Abstract
A laser-based hydrogel degradation technique was developed that allowed for local control over hydrogel porosity; fabrication of 3D vascular-derived, biomimetic, hydrogel-embedded microfluidic networks; and generation of two intertwining, yet independent microfluidic networks in a single construct.
Keywords: microfluidic, laser-based degradation, biomimetic, tissue engineering, vascularized constructs
Graphical abstract
The ability to fabricate multiple, high density, three-dimensional (3D) microfluidic networks in a small volume could significantly aid development of lab-on-a-chip,[1] organ-on-a-chip,[2–5] and human-on-a-chip devices.[3] Utilizing 3D microfluidic systems to generate vascularized tissue constructs has garnered significant interest, as these constructs have enormous potential in enhancing the use of in vitro models for drug screening;[1,4,6] elucidating disease mechanisms such as cancer metastasis;[7–10] and aiding the study, diagnosis, and treatment of cardiovascular disease.[11] Development of functional microtissues for use in on-a-chip devices necessitates microfluidics for transport of nutrients, oxygen, and waste to support cell function.[12] Two generalized methods exist to produce vasculature in hydrogels: self-assembly of pericyte-supported endothelial cell (EC) networks via angiogenic[13–16] or de novo vasculogenic[16–18] processes and implementation of microfabrication techniques to predefine vascular architecture followed by endothelialization.[19–23] While self-assembly methods are capable of forming dense vascular networks throughout large volume s, they often result in vessels with architecture[24] and permeability[9,15] characteristic of tumor vasculature; which may or may not be beneficial depending on the application. Additionally, as network formation relies on cell-mediated self-assembly, the vascular architecture cannot be controlled.
Conversely, microfabrication techniques allow for direct control over microfluidic architecture, but in general lack the resolution to recapitulate the highly tortuous and dense vascular networks formed in vivo. Lithographic molding,[25] computer-aided laser micromachining,[26,27] and direct-write assembly[28] have been used to create geometrically simple microfluidic systems consisting of planar, 3-sided channels on the surface of a matrix. Planar microfluidic networks completely embedded in polymer have been fabricated using advanced lithographic molding,[17] injection molding,[20,29] strategic placement and removal of needles,[19,22] and printing of sacrificial materials[21] in self-healing hydrogels.[30] While these approaches allow for fabrication of planar microfluidic systems, they do not provide a means to fabricate complex 3D networks.
A number of techniques to generate non-planar, 3D microfluidic networks have been developed including electrostatic discharge,[31] modular assembly,[32] molding to spring structures,[33] direct-write assembly,[34] 3D printing of sacrificial materials,[23] and omnidirectional printing.[35] While much progress has been made in fabricating 3D microfluidic networks, there are still a number of limitations with existing methods including the inability to: create capillary -sized structures; replicate the dense, tortuous architecture of in vivo vasculature; and generate multiple microfluidic networks within a single construct to mimic the close interaction and transport between lymphatic and cardiovascular systems for example.[10,36–38]
To overcome many of the limitations with existing microfluidic fabrication strategies, we combined laser-based hydrogel degradation[39–42] with image-guided laser control to generate complex 3D, highly tortuous, and dense biomimetic microfluidic networks embedded in poly(ethylene glycol) diacrylate (PEGDA) hydrogels. We demonstrate that these vascular-derived microfluidic systems accurately recapitulate the architecture, size, and density of in vivo microvasculature and that they can be lined with ECs to generate capillary-like networks. We also demonstrate the ability to generate two independent, yet closely intertwining networks that fill the same PEGDA volume. The two microfluidic networks never directly connect, yet allow for inter-network transport via their close proximity. The ability to generate 3D microfluidic networks that accurately recapitulate in vivo vascular architecture, as well as transport between independent networks, has the potential to open new avenues for the implementation of vascularized tissue constructs for a multitude of in vitro applications.
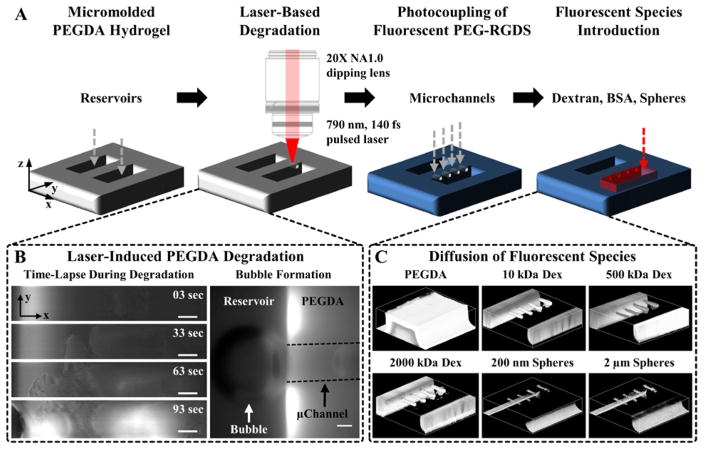
Figure 1 demonstrates the laser-based degradation process. A 790 nm, 140 fs pulsed Ti:S laser operating at a fluence of 37.7 nJ μm−2, focused through a 20X(NA1.0) water immersion objective, was raster scanned in desired geometric volumes in a PEGDA hydrogel to induce localized degradation (Figure 1A). The volume of PEGDA degraded was defined by digital, virtual masks that contained tens to thousands of single pixel wide regions-of-interest (ROIs) that guided the position of the laser.[18,43–45] Simple microchannel geometries, rectangular (Figure 1C and 2A) or cylindrical (Figure 3D), utilized ROIs manually defined in Zeiss Zen software. ROIs derived from more complex 3D geometries generated using computer aided design (CAD) software (Figure 4), or derived from confocal image stacks (Figure 3A,C), were produced using a MatLab algorithm.[18] The use of virtual masks eliminates the need for physical masters utilized for photolithographic techniques and provides the ability to quickly change network design “on-the-fly” through digital manipulation.
