Abstract
A-to-I RNA editing is an essential gene regulatory mechanism. Once thought to be a rare phenomenon only occurring in a few transcripts, the emergence of high-throughput RNA sequencing has facilitated the identification of over 2 million RNA editing sites in the human transcriptome. In this review, we survey the current RNA-seq based methods as well as historical methods used to identify RNA editing sites.
Keywords: RNA editing, post-transcriptional gene regulation, RNA sequencing
1. Introduction
Gene regulation occurs at every step along the central dogma of molecular biology. In addition to the myriad of regulatory mechanisms acting on DNA and proteins, RNA transcripts undergo a host of diverse processing mechanisms such as alternative splicing, regulated localization and nucleotide modifications. One such process acting on RNA is RNA editing, in which a base in RNA is modified enzymatically to form a different base. In metazoans, the most common type of RNA editing is Adenosine-to-Inosine (A-to-I), catalyzed by the adenosine deaminase acting on RNA (ADARs), a family of double stranded RNA binding enzymes [1]. Inosine, which is read as Guanosine by the cellular machinery, contributes to the diversity of the transcriptome by changing the amino acid sequences of proteins, influencing alternative splicing patterns, and affecting the ability of miRNAs to bind to their target sites. The identification of RNA editing sites has largely tracked with the development of DNA sequencing technologies (Figure 1) and the emergence of next-generation sequencing technologies has facilitated the identification of over 2 million RNA editing sites in humans, bringing attention to the critical role of RNA editing in gene regulation. In this review, we will provide a historical perspective on the identification of human RNA editing sites.
Figure 1.
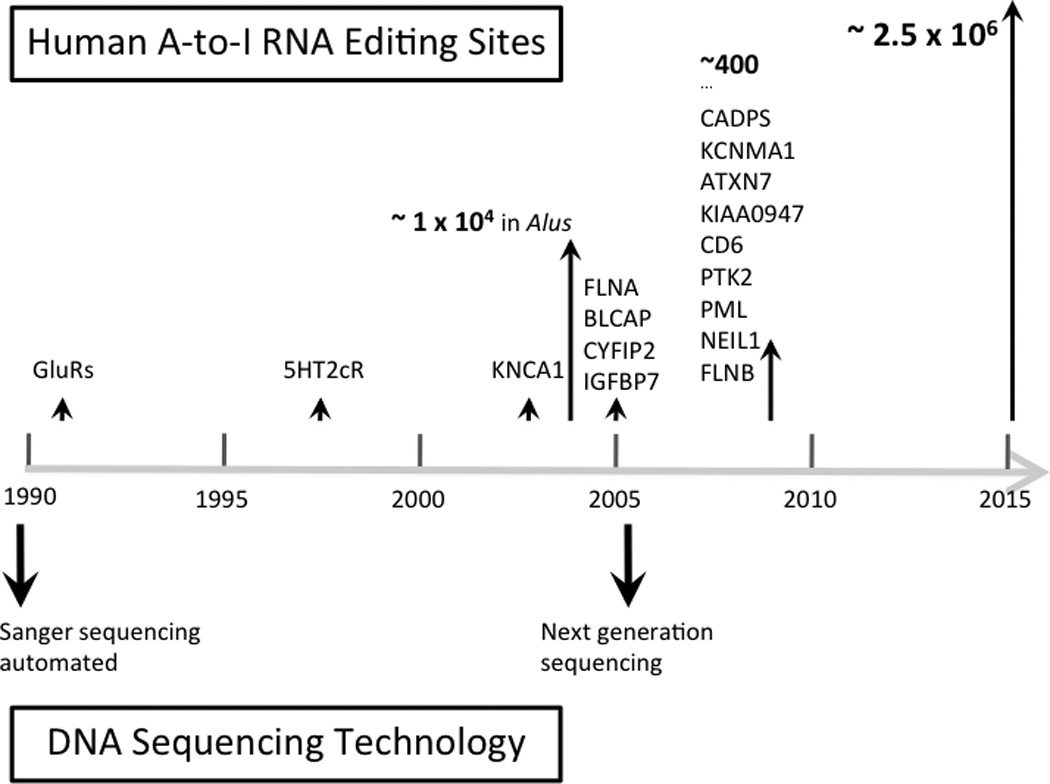
Timeline of A-to-I RNA editing site discovery
2. Historical methods
2.1 Sanger sequencing of ion channel receptors
The first human A-to-I RNA editing sites were discovered serendipitously using Sanger sequencing [2] to carefully identify nucleotide differences between DNA and cDNA sequences of ion channel receptors in the brain. The first examples of edited ion channel proteins were the glutamate receptor family subunits identified in 1991: GluR2, GluR5, and GluR6 [3]. The role of RNA editing in these glutamate receptor ion channels was shown to control their calcium permeability [4], and furthermore editing at the Q/R site in GluR2 is essential for brain function as removal of editing at this site in mice causes lethality resulting from severe seizures [5]. In 1997, the next gene identified to undergo RNA editing was the serotonin receptor 5-HT(2c)R which harbors 5 RNA editing sites within its transcript [6]. Editing within the 5-HT(2c)R gene was shown to reduce the potency of serotonergic agonists [7] and abnormalities in editing have been observed in suicide victims [8, 9]. The rate of discovery for RNA editing sites was quite slow in these first years, mainly due to technological shortcomings. Sanger sequencing, although very accurate, is an extremely tedious technique when used to screen entire human genes for RNA to DNA differences.
