Abstract
INTRODUCTION
We investigated change in DNA methylation in peripheral blood CD4+ lymphocytes over time, examined the relation between CD4+ lymphocytes and brain methylation, and compared their associations with AD pathology.
METHODS
Genome-wide methylation was measured three times in 41 older persons using Illumina InfiniumHumanMethylation450 array. The two CD4+ lymphocytes measures were at study baseline and proximate to death. Brain tissue came from frozen dorsolateral prefrontal cortex.
RESULTS
Global methylation features were conserved across tissue. At individual CpG sites, methylation level was concordant between the two CD4+ lymphocytes but more diffuse between CD4+ lymphocytes and brain. Previous associations of brain methylation with neuritic plaques at target methylation sites were not replicated in CD4+ lymphocytes.
DISCUSSION
There is no strong evidence of change in CD4+ lymphocytes methylation among older persons over an average of 7.5 years. Methylation associations with AD pathology found in neocortex is not directly reflected in CD4+ lymphocytes.
Keywords: DNA methylation, CD4+ lymphocytes, dorsolateral prefrontal cortex, Alzheimer’s disease
INTRODUCTION
DNA cytosine methylation is an epigenetic mechanism that is pivotal in regulating gene expression, cell development and differentiation [1]. Aberrant DNA methylation alterations are implicated in a number of neurodegenerative disorders including Alzheimer’s disease (AD) [2-8]. Dysregulations of DNA methylation in relation to AD pathology are observed in older persons free of clinical symptom, suggesting that it is an early indication of the disease onset. This, together with unavailability of brain samples until postmortem, leads to great interest of exploring methylation markers of AD in ante-mortem peripheral blood cells[9-13]. However, DNA methylation patterns are tissue specific [14-17], and the utility of interrogating peripheral tissue such as blood cells for a role of DNA methylation in neurodegeneration is unclear. While preliminary evidence suggests that peripheral blood may provide surrogate information for phenotypic variation due to methylation alteration in brain [18], findings on blood methylation with AD susceptibility have been largely conflicting [3, 10-12].
Another challenge of having blood methylation serve as surrogate for brain methylation is that DNA methylation patterns change over lifetime. Cross-sectional studies suggest that change in DNA methylation is fastest in prenatal period [19]; and that there are fewer methylated sites and lower methylation content in older age [20]. One longitudinal study shows both increase and decrease of DNA methylation in blood samples between two visits an average of 11 years apart [21]. These findings raise the question whether blood samples collected at different time period may influence the relation of methylation profiles in peripheral blood to postmortem brain.
In this study, we are interested in the potential utility of peripheral blood CD4+ lymphocytes DNA methylation to serve as a biomarker for the associations found between brain DNA methylation and AD pathology. CD4+ lymphocytes are selected because it represents a cell type related to immune function. Genome-wide DNA methylation was profiled three times from each of the 41 autopsied older persons. The CD4+ lymphocytes methylation was measured at study baseline and proximate to death. The postmortem brain methylation was obtained from frozen dorsolateral prefrontal cortex tissue. We compared DNA methylation profiles between the two CD4+ lymphocytes measures, and separately between CD4+ lymphocytes and brain, both in terms of global features as well as average methylation level at individual CpG sites. Further, using data from CD4+ lymphocytes measures we attempted to replicate the associations of target methylation sites with AD pathology that have previously been reported in post-mortem brain [4]. Notably, the sample size is not powered to discover genome-wide significant methylation signals of AD. Instead, the primary purpose of this study is to provide an essential cross walk between CD4+ lymphocytes and brain methylation as well as their relation to AD pathology. We are not aware of any other extant dataset in which this is possible. The data presented will provide important information to other researchers considering pursuing similar work limited to peripheral blood, clinical diagnoses, and in some cases neuroimaging.
METHODS
Study subjects
Data came from the Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP), the two community based clinical pathologic cohort studies of aging and dementia [22, 23]. Both studies are approved by the Institutional Review Board of Rush University Medical Center. Participants enroll without known dementia, and agree to annual clinical evaluations and brain autopsy at the time of death. A written informed consent and an anatomical gift act are obtained from each participant.
The DNA methylation data collection was performed in January 2010. Genome-wide DNA methylation data are available from approximately 750 postmortem dorsolateral prefrontal cortices. These data were previously used for a genome-wide association study of neuritic plaque burden, a key pathologic index of AD [4]. Because we are interested in comparing DNA methylation between peripheral blood and brain, and separately between blood measures over time, we performed this study on a subset of 41 subjects on whom two peripheral blood samples were available several years apart. The first CD4+ lymphocytes measure was at baseline (T1) to evaluate methylation profiles many years prior to death. The second CD4+ lymphocytes measure was proximate to death (T2) to investigate whether blood methylation was different between the two time points that were years apart, and whether their relation to brain methylation and AD pathology were also different. Considering the limited availability of the biospecimen resource of blood distal and proximate to death from persons with and without dementia who had come to autopsy, we elected for this small scale study so that it would inform the cost and benefit for a larger study.
Blood and brain DNA methylation measures
Blood DNA methylation was measured in CD4+ T cells isolated from frozen peripheral blood mononuclear cells (PBMCs). The PBMCs were washed with RPMI1640 medium to remove Dimethyl sulfoxide (DMSO) exposure. We chose to interrogate CD4+ lymphocytes as it is a single cell type related to immune function. CD4+ T-cells were isolated using magnetic-activated cell sorting (MACS), and reached the purity of at least 95% as assessed by flow cytometry. Blood DNA isolation was performed using AllPrep DNA/RNA Micro kit according to manufacturer’s instructions. Brain DNA methylation was measured in dorsolateral prefrontal cortex tissue, as previously described [4]. Briefly, 100-mg frozen sections were thawed on ice, with the gray matter dissected from the white matter. The Qiagen QIAamp DNA mini protocol was used for DNA isolation. DNA methylation profiles were generated using Illumina InfiniumHumanMethylation450 platform. The same processing and quality control (QC) pipeline was implemented to both CD4+ lymphocytes and brain methylation data, as described in the Supplemental Methods. At the end of QC pipeline, data on 420,132 CpG sites are retained across the 22 autosomes. At each site DNA methylation level was presented as a β value, that is, the ratio of the methylated probe intensity to the sum of methylated and unmethylated probe intensities [24]. The values range from 0 to 1, where a larger value indicates higher methylation.
