Abstract
Influenza viruses cause a highly contagious respiratory disease in humans. The NS1 proteins of influenza A and B viruses (NS1A and NS1B proteins, respectively) are comprised of two domains, a dimeric N-terminal domain and a C-terminal domain, connected by a flexible polypeptide linker. Here we report the 2.0-Å X-ray crystal structure and NMR studies of the NS1B C-terminal domain that reveal a novel and unexpected basic RNA-binding site that is not present in the NS1A protein. We demonstrate that single-site alanine replacements of basic residues in this site lead to reduced RNA-binding activity, and that recombinant influenza B viruses expressing these mutant NS1B proteins are severely attenuated in replication. This novel RNA-binding site of NS1B is required for optimal influenza B virus replication. Most importantly, this study reveals an unexpected RNA-binding function in the C-terminal domain of NS1B, a novel function that distinguishes influenza B viruses from influenza A viruses.
Graphical Abstract
eTOC Blurb
Ma et al. describe a novel RNA-binding site in the non-structural protein 1 (NS1B) from influenza B virus. Mutations of basic residues in this site reduce RNA-binding activity, and recombinant influenza B viruses expressing these mutant NS1B proteins are severely attenuated in virus replication.
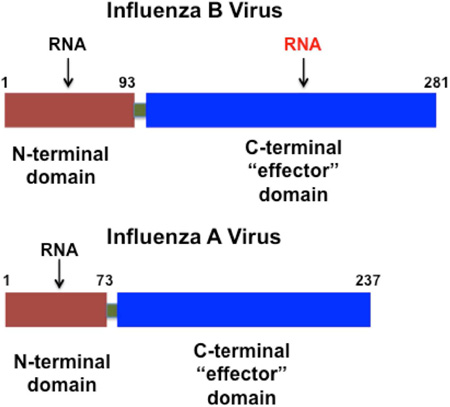
Critical questions in the field of influenza virus research involve elucidating the features of influenza A and influenza B viruses that distinguish their infection mechanism(s) and virulence. The influenza A and B virus genomes are both comprised of eight segments of viral RNAs (vRNAs) of negative polarity (Krug and Fodor, 2013). The smallest vRNA segment of both viruses encodes the nonstructural NS1 protein (Krug and Garcia-Sastre, 2013). The NS1 protein of influenza A virus (NS1A protein) has been extensively studied and shown to have multiple functions that counter host antiviral responses and regulate other cellular and viral functions (Krug and Garcia-Sastre, 2013). In contrast, the function(s) of the NS1 protein of influenza B virus (NS1B protein) are not well understood. The C-terminal domain (CTD) of NS1B inhibits activation of IRF3 and transcription of interferon-α/β(IFN-α/β) genes (Dauber et al., 2006; Donelan et al., 2004), but it is not known how NS1B causes this inhibition. No other functions of the NS1B CTD have been identified. The functions of the NS1A protein of influenza A viruses stem from its ability to bind RNA and several different host proteins (Krug and Garcia-Sastre, 2013). The N-terminal domain (NTD) of NS1A binds dsRNA, and adopts a 6-helical interwoven dimeric fold with three helices from each chain (Chien et al., 1997; Liu et al., 1997). The C-terminal domain (CTD) of NS1A binds several host proteins, including the cellular polyadenylation specificity factor 30 (CPSF30) (Das et al., 2008; Nemeroff et al., 1998), the p85β subunit human of phosphatidylinositol 3-kinase (Hale et al., 2006), polyA binding protein II (Chen et al., 1999), and protein kinase R (Li et al., 2006).
The host binding partners of the NS1B protein of influenza B viruses are less well studied. Like NS1A, the NTD of NS1B is also a 6-helical dimeric fold that binds dsRNA (Yin et al., 2007). A somewhat larger 103-residue N-terminal segment of NS1B binds the interferon-induced antiviral human ISG15 protein, and the structure of this complex has been determined (Guan et al., 2011). The NS1A protein of influenza A viruses does not share this ability to bind ISG15. Conversely, the CTD of the NS1B protein of influenza B virus has not been reported to bind the known host protein partners of the CTD of the NS1A protein of influenza A viruses.
Here we uncover a additional new and important difference between the NS1A and NS1B proteins of influenza viruses A and B, discovered by our determination of the 3D structure of the C-terminal domain of the NS1B protein of influenza B viruses. Despite low sequence identity with the CTD of NS1A, which prevents reliable homology modeling, the overall fold of the NS1B CTD is similar to that of the NS1A CTD. However, the structure of the NS1B CTD reveals an unanticipated conserved basic surface. This basic surface functions to bind RNA. Disruption of this RNA binding surface in the NS1B CTD by single site mutations of key basic residues substantially attenuates the replication of influenza B viruses in human cells, demonstrating that that this RNA-binding activity is required for optimal influenza B virus replication. The RNA-binding activity of the C-terminal domain of the NS1B protein of influenza B viruses is a novel, unexpected function that is absent from the NS1A proteins of influenza A viruses.
RESULTS
Construct design and optimization
To determine the 3D structure of the NS1B CTD, we expressed several constructs in E. coli expression hosts, guided by amide hydrogen deuterium exchange data on the full-length NS1B protein (Sharma et al., 2009) and disorder predictions (Huang et al., 2014). Several constructs, differing by varying the length of the N-terminal segment of the CTD, were produced and assessed by small-scale expression and NMR studies. The CTD construct containing amino-acid residues 141–281 behaved the best in expression, purification, and [15N-1H]-HSQC NMR experiments.
