Abstract
Background
The incidence and outcomes for patients with colorectal cancer (CRC) varies by age. Younger patients tend to have sporadic cancers not detected by screening and worse survival. To understand if genetic differences exist between age cohorts we sought to characterize unique genetic alterations in patients with CRC.
Methods
We identified 283 patients with sporadic CRC between 1998 and 2010 and divided them by age into two cohorts: ≤45 years old (younger) or ≥65 years old (older) and performed targeted exome sequencing. Fisher’s Exact test was used to detect differences in mutation frequencies between the two groups. Whole exome sequencing was performed on 21 additional younger patient samples for validation. Findings were confirmed in The Cancer Genome Atlas CRC dataset.
Results
246 samples were included for final analysis (195 older, 51 younger). Mutations in FBXW7 were more common in the younger cohort (27.5% vs. 9.7%, p=0.0022) as were mutations in the proofreading domain of POLE (9.8% vs. 1.0%, p= 0.0048). There were similar mutation rates between cohorts with regards to TP53 (64.7% vs. 61.5%), KRAS (43.1% vs. 46.2%), and APC (60.8% vs 73.8%). BRAF mutations were numerically more common in the older cohort, though not statistically significant (2.0% vs 9.7%, p=0.082).
Conclusions
In this retrospective study, we identified a unique genetic profile for younger CRC patients as compared to patients diagnosed at an older age. These findings should be validated in a larger study and could have an impact on future screening and treatment modalities for younger CRC patients.
Keywords: iomarkers, colorectal cancer, AYA
Condensed Abstract
In this manuscript, we describe our original research involving exome sequencing of patients with colorectal cancer in which we compared younger patients to older patients. We identified a unique genetic profile for younger patients which may have implications on future screening and treatment paradigms for these patients.
Introduction
CRC is a major cause of morbidity and mortality, both in the US and worldwide,1, 2 and is primarily a disease of older adults, with a median age at diagnosis of 65. Improvements in screening, increasingly effective therapies, and optimization of supportive care have contributed to the trend in improved survival for older patients with colorectal cancer (CRC).3, 4 Though the incidence of disease is declining in the over 50 population, data suggest that the cancer incidence is actually rising in younger patients.5, 6 Younger patients have worse outcomes when matched stage for stage, but also overall for multifactorial reasons such as higher stage at presentation, higher rate of mucinous histology, and intrinsic biologic differences in their disease.7–11 In addition, younger patients with CRC have not seen a significant improvement in survival over the past three decades.4, 12 One possible explanation for this discrepancy is that cancer in this younger cohort of CRC patients is biologically distinct from older adults and to date has not been the focus of large research initiatives.4 Though there are several genetic syndromes that predispose younger patients to CRC, the majority of CRC in younger patients is sporadic.13
Elucidating the reasons behind these disparities in outcome and survival improvement may reveal opportunities to improve cancer care for adolescents and young adults. Preliminary research has suggested that cancer incidence and outcome differences in young adults could relate to fundamental age-related differences in tumor biology.7, 8, 14–19
Recent efforts in molecular profiling have allowed great strides to be made in understanding underlying tumor pathophysiology.20, 21 We performed molecular characterization of tumors from younger and older sporadic CRC patient cohorts in an effort to determine potentially distinct genetic signatures. We hypothesized that CRC in young patients would harbor biologically distinct alterations and that these differences could ultimately play a role in developing personalized screening and management strategies.
Materials and Methods
Institutional Review Board approval was obtained to conduct this multi-institutional, observational study. Tumor samples from fresh frozen surgical biopsy specimens collected from 468 colon and rectal cancer patients between 1998 and 2010 at Moffitt Cancer Center and affiliated community hospitals as part of the Moffitt Total Cancer Care™ (TCC) protocol.22 Tissue is collected at the time of surgery or biopsy and all patients with cancer are eligible for inclusion.