Neuropathology of AD
Multiple indices of AD pathology are assessed in ROS and MAP postmortem brain [25]. In the recent genome-wide DNA methylation scan of more than 700 brains, we reported primary findings with neuritic plaque pathology [4]. For consistency, our replication analysis was restricted to the same measure. Bielschowsky silver stain was used to visualize neuritic plaques in frontal, temporal, parietal, entorhinal, and hippocampal cortices. In each region, neuritic plaques were counted in a 1 mm2 area of the area with highest density. These counts were scaled and averaged across the five regions, as previously described [26]. The measure was right skewed, and square root transformation was applied prior to the analysis. A higher score refers to a greater burden of neuritic plaque pathology. National Institute on Aging (NIA) Reagan criteria were used for sample description [27].
Statistical analysis
To capture the global features of DNA methylation across tissues, average methylation levels (across subjects) at individual CpG sites were obtained for each of the T1 and T2 CD4+ lymphocytes, and brain methylation profiles. Using histograms and box plots, we described the overall distributions of methylation. We compared the distributions by annotated and functional feature types including CpG island features, gene features and chromatin states [17]. At individual CpG level, we estimated pairwise correlations among T1 and T2 CD4+ lymphocytes, and brain methylation for each of the 420,132 CpG sites. Next, mean methylation levels at each site are compared using paired t-tests between T1 and T2 CD4+ lymphocytes, and separately T2 CD4+ lymphocytes and brain. Finally, in a series of multivariable linear regression models, we examined CD4+ lymphocytes methylation associations with neuritic plaque pathology at 14 CpG sites which show brain methylation alterations in AD. In these models, the measure of neuritic plaques was the continuous outcome, and methylation level at each target CpG site was the predictor. The models were adjusted for age and sex.
The analyses were performed using SAS/STAT software, version 9.3 [SAS Institute Inc., Cary, NC] and R statistical software, version 3.1.2 [www.R-project.org].
RESULTS
Subjects characteristics
All of the subjects (N=41) included in the analyses were of European ancestry, and 65.9% (N=27) were females, and they had an average of 15.2 (standard deviation (SD)=3.5) years of education. The mean age was about 81 years at T1, and about 89 years at T2 (Table 1). The mean interval from T1 to T2 was 7.5 years (SD=4.1), whereas from T2 to death was about 1 year (SD=0.7) such that mean age at death was about 90 years. At T1, a majority of the participants (95.1%) were dementia free. At T2, almost half (46.3%) had dementia. Similarly, the average score for Mini Mental State Examination (MMSE) at T1 was about 27 whereas it was 19 at T2 suggesting substantial cognitive deterioration among some participants. 58.5% of the subjects had AD or other dementia perimortem, and nearly three quarters (73.2%) had pathological AD (intermediate or high likelihood AD by NIA-Reagan criteria). The average burden of neuritic plaque pathology was about 0.94 (SD=0.57), similar to that of the overall cohorts.
Table 1.
Basic characteristics of the subjects (N=41)
| T1 | T2 | Death | |
|---|---|---|---|
| Age (Mean, SD) | 81.2 (6.2) | 88.7 (4.7) | 89.6 (4.9) |
| MMSE (Mean, SD) | 27.4 (2.7) | 19.4 (8.7) | - |
| NCI (N, %) | 21 (51.2%) | 9 (22.0%) | 9 (22.0%) |
| MCI (N, %) | 18 (43.9%) | 13 (31.7%) | 8 (19.5%) |
| Dementia (N, %) | 2 (4.9%) | 19 (46.3%) | 24 (58.5%) |
| Postmortem interval in hours (Mean, SD) | - | - | 7.3 (3.3) |
| Pathologic AD1 (N, %) | - | - | 30 (73.2%) |
| Neuritic plaque2 (Mean, SD) | - | - | 0.94 (0.57) |
NIA-Reagan intermediate or high likelihood AD,
square-root transformed
Global methylation features across tissues
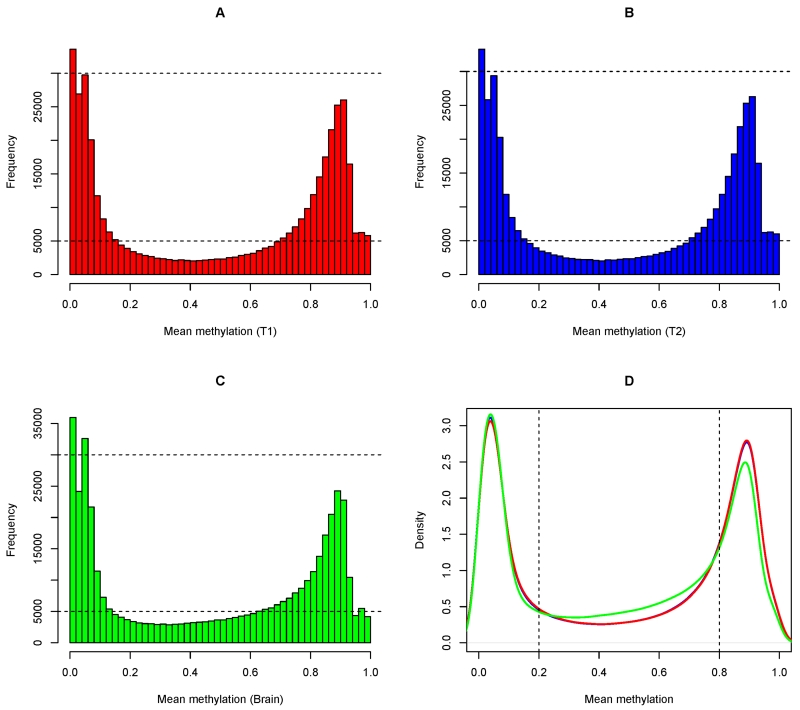
Despite tissue difference, genome-wide DNA methylation captured by InfiniumHumanMethylation450 array features a distinct bimodal distribution for both CD4+ lymphocytes and brain (Figure 1A-1C). A majority of the CpG sites were either extremely hypo-methylated (β value ≤0.2) or extremely hyper-methylated (β value ≥0.8). We further observed that the density distributions of T1 and T2 CD4+ lymphocytes are virtually on top of each other (Figure 1D). On the other hand, comparing brain with CD4+ lymphocytes, they differed at the upper end of the distribution where fewer hyper-methylated CpGs in brain were seen. Indeed, the proportions of extremely hyper-methylated CpGs were 37.2% and 37.4% in T1 and T2 CD4+ lymphocytes respectively, and the proportion (32.0%) was lower in brain.