Three-dimensional structure of NS1B-CTD
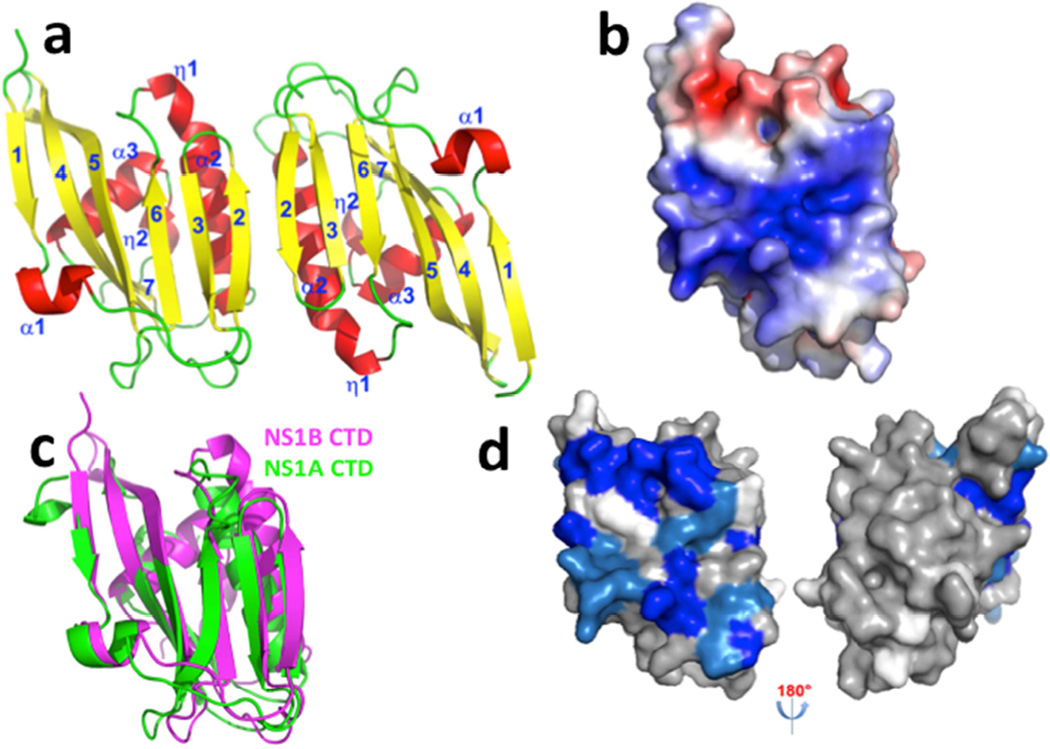
The NS1B CTD construct containing amino-acid residues 141–281 was crystallized and its structure determined to 2.0-Å resolution by X-ray crystallography using the iodide SAD method (Abendroth et al., 2011) (Table 1). There are two NS1B CTD molecules in the asymmetric unit, which form a homodimer in the crystal (Figure 1a). Each protomer has three α-helices, two 310 helices, and an antiparallel twisted β-sheet comprising seven β strands. The largest helix begins as a 310 helix (η1) and then forms an α-helix α2. The final β-stand β7 interacts with the C-terminal 310 helix η2. The β-sheets from the two monomers interact with each other to form an interfacial, extended β-sheet, which together with two interacting helices from each monomer form the dimer interface. There are multiple interactions at the dimer interface, including 19 salt bridges and/or hydrogen bonds as well as 64 van der Waals contacts. An unanticipated feature of the NS1B CTD structure is that it contains a broad basic surface (Figure 1b), including Arg and Lys residues that are highly conserved across NS1B proteins of influenza B virus strains (Supplemental Figure S1). Such conserved basic surfaces are typical of nucleic acid binding sites, a prediction that we verify below.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 34.69, 41.67, 49.68 |
| α, β, γ (°) | 68.72, 82.83, 78.12 |
| Wavelength (Å) | 1.5418 |
| Resolution (Å) | 2.0 (2.04 – 2.01) |
| Rmerge (%) | 8.4 (33.4) |
| I/σI | 12.22 (2.84) |
| Completeness (%) | 95.0 (90.1) |
| Redundancy | 3.9 (3.8) |
| Refinement | |
| Resolution (Å) | 2.0 |
| No. reflections | 31913 |
| Rwork / Rfree (%) | 19.36 / 24.25 |
| No. atoms | |
| Protein | 2184 |
| Iodide ion | 13 |
| Water | 200 |
| B-factors (Å2) | |
| Protein | 23.56 |
| Ligand/ion | 31.82 |
| Water | 28.48 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.775 |
| Bond angles (°) | 0.003 |
Values in parentheses are for highest-resolution shell.
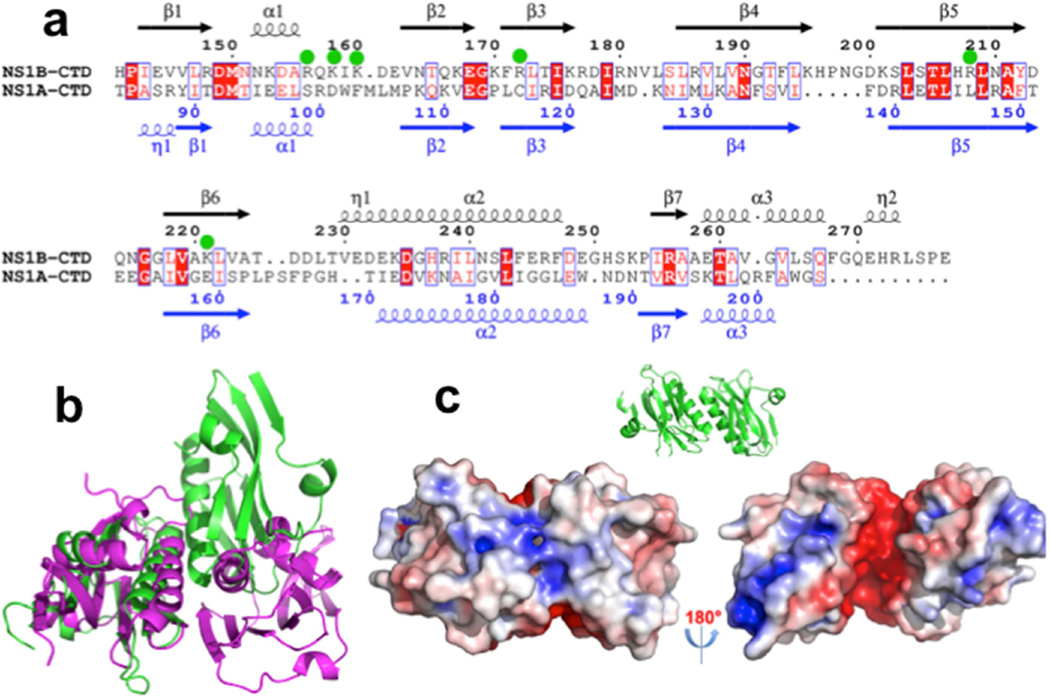
Fig. 1. The X-ray crystal structure of the CTD of NS1B protein reveals a broad basic surface.
(a) Ribbon diagram showing the 2.0-Å X-ray crystal structure of the dimeric NS1B CTD. β-strands are numbered 1 – 7, α-helices α1 – α3, and 310 helices as η1 – η2. (b) The electrostatic surface of the NS1B-CTD calculated with APBS plugin (Baker et al., 2001) of Pymol (DeLano, 2002) at ± 5 kT/e (red, negative; blue, positive). (c) The structure of NS1B CTD (magenta) superimposed on the structure of NS1A-CTD (green, PDB ID: 3EE9) (Xia and Robertus, 2010). (d) 15N-1H chemical shift perturbations Δδcomp due to dsRNA-binding mapped onto the 3D structure of the NS1B CTD. Residues with backbone 15N-1H CSPs due to dsRNA binding Δδcomp ≤ 20 ppb, 20 < Δδcomp < 30 ppb, and Δδcomp ≥ 30 ppb are highlighted in grey, lt. blue, and dk. blue, respectively (see Supplementary Figure S2). Residues for which CSPs could not be determined, including Pro residues, are shown in white. The molecular orientations in panels (a), (b), (c), and (d-left) are the same. Panel d also shows an orientation rotated by 180 deg, revealing no significant CSPs on the opposite side of the molecule.
Although the C-terminal domains of NS1A and NS1B have low (<18%) sequence identity (Supplemental Figure S1), our crystal structures show that the overall folds of these two domains are quite similar (Figure 1c). The NS1B CTD has a longer helix α3 than NS1A CTD, and an extra 310 helix in its C-terminal region. Compared with NS1A CTD, helix α2 of the NS1B CTD also has a short 310 helix extension on its N-terminal side, and β-strand β1 of NS1B CTD is part β-strand and part 310 helix in the NS1A CTD. The pronounced basic surface of NS1B CTD is not observed in the structure of the NS1A CTD.
The C-terminal domain of the NS1B protein of influenza B virus binds RNA
To determine whether the basic surface observed in the X-ray crystal structure of the NS1B C-terminal domain is indeed a novel binding-site for RNA, we measured backbone NMR chemical shifts of NS1B CTD in the presence and absence of RNA. A 16-base dsRNA was mixed with the 15N-labeled NS1B CTD, and chemical shifts of the apo and RNA-bound NS1B CTD were compared (Supplemental Figure S2). Amino-acid residues in the basic surface and in an adjacent acid patch, identified in the X-ray crystal structure exhibit backbone 15N-1H chemical shift perturbations upon binding of this 16-mer dsRNA (Figure 1d), demonstrating that the RNA-binding site in the NS1B CTD involves the conserved basic surface observed in the X-ray crystal structure (Figure 1b). Less extensive chemical shift perturbations are also observed on this same face of the NS1 CTD molecule upon binding a single-strand 16-nt RNA substrate (data not shown).