Figure 1. Fabrication of Microfluidic Channels in PEGDA Hydrogels.
(A) A 5% 3.4 kDa PEGDA hydrogel is photopolymerized against a PDMS master to create reservoirs. A 790 nm, 140 fs pulsed laser focused through a 20X(NA1.0) water immersion objective is raster scanned in desired 3D configurations to locally degrade the hydrogel. The hydrogel is fluorescently labeled via photocoupling of a monoacrylate fluorescent PEG-RGDS to visualize degraded volumes. The reservoirs are filled with a fluorescent species (dextran (Dex), bovine serum albumin (BSA), nano- or microspheres) and the microchannels imaged via confocal microscopy. (B: left column) Time-lapse images of PEGDA during laser-induced degradation of a 500×100×100 μm (x,y,z) channel. (B: right column) As microbubbles form they migrate to the reservoir and coalesce to form a large bubble. The microchannel walls are depicted by dashed black lines. (C) 3D renderings of a micromolded hydrogel and microchannels filled with 10, 500, and 2000 kDa fluorescent dextran and 200 nm and 2 μm fluorescent spheres. (B) SB=50 μm.
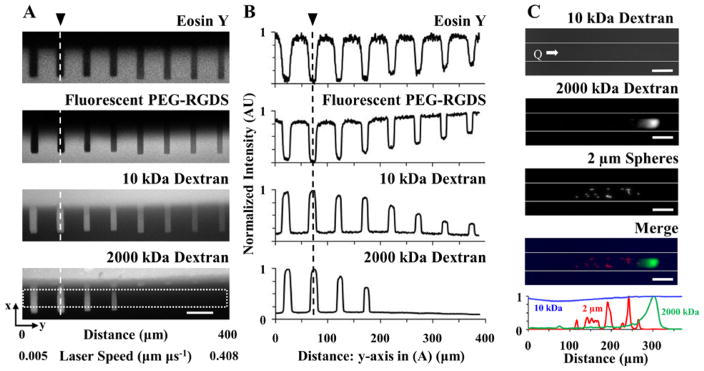
Figure 2. Controlled Local Hydrogel Porosity and Size-Based Separation of Fluorescent Species.
(A) The local hydrogel porosity was controlled by altering the laser scan speed while degrading at a constant fluence of 21.73 nJ μm−2. (A: top two rows) Z-projections of a 3D image stack depicting the eosin Y and fluorescent PEG-RGDS show where the hydrogel (lighter regions) was degraded to form microchannels (darker regions). (A: bottom two rows) Z-projections of fluorescent 10 and 2000 kDa dextran perfused into the microchannels are also shown. (A,B: channel on far left side) A slow scan speed, 0.005 μm μs−1, induced total hydrogel degradation resulting in a completely open microchannel as observed in the eosin Y and fluorescent PEG-RGDS (A) images and (B) intensity profiles. (A,B: channels from left to right) Increasing the scan speed induced partial hydrogel degradation resulting in microchannels containing some polymer but with an increased pore size relative to the unmodified base hydrogel as indicated by the (A,B: 4th row) inability for 2000 kDa dextran to perfuse into these channels and (A,B: 3rd row) decreased intensity of 10 kDa dextran. (A: 4th row) The white box (389×37.4 μm) indicates where the (B) intensity measurements were acquired for each z-projection. (A,B) The vertical dashed lines, indicated by the black arrows, are in the same location for orientation purposes. (C) The degradation parameters used to create the microchannels in (A) were applied in a gradient fashion by increasing the scan speed from left to right to create a long rectangular channel (white lines indicate channel walls) that allowed for size-based separation of fluorescent species (2000 kDa dextran and 2 μm spheres) during flow; Q = 10 μL min−1. Normalized intensity profiles measured for each fluorescent species were plotted as a function of distance from the channel entrance. (A,C) SB=50 μm.