Figure 1.
illustrates the global distribution of DNA methylation for T1 CD4+ lymphocytes (Panel A), T2 CD4+ lymphocytes (Panel B), and brain (Panel C). Each panel (A, B and C) shows the histogram of methylation of 420,132 autosomal CpG sites, which were averaged across 41 subjects. Panel D shows the density distributions of T1, T2, and brain, overlaying each other.
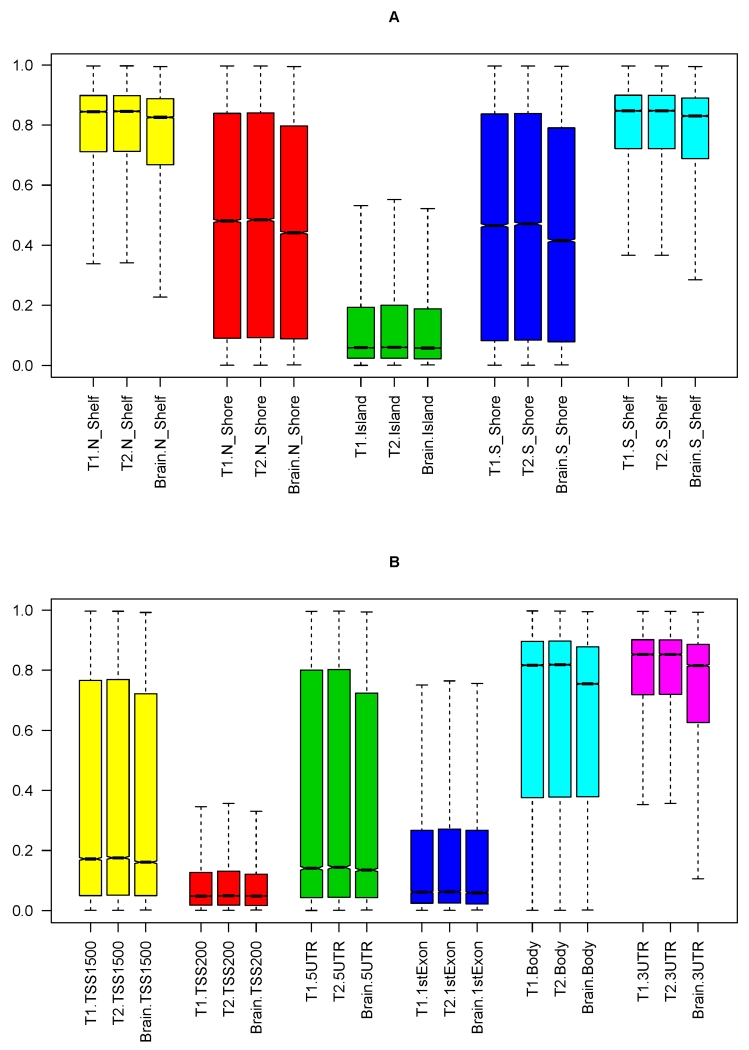
Since distributions of DNA methylation varied across island features, sub-genic regions, as well as chromatin states [17, 28], we examined whether these distinct distributions were conserved across tissues. We first examined methylation in CD4+ lymphocytes versus brain by island features (Figure 2A). As we reported earlier [28], most CpGs in islands were extremely hypo-methylated and a majority of CpGs in distal shelves were extremely hyper-methylated. These differences in distributions by island features were well conserved across tissues. Nonetheless, the median methylation levels were lower in brain (Table 2).
Figure 2.
illustrates global distribution for CD4+ lymphocytes and brain methylation by island features and gene features. Panel A shows the boxplot of methylation levels between T1 CD4+ lymphocytes, T2 CD4+ lymphocytes, and brain, stratified by islands, shores and shelves. Panel B shows the boxplot of methylation levels between T1 CD4+ lymphocytes, T2 CD4+ lymphocytes, and brain, stratified by sub-genic regions including TSS1500, TSS200, 5’UTR, 1stExon, gene body and 3’UTR.
Table 2.