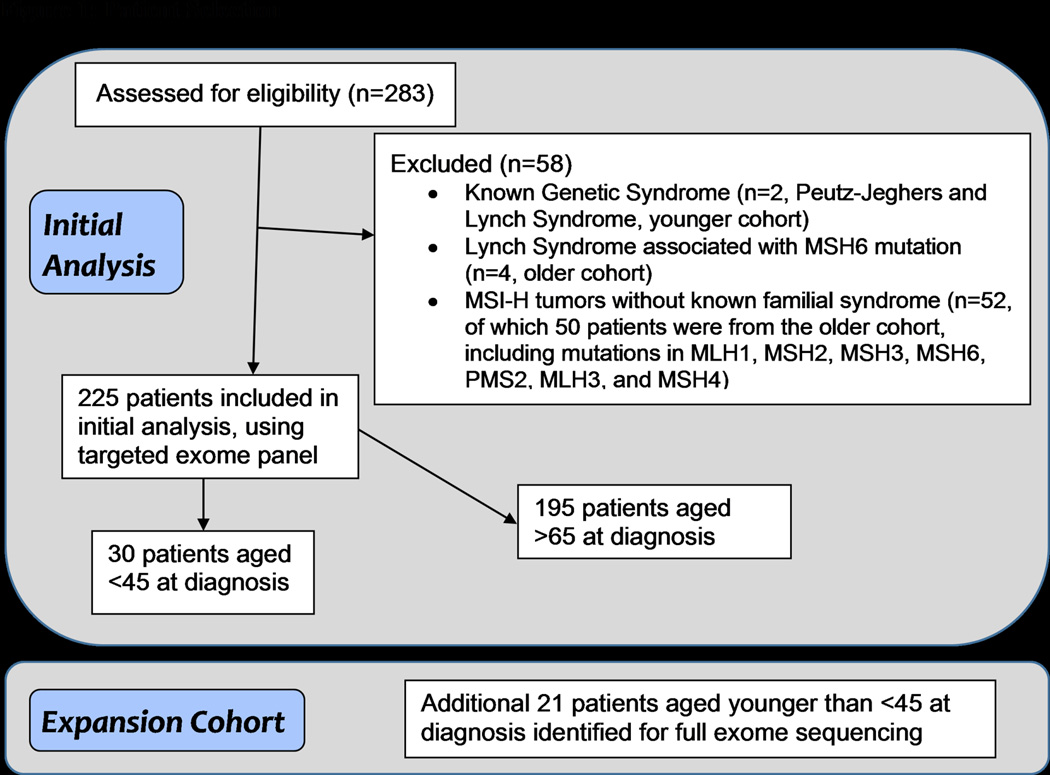
Our initial analysis included 283 patients. Following initial observations, we refined the cohort to exclude patients with known familial cancer syndromes as well as MSI high (MSI-H) tumors in order to minimize bias and capture those patients who truly had sporadic tumors. Out of the initial 283 patients, 58 were excluded for familial cancer syndromes or having MSI-H tumors (see Figure 1). Out of these 58 patients, 2 patients from the young cohort had a known genetic syndrome, 4 patients from the older cohort had Lynch syndrome associated with an MSH6 mutation, and 52 patients without a known familial syndrome had MSI-H tumors (50 from the older cohort and 2 from the younger cohort). Of the 225 patients remaining, 195 patients were 65 or older and 30 patients were 45 or younger at diagnosis.
Figure 1. Patient selection criteria.
Experimental design highlighting patient selection criteria in discovery and expansion cohorts.
MSI-H: Microsatellite instability; MLH1: MutL homolog 1 protein; MLH3: MutL homolog 3 protein; MSH2: MutS protein homolog 2; MSH3: MutS protein homolog 3; MSH4: MutS protein homolog 4; MSH6: MutS protein homolog 6; PMS2: Mismatch repair endonuclease 2
Tissue collection/analysis
The tumors were collected using a snap frozen technique in liquid nitrogen within 15–20 minutes of extirpation. Macrodissection was performed to ensure >80% tumor was present in the specimens that underwent sequence analysis. Normal tissue, necrotic tissue and excessive stromal tissues were dissected away from the specimen under frozen section control. DNA was then extracted for targeted gene sequencing which was performed by Beijing Genomic Institute (Beijing, China). 1,321 genes were targeted using the Agilent SureSelect technology (Agilent Technologies, Inc. Santa Clara CA), followed by 90 base pair, paired-end sequencing on GAIIx instruments (Illumina, Inc. San Diego, CA) to achieve 138× average depth of coverage across the target region.
Burrows-Wheeler Aligner (BWA23) was used to align sequence reads to the human reference (hs37d5). The Genome Analysis Toolkit (GATK24) was used for insertion/deletion realignment, quality score recalibration, and identification of single nucleotide and insertion/deletion variants. Matched normal samples were generally not available for comparison to identify somatic mutations, so filtering of normal variants was performed using the 1000 Genomes Project dataset25 and an internal pool of 238 normal samples. Variants identified in 1000 Genomes with an MAF ≥ 0.01 or identified in the normal pool with MAF ≥ 0.05 were removed. ANNOVAR26 was used to annotate variants, and VarSifter27, in-house web-based display applications, and custom Perl and R scripts were used for analysis.