2.2 Comparative genomics
In the late 1990s and early 2000s, the success of the human genome project [10, 11] together with the assembly of genomes from model organisms such as the mouse Mus musculus [12], the fruit fly Drosphila melanogaster [13], and the nematode worm Caenorhabditis elegans [14] enabled the use of comparative genomics to identify functional elements of the genome by evolutionary conservation. The first study to search for RNA editing sites using comparative genomics surveyed mRNAs from highly conserved genic sequences within 18 Drosophila species and identified 16 novel editing sites [15]. Intriguingly, one of the edited genes in Drosophila, the potassium channel Shaker (sh), was also edited in a human ortholog (KCNA1). Editing in KCNA1 substantially modifies the kinetic properties of the ion channel by speeding up the rate of recovery after inactivation [16]. The first studies to focus on comparative genomics in mammals searched within conserved genic loci between mouse and human to identify 4 novel editing sites in the genes: FLNA, BLCAP, CYFIP2, and IGFBP7 [17, 18]. Interestingly, none of these genes encode ion channel proteins and opened up possible roles for RNA editing outside of the nervous system.
2.3 High-throughput screening of expressed sequence tags (ESTs)
In addition to the assembly of whole genomes, the early 2000s saw the development of sequence databases and repositories to house a large collection of publically available sequences of cDNA transcripts from libraries of expressed sequence tags (ESTs). These collections of ESTs were a valuable resource for higher throughput identification of RNA editing sites. In 2004, four studies utilized systematic assessment of A-to-G mismatches, the signal for A-to-I editing, when aligning millions of human ESTs or full length cDNA transcripts to genomic sequences and in total identified greater than 10,000 RNA editing sites in over 1,000 genes [19–22]. The majority of these editing sites are clustered within Alu repeats which mostly reside in noncoding regions of genes. Mismatches from alignment of ESTs to genomic sequences were also used to populate databases of genomic polymorphisms such as dbSNP [23], however many of these variants were actually RNA editing sites. Computational strategies utilizing features of RNA editing sites such as clustering and RNA structural features were used to identify RNA editing sites that were incorrectly annotated within dbSNP [24–26]. Although thousands of editing sites were identified using alignment of ESTs to the genome, the clustering of editing sites in close proximity prohibited the alignment of “hyper-edited” (also known as ultra-edited) ESTs, sequences with extensive amounts of editing. To identify these hyper-edited transcripts, a pipeline was developed that masked potentially edited adenosines as guanosines in the genome before alignment of ESTs. Utilizing this genome masking pipeline allowed identification of greater than 15,000 editing sites in only 760 ESTs, an average of about 20 edits per EST [27].