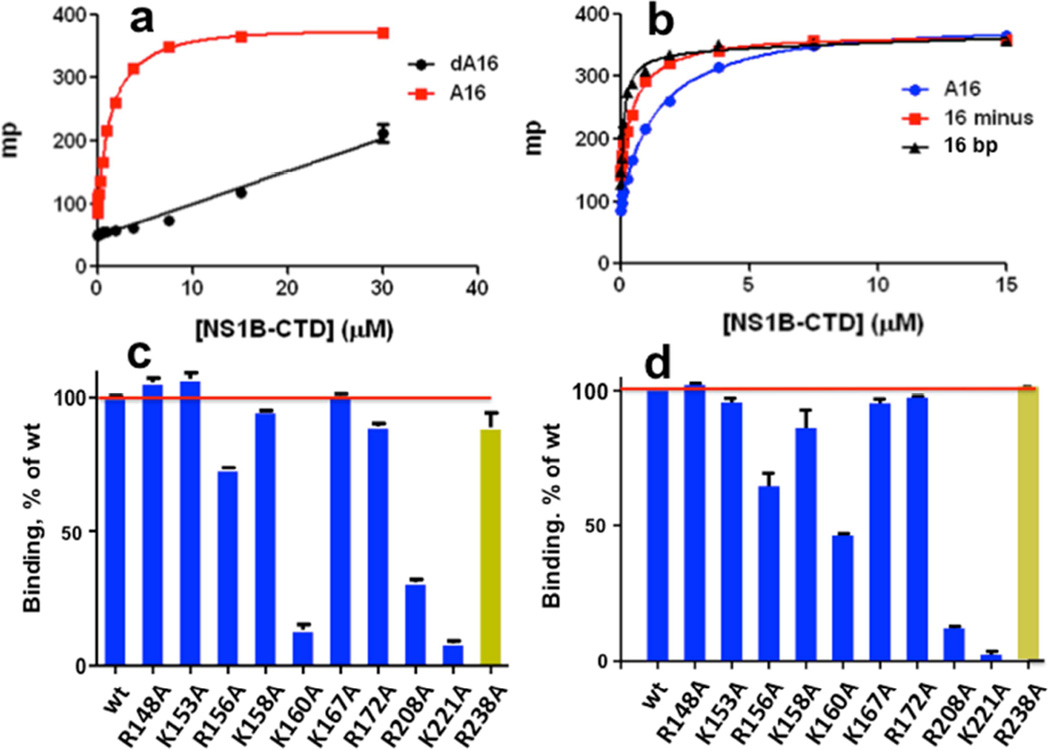
Polynucleotide binding was also assessed by fluorescence polarization (FP) assays using short synthetic DNA and RNA molecules containing a 5’ fluorescein label as substrates. Both polyA16 ssRNA (A16) and polydA16 ssDNA (dA16) exhibit FP due to interactions with NS1BCTD, but the affinity for A16 ssRNA (Kd = 1,400 ± 200 nM) is much tighter than for dA16 ssDNA, for which the binding is too weak to measure reliably (Figure 2a). These data indicate a preference for ssRNA versus ssDNA. We then compared the binding of A16 ssRNA to a 16-mer ssRNA with a sequence containing all four bases (CGCCUGUAGAGGAUGG). This 16-mer ssRNA (called ‘16 minus’) binds even more tightly (Kd = 580 ± 50 nM) than the A16 homopolymer (Figure 2b). A dsRNA substrate was then prepared by annealing this 16-mer ssRNA to its complementary RNA strand. The binding affinity of this dsRNA substrate (Kd = 130 ± 30 nM) is ~ 4 times tighter than the 16-mer ssRNA. Hence the NS1B CTD binds both ssRNA and dsRNA, with some preference for dsRNA.
Fig. 2. Fluorescence polarization (FP) assays of nucleic acid binding activity.
FP, in units of millipolarization (mP), resulting from binding of fluorescein-labeled DNA and RNA molecules to NS1B-CTD. (a) Binding of poly-A16 ssRNA (A16, red) versus poly-dA16 ssDNA (dA16, black). (b) Binding of poly-A16 ssRNA (A16, blue), minus-strand of a 16-nt ssRNA (16 minus, red), and 16-bp dsRNA (16 bp, black). The effects of alanine replacements of individual Lys and Arg residues on the (c) ssRNA- and (d) dsRNA-binding activity of NS1B CTD, assayed by FP. The yellow histogram bar in each plot designates data for dimer-disrupting mutant R238A at the protein-protein interface formed at high protein concentrations and observed in the X-ray crystal structure.
Mutation of surface basic residues of the NS1B CTD reduces RNA-binding activity
NMR chemical shift changes are not precise enough to identify exactly which residues of the NS1B CTD contribute to its RNA-binding activity. CSPs may arise simply by proximity to bound nucleic acid, rather than due to specific interactions. In addition, some residues (shown in white in Figure 1b) within the apparent binding site did not provide reliable CSP data, or are prolines which lack 15N-1H resonances. To identify which of the basic amino-acid surface sidechains contribute to the RNA-binding function, we cloned, expressed, and purified nine NS1B CTD constructs that each contained one single alanine substitution for each basic residue in the RNA binding site identified by X-ray crystallography and NMR studies; specifically R148A, K153A, R156A, K158A, K160A, K167A, R172A, R208A, and K221A mutants. As a control experiment to determine whether each of these alanine substitutions disrupt the 3D structure of the CTD, the mutant CTD proteins were also prepared with 15N enrichment, and were analyzed by [15N-1H]-HSQC NMR. These NMR data (Supplemental Figure S3) demonstrate that all of these mutant NS1B CTD molecules have native-like 3D structures similar to the wt NS1B CTD. Hence, any significant reductions in RNA binding affinity can be attributed to altered protein-RNA interactions, rather than to disruption of the overall NS1B CTD protein structure.
FP assays for dsRNA-binding showed that four of these NS1B CTD variants (R156A, K160A, R208A, K221A) have significantly reduced binding activity for both dsRNA (Figure 2c) and ssRNA (Figure 2d). Interestingly, the same four key surface residues are important for binding both the ssRNA and dsRNA substrates. Two other variants, K158A and R172A, have slight attenuating effect on dsRNA and ssRNA binding affinity. Not all of the basic amino-acid side chains on this surface affect RNA binding under these assay conditions (Figure 2c,d); the R148A, K153A, and K167A mutant NS1B CTD proteins each bind both ssRNA and dsRNA as efficiently as the wt protein.
