Abstract
Female members of many cephalopod species house a bacterial consortium in the accessory nidamental gland (ANG), part of the reproductive system. These bacteria are deposited into eggs that are then laid in the environment where they must develop unprotected from predation, pathogens, and fouling. In this study, we characterized the genome and secondary metabolite production of Leisingera sp. JC1, a member of the roseobacter clade (Rhodobacteraceae) of Alphaproteobacteria isolated from the jelly coat of eggs from the Hawaiian bobtail squid, Euprymna scolopes. Whole genome sequencing and MLSA analysis revealed that Leisingera sp. JC1 falls within a group of roseobacters associated with squid ANGs. Genome and biochemical analyses revealed the potential for and production of a number of secondary metabolites, including siderophores and acyl-homoserine lactones involved with quorum sensing. The complete biosynthetic gene cluster for the pigment indigoidine was detected in the genome and mass spectrometry confirmed the production of this compound. Furthermore, we investigated the production of indigoidine under co-culture conditions with Vibrio fischeri, the light organ symbiont of E. scolopes, and with other vibrios. Finally, both Leisingera sp. JC1 and secondary metabolite extracts of this strain had differential antimicrobial activity against a number of marine vibrios, suggesting that Leisingera sp. JC1 may play a role in host defense against other marine bacteria either in the eggs and/or ANG. These data also suggest that indigoidine may be partially, but not wholly, responsible for the antimicrobial activity of this squid-associated bacterium.
Keywords: symbiosis, Euprymna, roseobacter, Rhodobacteraceae, indigoidine, Leisingera, DART-MS, secondary metabolite regulation
Introduction
It is becoming increasingly evident that many animals and plants use compounds produced by symbiotic bacteria for protection against pathogens and other fouling organisms (reviewed in Flórez et al., 2015). In marine and aquatic environments a number of invertebrates (including sponges, tunicates, bryozoans, and molluscs) host microorganisms that produce compounds used for such protection. These groups have served as an important source for studying defensive symbioses and for the discovery of novel bioactive natural products (see example in Schmidt and Donia, 2010).
Among molluscs, one common yet poorly understood animal–bacterial association occurs between members of squid and cuttlefish species and bacterial consortia that reside within a reproductive gland of female hosts called the accessory nidamental gland (ANG; Kaufman et al., 1998; Grigioni et al., 2000; Barbieri et al., 2001; Pichon et al., 2005; Collins et al., 2012). This organ harbors a dense consortium of bacteria housed in epithelium-lined tubules that are attached to the nidamental gland, the organ that secretes the jelly coat (JC) surrounding fertilized eggs. Bacteria from the ANG are deposited into the JC where they have been hypothesized to help protect developing eggs from fouling microorganisms, pathogens, and/or predation (Barbieri et al., 1997, 2001; Collins et al., 2012, 2015).
The Hawaiian bobtail squid, Euprymna scolopes, has been used as a model organism to study bacteria–host interactions, mainly due to the host’s relationship with the bioluminescent bacterium Vibrio fischeri (McFall-Ngai, 2014). Recent studies have also focused on a second association found within the ANG of this species (Collins and Nyholm, 2011; Collins et al., 2012, 2015). These studies demonstrated that the ANG consortium in E. scolopes is dominated by members of the Rhodobacteraceae (roseobacters) within the Alphaproteobacteria, a common group of marine bacteria. A number of roseobacter-clade organisms are known to produce unique antimicrobial molecules and other secondary metabolites. For example, the antibiotic tropodithietic acid (TDA) and the algicidal roseobacticides are produced by Phaeobacter species and the antibacterial compound indigoidine is produced by Leisingera (formerly Phaeobacter) sp. Y4I (Geng et al., 2008; Seyedsayamdost et al., 2011; Cude et al., 2012). Most of these studies have focused on either free-living or plankton-associated roseobacters and the potential antimicrobial activity of the ANG strains has not been explored. A study that analyzed the genomes of 13 ANG roseobacter strains from E. scolopes did reveal the potential for secondary metabolite production (Collins et al., 2015) and Gammaproteobacteria from the ANG of another squid species have been shown to inhibit other bacteria (Barbieri et al., 1997).