2.4 Chemical mapping
In addition to direct nucleotide sequencing, inosines in messenger RNAs can also be detected by biochemical methods. The first biochemical method to detect inosines, developed in 1997, utilized inosine-specific cleavage of RNA by RNAse T1 after treatment with glyoxal, and demonstrated the specificity of this method using the two recoding editing sites in the GluR2 transcript [28]. In 2002, inosine-specific cleavage was used to identify 19 editing sites in human brain, 15 of which were located within repetitive elements [29]. A second chemical method to identify inosines was developed called inosine chemical erasing (ICE), which utilizes inosine cyanoethylation after acrylonitrile treatment to prevent extension of reverse transcriptase at inosine nucleotides [30]. By comparing cDNA sequence traces from untreated, which should have both adenosine and guanosine signal, against acrylonitrile treated transcripts, which should only have adenosine signal, this study identified over 5,000 RNA editing sites.
3. High-throughput RNA sequencing based methods
3.1 Targeted RNA sequencing
Within the past 10 years, the emergence of next-generation sequencing technologies has facilitated affordable sequencing for millions of DNA fragments in a single experiment [31]. In contrast to the ~5 million total publically available ESTs present in the year 2004, current RNA sequencing methods typically process around 50 million reads in a single experiment. These technologies have enabled powerful approaches to high-throughput identification of RNA editing sites. The effectiveness of next-generation sequencing to study RNA editing was demonstrated in 2009 by one of us (J.B.L.) in utilizing an effective combination of mining the EST database with next-generation sequencing. In this study, a thorough search of the EST database identified over 36,000 potential RNA editing sites that were subsequently captured using padlock probes and deeply sequenced to identify several hundred human RNA editing sites in non-repetitive sequences, a 10-fold expansion upon previously identified sites [32].
3.2 Matched RNA and DNA sequencing
Although next-generation sequencing was shown to be a potent tool in validation of editing sites predicted using ESTs, the most promising application of next-generation sequencing is ability to de novo identify editing sites without requiring any prior information. The most straightforward approach for de novo identification of editing sites is to sequence the genome and transcriptome of a single sample, call mismatches in both independently, and identify RNA specific variants. The first study to identify de novo RNA editing sites was published in 2011 and identified over 10,000 exonic RNA-DNA differences (RDDs) of all 12 possible mismatch types [33]. In 2014, the same authors go on to show that all of these RDDs are generated co-transcriptionally, within seconds after transcription [34]. These result were very surprising, because they implied the existence of numerous unknown RNA modification mechanisms in addition to canonical A-to-I and C-to-U editing. However, upon closer examination it was shown that most of the RDDs identified in the 2011 study were false positives caused by technical artifacts such as errors introduced during reverse transcription and inaccurate sequencing read alignments [35–38]. The main lesson learned from these studies was that accurate de novo identification of RNA editing sites using next-generation sequencing required meticulous methods to separate bona fide editing sites from false positives.
Keeping this lesson in mind, in 2012 and 2013 numerous groups developed computational pipelines for de novo identification of RNA editing sites from matched DNA and RNA sequencing. In stark contrast to the first study, all of these studies found that after applying rigorous filters to remove false positives, the largest type of editing sites identified were canonical A-to-I editing. Bahn et al. identified over 9,000 RNA specific variants from a human glioblastoma cell line U87MG with a majority (62%) of them indicative of A-to-I editing [39]. Peng et al. identified over 22,000 RNA specific variants from a lymphoblastoid cell line YH with a vast majority (93%) of them indicative of A-to-I editing. We identified over 500,000 RNA specific variants from two lymphoblastoid cell lines GM12878 and YH with a vast majority (95%) of them indicative of A-to-I editing. Park et al. identified over 10,000 RNA specific variants from 14 different ENCODE cell lines with the largest fraction (43%) indicative of A-to-I editing [40]. Kleinman et al. identified over 10,000 RNA specific amino acid recoding variants from two lymphoblastoid cell lines NA12891 and NA12892 with a vast majority (80%) of them indicative of A-to-I editing. Chen identified over 300,000 RNA specific variants from 7 different ENCODE cell lines with a vast majority (94–99%) of them indicative of A-to-I editing. The overlap in editing sites identified between these studies is quite low, suggesting that each study is only querying a fraction of the total editing repertoire. Although all of these studies conclude that A-to-I is the predominant RNA editing mechanism in human cells, they did not rule out the possibility that the small fraction of noncanonical mismatches remaining are caused by novel editing mechanisms. To follow up, a careful examination of these noncanonical mismatches demonstrated that all of them are likely false positives [41].