The monomeric form of the NS1B CTD is sufficient to bind RNA
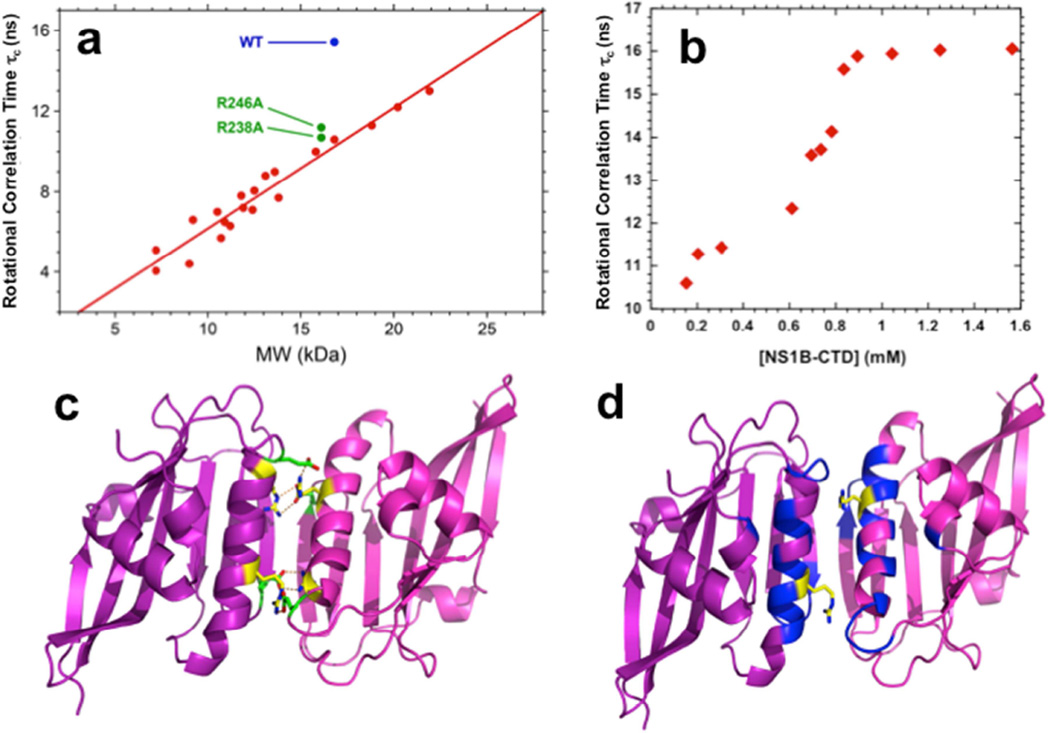
Dimeric interactions observed in crystal structures may or may not occur in solution, particularly under dilute protein conditions. Analytical gel filtration with multi-angle static light scattering (AGF-MALS), analytical ultracentafugation (AUC), and NMR were used to assess whether the NS1B CTD dimer observed in the crystal also forms in solution. Using an initial protein concentration of 200 µM, AFG-MALS demonstrates a monodisperse monomer with MW ~ 17 kDa (Supplemental Figure S4), consistent with expected MW of 15.9 kDa. AUC studies using initial protein concentrations ranging from 13 to 100 µM also demonstrate a monodisperse monomer with MW 15.2 ± 0.3 kDa (Supplemental Figure S5). However, at significantly higher protein concentrations (> 1 mM protein concentration), NMR rotational correlation time estimates, based on 15N T1/T2 measurements, demonstrate that dimeric NS1-CTD is formed (Figure 3a,b), with an estimated dimer self-association constant Kd of ~ 700 µM (Figure 3b).
Fig. 3. NMR studies demonstrate that the dimer interface in solution is the same as in the X-ray crystal structure.
(a) Plot of rotational correlation times, τc (ns), obtained from 1D 15N T1/T2 relaxation data versus protein molecular weight (kDa) for wt NS1B CTD (blue), interfacial mutants [R238A]- and [R246A]-NS1B CTD (green), and monomeric proteins of known molecular weight and oligomerization state. (b) Plot of rotational correlation times, τc (ns), obtained from 1D 15N T1/T2 relaxation data at different protein concentrations of NS1B CTD, providing an estimate the self-dissociation equilibrium constant Kd ~ 700 µM. (c) The homodimer structure of NS1B CTD determined by X-ray crystallography, highlighting key Arg238 – Glu249 and Arg246 – Glu168 interchain salt bridges (yellow: Arg; green: Glu). (d) Backbone amide 15N-1H chemical shift differences between wt NS1B CTD dimer and its R238A mutant monomer mapped onto the X-ray crystal structure of NS1B CTD dimer. Residues are colored as follows: blue, Δδcomp > 50 ppb; magenta, Δδcomp < 50 ppb; white, missing amide NH assignments. The sidechains of residue Arg-238, mutated to Ala to form a monomer, are shown in yellow. The protein-protein interface of the weak homodimer that is formed in solution at high (> 1 mM) protein concentrations is the same as that observed in the crystal (Fig. 1a). All data were obtained on a Bruker 600 MHz spectrometer at 298 K.
In the crystal structure, residues R238 and R246 are involved in intermolecular salt bridges with residues N249 and E168, respectively, at the dimer interface (Figure 3c). To determine whether these same interactions stabilize the weak dimer observed in solution, NMR rotational correlation time estimates were also made for R238A and R246A NS1-CTD mutants. Each of these mutations results in NS1 CTD species with monomeric rotational correlation times under conditions (~ 1 mM protein concentration) at which wt NS1 CTD is largely dimeric (Figure 3a), demonstrating the protein-protein interface that is formed in solution at high protein concentrations is the same as that observed in the crystal. [15N-1H]-HSQC spectra of wt NS1B CTD, recorded over a range of protein concentrations, also exhibit chemical shift changes due to the dimer / monomer equilibrium (Kd ~ 600 µM, Supplemental Figure S6), located in the same dimer interface observed in the X-ray crystal structure. Chemical shift differences between dimeric wt and monomeric [R238A]-NS1B CTD are also observed for this same dimer interface. These biophysical studies demonstrate that the NS1B CTD has a weak propensity to form a homodimeric structure with the same interface observed in the X-ray crystal structure. However, this homodimer is predominant in solution only at relatively high (> 500 to 1,000 µM) protein concentrations. The RNA-binding studies described above (Figure 2) were carried out using protein concentrations of 5 µM, well below the Kd for NS1 CTD self association (~ 600–700 µM), indicating that the CTD monomer (rather than the dimer) binds dsRNA. In order to verify this conclusion, and to assess the possibility that dimer formation is driven by RNA binding, we also measured ssRNA and dsRNA binding activity of the Arg238Ala mutation at the homodimer interface, which reduces the homodimer self association (Figure 3a). R238A mutation has little or no effect on ssRNA binding, and no effect on dsRNA binding affinity (yellow histogram bars in Figs. 2c,d). From these measurements we conclude that homodimer formation of the NS1B CTD, while potentially important in cooperative binding to large RNA substrates in vivo, is not required for binding these short RNA substrates.