Mutation rate differences were assessed in two ways: mutation frequency at specific positions to focus on likely recurrent activating oncogenic mutations and entire genes when a truncating mutation was present to account for more diverse inactivating tumor suppressor mutations.
In the validation phase of this study, a focused search through TCC revealed an additional 21 CRC samples from patients diagnosed at age 45 or younger with sporadic disease. Retrospective chart review was performed to capture information on age, sex, gender, stage, pathology, and survival outcome.
These tumors underwent whole exome sequencing in order to identify somatic mutations in the coding regions of the human genome. Two micrograms of DNA was used as input into the Agilent SureSelect XT Clinical Research Exome kit, which includes the exon targets of Agilent’s v5 whole-exome kit, with increased coverage at 5000 disease-associated targets. For each tumor DNA sample, a genomic DNA library was constructed according to the manufacturer’s protocol and the size and quality of the library was evaluated using the Agilent BioAnalzyer. An equimolar amount of library DNA was used for a whole-exome enrichment using the Agilent capture baits and after quantitative PCR library quantitation and QC analysis on the BioAnalzyer, approximately 150 million 75-base paired-end sequences were generated using v2 chemistry on an Illumina NextSeq 500 sequencer. Average depth of coverage across the target regions was 116×. Data were analyzed as described for the original TCC cohort.
Candidate mutation differences were detected in the original cohort by testing for frequency differences using a two-sided Fisher’s exact test at the position level and at the truncating gene level (counting the presence of any truncating mutations in a gene). Marginally significant findings (p<0.05 uncorrected) in the discovery set were then retested including the results from 21 additional younger patients, resulting in an expanded cohort with 51 younger and 195 older patients. Multiple test correction was performed using the Benjamini and Hochberg method.28 Gene and mutations known to drive colorectal cancer were specifically tested to understand differences in colorectal cancer driver genes.
The Cancer Genome Atlas (TCGA) data were used for comparison and external validation.29 Sequencing data was downloaded for both colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ). Individuals with the clinical indicator “microsatellite instability” of YES were excluded, resulting in 29 young individuals and 220 old individuals. MAF files were converted to VCF, and re-annotated as described above. Differential mutation rates in specific genes, including known CRC driver genes and differentially mutated genes in our cohort were retested in this independent dataset. Lollipop plots were prepared using the cBioPortal MutationMapper tool.30
Results
283 CRC patient specimens underwent targeted gene sequencing covering 1,321 genes. We observed a marginally significant imbalance in MSI frequency between groups (younger= 6.2%, older = 20.7%, p=0.055). MSI patients as well as patients with familial syndromes were excluded from further analysis to avoid introduction of mutational bias potentially caused by this baseline difference. Our final discovery cohort of sporadic, mismatch repair sufficient CRC (30 younger, 195 older) was then analyzed to identify mutation differences between groups. Baseline patient characteristics of the expanded cohort are described in Table 1.
Table 1.
Baseline Characteristics
| Clinical Characteristics of patients ≥65 years | Number of patients 195 (%) | |
|---|---|---|
| Age at diagnosis | Median | 73 |
| Range | 65–93 | |
| Gender | Male | 95 (49%) |
| Stage at diagnosis | I | 29 (15%) |
| II | 53 (27%) | |
| III | 66 (34%) | |
| IV | 44 (23%) | |
| Unknown | 3 (1%) | |
| Clinical Characteristics of patients ≤45 years | Number of patients 51 (%) | |
|---|---|---|
| Age at diagnosis | Median | 42 |
| Range | 30–45 | |
| Gender | Male | 28 (55%) |
| Stage at diagnosis | I | 3 (6%) |
| II | 9 (18%) | |
| III | 16 (31%) | |
| IV | 23 (45%) | |
| Unknown | 0 | |
In the older cohort (age ≥65), the median age at diagnosis was 73 with a range from 65–93 and 49% were male. In the younger cohort (age ≤45, the median age at diagnosis was 42 with a range from 30–45 and 55% were male. There was a higher frequency of advanced stage cancers in the younger cohort.