Median DNA methylation levels by global features across tissues
| # CpGs (Blood/Brain) | T1 | T2 | Brain | p Δ | |
|---|---|---|---|---|---|
| Island features | |||||
| Island | 135,303/135,303 | 0.059 | 0.060 | 0.058 | <0001 |
| North shore | 55,328/55,328 | 0.481 | 0.485 | 0.442 | <0001 |
| South shore | 43,325/43,325 | 0.466 | 0.471 | 0.415 | <0001 |
| North shelf | 20,732/20,732 | 0.845 | 0.846 | 0.826 | <0001 |
| South shelf | 18,409/18,409 | 0.847 | 0.848 | 0.830 | <0001 |
| Gene features | |||||
| TSS1500 | 60,703/60,703 | 0.172 | 0.176 | 0.161 | <0001 |
| TSS200 | 47,242/47,242 | 0.048 | 0.049 | 0.048 | .031 |
| 5’UTR | 37,987/37,987 | 0.141 | 0.144 | 0.135 | <0001 |
| 1stExon | 20,239/20,239 | 0.062 | 0.063 | 0.059 | .002 |
| Gene body | 141,917/141,917 | 0.817 | 0.819 | 0.755 | <0001 |
| 3’UTR | 15,188/15,188 | 0.853 | 0.853 | 0.815 | <0001 |
| Chromatin structure* | |||||
| 1_TssA | 60,614/125,965 | 0.039 | 0.040 | 0.048 | <0001 |
| 2_TssAFlnk | 41,806/21,400 | 0.093 | 0.095 | 0.187 | <0001 |
| 3_TxFlnk | 29,974/872 | 0.037 | 0.038 | 0.497 | <0001 |
| 4_Tx | 28,060/23,942 | 0.895 | 0.895 | 0.863 | <0001 |
| 5_TxWk | 33,593/55,988 | 0.876 | 0.877 | 0.835 | <0001 |
| 6_EnhG | 4,486/4399 | 0.856 | 0.858 | 0.701 | <0001 |
| 7_Enh | 23,797/24,161 | 0.741 | 0.746 | 0.586 | <0001 |
| 8_ZNF/Rpts | 894/702 | 0.852 | 0.851 | 0.673 | <0001 |
| 9_Het | 2,119/1,528 | 0.805 | 0.805 | 0.806 | 0.775 |
| 10_TssBiv | 3,872/12,473 | 0.064 | 0.066 | 0.087 | <0001 |
| 11_BivFlnk | 9,281/4,212 | 0.090 | 0.094 | 0.091 | 0.008 |
| 12_EnhBiv | 9,381/3,645 | 0.146 | 0.151 | 0.186 | <0001 |
| 13_ReprPC | 15,708/17,536 | 0.254 | 0.263 | 0.264 | <0001 |
| 14_ReprPCWk | 39,455/36,929 | 0.684 | 0.683 | 0.804 | <0001 |
| 15_Quies | 117,092/86,380 | 0.841 | 0.841 | 0.848 | <0001 |
1: Active TSS; 2: Flanking Active TSS; 3: Transcription at gene 5’ and 3’; 4: Strong transcription; 5: Weak transcription; 6: Genic enhancers; 7: Enhancers; 8: ZNF genes & repeats; 9: Heterochromatin; 10: Bivalent/Poised TSS; 11: Flanking Bivalent TSS/Enh: 12: Bivalent Enhancer; 13: Repressed PolyComb; 14: Weak Repressed PolyComb; 15: Quiescent/Low. Detailed description is reported previously [17].
Each p-value was derived from a non-parametric Kruskal-Wallis test which compares the difference of median methylation levels between T1, T2 and brain.
Next, we examined the distribution of methylation by gene features (Figure 2B). On average, CpGs in sub-genic regions including 1stExon, 5’UTR, TSS200 and TSS1500 (200 and 1500 base pairs from transcription starting site) were hypo-methylated, and CpGs in 3’UTR and gene bodies were hyper-methylated. Similar to the findings for island features, the differences in distributions by gene features were also conserved across tissues, and we also found that the median methylation levels were lower in brain for these sub-genic regions (Table 2).
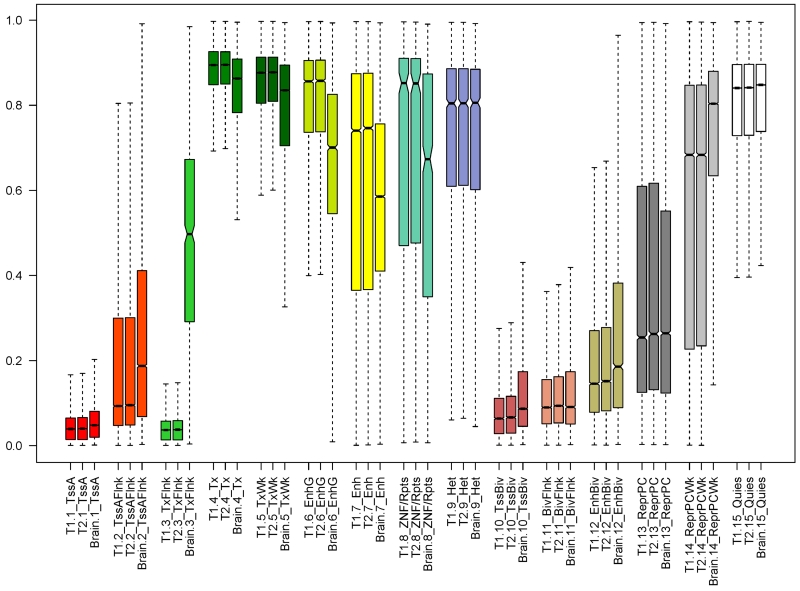
Finally, we examined the distribution of methylation by chromatin structure. Notable differences were observed across the 15 previously identified chromatin states [17] (Figure 3). As expected, CpGs in active promoters (i.e. TssA) were extremely hypo-methylated; and CpGs in strong transcription state (i.e. Tx) were extremely hyper-methylated. We observed overall similarity across tissues in methylation distribution by chromatin state. However, there was evidence that the average methylation level varied between CD4+ lymphocytes and brain in select states. In particular, the median DNA methylation level in transcribed state at 3’UTR and 5’UTR of genes (i.e. TxFlnk) was over 12 times higher in brain than in CD4+ lymphocytes proximate to death (Table 2). The difference between CD4+ lymphocytes and brain was also observed in other states including enhancer states and a state associated with zinc finger protein genes. Previous report shows that H3K4mel-associated states, including TxFlnk, EnhG, Enh and EnhBiv, are the most variable states across tissues [17].