Mutation of key basic residues in the RNA-binding surface on the Cterminal domain of full-length NS1B protein results in attenuated influenza B viruses
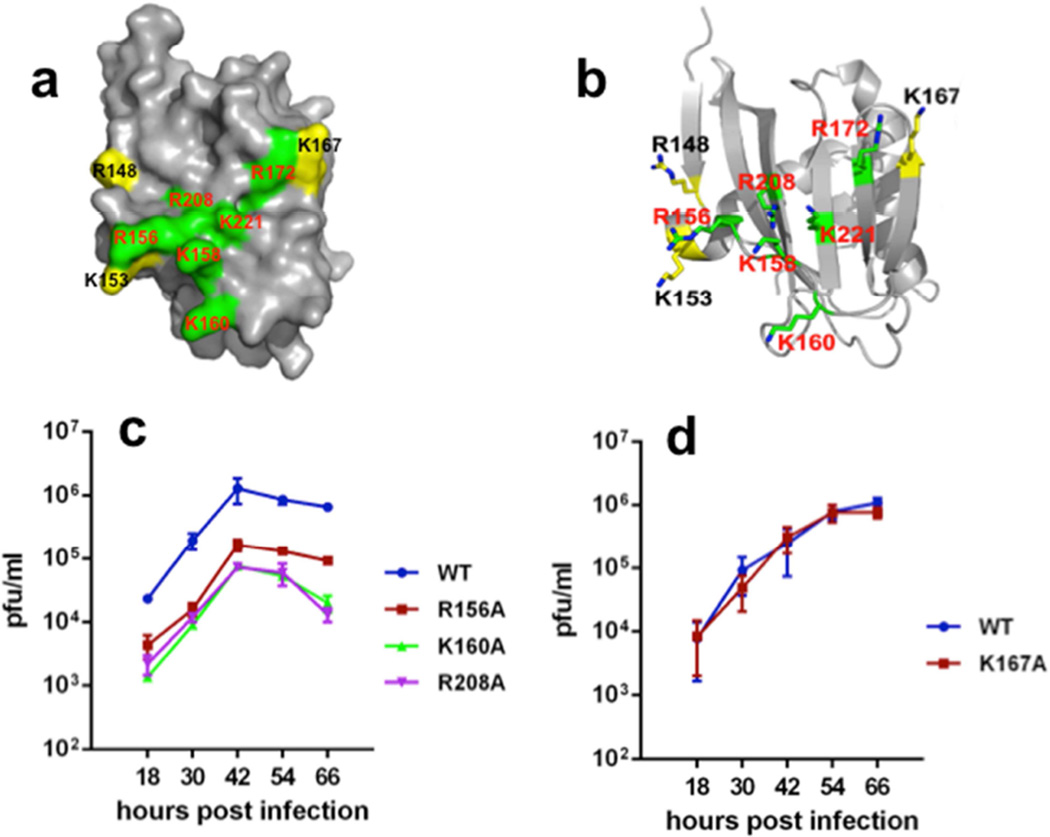
The structural and biophysical results outlined above reveal a novel, unanticipated RNA binding function in the C-terminal domain of the NS1 protein of influenza B viruses. The key basic residues responsible for RNA-binding activity are shown in green in Figs. 4a,b. In order to determine whether RNA-binding by the C-terminal domain of full-length NS1B is required for influenza B virus replication, we generated recombinant influenza B viruses that express a full-length NS1B protein containing either the R156A, K160A, or R208A mutations, each of which significantly inhibits RNA-binding activity in the biophysical FP assays. The replication of these mutant viruses was assayed in human A549 cells using a multiplicity of infection of 0.10 plaque-forming units (pfu) / cell. All three mutant viruses were highly attenuated in replication (Figure 4c). The R156A virus yielded approximately 20-fold less virus than the wt virus, and the K160A and R208A viruses were even more attenuated, yielding ~50–100-fold less virus. These different degrees of viral attenuation parallel the different extents of in vitro RNA binding activity of these mutant NS1B CTDs (compare panels 2c,d and 4c). We were not able to generate a recombinant virus expressing an NS1B protein with a K221A mutation, which has the weakest RNA binding activity in the FP assay (Figure 2c,d), suggesting that this mutation renders the virus even more attenuated. As a control, we generated a virus that expresses a full-length NS1B protein with the K167A mutation that does not reduce the RNA binding activity of the CTD in the FP measurements. Consistent with the RNA binding results obtained in the biophysical studies, the K167A mutant virus is not attenuated and replicates like the wt virus (Figure 4d). These results demonstrate that the RNA binding activity of the NS1B CTD is required for optimal replication of influenza B virus.
Fig. 4. The RNA-binding activity of the RNA-binding epitope in the Cterminal domain of NS1B is required for influenza B virus replication.
The data of Fig. 2c,d are mapped onto the 3D structure of the NS1B ED dimer, shown in both (a) space-filling and (b) ball-and-stick representations, revealing the pattern of surface basic residues contributing to RNA binding (highlighted in green) and those surface basic residues which apparently do not participate in RNA binding (highlighted in yellow). (c) Growth curves of wt, R156A, K160A and R208A mutant viruses in A549 cells infected with a low multiplicity of infection, 0.1 pfu / cell. (d) Growth curves of wt and K167A mutant viruses in A549 cells infected with 0.1 pfu / cell.
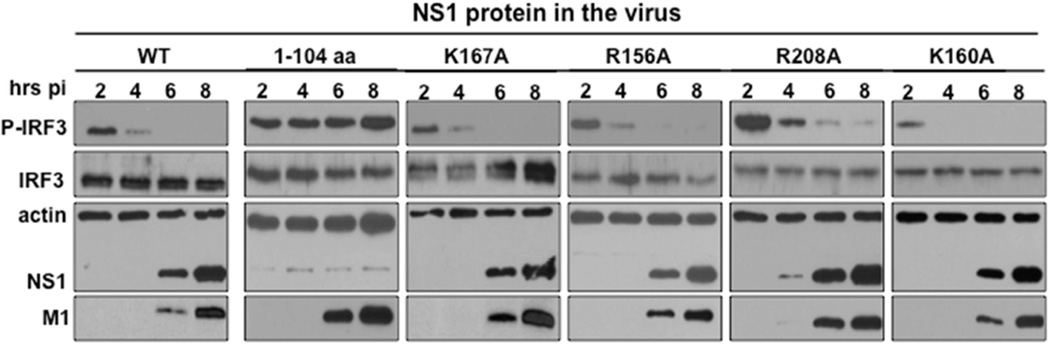
The NS1B CTD of influenza B is required for the inhibition of the IFN-α/β induction pathway, which includes activation of the IRF3 transcription factor (Dauber et al., 2006). Because dsRNA binding by the N-terminal domain of influenza B virus NS1B protein does not antagonize production of IFN-α/β (Dauber et al., 2006), we hypothesized that the RNA-binding activity of the NS1B CTD might sequester key RNAs required for suppression of this IFN induction pathway. Accordingly, we assayed IRF3 activation (phosphorylation) in cells infected by (i) the wt virus, (ii) a recombinant virus expressing a NS1B protein encoding only N-terminal amino acids (1–104 NS1B amino acids), or (iii) the recombinant viruses expressing NS1B with single Ala mutations in the CTD RNA-binding domain (Figure 5). Infection by each of these influenza B viruses caused activation of IRF3 at early times (2–4 hours postinfection), triggered directly by the incoming virus (Makela et al., 2015). As soon as significant levels of the full-length wt or mutant NS1B proteins were synthesized, starting at 6 hours postinfection, activated IRF3 disappeared. In contrast, activated IRF3 remained at high levels after 4 hours postinfection in cells infected with the virus expressing only the 104 N-terminal residues of NS1B, verifying that the NS1B CTD is required for the suppression of IRF3 activation. Hence, like the wt virus, all of the recombinant viruses with a point mutation in the CTD RNA-binding surface block IRF3 activation. These results indicate that the function of this RNA-binding activity does not involve suppression of IRF3 activation.
Fig. 5. Recombinant influenza B viruses with mutations in the CTD RNA-binding domain of NS1 all inhibit IRF3 activation.
A549 cells were infected with 5 pfu / cell of wt influenza B virus, a mutant virus encoding the N-terminal 104 amino acids of the NS1 protein, or a mutant virus expressing a NS1 protein with one of the indicated mutations in the NS1 protein. At the indicated times after infection, cell extracts were immunoblotted with antibodies against activated (phosphorylated) IRF3 (Cell Signaling), total IRF3, actin, viral NS1, or M1 protein.
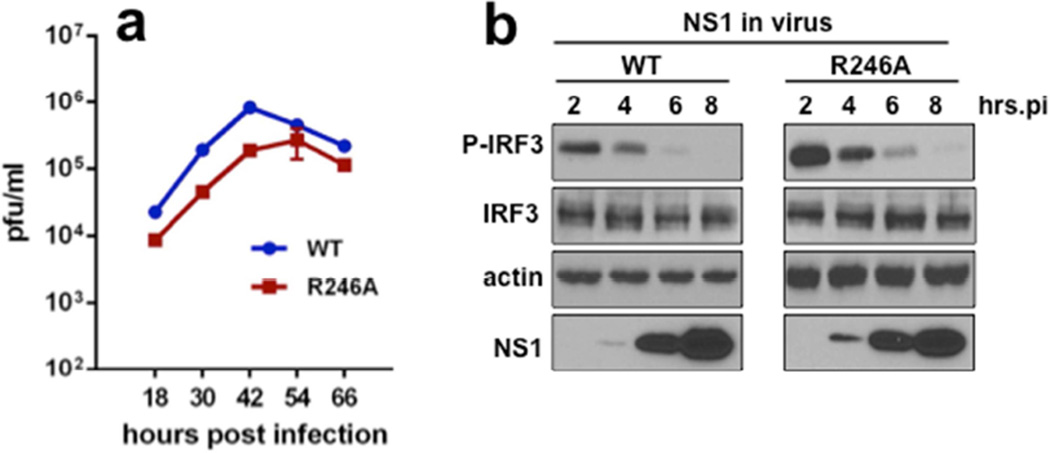
Residue Arg-246 in the CTD homo-dimerization interface is also important for optimal viral replication
Both wt NS1B CTD and [R238A]-NS1B CTD bind both 16-nt ssRNA and 16-bp dsRNA (Figure 2), demonstrating that monomeric NS1B CTD binds short RNA substrates. However, as is the case for the NS1A protein, dimerization of the NS1B CTD in full-length NS1B may contribute to binding larger RNA substrates in virus-infected cells (Aramini et al., 2014; Aramini et al., 2011; Ayllon et al., 2012; Kerry et al., 2014). To determine whether the homodimerization interface of the CTD plays any role in viral replication, we generated a recombinant virus expressing a NS1B protein with the R246A mutation that inhibits CTD dimerization. This mutant virus is attenuated in replication approximately 10-fold (Figure 6a). This attenuation is not due to the loss of the CTD ability to inhibit activation of IRF3 (Figure 6b). Although we cannot exclude alternative interactions with residue R246 that have other functions in viral replication, these data suggest NS1B dimerization mediated by its CTD plays some role in providing optimal viral replication in influenza B virus-infected cells.