Following a test for proportional differences we identified 27 candidate positions and 17 truncated genes differentially mutated between younger and older patients (Supplemental Table 1) (p<0.05 uncorrected). Mutated positions were found in many genes, including specific mutations in the recurrently mutated APC, KRAS, and TP53 genes. Top hits included a stop mutation in FBXW7 and a point mutation in MAP2K4 (both seen in 3/30 younger and 0/195 older patients). Both mutations are observed in published data of colorectal cancer (cBioPortal29). Differentially truncating genes included several known tumor suppressors, including WRN, FBXW7, ATR, and NF1.
To confirm these results, an additional 21 younger patient tumor samples were sequenced with whole exome sequencing, and analyzed and enriched for somatic mutations as described (Supplemental Table 2). 16 positions remained significant (p<0.05, uncorrected), including mutations observed in FBXW7 and MAP2K4. Well characterized mutations in KRAS, TP53, and APC were no longer significant with the increased sample size. A multiple test correction was applied using the Benjamini-Hochberg method and resulted in two significant mutations: a stopgain in FBXW7 (4/51 young, 0/195 old, q=0.042) and a non-frameshifting insertion in CBX4 (0/39 young, 30/151 old, q=0.042, total sample counts reduced as low read depth resulted in some instances of missing data.) A similar non-frameshifting insertion is seen in the Exome Variant Server (http://evs.gs.washington.edu/EVS/) at overall allele frequency of 5.8%, suggesting this variant may be inherited. 11 truncated genes remained significant (p<0.05, uncorrected) but only FBXW7 survived multiple test correction (6/51 young, 2/195 old, q=0.021).
Given the significant results observed in FBXW7 using both specific mutation and truncating gene approaches, we compared the overall gene mutation rate in the expanded cohort. FBXW7 was about 2.5-fold more frequently mutated in younger samples compared to older (14/51 = 27.5% in young, 19/195 = 9.7% in older, p=0.0022) (Table 2). Mutations in this gene included the stop gain mutation we observed as more frequently mutated in young patients, as well as other recurrent and rarer mutations. The most highly mutated positions in our cohort were R578X (4/51 = 7.8% in young, 0/195 = 0% in old; R658X in cBioPortal), R425C (2/51 = 3.9% in young, 3/195 = 1.5% in old; R505C in cBioPortal) and R385C (4/51 = 7.8% in young, 4/195 = 2.1% in old; R465C in cBioPortal). Although not always statistically significant, these recurrent positions were each more frequently mutated in young patients, contributing to a significantly higher overall mutation rate.
Table 2.
Mutation Frequencies in TCC and TCGA
| Gene | TCC Extended | TCGA | combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| younger n=51 |
older n=195 |
P-value | younger n=29 |
older n=220 |
P-value | younger n=80 |
older n=415 |
P-value | |
| APC | 60.8% | 73.8% | 0.66 | 51.7% | 73.6% | 0.027 | 57.5% | 73.7% | 0.0046 |
|
BRAF (V600E) |
2.0% | 5.6% | 0.47 | 0% | 8.2% | 0.24 | 1.3% | 7.0% | 0.043 |
| FBXW7 | 27.5% | 9.7% | 0.0022 | 20.7% | 11.4% | 0.22 | 25.0% | 10.6% | 0.0016 |
| KRAS | 43.1% | 46.1% | 0.33 | 41.4% | 33.6% | 0.41 | 42.5% | 39.5% | 0.62 |
|
POLE proofre ading |
9.8% | 1.0% | 0.0048 | 10.3% | 0.91% | 0.012 | 10% | 0.96% | 9.7e–5 |
| TP53 | 64.7% | 61.5% | 0.42 | 51.7% | 52.7% | 1 | 60% | 56.9% | 0.62 |
TCC: Total Cancer Care; TCGA: The Cancer Genome Atlas; APC: Adenomatous polyposis coli gene; BRAF: Proto-oncogene B-Raf; FBXW7: F-box/WD repeat containing protein 7; KRAS: Kirsten rat sarcoma viral oncogene homolog; POLE: Polymerase epsilon catalytic subunit; TP53: Tumor protein p53
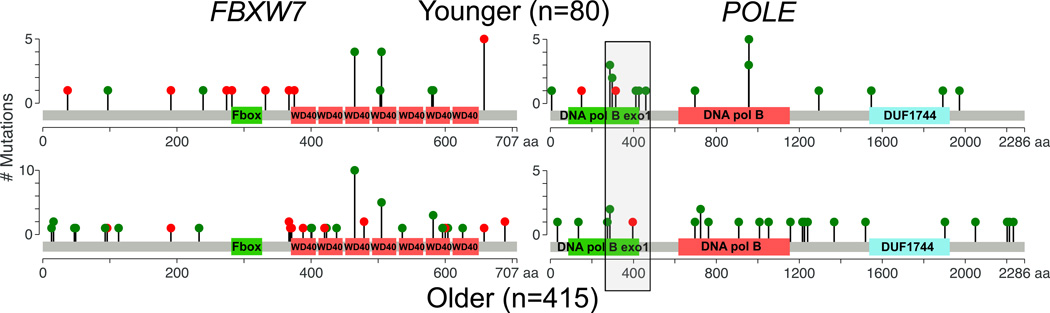