Figure 3.
illustrates global distribution for CD4+ lymphocytes and brain methylation by chromatin structure. The boxplot compares the methylation levels between T1 CD4+ lymphocytes, T2 CD4+ lymphocytes, and brain, stratified by the 15 chromatin states. Abbreviations: TssA: Active transcirption stating site (TSS); TssAFlnk: Flanking Active TSS; TxFlnk:Transcription at gene 5’ and 3’; Tx: Strong transcription; TxWk: Weak transcription; EnhG: Genic enhancers; Enh: Enhancers; ZNF/Rpts: ZNF genes & repeats; Het: Heterochromatin; TssBiv: Bivalent/Poised TSS; BivFlnk: Flanking Bivalent TSS/Enhancer; EnhBiv: Bivalent Enhancer; ReprPC: Repressed PolyComb; ReprPCWk: Weak Repressed PolyComb; Quies: Quiescent. Detailed description of Chromatin states is reported previously [17].
CpG specific methylation correlation and difference across tissues
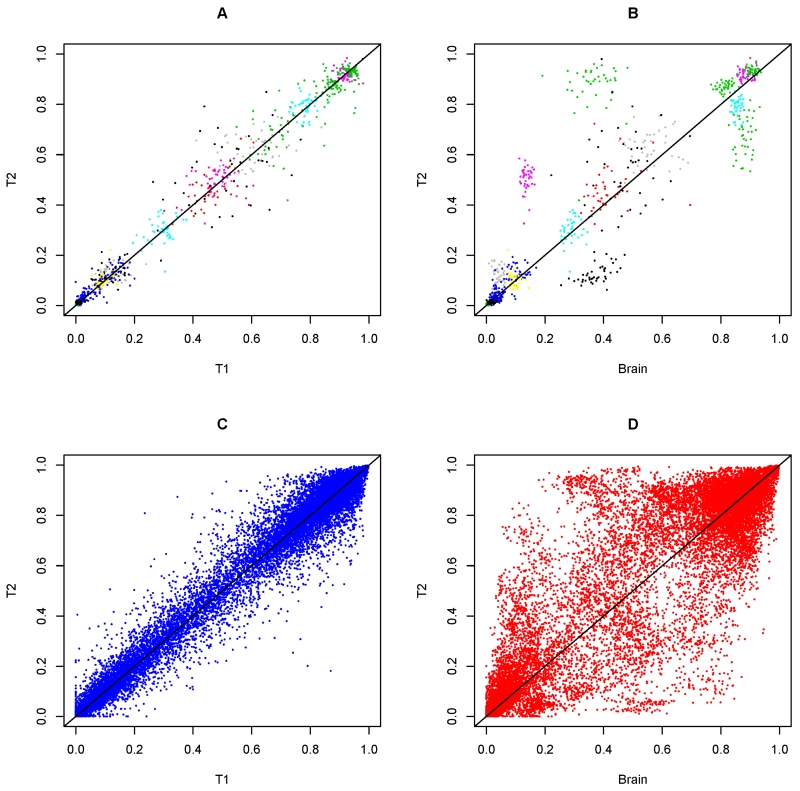
Next, we compared the DNA methylation at individual CpG sites. In contrast to the global distribution, methylation at individual CpG sites was mostly unimodal (Figure S1). Further, distributions for T1 and T2 CD4+ lymphocytes methylation were largely overlapping with one another. Comparing CD4+ lymphocytes with brain, the distribution was similar at some sites, but differed markedly at others. Pair plots of methylation across tissues show that T1 and T2 CD4+ lymphocytes methylation were highly concordant, but that of CD4+ lymphocytes and brain were more diffuse (Figure 4). For each of the 420,132 CpGs, we obtained the Pearson correlation coefficient (r) for methylation between T1 and T2 CD4+ lymphocytes, and separately between T2 CD4+ lymphocytes and brain. We observe mild to moderate correlations between CD4+ lymphocytes methylation (mean Pearson r =0.23, SD=0.23), and the correlations for T2 CD4+ lymphocyte and brain was much weaker (mean Pearson r =0.06, SD=0.18).
Figure 4.
illustrates the concordance and discordance of DNA methylation across tissues. Panel A shows the pair plot of T1 and T2 CD4+ lymphocytes methylation at 10 random selected CpG sites. Each colored cloud represents methylation measure for a particular CpG. Panel B shows the pair plot of T2 CD4+ lymphocytes and brain for the same 10 CpGs. Panel C shows the pair plot of T1 and T2 CD4+ lymphocytes methylation at 500 random selected CpG sites. Panel D shows the pair plot of T2 CD4+ lymphocytes and brain for the same 500 CpGs.
Using paired t-tests, we scanned for difference of average DNA methylation between T1 and T2 CD4+ lymphocytes (Figure S2a). Only 1 CpG (cg24912310, p=8.78×10−8) was significant after Bonferroni correction for multiple testing (α=10−7). The CpG is located in a strong transcription region of the WDR88 gene in chromosome 19. By contrast, in a separate scan for difference between T2 CD4+ lymphocytes and brain, 46.8% of the 420,131 interrogated CpGs showed significant difference (Figure S2b). A majority (59%) of the significant CpGs had lower average methylation levels in brain, compared to T2 CD4+ lymphocytes.
