, Supplementary Fig. 25 and Method 6.2 for details). The high-precision visualisation of individual targets (down to angstrom level, as shown above) allows highly-accurate alignment across all channels (<1 nm), enabling DMI imaging (Fig. 6f).
We then imaged a three-colour mixture structure of the 5 nm grid with multiplexed DMI, with average DAFL localisation precision 2.0 nm, and TLS resolution 5.9 nm in FWHM (Fig. 6g,h, Supplementary Fig. 25). Automatic target fitting to single-molecule structures revealed the regular grid pattern as designed.
Conclusion
Discrete molecular imaging (DMI) contrasts to current super-resolution fluorescence microscopy techniques in a similar way as digital to analogue signal processing. Whereas current super-resolution demonstrations typically focus on depicting continuous morphology of macromolecular structures and biomolecule spatial distributions (analogue)22, 26, 37-39 and have sampling density-limited resolution (Nyquist sampling criterion), DMI imaging aims at discrete visualisation of each individual molecular components within these structures. Combined with the high multiplexing power of Exchange-PAINT, DMI could potentially enable determination of the position and identity of each molecular component in a complex biological or synthetic nanoscale system, and thus offers a complementary method to electron microscopy and crystallography with single-molecule sensitivity. Furthermore, the angstrom-level precision (<2 Å) structural study on DNA nanostructures suggests that DMI could be applicable to studying sub-molecular and even potentially atomic level features (e.g. macromolecular composition and component positions, geometric distortions and conformational changes).
We see two challenges in the further development of DMI. The first comes from the physical tradeoff between spatial and temporal resolution13,26. Specifically, achieving higher spatial resolution requires longer blinking on-time, larger number of blinking events, and lower blinking on-off duty cycle, all of which necessarily increases the total image acquisition time. Potential ways to shorten the imaging time include engineering brighter fluorophores to shorten the blinking on-time required to collect enough photons. The second challenge is from imperfect labeling of biomolecular targets. Although our method is not limited by Nyquist sampling criterion (number of probes labelled per unit space), it is still limited by the absolute labeling efficiency (average number of probes labelled per molecular target). Conventional immunostaining method with IgG antibodies both introduces a large offset from the target epitope to the chemically conjugated probe (~10 nm) and often has limited labeling efficiency (due to the large size and sometimes the low affinity of antibodies). Several approaches could potentially address these challenges: genetically labelled tags (e.g. SNAP-tag, unnatural amino acids) can provide smaller probe size and higher labeling efficiency; aptamers, small-molecule labels, single-chain antibody fragments and camelid single-chain antibodies (nanobodies) could also provide effective alternatives of smaller affinity probes.
DMI allows direct visualisation of each individual component in densely packed biomolecular environment. In the future, we expect the development of DMI to eventually allow studying quantitative molecular features in diverse biological systems, such as (1) molecular composition and architecture of diverse cellular systems (e.g. cell membrane receptor clusters, neuronal synapses), (2) molecular states of individual protein components within macromolecular context (e.g. binding and rotational states of ring ATPase complexes, patterns of histone modifications), and (3) 3D architecture of chromosomes with high spatial and genomic resolution.
Supplementary Material
Acknowledgements
We thank M. Avendaño, J. Wöhrstein, F. Schüder, G. Church, W. Shih, D. Zhang, S. Agasti, E. Winfree, P. Rothemund for helpful discussions, and C. Myhrvold, N. Liu, W. Sun, X. Chen, F. Vigneault, and H. Tang for helpful comments on the manuscript.
This work is supported by a National Institutes of Health (NIH) Director's New Innovator Award (1DP2OD007292), an NIH Transformative Research Award (1R01EB018659), an NIH grant (5R21HD072481), an Office of Naval Research (ONR) Young Investigator Program Award (N000141110914), ONR grants (N000141010827 and N000141310593), a National Science Foundation (NSF) Faculty Early Career Development Award (CCF1054898), an NSF grant (CCF1162459) and a Wyss Institute for Biologically Engineering Faculty Startup Fund to P.Y.. M.D. acknowledges support from the HHMI International Predoctoral Fellowship. R.J. acknowledges support from the Alexander von Humboldt-Foundation through a Feodor-Lynen Fellowship.