To confirm the observation of increased FBXW7 mutations in an independent dataset, we examined TCGA colon and rectal adenocarcinoma somatic mutation data. Although the number of young patients was much smaller than in our study, we observed increased mutation rates at positions and in genes we have observed in this study. FBXW7 R578X was observed 1/29 in young, and 1/220 in old (p=0.22). FBXW7 truncating mutations were significantly higher in young patients (5/29 young, 11/220 old, p=0.026). Overall FBXW7 mutation rate was higher in young patients, but was not significant: (6/29 = 20.7% in young, 25/220 = 11.4% in old, p=0.22) (Table 2). Combining the extended TCC data set with published TCGA data resulted in a significantly higher overall FBXW7 mutation rate in younger patients: 20/80 = 25.0% in young, 44/415 = 10.6% in old, p=0.0016 (Table 2). CBX4 H398delinsHH was not observed in TCGA somatic mutation data, further suggesting it was an inherited variant. Truncating and recurrent mutations observed in the combined cohort were spread across the gene, but R578X (R658X in cBioPortal) was much more common in younger patients (Figure 2).
Figure 2. Distribution of FBXW7 and POLE mutations in combined CRC cohorts.
Lollipop plots depict the number and location of somatic coding mutations in these two genes. Note that patients may have more than one mutation. Color indicates functional consequence of mutation: truncating (nonsense, frameshifting, splicing) (red) and missense (green). In both cohorts, FBXW7 mutations are spread throughout the protein and are enriched for truncating mutations, a typical pattern for tumor suppressor genes. The grey box overlapping POLE indicates the NCBI defined exonuclease domain used to define proofreading mutations. Younger patients have enrichment of mutations in the proofreading domain of POLE as compared to older patients.
Mutational landscape studies have identified a number of recurrently mutated genes.29 We specifically examined mutation frequencies between young and old patient tumors across eight of the most commonly mutated genes in CRC as well as FBXW7: APC, KRAS, TP53, BRAF, FBXW7, NRAS, PIK3CA, SMAD4, and TCF7L2. We observed a significantly different mutation rate in FBXW7, but not in any of the other genes (Supplemental Table 3). Specific positions in these genes showed potential significance (Supplemental Table 1a), but did not survive multiple test correction, even after expanding the number of younger patients (Supplemental Table 2a). Interestingly, an initial analysis performed before removal of MSI-H samples indicated BRAF V600E was more frequently observed in older patients (0/34 young, 46/248 old, p=0.0023 uncorrected). Additional mutations related to MSI-H status were also more frequently observed in older samples. The difference was no longer significant after removing MSI-H samples, but BRAF V600E was still more frequent in older patients in the expanded cohort (1/51 young, 11/195 old, p=0.47) (Table 2).
Mutation rates in these genes were generally not significantly different in the TCGA dataset (Supplemental Table 4). APC was significantly more mutated in older patients when considering truncating mutations (13/29 in young, 158/220 in old, p=0.0051 uncorrected) or all mutations (15/29 in young, 162/220 in old, p=0.027, Supplemental Table 5). As APC trended to higher mutation rates in older TCC patients, we combined datasets: 46/80 (57.5%) younger, 306/415 (73.7%), p=0.0046 (Table 2). BRAF V600E was observed in older patients even after removal of MSI-H cases, but not in young (0/29 young, 18/220 old, p=0.24) Combining counts from our expanded TCC cohort with the TCGA cohort results in a significantly lower incidence in young patients: 1/80 (1.3%) younger, 29/415 (7.2%) older, p=0.043 (Table 2).