Blood methylation and AD pathology
More than 70% of the subjects included in the analysis were diagnosed with pathologic AD at autopsy. The limited sample size (N=41) prohibited conducting a genome-wide comparison of CD4+ lymphocytes and brain methylation for association with AD traits. Therefore, we first describe the global features of methylation level by disease status. Interestingly, there was only modest difference in global methylation distribution between subjects with (N=30) and without (N=11) pathologic AD. Specifically, the percent difference in median methylation level between the two groups was mostly under 5%, regardless of island features, gene features or chromatin states (Table S1). This was true for both CD4+ lymphocytes and brain, and the corresponding density distributions were visually identical.
Next, we targeted 12 validated CpGs and 2 CpGs in known AD loci that we previously published with a genome wide brain DNA methylation association with neuritic plaque burden [4]. Using multivariable linear regression, we examined the association of brain methylation with neuritic plaque pathology, and repeated the models for T1 and T2 CD4+ lymphocytes methylation (Table 3). Notably, even with this markedly smaller sample (prior N>700), we captured most of the signals using brain methylation measure, such that 3 CpG sites survived Bonferroni correction for multiple testing (α=0.0036) and additional 8 CpGs reached nominal significance (α=0.05). Further, consistent with the prior report, all the regression coefficients showed positive signs, suggesting that higher methylation level at these sites were associated with more burden of neuritic plaque pathology. By contrast, none of the associations was replicated with either CD4+ lymphocytes measure, and many of the associations were in the opposite direction from that found with brain.
Table 3.
DNA methylation association with neuritic plaque pathology at target CpGs
| CpG | Chr | Position (bp) | Brain (Est, SE, p) | T1 (Est, SE, p) | T2 (Est, SE, p) | Genes* |
|---|---|---|---|---|---|---|
| cg11724984 | 12 | 121890864 | 4.484, 1.611, 0.008 | −4.476, 14.620, 0.761 | −37.329, 16.193, 0.027 | RNF34, KDM2B |
| cg23968456 | 10 | 73521631 | 8.923, 3.474, 0.014 | 0.292, 15.042, 0.985 | 3.861, 17.455, 0.826 | CDH23, C10orf105, C10orf54 |
| cg15821544 | 1 | 43473840 | 6.054, 2.591, 0.025 | −18.283, 7.730, 0.023 | 0.629, 7.262, 0.931 | SLC2A1, FLJ32224 |
| cg16733298 | 16 | 19127132 | 4.533, 1.546, 0.006 | 5.073, 5.680, 0.378 | 1.841, 4.879, 0.708 | COQ7, ITPRIPL2 |
| cg22962123 | 7 | 27153605 | 2.433, 1.118, 0.036 | 12.066, 8.077, 0.144 | −6.713, 4.742, 0.165 |
HOXA1, HOTAIRM1, HOXA2, AK291164, HOXA3, AK311383, BC035889, HOXA4, LOC100133311, HOXA5, HOXA6, DQ655986, HOXA7, HOXA9, HOXA10 |
| cg13076843 | 17 | 74475294 | 2.664, 1.204, 0.033 | −3.627, 6.847, 0.599 | −7.978, 6.577, 0.233 |
UBE2O, AANAT, RHBDF2, AX747521, CYGB, PRCD |
| cg25594100 | 7 | 4786943 | 5.983, 1.679, 0.001 | −14.313, 8.775, 0.111 | −2.784, 9.799, 0.778 |
FOXK1, AP5Z1 (KIAA0415), RADIL |
| cg00621289 | 21 | 47855916 | 7.654, 2.446, 0.003 | 2.541, 3.762, 0.504 | −9.643, 4.714, 0.048 | PCNT, DIP2A |
| cg19803550 | 17 | 1637391 | 7.797, 2.310, 0.002 | 3.197, 9.073, 0.727 | 2.193, 12.778, 0.865 |
PRPF8, TLCD2, MIR22HG, AF070569, MIR22, WDR81, SERPINF2, SERPINF1, SMYD4 |
| cg03169557 | 16 | 89598950 | 7.012, 2.428, 0.006 | −5.847, 6.425, 0.369 | −25.303, 18.51, 0.180 |
ANKRD11, SPG7, SNORD68, RPL13, CPNE7 |
| cg05066959 | 8 | 41519308 | 2.357,1.360, 0.091 | −4.726, 2.455, 0.062 | 0.109, 2.765, 0.969 |
AGPAT6, NKX6-3, JA429246, ANK1 |
| cg05810363 | 17 | 74475270 | 3.502,1.744, 0.052 | −11.120, 15.853, 0.487 | −13.722, 24.318, 0.576 |
UBE2O, AANAT, RHBDF2, AX747521, CYGB, PRCD |
| cg22883290 | 2 | 127800646 | 8.232, 2.802,0.006 | −11.023,12.589, 0.387 | 4.795, 11.464, 0.678 | BIN1 |
| cg02308560 | 19 | 1071176 | 0.895, 1.293, 0.493 | 1.249, 2.399, 0.606 | 2.389, 2.691, 0.380 |
CNN2, ABCA7, HMHA1, POLR2E, GPX4, SBNO2 |
Genes within 50 kb of associated CpG [4]
DISCUSSION
Increasing evidence suggests that alteration of DNA methylation is involved in AD [29]. Challenges in interrogating methylation in AD includes searching for suitable neuroepigenomic surrogates [30] and understanding the dynamics of change in DNA methylation [31]. In this study, by leveraging three sets of genome-wide DNA methylation data from 41 autopsied older persons, we characterized and compared the methylation profiles from two CD4+ lymphocytes measures on average 7.5 years apart, including one proximate to death, as well as post-mortem brain tissue. We then explored the association of CD4+ lymphocytes and brain methylation with neuritic plaque pathology.