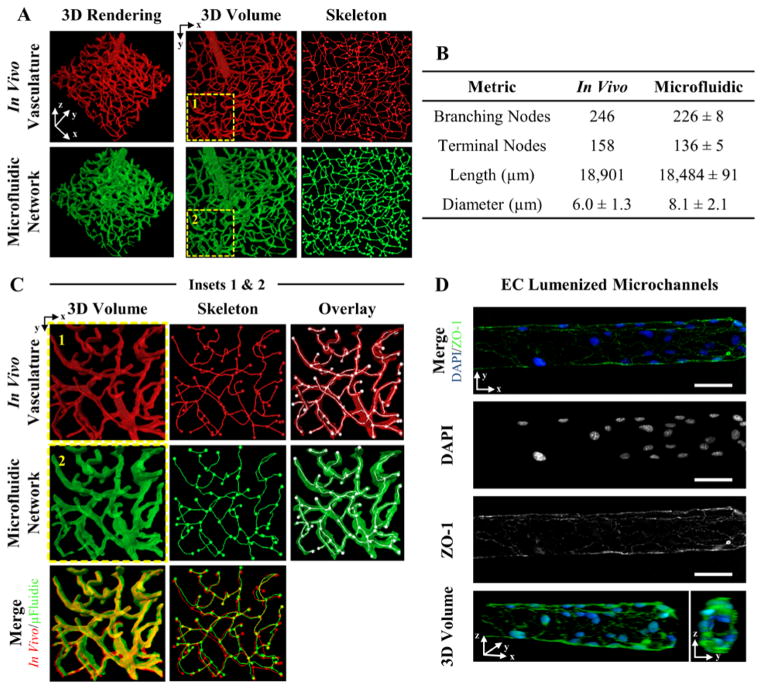
Figure 3. 3D Vascular -Derived Microfluidic Networks.
(A) A confocal image stack of (red) cerebral cortex vasculature was used to fabricate a (green) biomimetic microfluidic network in a PEGDA hydrogel. The in vivo and in vitro microfluidic networks were skeletonized for quantitative analysis. (B) Four metrics were used to quantity the microfluidic networks. (C) The insets in (A) are expanded to demonstrate the ability to recapitulate the dense, tortuous in vivo vascular network in PEGDA. (D) Microchannels were seeded with mouse brain endothelial cells, fluorescently labeled with DAPI (blue: nucleus) and ZO-1 (green: tight junctions), and imaged via confocal microscopy. (D) SB=50 μm.
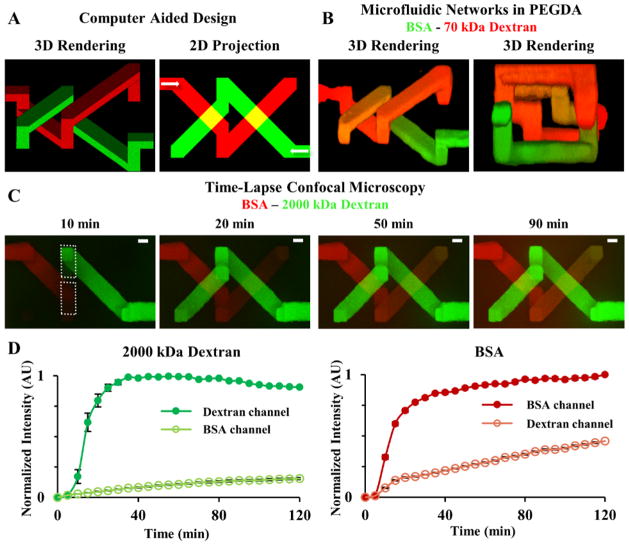
Figure 4. Transport Between 3D Intertwining Microchannels.
(A) A 3D model containing two independent yet intertwining microchannels (20×20 μm: x,y) was designed in SolidWorks; the white arrows in the 2D projection indicate the flow direction. (B) The 3D model was used to fabricate two microfluidic networks in PEGDA and the microchannels were exposed to (red) 70 kDa dextran and (green) BSA. Orange in the 3D renderings indicates transport of the two fluorescent species from their respective microchannel, through the hydrogel, into the adjacent channel. (B: right panel) Looking down the central axis of the intertwining channels shows that the two networks come within 15 μm of each other but never directly connect. (C) Time-lapse confocal images of species movement show (green) 2000 kDa dextran and (red) BSA filling their respective microchannel and transport through the hydrogel into the adjacent channel. (D) White dotted boxes in (C) indicate where intensities of 2000 kDa dextran and BSA were measured over time. (D: left graph) Normalized intensity of 2000 kDa dextran in its own channel and diffusion into the BSA channel (D: right graph). Normalized intensity of BSA in its own channel and diffusion into the 2000 kDa channel. (C) SB=20 μm.
Laser-based hydrogel degradation has previously been implemented to create microchannels to guide neuronal outgrowth in PEGylated fibrinogen[39,40] or silk protein hydrogels[42] and to direct cancer cell migration in collagen,[41] but has not been utilized to create perfusable 3D, biomimetic microfluidic networks as demonstrated here. Hydrogels composed entirely of protein (silk[42] or collagen[41]) are degraded via multiphoton absorption.[42] The presence of proteins with large multiphoton cross-sections allows for laser-based degradation through multiphoton absorption induced chemical breakage.[42] In this study, PEGDA hydrogels free of protein were utilized. We speculate that localized degradation of PEGDA is mediated by laser-induced optical breakdown of water which generates localized plasma formation leading to a rapid thermoelastic expansion of water.[46] Rapid thermoelastic expansion can induce high levels of stress on the surrounding polymer leading to breakage of covalent bonds and the formation of open voids.[46] PEGDA degradation only occurred when enough energy was delivered to induce cavitation (Figure 1B); if microbubbles did not form, then polymer breakage did not occur. This supports the idea that thermoelastic expansion induced breakage of covalent bonds is the primary mechanism of degradation for hydrogels without a large multiphoton cross-section.