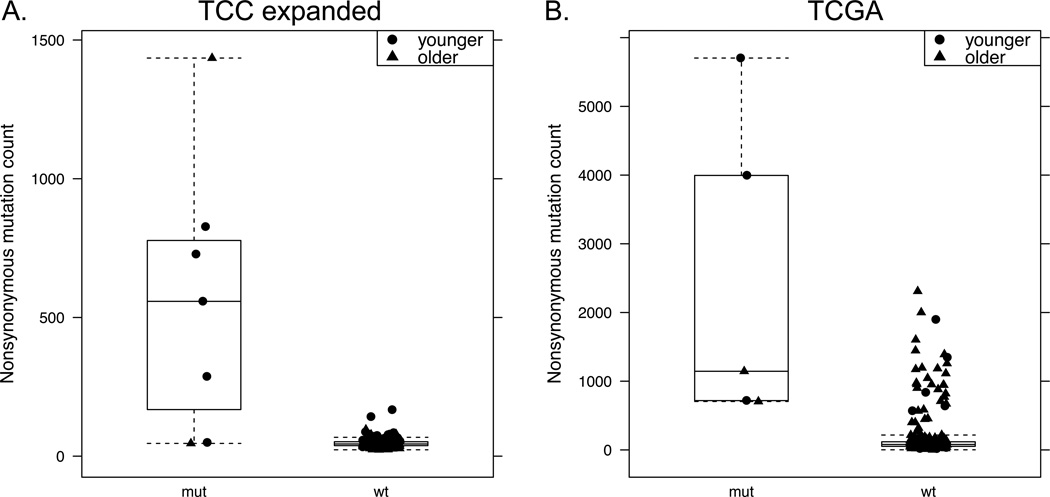
Recurrent polymerase episilon (POLE) mutations specific to the proofreading domain have been demonstrated in several different tumor types, including colorectal cancer.21, 31 We therefore examined our cohorts for mutational differences in the POLE proofreading domain, from amino acids 268–471.32 We observed such mutations in 5/51 young and 2/195 old patients (p-value 0.0048). Significantly increased proofreading domain mutation rates in young patients were also observed in TCGA colorectal data: 3/29 young, 2/220 old, p=0.012. Combining the datasets results in a significantly higher POLE proofreading domain mutation rate in younger patients: 8/80 (10%) younger, 4/415 (0.96 %) older, p=9.7×10−5 (Table 2, Figure 2). POLE proofreading domain mutations have previously been associated with high somatic mutation rates.33 We examined mutation counts in POLE mutated tumors in the TCC expanded and TCGA cohorts, and observed higher mutation counts in these samples within each cohort (Figure 3, Supplemental Table 5).
Figure 3. Mutation counts with POLE proofreading domain mutations.
Total nonsynonymous mutation counts are plotted by POLE proofreading domain mutation status in A) TCC discovery and expansion cohorts (1,321 genes sequenced) and B) TCGA whole exome sequencing. Younger patients are noted with circles, older patients with triangles. All data points are plotted for clarity.
Discussion
We have investigated differences in mutation rates between younger and older CRC patients that may contribute to variations in disease progression and outcome. While the overall genetic landscape of colorectal cancer is similar, we have shown that FBXW7 truncating mutations and POLE proofreading domain mutations are significantly more common in younger patients, with approximately 10-fold enrichment of each in this population.
FBXW7 is an F-box member of the Skp1-cullin-F-box (SCF) complexes responsible for phosphorylation dependent ubiquitination of specific proteins. Target substrates include cyclin-E, JUN, MYC, and others. The FBXW7 protein interacts with p53 and has been established as a tumor suppressor in several human cancers, including cholangiocarcinomas and endometrial cancers.34 Genetic alterations in the FBXW7 gene have been associated with poorer prognosis in CRC patients.35
Following differential mutation discovery, expansion, and comparison to independent external datasets, we found that FBXW7 is more frequently mutated in younger patients. The difference is seen in our patient samples when considering any mutation in the gene (27.5% in young, 10.7% in old) and when considering only truncating mutations (11.8% young, 1.0% old). Truncating mutations are significantly enriched in younger samples in the TCGA data as well. Future investigation of FBXW7 alterations may reveal contributions to clinical outcome (drug response, survival).