All of the RNA editing pipelines share common features to filter false positive mismatches inherent to RNA sequencing data (Figure 2). A major concern is the accuracy of mapping RNA-seq reads to their correct position in the genome. Due to the spliced nature of mRNA transcripts, many reads spanning exon-exon splicing boundaries are incorrectly mapped onto processed pseudogenes or the read ends are incorrectly mapped into adjacent introns. We recommend mapping reads in a splice-aware manner by including sequences spanning known splicing junctions as “pseudo-chromosomes” in addition to the reference genome [42] and verifying the correct alignment of mismatched reads using a highly sensitive aligner such as BLAT [43]. We also recommend filtering out mismatches in introns that reside close to the intron-exon boundary. Another major concern is artificial mismatches introduced during reverse transcription priming using random hexamers. We recommend filtering out mismatches in the first 6 base pairs of each sequencing read.
Figure 2.
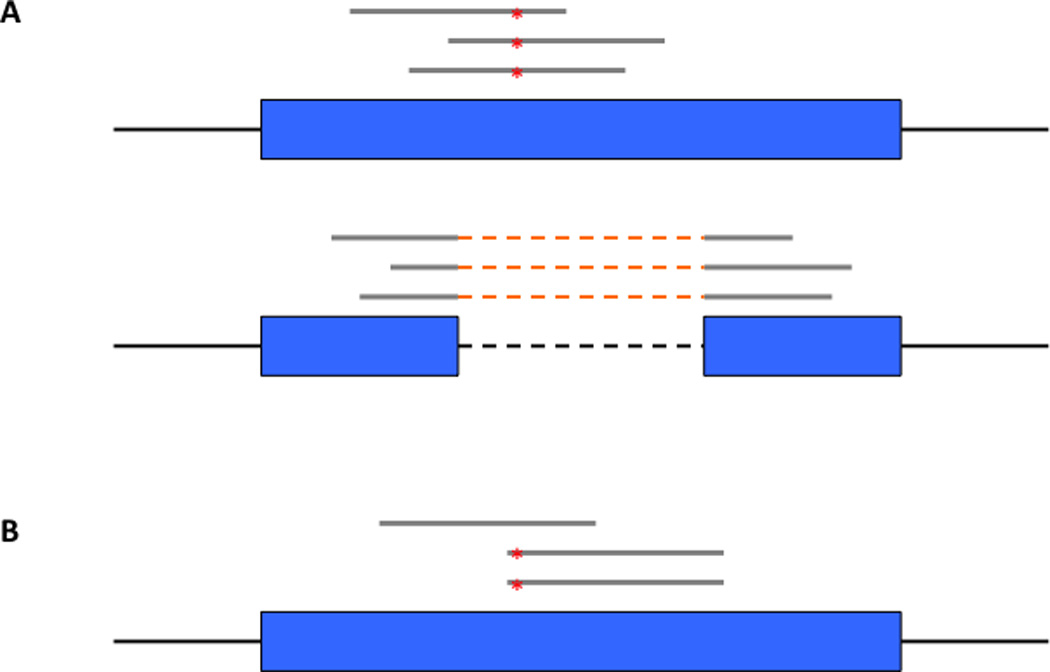
Common false positive mismatches in RNA-seq. (a) If RNA-seq reads are not mapped in a splice-aware manner, false positive mismatches in pseudogenes (top) are caused by misalignment of reads spanning splice junctions (bottom). (b) Putative variants supported only by mismatches at read ends are likely artifacts caused by errors during reverse transcription.