We found one study that examined the change in DNA methylation in unfractionated peripheral blood cells among 111 older persons, where both increase and decrease in global methylation level are observed between 2 visits that were on average 11 years apart [21]. However, the comparison is primarily restricted to global methylation, and the Illumina chip used for analysis targets less than 2,000 CpG sites. In our study, there is no strong evidence of such change in CD4+ lymphocytes over 7.5 years despite significant changes in phenotype, e.g., the development of AD dementia. Specifically, global methylation distributions are almost identical between T1 and T2, and this holds for distributions stratified by several different epigenomic feature types. Numeric comparison indicates that methylation in CD4+ lymphocytes shows only slight elevation in T2. At the individual CpG level, the pairwise correlations are not strong, but methylation levels are largely concordant between the two time points. We found only one differentially methylated CpG site between the two CD4+ lymphocytes measures, after correction for multiple comparisons. Taken together, these results indicate that there is little change of large magnitude in CD4+ lymphocytes DNA methylation over several years in late life.
Until recently, the tissue specific methylation landscape was not well understood. As part of the NIH Roadmap Epigenomics Mapping Consortium (http://www.roadmapepigenomics.org) [32], next-generation sequencing approaches were recently used to characterize distinct patterns of genome-wide DNA methylation between whole blood and multiple brain regions. Here, we compare our results with findings in a study from the Epigenomics Mapping Consortium [18]. First, the gene methylation profiles captured in our study using Illumina array are overall consistent. Briefly, both studies show that CpGs around the transcription start sites are on average hypo-methylated and those in the gene body are hyper-methylated. The Mapping Consortium suggested a subtle decrease in methylation level at the 3’ end of the gene. By contrast, our data suggest that CpGs in the 3’ remain extremely hyper-methylated. Most importantly, both studies confirm that these gene features are generally conserved across tissues. Further, we show that methylation profiles by other epigenomic features including island features and chromatin structure are also generally conserved between CD4+ lymphocytes and brain.
Second, we confirm that there are significant difference in average methylation level between CD4+ lymphocytes and brain within the same individuals. The Mapping Consortium identified a number of tissue-specific differentially methylated regions (TS-DMRs). Among the top TS-DMRs, a majority have lower methylation level in brain. They further reported that 74% of the 206 CpG sites covered by these TS-DMRs show methylation difference between tissues. In this study, we find that DNA methylation levels at approximately half of the autosomal CpG sites differ between CD4+ lymphocytes proximate death and brain, a majority of which have lower methylation in brain. Notably, in our data, 73% of the same 206 CpGs reported in the roadmap study also show significant difference.
The results from the two studies also have differences. In particular, the Mapping Consortium found that between-subject differences in methylation are correlated across tissue, implicating potential utility of using methylation in peripheral tissues as a surrogate marker for brain methylation. Our results show that, on average, the pairwise correlations of CD4+ lymphocytes and brain methylation at the individual CpG level are minimal, and the lack of concordance is striking. More importantly, we target DNA methylation at 14 CpG sites that we previously reported to be associated with neuritic plaque pathology. While we replicate the finding with brain at most sites with a sample size barely 1/20th of the size, we are not able to replicate any of the associations using CD4+ lymphocytes measures from the same individuals. These findings suggest that the brain methylation associations with AD pathology at target sites are not directly reflected in CD4+ lymphocytes from the identical sample. We appreciate that these results only apply to CD4+ lymphocytes and we cannot extrapolate to other peripheral blood cells. It is possible that alterations in the methylation of other peripheral blood cells (e.g. monocytes) are involved in the disease process through different systems or may reflect ongoing processes in the central nervous system (CNS) [33]. Future studies are warranted to explore the utility of peripheral measures of methylation. Overall, the availability of blood distal and proximate to death allows us to associate with brain methylation as well as AD pathology from the same subjects. The new data reported here reflect findings that are different and complementary to those of the Consortium’s.
To our knowledge, this is the first study to cross-examine the genome-wide methylation profiles in CD4+ lymphocytes and postmortem brain from same subjects. Given the considerable interest in peripheral blood biomarkers for AD, our study offers insights into change of DNA methylation in CD4+ lymphocytes over several years in late life, as well as the potential for DNA methylation of CD4+ lymphocytes to recapitulate what can be identified via DNA methylation of neocortical homogenates. Given the rare biospecimens and costs, we intentionally did not power the study for genome-wide discovery of methylation signals with CD4+ lymphocytes. The lack of association with AD pathology in CD4+ lymphocytes may only apply to the target sites interrogated in the study. Nonetheless, we believe that the sample size is sufficient for the intended purpose, namely to inform go-no go decisions regarding larger studies. Indeed, one of our take home messages is that brain methylation associations with AD pathology are not directly reflected in peripheral blood CD4+ lymphocytes. Based on our finding, we elected not to proceed with a well-powered study given the virtually irreplaceable blood biospecimen in our autopsied subjects. We believe these data will be of great value to researchers when considering the design of future studies in this space.
Other limitations are noted. First, due to the design of microarray, the CpGs interrogated in the study, although genome-wide, only cover a small proportion of the methylation sites and are primarily located in promoters and CpG islands. As a result, the findings may not be generalizable to the entire methylome. Second, the time interval between the two blood collections is on average 7.5 years, which may not be long enough to capture the change of age related DNA methylation in CD4+ lymphocytes. Pathologic measures are not available at the time of blood sampling, and this presents a challenge in directly comparing the blood to brain methylation in association with AD pathology.
Supplementary Material
Research in context.