After degradation, the hydrogel was functionalized with a PEGylated, fluorophore-labeled cell adhesive ligand, RGDS, via photocoupling (Figure 1A), for both hydrogel visualization (Figure 1B,C and 2A) and for cell adhesion (Figure 3D). The resulting microfluidic channels were visualized by perfusing a fluorescent species (dextran, bovine serum albumin (BSA), or spheres) into the channels (Figure 1A,C). In Figure 1C, the degraded microchannels were filled with fluorescent spheres up to 2 μm in diameter. The spheres were able to enter the degraded microchannels, but unable to enter the bulk hydrogel, indicating extensive degradation in the desired volume defining the channels. To better understand the degradative process, we investigated the influence of laser energy on the extent of PEGDA degradation (Figure 2).
In addition to fabricating microfluidic channels that are completely void of hydrogel (Figure 1C and 2A), the total energy delivered to a chosen volume can be reduced by decreasing the laser fluence and/or increasing the laser scan speed, thus reducing the exposure time, resulting in partial degradation (Figure 2). To demonstrate the ability to locally control the extent of hydrogel degradation, eight microchannels were fabricated by scanning the laser at a constant fluence, 21.7 nJ μm−2, with eight different scan speeds ranging from 0.005–0.409 μm μs−1 (left to right in Figure 2A,B). The extent of PEGDA degradation was quantified by measuring the intensity of the hydrogel autofluorescence provided by the eosin Y photoinitiator (Figure 2A,B: 1st row). A clear trend relating eosin Y intensity to scan speed was observed, with higher eosin Y intensity correlating to a faster scan speed, indicating that more hydrogel remained in the scanned volume when less energy was delivered (Figure 2A,B: 1st row). To verify that eosin Y intensity differences were not due to photobleaching during degradation, a fluorescently labeled, monoacrylate PEG-RGDS was photocoupled into the hydrogel after degradation, and its intensity measured (Figure 2A,B: 2nd row). The monoacrylate PEG-RGDS will only couple to free acrylates, acting as a fluorescent reporter for the amount of PEGDA present after degradation. The eosin Y and fluorescent PEG-RGDS signals corroborated each other, verifying that the hydrogel was fully degraded at slow laser scan speeds and partially degraded at faster speeds (Figure 2A,B: 1st and 2nd rows).
The ability to regulate the extent of hydrogel degradation was further verified by quantifying the intensity of two molecular weight dextrans, 10 and 2000 kDa, with Stokes radii of 2.36 nm and 27 nm, respectively, that were perfused into the microchannels (Figure 2A,B: 3rd and 4th rows). The smaller 10 kDa dextran was able to fill all of the channels, as indicated by the intensity profile, but displayed a gradual decrease in intensity as the laser scan speed increased (Figure 2A,B: 3rd row). This indicated that the concentration of the 10 kDa dextran in the microchannels decreased as more hydrogel was left in the degraded volumes using faster scan speeds (Figure 2A,B: 3rd row), corroborating what was observed for the eosin Y and PEG-RGDS measurements (Figure 2A,B: 1st and 2nd rows). Similarly, the larger 2000 kDa dextran was able to fill microchannels totally devoid of hydrogel that were fabricated at slow laser scan speeds (<0.010 μm μs−1) and the fluorescence intensity tapered off as the extent of degradation decreased (Figure 2A,B: 4th row). A clear cut-off where perfusion of the 2000 kDa dextran into microchannels no longer occurred was observed, indicating that when the laser was scanned at faster speeds (> 0.082 μm μs−1), the hydrogel was not degraded enough to increase the local porosity to facilitate perfusion of the 2000 kDa dextran into the channels (Figure 2A,B: 4th row). This data demonstrates that the extent of hydrogel degradation in a desired volume can be tuned by controlling the energy delivered.
To demonstrate an application of controlled degradation, we performed size-based separation of fluorescent biomolecules in a microchannel (Figure 2C). A PEGDA hydrogel was photopolymerized within a microfluidic device. The scan speeds used to create the eight differentially degraded microchannels in Figure 2A were applied in a gradient fashion to generate a long microchannel that contained a gradual increase in polymer density from left to right. A solution of 2 μm spheres, and 2000 and 10 kDa dextran, was perfused through the hydrogel at a flowrate of 10 μL min−1. The movement of varying molecular weight fluorescent species, in the direction of flow through the microchannel, was halted at different positions along the flow axis resulting in size-based separation (Figure 2C). While the 10 kDa dextran was able to perfuse the entire channel (Figure 2C: 1st row, blue in 4th row and intensity plot), the 2 μm spheres were clustered 125–250 μm from the channel entrance (Figure 2C: 3rd row, red in 4th row and intensity plot), while the 2000 kDa dextran was able to travel further down the channel, due to its smaller size, until being halted approximately 300 μm from the channel entrance (Figure 2C: 2nd row, green in 4th row and intensity plot). The ability to separate biomolecules based on size indicates that the polymer density increased, and thus porosity decreased, from left to right along the channel length (Figure 2C).