Fig. 6. A recombinant influenza B virus with a R246A NS1B mutation that suppresses CTD homo-dimerization is attenuated in viral replication.
(a) Growth curves of wt and R246A mutant viruses in A549 cells infected with 0.1 pfu / cell. (b) A549 cells were infected with 5 pfu / cell of wt or R246A mutant virus, and at the indicated times after infected cell extracts were assayed by immunoblots for activated (phosphorylated) IRF3, total IRF3, and NS1B levels, as described in the legend of Figure 5.
The NS1A protein of influenza A does not have an RNA-binding site in its C-terminal domain
The results outlined above demonstrate that specific basic amino-acid side chains on the surface of the NS1B CTD participate in RNA binding, and that this RNA-binding activity is required for optimal influenza B virus replication. Conversely, the CTD of NS1A exhibits no RNA binding activity under these same conditions (Supplemental Figure S7). The six basic residues in NS1B CTD exhibiting significant or minor RNA-binding function attenuation are colored green in the 3D structure (Figure 4a,b) and indicated by green dots in the protein sequence (Figure 7a). They form a largely basic patch on the surface of the protein structure, and are highly conserved across influenza B NS1B proteins (Supplemental Figure S1b). However, using a structure-based sequence alignment, the corresponding residues in influenza A NS1A proteins are neither basic nor conserved (cf. Figure 7a and Supplemental Figure S1), explaining its inability to bind RNA substrates Hence, the RNA-binding activity of the CTD of the NS1B protein of influenza B viruses is a newly identified function that distinguishes influenza A and influenza B viruses.
Fig. 7. NS1A proteins from influenza A viruses do not share conserved surface features that form and RNA-binding surface of NS1B proteins from influenza B viruses.
(a) A structure-based sequence alignment of the CTD domains of NS1 from influenza A and influenza B strains. Basic residues of the NS1B CTD that affect RNA binding activity when mutated to Ala are indicated with green dots. (b) Comparison of protomer orientations in the X-ray crystal structure of NS1B CTD and in one of the published X-ray crystal structures (PDB ID: 3EE)(Xia and Robertus, 2010) of the NS1A CTD. (c) The same dimer structure of the NS1A CTD (PDB ID: 3EE) (Xia and Robertus, 2010), which has been confirmed in solution by NMR studies (Aramini et al., 2014; Aramini et al., 2011), with electrostatic surface calculated with APBS (Baker et al., 2001) plugin of Pymol (DeLano, 2002) at ± 5 kT/e (left – same orientation as illustrated by the ribbon diagram inset, right – rotated by 180 deg). The structure of NS1A C-terminal domain does not include a basic surface needed to support RNA binding.
DISCUSSION
Our combined biophysical and virology studies of the CTD of the influenza B NS1B protein have led to an unexpected and novel finding regarding differences in the mechanisms of infection by A and B strains of influenza viruses. We have identified an unanticipated and essential RNA-binding site in the C-terminal domain of the NS1B protein of influenza B virus. This RNA-binding site is not present in the influenza A virus NS1A protein. Remarkably, even though the C-terminal domains of NS1A and NS1B have the same overall fold, the newly-discovered RNA-binding function of the NS1B, localized to its C-terminal domain, is a key difference between the functions of these two NS1 proteins. This result highlights differences in the replication strategies of influenza A and B viruses. This is a classic example in which the three-dimensional protein structure provides evidence for an unexpected and important biological function, which was then validated first by biophysical studies with purified protein samples, followed by the generation of viruses bearing specific mutations.
The NS1 proteins of influenza A and B strains bind some common host factors; e.g. dsRNA binding by their N-terminal domains (Krug and Garcia-Sastre, 2013; Qian et al., 1995; Yin et al., 2007). However, they also interact with some different host factors. These distinct binding-partner networks underlie the multiple differences in the mechanisms used by A and B viral strains in host infection. In particular, the NS1B protein of influenza B viruses does not bind at least some of the host proteins bound by the NS1A of influenza A, but does have an essential RNA binding site it its C-terminal domain that is not present in the NS1A proteins of influenza A viruses.
The NS1A CTD, unlike the NS1B CTD, does not have a basic surface (cf. Figure 1b and Figure 7c). Instead, the conserved basic residues in the C-terminal domain of the NS1B proteins of influenza B viruses are substituted by acidic, polar, and/or non-polar residues in the structure-based sequence alignment with the NS1A proteins of influenza A viruses (Figure 7a and Supplemental Figure S1). The CTD of NS1A also does not bind these RNA substrates (Supplemental Figure S8). The novel RNA-binding function in the C-terminal domain of NS1B proteins described in this study, arising from its conserved, broad, basic surface, is a unique property of NS1B proteins of influenza B viruses, that is not shared by NS1A proteins of influenza A viruses.
The C-terminal domain of NS1A from influenza A viruses also forms weak dimers in solution, with a helix α2 – helix α2’ interaction that includes residues Trp-187 (Aramini et al., 2014; Aramini et al., 2011; Kerry et al., 2014). This dimerization interaction contributes to cooperativity in binding long dsRNA molecules by full-length NS1A proteins (Aramini et al., 2011). In the crystal structure, and at high concentrations in solution, the NS1B CTD dimer structure also has a helix α2 – helix α2’ interaction at its interface. However, this dimer structure differs significantly in the relative orientations of protomers from those reported for the influenza A virus NS1A CTD (Aramini et al., 2011; Bornholdt and Prasad, 2006, 2008; Xia and Robertus, 2010) (Figure 7b). The details of interchain packing at this interface varies across these published crystal structures of NS1A CTD, and the interface exhibits intrinsic dynamics which have been studied by 19F NMR nuclear relaxation measurements (Aramini et al., 2014).
However, the range of protomer orientations reported in the published structures of NS1A CTD dimers is much narrower than the large differences between interdomain orientations in NS1A CTD and NS1B CTD dimers illustrated in Figure 7b. While dimerization mediated by the CTD of NS1B is not required for binding small RNA substrates, our observation that virus replication is attenuated 10-fold by a dimer-disrupting R246A mutation suggests that dimerization of full-length NS1B mediated by its CTD may none-the-less contribute to the efficiency of viral replication. Alternatively, this mutation may be affecting other functions of NS1B in influenza B virus-infected cells.