Footnotes
Author contribution
M.D. conceived of and designed the study, designed and performed the experiments, developed the software, analysed the data, and wrote the manuscript. R.J. conceived of and designed the study, interpreted the data, and critiqued the manuscript. P.Y. conceived of, designed and supervised the study, interpreted the data and wrote the manuscript. All authors reviewed and approved the manuscript.
Competing financial interest
The authors declare conflict of interests, and have filed a patent application. P.Y. and R.J. are co-founders of Ultivue, Inc., a startup company with interest to commercialise the reported technology.
References
- 1.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Optics Letters. 1994;19 doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 2.Klar TA, Hell SW. Subdiffraction resolution in far-field fluorescence microscopy. Optics letters. 1999;24:954–956. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- 3.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Journal of microscopy. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 4.Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science (New York, N.Y.) 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical journal. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angewandte Chemie (International ed. in English) 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 9.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nature methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hell SW. Microscopy and its focal switch. Nature methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- 12.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annual review of biochemistry. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahl SJ, Moerner WE. Super-resolution fluorescence imaging with single molecules. Current opinion in structural biology. 2013;23:778–787. doi: 10.1016/j.sbi.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moerner WE, Kador L. Optical detection and spectroscopy of single molecules in a solid. Physical Review Letters. 1989;62:2535–2538. doi: 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- 15.Russell ET, Daniel RL, Watt WW. Precise nanometer localization analysis for individual fluorescent probes. Biophysical journal. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Löschberger A, et al. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. Journal of cell science. 2012;125:570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- 17.Szymborska A, et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science (New York, N.Y.) 2013;341:655–658. doi: 10.1126/science.1240672. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz A, et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science (New York, N.Y.) 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 19.Rittweger E, Han K, Irvine SE, Eggeling C, Hell SW. STED microscopy reveals crystal colour centres with nanometric resolution. Nature Photonics. 2009;3 [Google Scholar]
- 20.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pertsinidis A, Zhang Y, Chu S. Subnanometre single-molecule localization, registration and distance measurements. Nature. 2010;466:647–651. doi: 10.1038/nature09163. [DOI] [PubMed] [Google Scholar]
- 22.Vaughan JC, Jia S, Zhuang X. Ultrabright photoactivatable fluorophores created by reductive caging. Nature methods. 2012;9:1181–1184. doi: 10.1038/nmeth.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mario R, Jürgen JS, Ija J, Carsten F, Philip T. Fluorescence microscopy with 6 nm resolution on DNA origami. Chemphyschem : a European journal of chemical physics and physical chemistry. 2014;15:2431–2435. doi: 10.1002/cphc.201402179. [DOI] [PubMed] [Google Scholar]
- 24.Charlotte K, et al. Absolute Arrangement of Subunits in Cytoskeletal Septin Filaments in Cells Measured by Fluorescence Microscopy. Nano letters. 2015 doi: 10.1021/acs.nanolett.5b00693. [DOI] [PubMed] [Google Scholar]
- 25.Fernández-Suárez M, Chen TS, Ting AY. Protein-protein interaction detection in vitro and in cells by proximity biotinylation. Journal of the American Chemical Society. 2008;130:9251–9253. doi: 10.1021/ja801445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nature methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nature methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nature methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannone G, et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophysical journal. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-activated localization microscopy of DNA structures. Nano letters. 2011;11:4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- 31.Jungmann R, et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano letters. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 32.Johnson-Buck A, et al. Super-resolution fingerprinting detects chemical reactions and idiosyncrasies of single DNA pegboards. Nano letters. 2013;13:728–733. doi: 10.1021/nl304415b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iinuma R, et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science (New York, N.Y.) 2014;344:65–69. doi: 10.1126/science.1250944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul WR. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 35.Gregory MA, et al. High-Resolution Microtubule Structures Reveal the Structural Transitions in αβ-Tubulin upon GTP Hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertsinidis A, et al. Ultrahigh-resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20. doi: 10.1073/pnas.1310654110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andriy C, et al. Nanoscopy with more than 100,000 'doughnuts'. Nature Methods. 2013 doi: 10.1038/nmeth.2556. [DOI] [PubMed] [Google Scholar]
- 38.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science (New York, N.Y.) 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong L, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349 doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.