While many methods exist for adjusting bulk porosity of photopolymerized hydrogels pre-synthesis (monomer/macromer length, weight percent, photoinitiator type and concentration, light intensity and exposure time),[47] or mechanically during synthesis,[48] few exist for locally adjusting porosity post-synthesis, especially in specific 3D geometries. Laser-based hydrogel degradation provides enhanced control over local porosity within desired 3D geometries, which could be useful in directing transport within on-a-chip devices.
In vivo, for vascularized tissue, cells are rarely more than 50 to 100 μm from a blood vessel, as proximity to vasculature is necessary for oxygen and nutrient delivery.[49] The ability to replicate dense, tortuous vasculature in vitro is important in creating engineered tissue constructs that recapitulate transport of oxygen and nutrients essential to cell function. While many fabrication techniques exist for creating 3D microfluidic networks, no current technology is able to recapitulate the native architecture of in vivo vasculature. To address this limitation, we demonstrate the ability to recapitulate the native architecture, size, tortuosity, and density of in vivo blood vessels in a hydrogel using image-guided,[18,43–45] laser-based degradation of vascular -derived microfluidic networks (Figure 3A–C).
A confocal image stack of a microvascular network in the cerebral cortex of a mouse injected with fluorescent dextran (Figure 3A,C: 1st row) was converted to a series of virtual masks using a MatLab algorithm.[18] The virtual masks guided the position of the laser[18,43–45] during feature formation, resulting in highly localized PEGDA degradation in the vascular-derived configuration. The resulting biomimetic microfluidic network was perfused with 2000 kDa dextran and imaged (Figure 3A,C: 2nd row). Visual comparison of the 3D renderings (Figure 3A: 1st column) and volumes (Figure 3A: 2nd column) indicate a high level of fidelity between the in vivo and in vitro microfluidic architectures. For further validation, a volume of the network was chosen as indicated by the dashed yellow lines (Figure 3A: 2nd column), colored red for the in vivo vasculature (Figure 3C1), and green for the vascular-derived microfluidic network (Figure 3C2), and the two renderings merged (Figure 3C: 1st column, 3rd row). The dominate yellow in the merged image (Figure 3C: 1st column, 3rd row) visually verifies the ability to generate highly tortuous, biomimetic microfluidic networks that closely match the complex architecture of in vivo vasculature. To quantify the visual observation, the image stacks were skeletonized (Figure 3A: 3rd column and 3C: 2nd column) and analyzed to measure the number of branching and terminal nodes, network length, and vessel diameter (Figure 3B). Visual inspection of the 3D renderings displaying the skeletonized structures (Figure 3A: 3rd column and 3C: 2nd column) with branching and terminal nodes (Figure 3A: 3rd column and 3C: 2nd and 3rd columns) indicates a high degree of alignment.
The vessel diameters were quantitatively compared by acquiring measurements at the same 125 locations for both the in vivo and in vitro networks (Figure 3B). The in vivo network had an average diameter of 6.0 ± 1.3 μm, while the vascular-derived, in vitro networks were slightly larger at 8.1 ± 2.2 μm (Figure 3B). A t-test for populations with unknown variance was performed and the difference between the in vivo and in vitro diameters was not significant within 95% confidence. However, the mean diameters of the in vitro networks were slightly larger, occasionally resulting in merging of vessels in close proximity in high density regions. This prompted us to investigate the resolution achievable with laser-based PEGDA degradation. Using a 790 nm laser focused through a 20X(NA1.0) water immersion objective operating at 37.7 nJ μm−2 with a scan speed of 0.010 μm μs−1, microchannels with widths as small as 3.28 μm were achieved (Supplemental Figure 1). The corresponding height, in the z-direction along the axis of the focused beam, was 8.86 μm (Supplemental Figure 1). Elongation of channel features in the z-direction is a consequence of the shape of the focal volume of the focused laser[50] and also from the use of at least two z-planes to define the channel. Microchannels fabricated using a single z-plain were not able to support flow and therefore two z-planes spaced by 1 μm were implemented resulting in elongation of channel features in the z-direction. Elongatoin induced occasional merging of vessels in close proximity, and thus a reduction in the number of branching nodes and terminal nodes, and total network length for the in vitro architecture leading to significant differences compared to the in vivo network.
Occasional merging of two vessels in consolidated branching segments decreased the number of branching nodes and segments contributing to the total vessel length. Consequently, the number of branching nodes decreased by 8% from 246 to 226 and network length by 2% from 18,901 to 18,484 μm from the in vivo to the in vitro networks respectively (Figure 3B). The slightly increased diameter also consolidated terminal segments resulting in less defined surface protrusions, reducing the number by 14% from 158 to 136 (Figure 3B). Despite these small discrepancies, the vascular-derived, biomimetic microfluidic networks more accurately recapitulate the architecture of native in vivo vasculature than networks fabricated with any existing method. This data demonstrates the ability to recapitulate the complex architecture of in vivo vasculature in vitro with high resolution and accuracy.