Both the NS1A and NS1B proteins have structurally-similar NTDs which bind to dsRNA in vitro (Krug and Garcia-Sastre, 2013; Qian et al., 1995; Yin et al., 2007), and this dsRNA binding is essential for optimal replication of both influenza A and B viruses. Here we show that the influenza B virus NS1B protein has a second RNA-binding domain in its CTD that also binds RNA in vitro and is essential for optimal virus replication. Why would influenza B virus, but not influenza A virus, require a second RNA-binding domain in its NS1 protein that binds RNA in vitro? We tested one possible function of the NS1B CTD RNA-binding domain, namely, that it sequesters virus-specific RNAs that would otherwise activate the IFN-α/β induction pathway. We showed that this is not the case. To determine the function of the NS1B CTD RNA-binding activity, it will be necessary to next identify the RNA species that bind to this domain in infected cells, followed by a determination of the mechanism(s) by which this RNA inhibits specific steps in virus replication.
EXPERIMENTAL PROCEDURES
Cloning, expression, purification, and sample preparation
A DNA fragment encoding the effector domain of influenza B (B/lee/1940) NS1B-CTD (residues 141–281 of NS1B) was generated by PCR from the full-length gene and cloned into the modified pSUMO vector (LifeSensors), as described previously (Acton et al., 2011; Aramini et al., 2014; Aramini et al., 2011; Panavas et al., 2009) The NS1B-CTD construct was expressed as a N-terminal (6xHis)-tagged SUMO fusion protein in order to produce native NS1B-CTD following cleavage with SUMO protease. Details of sample preparation for crystallization, nucleic acid binding, and NMR studies, including procedures for production of isotope-enriched samples, are presented as Supplemental Information.
Crystallization and data collection
High-throughput robotic crystallization screening of purified NS1B-CTD was carried out at the High Throughput (HTP) Crystallization Facility of the Hauptman-Woodward Medical Research Institute using the microbatch-under-oil method (Luft et al., 2003). Further crystallization optimization was carried out using hanging drop evaporation method. The best crystals were obtained in 20% PEG 3350 with 0.2 M ammonium iodide, pH 5.6.
Structure determination and refinement
At the home X-ray source wavelength (1.54178 Å), the anomalous signal of iodide is very strong. The structure of NS1B-CTD was determined using the iodide SAD method with program AutoSol (Terwilliger et al., 2009) in Phenix (Adams et al., 2011). With the phases and initial map from AutoSol, AutoBuild was used to build a model with more than 50% of residues assigned. Structure refinement was carried out with program PHENIX (Adams et al., 2011). Manual model building and adjusting was done using COOT(Emsley and Cowtan, 2004), and the final model validated by the Protein Structure Validation Software Suite PSVS (Bhattacharya et al., 2007) and MOLPROBITY (Davis et al., 2007). Crystallographic statistics and final structure refinement and geometry statistics are summarized in Table 1.
NMR spectroscopy
NMR spectra of NS1B CTD and its mutants were carried out in buffered solutions containing 25 mM NH4OAc (pH 5.5), 450 mM NaCl, 5 mM CaCl2, 50 mM Arg, 0.02% NaN3, 10% v/v 2H2O, using protein concentrations of 0.1 to 0.3 mM, excepted where noted otherwise. Spectra were acquired at 298 K on Bruker AVANCE 800 and 600 MHz spectrometers equipped with a 5-mm TXI and 1.7-mm TCI cryoprobes, respectively, and referenced to internal DSS (2,2-dimethyl-2-silapentane-5-sulfonic acid). All multi-dimensional NMR spectra were processed with NMRPipe (Delaglio et al., 1995) and visualized using the programs Sparky (Goddard and Kneller, 2000) or CCPN-NMR (Vranken et al., 2005). Sequence-specific resonance assignments and chemical shift perturbation studies were carried out using standard methods, outlined in Supplemental Information. Near complete backbone 1H, 13C, and 15N resonance assignments for wt and [R238A]-NS1B CTD (85–215) were determined using conventional triple resonance NMR methods (Supplementary Figure S8).
Composite changes in 1H and 15N backbone amide chemical shifts, Δδcomp, were computed from assigned 1H-15N TROSY-HSQC spectra of wild type and [R238A]-NS1B CTD using Eqn. 1 (Farmer et al., 1996):
| (1) |
A threshold of 20 ppb (0.020 ppm) was used to define chemical shift perturbations in the presence of RNA, as outlined in the legend to Supplemental Figure S2.
Rotational correlation time measurements
To assess the oligomerization states of wt and mutant NS1B CTD in solution, rotational correlation times (τc) were computed from 15N T1 and T2 relaxation data acquired T 25 °C using pseudo-2D experiments, as described previously (Aramini et al., 2011). For each protein, the τc was then calculated from the 15N T1/T2 ratio using the following approximation given in Eqn. 2 (Fushman et al., 1994; Kay et al., 1989):
| (2) |
where νN is the resonance frequency of 15N in Hz. Finally, values of τc for wt NS1B CTD and mutants were plotted against protein molecular weight and compared to data for known monomeric proteins.
Analytical gel filtration with multiangle static light scattering detection
AGF-MALS was carried out as previously described (Acton et al., 2005; Xiao et al., 2010). Sedimentation velocity and equilibrium ultracentrifugation analyses were carried out by the University of Connecticut Analytical Ultracentrafugation Facility, directed by Drs. J. L. Cole and J. W. Lary. Details of sample preparation and methods used in these biophysical studies are presented as Supplemental Information.
RNA binding assays
For the fluorescence polarization (FP) experiments, the 5’- end of each oligonucleotide was fluorescein (FAM) labeled. Three single-stranded (ss) 16-nucleotide RNAs, FAM-poly-A16, FAM-CCAUCCUCUACAGGCG (sense) and FAM-CGCCUGUAGAGGAUGG (antisense) were purchased from Dharmacon Inc. or Integrated DNA technologies (IDT). FAM-labeled ssDNA poly-dA16 and the FAM-labeled 16-bp dsRNA, the duplex of the sense and antisense FAM-labeled ssRNAs, were purchased from IDT. Fluorescence polarization was measured on a Tecan GENios-Pro plate reader with excitation at 485 nm and emission at 535 nm. FP values were reported in millipolarization units (mP). Details of these FP measurements are presented as Supplemental Information. To estimate the affinity of NS1B CTD to the various nucleic acids described in the text, 20 µL of FAM-dsRNA was titrated with 20 µL of increasing protein concentration and the FP data was analyzed to determine dissociation constants (Kd) for binding NS1B CTD. These measurements were performed in a buffer containing 25 mM NH4OAc, pH 5.5, 225 mM NaCl, 5 mM CaCl2, 1 mM EDTA, 1 mM TCEP, 25 mM Arg, and 0.02% NaN3. For all FP-binding assays, fluorescence intensity (FI) and FP were measured after 50 minutes of incubation at room temperature to ensure equilibrium measurements. The data was fitted using nonlinear regression in PRISM 5.0 (GraphPad Software Inc., La Jolla, CA). To evaluate the effects of single-site mutations of NS1B CTD on its RNA-binding activities, the change in fluorescence polarization (FP) of RNA with or without the protein was measured. For wt NS1B CTD and each NS1B CTD mutant variant, measurements were carried out in a solution containing 5 µM of protein and 20 nM of RNA, in a buffer containing 25 mM NH4OAc, pH 5.5, 275 mM NaCl, 5 mM CaCl2, 1 mM EDTA, 1 mM TCEP, 25 mM Arg, and 0.02% NaN3. The data are normalized so that ΔmP = 100% for the difference in FP measured for the complex formed with wt NS1B CTD and the FP measured for the negative control (RNA alone). Each measurement was made in triplicate.