We also observed an increased rate of polymerase epsilon (POLE) proofreading domain mutations in younger patients. POLE mutations have been identified in highly mutated samples across cancer types. These mutations have been described in early onset CRC in individuals with no known familial syndrome.33 The tumors have been found to be microsatellite stable but hypermutated because of a tendency to develop base substitution mutations.
The recent advent of immunotherapy treatments in cancer could have clinical implications for POLE mutant patients. A link between high mutation rates and response to immune checkpoint inhibitors has been established.36 POLE mutations have been associated with increased neoantigen load, as well as increased immune cell infiltrate and expression of PD1 and PD-L1 in endometrial cancer, suggesting a potential therapeutic approach to these specific tumors.29, 37, 38 In a recent study of patients with advanced colorectal cancer, it was found that only patients with mismatch repair deficient tumors showed a response to an anti-PD1 agent.39 The mechanism by which this was hypothesized to occur was through higher tumor neoantigen load allowing for increased recognition by the immune system.
We specifically focused on POLE mutations in the proofreading domain, and observed significantly elevated mutation rates among young patients in our expanded cohort as well as the TCGA data set. These mutations are observed in young patients at around 10% frequency in both data sets versus only 1% in the older population. Furthermore, we observed that POLE proofreading domain mutations were associated with higher tumor mutational burden in both our cohort and TCGA datasets. Given the high rate of POLE mutation, the efficacy of immune checkpoint inhibitors merits specific study in these young CRC patients.
Several of the most common classically mutated genes in CRC, including KRAS and TP53, did not show a significant difference between cohorts in our discovery and expanded datasets, nor in the independent TCGA dataset, suggesting that there is molecular overlap among younger and older individuals with CRC. BRAF V600E mutations were more commonly observed in older patients. Although BRAF mutations are more associated with MSI-H CRC, this difference was observed in the context of MSS tumors. APC mutations were also somewhat more common in older patients. Although these differences were subtle, further study is warranted to confirm and understand such discrepancies.
Though there are overall similarities in commonly mutated genes between older and younger patients, distinct characteristics exist. Most strikingly, we noted significantly increased mutation rate in FBXW7 and in the POLE proofreading domain in younger patients. Further study is warranted to confirm our findings and to investigate the translational impact of FBXW7 and POLE in younger CRC patients, including the potential of immunotherapy to benefit a subset of younger patients.
Supplementary Material
Acknowledgments
Total Cancer Care® is enabled, in part, by the generous support of the DeBartolo Family, and we thank the many patients who so graciously provided data and tissue to the Total Cancer Care Consortium. Our study also received valuable assistance from the Cancer Informatics, Collaborative Data Services, and Tissue Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center, supported under NIH grant P30-CA76292. Funding was also provided by the Lewis Family Cancer Fund and the Gonzmart Family Foundation.
Footnotes
Conflict of Interest Disclosures: The authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr. Teer reports personal fees from Philips, outside the submitted work. In addition, Dr. Teer has a patent pending for a large data storage model. The other authors have nothing to disclose.
Author Contributions:
Conceptualization: Kothari, Teer, Abbott, Kim, Reed, Shibata.
Methodology: Teer, Reed, Shibata.
Software: Teer, Yoder, Zhang.
Validation: Teer, Yoder.
Formal analysis: Brohl.
Investigation: Kothari, Teer, Abbott, Srikumar, Yoder, Zhang.
Resources: Shibata.
Data Curation: Kothari, Teer, Abbott, Srikumar.
Writing – original draft: Kothari, Teer, Reed.
Writing – review and editing: Kothari, Teer, Abbott, Brohl, Kim, Reed, Shibata.
Visualization: Teer, Abbott, Srikumar, Reed.
Supervision: Kim, Reed, Shibata.
Project Administration: Yoder, Reed, Shibata.