While the ability to generate biomimetic microfluidic structures overcomes many of the limitations of existing fabrication methods, it is important to create EC linings in these microchannels. After degradation, monoacrylate PEG-RGDS was photocoupled into the hydrogel to enable cell adhesion to the channel surfaces. ECs were perfused into the microchannels and cultured for 11 days, after which the ECs were fixed, fluorescently labeled to visualize the nuclei (Figure 3D: 2nd row, blue in 1st row) and tight junctions (zona occludens protein-1: (ZO-1)) (Figure 3D: 3rd row, green in 1st row), and imaged. A 3D rendering of the fluorescently labeled ECs demonstrates complete coverage of the channel surface and formation of tight junctions (ZO-1) between adjacent ECs (Figure 3D: 4th row). The ability to generate 3D, biomimetic microfluidic networks that recapitulate in vivo vascular architecture, and that can be endothelialized, opens new avenues for generating vascularized engineered microtissues.
Most research concerning the formation of 3D microfluidic networks has focused on generation of single networks. In vivo, tissues contain multiple microfluidic networks in close proximity: lymphatic, cardiovascular, and biliary, for example. Interstitial flow facilitates cell function via transport of soluble factors,[10,36] and incorporation of mimetic lymphatic systems, 9 along with cardiovasculature, stabilizes vascular structures.[22] Using multiple microfluidic networks, one can begin to impart interstitial flow within in vitro constructs and begin mimicking in vivo-like transport. Toward this goal, we demonstrate the ability to fabricate two independent and closely intertwining microfluidic networks that never directly connect, yet allow for inter-network communication via transport through the hydrogel (Figure 4). While there is a long way to go to recapitulate in vivo transport in vitro, the data presented here is a first attempt demonstrating feasibility.
Two independent microfluidic networks filling the same hydrogel volume by intertwining, without directly connecting, were fabricated (Figure 4B), and the inter-network transport of fluorescent biomolecules quantified (Figure 4C,D). In vivo, lymphatic and cardiovascular networks come within 10–20 μm of each other.[51] To mimic this distance, the two intertwining networks were designed to be separated by 15 μm at their closest point (Figure 4A,B). The microchannel volumes were converted to a series of virtual masks[18] and formed using laser-based hydrogel degradation. Each microchannel connected to its own reservoir to allow perfusion of two different fluorescent biomolecules, BSA and 2000 kDa dextran, into each microchannel independently (Figure 4C,D). Each fluorescent species was added to one of the reservoirs and the perfusion of these species into their own channels and inter-network transport to the adjacent microchannel was monitored (Figure 4C). Intensity measurements were collected in the area delineated by the dashed white boxes in Figure 4C. The intensity data for each species within its own channel and in the adjacent microchannel was normalized and plotted as a function of time (Figure 4D). The intensity profiles show that both microchannels filled quickly, reaching ~75% of their maximum intensity within 20 min, and transport of the BSA into the dextran channel, and vice versa, occured (Figure 4D). The BSA, 68 kDa, was able to diffuse through the hydrogel into the adjacent microchannel more readily than the 2000 kDa dextran (Figure 4D), as expected based on size. The transport of BSA into the adjacent dextran channel reached 46% of its maximum in-channel value, while the larger dextran only reached 15% intensity in the BSA channel (Figure 4D). While this is a simple proof-of-principle demonstration, it is the first, to our knowledge, observing transport between adjacent, 3D microfluidic networks in a hydrogel. Expanding upon this idea may provide the capability of recapitulating in vivo-like interstitial transport in in vitro tissue constructs.
We developed a new approach to fabricate 3D, hydrogel-embedded, biomimetic microfluidic networks by combining laser-based degradation with image-guided laser control and demonstrated that it can be used to: locally control the concentration and porosity of hydrogels in desired 3D geometries; fabricate truly 3D, biomimetic, capillary-like microfluidic networks; and generate two independent, yet interacting, microfluidic networks in close proximity. Using these unique hydrogel modification properties, we demonstrated: size-based separation of varying molecular weight biomolecules in a single microchannel; vascular-derived microfluidic networks that recapitulate the dense, tortuous architecture of in vivo vasculature; and interstitial transport between intertwining, yet independent, microfluidic systems. Although we utilized PEGDA here, laser-based degradation of PEG-fibrinogen[39,40] and collagen[41] hydrogels has been implemented for other applications, indicating the ability to generate microfluidic networks in many synthetic and natural hydrogels. Since this image-guided process relies on the use of virtual masks, design modifications are easily made through simple digital manipulation, removing the need to generate new masters for iterative design changes needed for photolithographic technologies. Furthermore, since virtual masks can be developed from both image- and CAD-derived architectures, this approach opens endless possibilities for microfluidic design. While photolithographic techniques usually require a cleanroom, the fabrication presented here was performed using a commercially available confocal microscope that is present on most university campuses, providing the possibility for virtually any academic researcher to apply this technology. As organ- and human-on-a-chip devices become more sophisticated and biomimetic, there is a need to transport nutrients and oxygen to, and waste away from cells via different microfluidic networks. The data presented here demonstrates proof-of-principle that recapitulating in vivo-like transport using multiple, high density microfluidic networks, that mimic the architecture of native tissues, may prove advantageous in fabricating advanced on -a-chip devices.