3.3 RNA sequencing alone
As RNA sequencing became a commonly used methodology, many publicly available datasets were deposited in sequence repositories. These datasets can be repurposed as valuable resources to identify RNA editing sites from a vast variety of human tissues and cell types. However, genome sequencing information is unavailable for most of these datasets and the pipelines mentioned in the previous section would not be able to distinguish RNA editing sites from rare genetic variants. We developed the first methods to identify RNA editing sites using RNA sequencing data without the need for matched genome sequencing. Our methods leverage the notion that RNA editing sites are common to multiple individuals while rare genetic variants reside mainly in single individuals. Using data from lymphoblastoid cell lines, brain tissues, and the Illumina Human BodyMap project, we identified over 500,000 editing sites [44]. Another method utilized the fact that A-to-I editing sites tend to be clustered together and identified over 2,000 editing sites in 266 clusters from 18 different human RNA-seq datasets [45]. Recently, a method was developed that can identify RNA editing sites in a single RNA-seq sample [46]. This method utilizes allelic linkage to distinguish between RNA editing sites and genetic variants and its utility was demonstrated using 38 samples from the GTEx project [47].
3.4 Hyper-editing and chemical mapping
Both the genome-masking technique to identify hyper-edited sequences as well as the inosine chemical erasing method (ICE) (described in Section 2 above) have been successfully coupled with high-throughput RNA sequencing. Using RNA-seq reads from the Illumina Human BodyMap project that did not map conventionally to the human reference genome, Porath et al. map these sequences onto an A-to-G masked genome and identify over 300,000 novel RNA editing sites [48]. These novel editing sites were all missed by a previous study identifying RNA editing sites on the same dataset [44], demonstrating the usefulness of the hyper-editing pipeline. The inosine chemical erasing method coupled with deep sequencing (ICE-seq) was applied to an adult human brain sample to identify almost 20,000 novel editing sites [49].
4. Databases of RNA editing
There are two main databases of RNA editing: DARNED [50, 51] and RADAR [52]. Both offer searching features to identify RNA editing sites in a particular genomic location or gene. Features unique to DARNED include sequence-based searches, dbSNP identifiers and links to Wikipedia annotations, while features unique to RADAR include annotation based searching by genic location (UTR/CDS/introns), repetitive elements and editing conservation as well as a curated list of tissue specific editing levels. The latest update to either database was to RADAR in December 2014 and includes a listing of over 2,500,000 editing sites (Figure 3).
Figure 3.
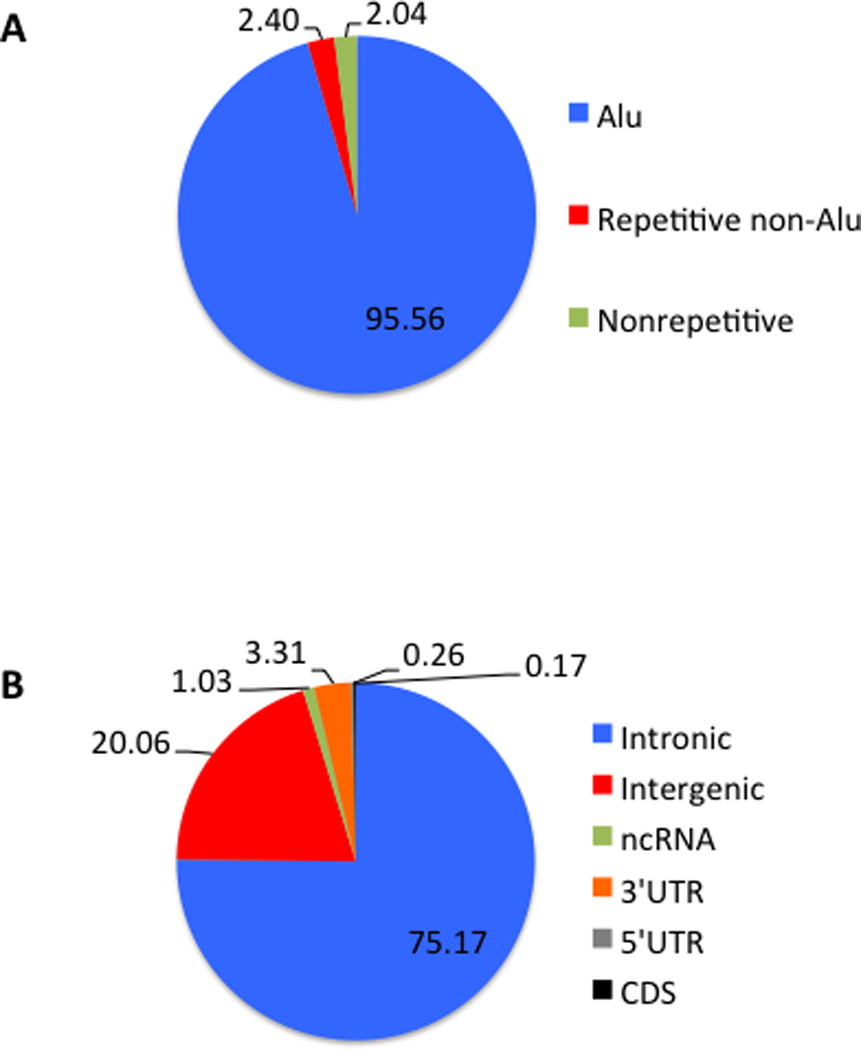
Current classifications of 2,576,459 human A-to-I RNA editing sites, downloaded from RADAR [52] release version 2. (a) Breakdown of RNA editing sites by location in Alu, repetitive non-Alu, or Nonrepetitive regions of the genome. (b) Breakdown of RNA editing sites by genic location. Numbers displayed in (a–b) are percentages.