RNA binding to the CTD of the NS1A protein from influenza A was also assessed by FP (Supplementary Figure S7). A DNA fragment encoding the effector domain of influenza A (Udorn) NS1A-CTD (residues 85–215 of NS1A) was generated by PCR from the full-length gene and cloned into the modified pMBP vector, pET15Avi8HMbpTEV_NESG, as described previously (Acton et al., 2011; Aramini et al., 2014). The NS1A-CTD construct was expressed as a N-terminal Avi(8xHis)-tagged MBP fusion protein, and the N-terminal tag was then removed by cleavage with TEV protease. FP measurements with FAM-dsRNA were then carried out as described above.
Viruses and Cells
MDCK and A549 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum. Influenza B/Yamanashi/1998 viruses encoding NS1 proteins with alanine substitutions for amino acids at the indicated positions were generated by plasmid-based reverse genetics (Hoffmann et al., 2002). Virus stocks were grown in 10-day-old fertilized eggs. For multiple cycle growth, A549 cells were infected with 0.1 pfu/cell of wt or mutant virus, and virus titers at different times after infection were determined by plaque assays in MDCK cells. For single cycle growth, A549 cells were infected with 5 pfu/cell of wt or mutant virus. Assays for the activation (phosphorylation) of IRF3 were carried out as described previously (Kuo et al., 2010). Where indicated, infected cell extracts were analyzed by immunoblots using antibodies against the viral M1 protein (Southern Biotech) and NS1B protein. The rabbit anti-NS1B Ab was generated against the purified, E. coli-expressed NS1B CTD.
Database Depositions
Atomic coordinates and structure factors have been deposited in the Protein Data Bank PDB ID (PDB ID 5DIL). Backbone NMR resonance assignments for wt NS1B CTD (BMRB ID 25462) and [R238A]-NS1B CTD (BMRB ID 25463) have been deposited in the BioMagResDB.
Supplementary Material
HIGHLIGHTS.
X-ray crystal structure of the C-terminal domain of NS1 from influenza B
Conserved basic surface suggests nucleic-acid binding function
NS1B C-terminal (or ‘effector’) domain has novel RNA binding activity
This RNA-binding activity is required for optimal viral replication
Acknowledgments
We thank Drs. L. Cole and J. W. Lary of the University of Connecticut Analytical Ultracentrafugation Facility for providing analytical ultracentrifugation analysis, and Dr. R. Xiao for providing AGF-MALS data. We also thank Drs. T.B. Acton, G.V.T. Swapna, and R. Xiao for helpful discussions and comments on this manuscript. This work was supported by National Institute of Allergy and Infectious Disease grants R01-AI11772 (R.M.K.) and R21-AI117510 (G.T.M.), and the National Institute of General Medical Science grant U54 GM094597-05 (G.T.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions L. Ma, R.G., K.H., J.M.A., S.W., R.M.K., and G.T.M. designed the research, analyzed data, and wrote the paper. K.H. made the initial observation that NS1B binds nucleic acid. K.H. and J.M.A. carried out NMR studies. L. Ma, K.H. and L. Mao carried out sample preparation, crystallization, and biophysical studies. K.H. and R.G. carried out crystallization and X-ray crystallography experiments; and S.W. and R.M.K. carried out virology experiments.
REFERENCES
- Abendroth J, Gardberg AS, Robinson JI, Christensen JS, Staker BL, Myler PJ, Stewart LJ, Edwards TE. SAD phasing using iodide ions in a high-throughput structural genomics environment. J Stuct Funct Genomics. 2011;12:83–95. doi: 10.1007/s10969-011-9101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang Y-W, Climent T, Cooper B, Denissova NG, et al. Robotic cloning and protein production platform of the Northeast Structural Genomics Consortium. Meth Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini JM, Hamilton K, Ma LC, Swapna GV, Leonard PG, Ladbury JE, Krug RM, Montelione GT. (19)F NMR reveals multiple conformations at the dimer interface of the nonstructural protein 1 effector domain from influenza A virus. Structure. 2014;22:515–525. doi: 10.1016/j.str.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini JM, Ma LC, Zhou L, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, et al. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J Biol Chem. 2011;286:26050–26060. doi: 10.1074/jbc.M111.248765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayllon J, Russell RJ, Garcia-Sastre A, Hale BG. Contribution of NS1 effector domain dimerization to influenza A virus replication and virulence. J Virol. 2012;86:13095–13098. doi: 10.1128/JVI.02237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13:559–560. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008;456:985–988. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Li Y, Krug RM. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3'-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, et al. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci USA. 2008;105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Schneider J, Wolff T. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J Virol. 2006;80:11667–11677. doi: 10.1128/JVI.01142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 1504. Schrödinger, LLC: 2002. [Google Scholar]
- Donelan NR, Dauber B, Wang X, Basler CF, Wolff T, Garcia-Sastre A. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J Virol. 2004;78:11574–11582. doi: 10.1128/JVI.78.21.11574-11582.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- Fushman D, Weisemann R, Thuring H, Ruterjans H. Backbone dynamics of ribonuclease T1 and its complex with 2'GMP studied by two-dimensional heteronuclear NMR spectroscopy. J Biomol NMR. 1994;4:61–78. doi: 10.1007/BF00178336. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. San Francisco, CA: University of California; 2000. [Google Scholar]
- Guan R, Ma LC, Leonard PG, Amer BR, Sridharan H, Zhao C, Krug RM, Montelione GT. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus. Proc Natl Acad Sci USA. 2011;108:13468–13473. doi: 10.1073/pnas.1107032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci USA. 2006;103:14194–14199. doi: 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Mahmood K, Yang CF, Webster RG, Greenberg HB, Kemble G. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci USA. 2002;99:11411–11416. doi: 10.1073/pnas.172393399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Acton TB, Montelione GT. DisMeta: a meta server for construct design and optimization. Methods Mol Biol. 2014;1091:3–16. doi: 10.1007/978-1-62703-691-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kerry PS, Turkington HL, Ackermann K, Jameison SA, Bode BE. Analysis of influenza A virus NS1 dimer interfaces in solution by pulse EPR distance measurements. J Phys Chem B. 2014;118:10882–10888. doi: 10.1021/jp508386r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Fodor E. The virus genome and its replication. In: Webster RG, Braciale TJ, Lamb RA, editors. Influenza Textbook. 2nd. Wiley-Blackwell; 2013. pp. 57–66. [Google Scholar]
- Krug RM, Garcia-Sastre A. The NS1 protein: A master regulator of host and viral functions. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Textbook of influenza. 2nd. Wiley-Blackwell; 2013. pp. 114–132. [Google Scholar]
- Kuo RL, Zhao C, Malur M, Krug RM. Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-beta transcription. Virology. 2010;408:146–158. doi: 10.1016/j.virol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3'end formation of cellular pre-mRNAs. Molecular cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Panavas T, Sanders C, Butt TR. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2009;497:303–317. doi: 10.1007/978-1-59745-566-4_20. [DOI] [PubMed] [Google Scholar]
- Qian XY, Chien CY, Lu Y, Montelione GT, Krug RM. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1995;1:948–956. [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Zheng H, Huang YJ, Ertekin A, Hamuro Y, Rossi P, Tejero R, Acton TB, Xiao R, Jiang M, et al. Construct optimization for protein NMR structure analysis using amide hydrogen/deuterium exchange mass spectrometry. Proteins. 2009;76:882–894. doi: 10.1002/prot.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Cryst Section D, Biological Crystallography. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Xia S, Robertus JD. X-ray structures of NS1 effector domain mutants. Arch Biochem Biophys. 2010;494:198–204. doi: 10.1016/j.abb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Anderson S, Aramini J, Belote R, Buchwald WA, Ciccosanti C, Conover K, Everett JK, Hamilton K, Huang YJ, et al. The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J Struct Biol. 2010;172:21–33. doi: 10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Khan JA, Swapna GV, Ertekin A, Krug RM, Tong L, Montelione GT. Conserved surface features form the doublestranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J Biol Chem. 2007;282:20584–20592. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.