Funding Acquisition: Reed, Shibata.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Edwards BK, Noone AM, Mariotto AB, et al. Annual Report to the Nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2013 doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albritton KCM, Anderson B, Nichols C, Ulman D Adolescent and Young Adult Oncology Progress Review Group. Closing the gap: research and care imperatives for adolescents and young adults with cancer. National Cancer Institute. 2006:1–108. [Google Scholar]
- 5.Davis DM, Marcet JE, Frattini JC, Prather AD, Mateka JJ, Nfonsam VN. Is it time to lower the recommended screening age for colorectal cancer? Journal of the American College of Surgeons. 2011;213:352–361. doi: 10.1016/j.jamcollsurg.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 6.Bailey CE, Hu C-Y, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA surgery. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleyer A, Barr R, Hayes-Lattin B, Thomas D, Ellis C, Anderson B. The distinctive biology of cancer in adolescents and young adults. Nature reviews. Cancer. 2008;8:288–298. doi: 10.1038/nrc2349. [DOI] [PubMed] [Google Scholar]
- 8.Tricoli JV, Seibel NL, Blair DG, Albritton K, Hayes-Lattin B. Unique characteristics of adolescent and young adult acute lymphoblastic leukemia, breast cancer, and colon cancer. Journal of the National Cancer Institute. 2011;103:628–635. doi: 10.1093/jnci/djr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zbuk K, Sidebotham EL, Bleyer A, La Quaglia MP. Colorectal cancer in young adults. Seminars in oncology. 2009;36:439–450. doi: 10.1053/j.seminoncol.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Furman WL, Tricoli JV. Important considerations in treating children, adolescents and young adults with colorectal carcinoma. Colorectal Cancer. 2013;2:347–358. [Google Scholar]
- 11.Saab R, Furman WL. Epidemiology and management options for colorectal cancer in children. Pediatric Drugs. 2008;10:177–192. doi: 10.2165/00148581-200810030-00006. [DOI] [PubMed] [Google Scholar]
- 12.Bleyer A, Barr R. Introduction--impact of malignant diseases on young adults II. Seminars in oncology. 2009;36:380. doi: 10.1053/j.seminoncol.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Dozois EJ, Boardman LA, Suwanthanma W, et al. Young-onset colorectal cancer in patients with no known genetic predisposition: can we increase early recognition and improve outcome? Medicine. 2008;87:259. doi: 10.1097/MD.0b013e3181881354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood WA, Lee SJ. Malignant hematologic diseases in adolescents and young adults. Blood. 2011;117:5803–5815. doi: 10.1182/blood-2010-12-283093. [DOI] [PubMed] [Google Scholar]
- 15.Mullighan CG, Willman CL. Advances in the Biology of Acute Lymphoblastic Leukemia-From Genomics to the Clinic. Journal of adolescent and young adult oncology. 2011;1:77–86. doi: 10.1089/jayao.2011.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gramatges MM, Rabin KR. The adolescent and young adult with cancer: state of the art-- acute leukemias. Curr Oncol Rep. 2013;15:317–324. doi: 10.1007/s11912-013-0325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard JM, Grothey A. Adolescent and young adult colorectal cancer. J Natl Compr Canc Netw. 2013;11:1219–1225. doi: 10.6004/jnccn.2013.0144. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RH, Chien FL, Bleyer A. Incidence of breast cancer with distant involvement among women in the United States, 1976 to 2009. JAMA. 2013;309:800–805. doi: 10.1001/jama.2013.776. [DOI] [PubMed] [Google Scholar]
- 19.Keegan TH, Press DJ, Tao L, et al. Impact of breast cancer subtypes on 3-year survival among adolescent and young adult women. Breast Cancer Res. 2013;15:R95. doi: 10.1186/bcr3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fearon ER. Molecular genetics of colorectal cancer. Annual Review of Pathology: Mechanisms of Disease. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 21.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer journal (Sudbury, Mass.) 2011;17:528. doi: 10.1097/PPO.0b013e318238216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Autosomes Chromosome X. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:1. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: Visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995:289–300. [Google Scholar]
- 29.Institute NCINHGR. The Cancer Genome Atlas. [accessed 25 January, 2016]; Available from URL: https://tcga-data.nci.nih.gov/tcga/
- 30.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Church DN, Stelloo E, Nout RA, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. Journal of the National Cancer Institute. 2015;107:dju402. doi: 10.1093/jnci/dju402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Church DN, Briggs SE, Palles C, et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Human molecular genetics. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palles C, Cazier J-B, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nature genetics. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akhoondi S, Sun D, von der Lehr N, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer research. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 35.Iwatsuki M, Mimori K, Ishii H, et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. International Journal of Cancer. 2010;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- 36.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 37.van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE proofreading mutations elicit an anti-tumor immune response in endometrial cancer. Clinical Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-15-0057. clincanres.0057.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howitt BE, Shukla SA, Sholl LM, et al. Association of polymerase e–mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA oncology. 2015;1:1319–1323. doi: 10.1001/jamaoncol.2015.2151. [DOI] [PubMed] [Google Scholar]
- 39.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. New England Journal of Medicine. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.