5. Concluding remarks
The past few years have been an exciting time for the study of RNA editing. The development of powerful computational pipelines has made the study of RNA editing using RNA-seq datasets a routine endeavor. RNA editing is quite prevalent throughout the transcriptome, with almost all adenosine nucleotides edited to some degree within Alu repeats [53]. Analyses using RNA-seq datasets have already started to shed light on previously uncharacterized topics such as the evolutionary conservation of editing in primates [54], the tissue-specificity of particular editing sites [55], and the role of RNA editing in diseases such as cancer [56–58] and neurological disorders [59, 60]. Still there are many critical unanswered questions. How is the process of RNA editing regulated – what mechanisms are used by ADAR proteins to target specific adenosine nucleotides and which other proteins act in concordance with ADAR? Which of the editing sites have functional roles, and what are these functions? It will be an exciting challenge to uncover the roles of RNA editing in biology and disease.
Highlights.
Historical review of methods to identify A-to-I RNA editing sites
Our ability to identify editing sites is heavily dependent on the DNA sequencing technologies
Most likely we have identified majority of sites that are highly or moderately edited
Acknowledgments
This work was supported by a Stanford Graduate Fellowship (G.R.), NIH grant R01GM102484 (J.B.L.), and the Ellison Medical Foundation (J.B.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annual review of biochemistry. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 4.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266(5191):1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 5.Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270(5242):1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 6.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387(6630):303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 7.Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. The Journal of biological chemistry. 1999;274(14):9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 8.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34(3):349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 9.Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2001;24(5):478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Mouse Genome Sequencing C. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 13.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 14.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 15.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301(5634):832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 16.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nature structural & molecular biology. 2004;11(10):950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 17.Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic acids research. 2005;33(4):1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clutterbuck DR, Leroy A, O'Connell MA, Semple CA. A bioinformatic screen for novel A-I RNA editing sites reveals recoding editing in BC10. Bioinformatics. 2005;21(11):2590–2595. doi: 10.1093/bioinformatics/bti411. [DOI] [PubMed] [Google Scholar]
- 19.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS biology. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature biotechnology. 2004;22(8):1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 21.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome research. 2004;14(12):2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome research. 2004;14(9):1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenberg E, Adamsky K, Cohen L, Amariglio N, Hirshberg A, Rechavi G, Levanon EY. Identification of RNA editing sites in the SNP database. Nucleic acids research. 2005;33(14):4612–4617. doi: 10.1093/nar/gki771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gommans WM, Tatalias NE, Sie CP, Dupuis D, Vendetti N, Smith L, Kaushal R, Maas S. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. Rna. 2008;14(10):2074–2085. doi: 10.1261/rna.816908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. Rna. 2007;13(5):698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmi S, Borukhov I, Levanon EY. Identification of widespread ultra-edited human RNAs. PLoS genetics. 2011;7(10):e1002317. doi: 10.1371/journal.pgen.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morse DP, Bass BL. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36(28):8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- 29.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(12):7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nature chemical biology. 2010;6(10):733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- 31.Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 32.