Experimental Section
See Supporting Information for experimental details.
Supplementary Material
Acknowledgments
The authors thank Dr. Jim Culver and Dr. Mary Dickinson at the Baylor College of Medicine for graciously providing the confocal image stack of cerebral cortex vasculature. We also thank Dr. Jeff Caplan and Mr. Michael Moore at the Delaware Biotechnology Institute Bio-Imaging Center for assistance with confocal microscopy. Access to microscopy equipment was supported by the National Institutes of Health (NIH) shared instrumentation grants (S10 RR0272773) and (S10 OD016361). This research was supported by grants from the Institutional Development Award (IDeA) from the NIH National Institute of General Medical Sciences (P20GM103446), the American Cancer Society (14-251-07-IRG), the University of Delaware Research Foundation (14A00778), and the State of Delaware Federal Research and Development Grant Program (16A00471). Jennifer L. Mantle is supported by an Integrative Graduate Education and Research Traineeship Program from the National Science Foundation (1144726).
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Keely A. Heintz, Department of Biomedical Engineering, University of Delaware, 150 Academy Street, 161 Colburn Lab, Newark, DE 19716, USA
Michael E. Bregenzer, Department of Biomedical Engineering, University of Delaware, 150 Academy Street, 161 Colburn Lab, Newark, DE 19716, USA
Jennifer L. Mantle, Department of Chemical and Biomolecular Engineering, University of Delaware, 150 Academy Street, Colburn Lab, Newark, DE 19716, USA. Delaware Biotechnology Institute, 15 Innovation Way, Newark, DE 19711, USA
Prof. Kelvin H. Lee, Department of Chemical and Biomolecular Engineering, University of Delaware, 150 Academy Street, Colburn Lab, Newark, DE 19716, USA. Delaware Biotechnology Institute, 15 Innovation Way, Newark, DE 19711, USA
Prof. Jennifer L. West, Department of Biomedical Engineering, Duke University, 101 Science Drive, 1427 Fitzpatrick Center, Durham, NC 27708, USA
Prof. John H. Slater, Email: jhslater@udel.edu, Department of Biomedical Engineering, University of Delaware, 150 Academy Street, 161 Colburn Lab, Newark, DE 19716, USA. Delaware Biotechnology Institute, 15 Innovation Way, Newark, DE 19711, USA
References
- 1.Dittrich PS, Manz A. Nat Rev Drug Discov. 2006;5:210. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 2.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Lab Chip. 2013;13:3496. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung JH, Esch MB, Prot JM, Long CJ, Smith A, Hickman JJ, Shuler ML. Lab Chip. 2013;13:1201. doi: 10.1039/c3lc41017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esch EW, Bahinski A, Huh D. Nat Rev Drug Discov. 2015;14:248. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh D, Torisawa Y, Hamilton GA, Kim HJ, Ingber DE. Lab Chip. 2012;12:2156. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 6.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. Drug Discov Today. 2012;17:173. doi: 10.1016/j.drudis.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. Proc Natl Acad Sci. 2015;112:214. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MB, Whisler JA, Jeon JS, Kamm RD. Integr Biol. 2013;5:1262. doi: 10.1039/c3ib40149a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Proc Natl Acad Sci. 2012;109:13515. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisano M, Triacca V, Barbee KA, Swartz MA. Integr Biol. 2015;7:525. doi: 10.1039/c5ib00085h. [DOI] [PubMed] [Google Scholar]
- 11.He B, Baird R, Butera R, Datta A, George S, Hecht B, Hero A, Lazzi G, Lee RC, Liang J, Neuman M, Peng GCY, Perreault EJ, Ramasubramanian M, Wang MD, Wikswo J, Yang GZ, Zhang YT. IEEE Trans Biomed Eng. 2013;60:589. doi: 10.1109/TBME.2013.2244886. [DOI] [PubMed] [Google Scholar]
- 12.van Duinen V, Trietsch SJ, Joore J, Vulto P, Hankemeier T. Curr Opin Biotechnol. 2015;35:118. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmökel H, Bezuidenhout D, Djonov V, Zilla P, Hubbell JA. FASEB J. 2003 doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 14.Moon JJ, Saik JE, Poché RA, Leslie-Barbick JE, Lee SH, Smith AA, Dickinson ME, West JL. Biomaterials. 2010;31:3840. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saik JE, Gould DJ, Keswani AH, Dickinson ME, West JL. Biomacromolecules. 2011;12:2715. doi: 10.1021/bm200492h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, Lee H, Chung M, Jeon NL. Lab Chip. 2013;13:1489. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 17.Cuchiara MP, Gould DJ, McHale MK, Dickinson ME, West JL. Adv Funct Mater. 