Systematic review: Increasing literature suggests that dysregulations of DNA methylation is an early indication of AD, which leads to great interest in exploring methylation markers of AD in peripheral tissue such as blood cells. Yet findings on blood methylation with AD have been inconsistent.
Interpretation: Our data shed light on the potential for DNA methylation of CD4+ cells to recapitulate what can be identified via DNA methylation of neocortical homogenates. We observe that global methylation features are conserved in both CD4+ cells and brain. However, there is no strong evidence of change in CD4+ methylation over an average of 7.5 years in late life. The previously identified brain methylation association with neuritic plaques at target sites is not replicated in CD4+ cells.
Future directions: Methylation association with AD pathology found in neocortex is not directly reflected in CD4+ cells. Future studies are warranted to replicate these findings and further elucidate the utility of peripheral measures for a role of methylation in neurodegeneration.
ACKNOWLEDGEMENT
The study was supported by National Institutes on Aging grants P30AG10161, R01AG15819, R01AG17917, R01AG36042, and U01AG46152. We thank the staff of the Rush Alzheimer’s Disease Center of Rush University Medical Center and the De Jager Lab of the Brigham and Women’s Hospital and Broad Institute of Harvard and MIT. Special thanks to the participants in the Religious Orders Study and the Rush Memory and Aging Project for donating their data and biospecimen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- [2].Jowaed A, Schmitt I, Kaut O, Wullner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:6355–9. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nature neuroscience. 2014;17:1164–70. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nature neuroscience. 2014;17:1156–63. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiology of aging. 2014;35:1334–44. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- [6].Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2354–9. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis : official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases. 2009;10:418–29. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- [8].Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HW, Mastroeni D, et al. The epigenetics of aging and neurodegeneration. Progress in neurobiology. 2015 doi: 10.1016/j.pneurobio.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bollati V, Galimberti D, Pergoli L, Dalla Valle E, Barretta F, Cortini F, et al. DNA methylation in repetitive elements and Alzheimer disease. Brain, behavior, and immunity. 2011;25:1078–83. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Di Francesco A, Arosio B, Falconi A, Micioni Di Bonaventura MV, Karimi M, Mari D, et al. Global changes in DNA methylation in Alzheimer’s disease peripheral blood mononuclear cells. Brain, behavior, and immunity. 2015;45:139–44. doi: 10.1016/j.bbi.2014.11.002. [DOI] [PubMed] [Google Scholar]
- [11].Tannorella P, Stoccoro A, Tognoni G, Petrozzi L, Salluzzo MG, Ragalmuto A, et al. Methylation analysis of multiple genes in blood DNA of Alzheimer’s disease and healthy individuals. Neuroscience letters. 2015 doi: 10.1016/j.neulet.2015.06.009. [DOI] [PubMed] [Google Scholar]
- [12].Arosio B, D’Addario C, Gussago C, Casati M, Tedone E, Ferri E, et al. Peripheral blood mononuclear cells as a laboratory to study dementia in the elderly. BioMed research international. 2014;2014:169203. doi: 10.1155/2014/169203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hernandez HG, Mahecha MF, Mejia A, Arboleda H, Forero DA. Global long interspersed nuclear element 1 DNA methylation in a Colombian sample of patients with late-onset Alzheimer’s disease. American journal of Alzheimer’s disease and other dementias. 2014;29:50–3. doi: 10.1177/1533317513505132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jiang R, Jones MJ, Chen E, Neumann SM, Fraser HB, Miller GE, et al. Discordance of DNA methylation variance between two accessible human tissues. Scientific reports. 2015;5:8257. doi: 10.1038/srep08257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Human molecular genetics. 2009;18:4808–17. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R, et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome biology. 2014;15:r54. doi: 10.1186/gb-2014-15-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome biology. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics. 2012;90:260–72. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. Distinct DNA methylomes of newborns and centenarians. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. Jama. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Current Alzheimer research. 2012;9:628. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the Rush Memory and Aging Project. Current Alzheimer research. 2012;9:646. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome research. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schneider JA, Arvanitakis Z, Yu L, Boyle PA, Leurgans SE, Bennett DA. Cognitive impairment, decline and fluctuations in older community-dwelling subjects with Lewy bodies. Brain : a journal of neurology. 2012;135:3005–14. doi: 10.1093/brain/aws234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bennett DA, Schneider JA, Bienias JL, Evans DA, Wilson RS. Mild cognitive impairment is related to Alzheimer disease pathology and cerebral infarctions. Neurology. 2005;64:834–41. doi: 10.1212/01.WNL.0000152982.47274.9E. [DOI] [PubMed] [Google Scholar]
- [27].Bennett D, Arnold S, Valenzuela M, Brayne C, Schneider J. Cognitive and social lifestyle: links with neuropathology and cognition in late life. Acta neuropathologica. 2014;127:137–50. doi: 10.1007/s00401-013-1226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bennett DA, Yu L, Yang J, Srivastava GP, Aubin C, De Jager PL. Epigenomics of Alzheimer’s disease. Translational research : the journal of laboratory and clinical medicine. 2015;165:200–20. doi: 10.1016/j.trsl.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lord J, Cruchaga C. The epigenetic landscape of Alzheimer’s disease. Nature neuroscience. 2014;17:1138–40. doi: 10.1038/nn.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Satterlee JS, Beckel-Mitchener A, Little R, Procaccini D, Rutter JL, Lossie AC. Neuroepigenomics: Resources, Obstacles, and Opportunities. Neuroepigenetics. 2015;1:2–13. doi: 10.1016/j.nepig.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PloS one. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature biotechnology. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lunnon K, Ibrahim Z, Proitsi P, Lourdusamy A, Newhouse S, Sattlecker M, et al. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer’s disease blood. Journal of Alzheimer’s disease : JAD. 2012;30:685–710. doi: 10.3233/JAD-2012-111592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.