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324(5931):1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333(6038):53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang IX, Core LJ, Kwak H, Brady L, Bruzel A, McDaniel L, Richards AL, Wu M, Grunseich C, Lis JT, et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell reports. 2014;6(5):906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinman CL, Majewski J. Comment on "Widespread RNA and DNA sequence differences in the human transcriptome". Science. 2012;335(6074):1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 36.Lin W, Piskol R, Tan MH, Li JB. Comment on "Widespread RNA and DNA sequence differences in the human transcriptome". Science. 2012;335(6074):1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 37.Pickrell JK, Gilad Y, Pritchard JK. Comment on "Widespread RNA and DNA sequence differences in the human transcriptome". Science. 2012;335(6074):1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrider DR, Gout JF, Hahn MW. Very few RNA and DNA sequence differences in the human transcriptome. PloS one. 2011;6(10):e25842. doi: 10.1371/journal.pone.0025842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahn JH, Lee JH, Li G, Greer C, Peng G, Xiao X. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome research. 2012;22(1):142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park E, Williams B, Wold BJ, Mortazavi A. RNA editing in the human ENCODE RNA-seq data. Genome research. 2012;22(9):1626–1633. doi: 10.1101/gr.134957.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piskol R, Peng Z, Wang J, Li JB. Lack of evidence for existence of noncanonical RNA editing. Nature biotechnology. 2013;31(1):19–20. doi: 10.1038/nbt.2472. [DOI] [PubMed] [Google Scholar]
- 42.Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nature methods. 2012;9(6):579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent WJ. BLAT--the BLAST-like alignment tool. Genome research. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O'Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nature methods. 2013;10(2):128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu S, Xiang JF, Chen T, Chen LL, Yang L. Prediction of constitutive A-to-I editing sites from human transcriptomes in the absence of genomic sequences. BMC genomics. 2013;14:206. doi: 10.1186/1471-2164-14-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, Xiao X. Genome sequence-independent identification of RNA editing sites. Nature methods. 2015;12(4):347–350. doi: 10.1038/nmeth.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porath HT, Carmi S, Levanon EY. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nature communications. 2014;5:4726. doi: 10.1038/ncomms5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, Suzuki T. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome research. 2014;24(3):522–534. doi: 10.1101/gr.162537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26(14):1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 51.Kiran AM, O'Mahony JJ, Sanjeev K, Baranov PV. Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic acids research. 2013;41(Database issue):D258–D261. doi: 10.1093/nar/gks961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic acids research. 2014;42(Database issue):D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome research. 2014;24(3):365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Bammann H, Li M, Liang H, Yan Z, Phoebe Chen YP, Zhao M, Khaitovich P. Evolutionary and ontogenetic changes in RNA editing in human, chimpanzee, and macaque brains. Rna. 2013;19(12):1693–1702. doi: 10.1261/rna.039206.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picardi E, Manzari C, Mastropasqua F, Aiello I, D'Erchia AM, Pesole G. Profiling RNA editing in human tissues: towards the inosinome Atlas. Scientific reports. 2015;5:14941. doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nature medicine. 2013;19(2):209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HM, Eterovic AK, Yuan Y, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer cell. 2015;28(4):515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paz-Yaacov N, Bazak L, Buchumenski I, Porath HT, Danan-Gotthold M, Knisbacher BA, Eisenberg E, Levanon EY. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell reports. 2015;13(2):267–276. doi: 10.1016/j.celrep.2015.08.080. [DOI] [PubMed] [Google Scholar]
- 59.Eran A, Li JB, Vatalaro K, McCarthy J, Rahimov F, Collins C, Markianos K, Margulies DM, Brown EN, Calvo SE, et al. Comparative RNA editing in autistic and neurotypical cerebella. Molecular psychiatry. 2013;18(9):1041–1048. doi: 10.1038/mp.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khermesh K, D'Erchia AM, Barak M, Annese A, Wachtel C, Levanon EY, Picardi E, Eisenberg E. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer's disease. Rna. 2016;22(2):290–302. doi: 10.1261/rna.054627.115. [DOI] [PMC free article] [PubMed] [Google Scholar]