2012;22:4511. doi: 10.1002/adfm.201200976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culver JC, Hoffmann JC, Poché RA, Slater JH, West JL, Dickinson ME. Adv Mater. 2012;24:2344. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchanan CF, Voigt EE, Szot CS, Freeman JW, Vlachos PP, Rylander MN. Tissue Eng Part C Methods. 2014;20:64. doi: 10.1089/ten.tec.2012.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. Proc Natl Acad Sci. 2012;109:9342. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong KHK, Truslow JG, Khankhel AH, Chan KLS, Tien J. J Biomed Mater Res A. 2013;101A:2181. doi: 10.1002/jbm.a.34524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. Adv Mater. 2014;26:3124. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 24.Roudsari LC, West JL. Adv Drug Deliv Rev. 2015 doi: 10.1016/j.addr.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 25.He J, Mao M, Liu Y, Shao J, Jin Z, Li D. Adv Healthc Mater. 2013;2:1108. doi: 10.1002/adhm.201200404. [DOI] [PubMed] [Google Scholar]
- 26.Doraiswamy A, Narayan RJ. Philos Trans R Soc Math Phys Eng Sci. 2010;368:1891. doi: 10.1098/rsta.2010.0004. [DOI] [PubMed] [Google Scholar]
- 27.De Maria C, Grassi L, Vozzi F, Ahluwalia A, Vozzi G. Rapid Prototyp J. 2014;20:490. [Google Scholar]
- 28.Wu W, Hansen CJ, Aragón AM, Geubelle PH, White SR, Lewis JA. Soft Matter. 2010;6:739. [Google Scholar]
- 29.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Nat Mater. 2007;6:908. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 30.Highley CB, Rodell CB, Burdick JA. Adv Mater. 2015;27:5075. doi: 10.1002/adma.201501234. [DOI] [PubMed] [Google Scholar]
- 31.Huang JH, Kim J, Agrawal N, Sudarsan AP, Maxim JE, Jayaraman A, Ugaz VM. Adv Mater. 2009;21:3567. [Google Scholar]
- 32.He J, Zhu L, Liu Y, Li D, Jin Z. J Mater Sci Mater Med. 2014;25:2491. doi: 10.1007/s10856-014-5270-9. [DOI] [PubMed] [Google Scholar]
- 33.Huang G, Wang S, He X, Zhang X, Lu TJ, Xu F. Biotechnol Bioeng. 2013;110:980. doi: 10.1002/bit.24764. [DOI] [PubMed] [Google Scholar]
- 34.Therriault D, White SR, Lewis JA. Nat Mater. 2003;2:265. doi: 10.1038/nmat863. [DOI] [PubMed] [Google Scholar]
- 35.Wu W, DeConinck A, Lewis JA. Adv Mater. 2011;23:H178. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 36.Ng CP, Helm CLE, Swartz MA. Microvasc Res. 2004;68:258. doi: 10.1016/j.mvr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Swartz MA, Lund AW. Nat Rev Cancer. 2012;12:210. doi: 10.1038/nrc3186. [DOI] [PubMed] [Google Scholar]
- 38.Margaris KN, Black RA. J R Soc Interface. 2012;9:601. doi: 10.1098/rsif.2011.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarig-Nadir O, Livnat N, Zajdman R, Shoham S, Seliktar D. Biophys J. 2009;96:4743. doi: 10.1016/j.bpj.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livnat N, Sarig-Nadir O, Seliktar D, Shoham S. Neural Eng. 2009 NER09 4th Int. IEEEEMBS Conf. On, IEEE; 2009; pp. 116–119. [Google Scholar]
- 41.Ilina O, Bakker GJ, Vasaturo A, Hoffman RM, Friedl P. Phys Biol. 2011;8:015010. doi: 10.1088/1478-3975/8/1/015010. [DOI] [PubMed] [Google Scholar]
- 42.Applegate MB, Coburn J, Partlow BP, Moreau JE, Mondia JP, Marelli B, Kaplan DL, Omenetto FG. Proc Natl Acad Sci. 2015;112:12052. doi: 10.1073/pnas.1509405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slater JH, Culver JC, Long BL, Hu CW, Hu J, Birk TF, Qutub AA, Dickinson ME, West JL. ACS Nano. 2015;9:6128. doi: 10.1021/acsnano.5b01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shukla A, Slater JH, Culver JC, Dickinson ME, West JL. ACS Appl Mater Interfaces. 2015 doi: 10.1021/acsami.5b08978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slater JH, West JL. Methods Cell Biol. Elsevier; 2014. pp. 193–217. [DOI] [PubMed] [Google Scholar]
- 46.Vogel A, Noack J, Hüttman G, Paltauf G. Appl Phys B. 2005;81:1015. [Google Scholar]
- 47.Thomas AM, Shea LD. J Mater Chem B. 2014;2:4521. doi: 10.1039/C4TB00585F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F. Tissue Eng Part B Rev. 2010;16:371. doi: 10.1089/ten.teb.2009.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baish JW, Stylianopoulos T, Lanning RM, Kamoun WS, Fukumura D, Munn LL, Jain RK. Proc Natl Acad Sci. 2011;108:1799. doi: 10.1073/pnas.1018154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berkovitch Y, Yelin D, Seliktar D. Eur Polym J. 2015;72:473. [Google Scholar]
- 51.Schmid-Schoenbein GW. Physiol Rev. 1990;70:987. doi: 10.1152/physrev.1990.70.4.987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.