In this study, we characterized the genome and secondary metabolite production of a new bacterial strain, Leisingera sp. JC1, isolated from the JC of E. scolopes squid eggs. Whole genome sequencing and biochemical analyses revealed the potential for and production of a number of secondary metabolites, including siderophores and acyl-homoserine lactones involved with quorum sensing. The complete indigoidine biosynthetic gene cluster was detected in the genome and mass spectrometry confirmed the production of this compound. Furthermore, we investigated the regulation of indigoidine under co-culture conditions with V. fischeri, the light organ symbiont. Finally, both Leisingera sp. JC1 and extracts from this strain exhibited differential antimicrobial activity against a number of marine vibrios, suggesting that indigoidine may be partially, but not wholly, responsible for the antimicrobial activity of this squid-associated bacterium.
Materials and Methods
Bacterial Isolation
Hawaiian bobtail squid, E. scolopes, were obtained from sand flats in Oahu (Maunalua Bay, 21°16′51.42″ N, 157°43′33.07″ W), Hawaii and maintained in aquaria as previously described (Schleicher and Nyholm, 2011). Eggs laid in captivity from one adult female were collected, flash frozen on the 11th day of development, and stored at -80°C. Ten eggs were thawed for bacterial isolation and their outer capsules and embryos were removed and discarded with sterile forceps. The JCs were isolated, surface sterilized with 70% ethanol, and rinsed with filter-sterilized squid Ringers (FSSR, 530 mM NaCl, 25 mM MgCl2, 10 mM CaCl2, 20 mM HEPES, pH = 7.5). The 10 JCs were pooled and homogenized in FSSR, then serially diluted and plated on seawater tryptone (SWT) medium (5 g/L tryptone, 3 g/L yeast extract, 3 mL/L glycerol, 700 mL/L Instant Ocean sea salts, 15 g/L agar, 300 mL/L DI water). Leisingera sp. JC1 colonies appeared dark blue on this medium and were streaked to isolation.
Genomic Sequencing and Analysis
Genomic DNA was extracted using the MasterPure DNA Purification kit (Epicentre, Madison, WI, USA) from an overnight liquid culture of Leisingera sp. JC1 grown shaking at 30°C in SWT. DNA was quantified using a Qubit 2.0 fluorometer (Life Technologies, Agawam, MA, USA) and checked for quality on a 1% agarose gel and using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Agawam, MA, USA). A paired end library was prepared from 1 ng of genomic DNA using the Nextera XT DNA library kit (Illumina, Inc., San Diego, CA, USA) and quantified using the Qubit fluorometer and bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The library was sequenced on an Illumina MiSeq sequencer using 2 bp × 250 bp reads at the Microbial Analysis Resources and Services (MARS) facility at the University of Connecticut (Storrs, CT, USA).
Reads were trimmed using the CLC Genomic Workbench (Qiagen, Hilden, Germany) and a draft genome was assembled using the A5 assembler (Tritt et al., 2012). Coverage was determined by mapping trimmed reads to the draft genome assembly using CLC Genomic Workbench. The genome was annotated using the Rapid Annotation using Subsystem Technology (RAST, Aziz et al., 2008)1 server and analyzed with the Antibiotic and Secondary Metabolite Analysis Shell 3.0 (antiSMASH, Weber et al., 2015)2 for potential secondary metabolite biosynthesis gene clusters. The draft genome assembly has been deposited in DDBJ/EMBL/GenBank under accession LYUZ00000000. The version described in this paper is version LYUZ01000000.
Taxonomic Analysis and Whole Genome Comparison
Initial 16S identity suggested JC1 belonged to the genus Leisingera (data not shown). To validate this conclusion and to evaluate its relationship to the previously sequenced ANG isolates, a further taxonomic analysis was undertaken that used 17 previously described Leisingera genomes (Collins et al., 2015). A 33 gene multilocus sequence analysis was carried out following the methodology described in Collins et al. (2015). After generating alignments for each of the 33 genes using MUSCLE (Edgar, 2004), a concatenated alignment was generated using in-house python scripts. An optimal model of evolution was determined using the Akaike information criterion with correction for small sample size as implemented in jModelTest v2.1.4 (Darriba et al., 2012). The best-fitting model reported was GTR + Gamma estimation + Invariable site estimation. A maximum-likelihood (ML) phylogeny was generated from the concatenated multi-sequence alignment using PhyML v3.0_360-500M (Guindon et al., 2010). PhyML parameters consisted of GTR model, estimated p-invar, four substitution rate categories, estimated gamma distribution, sub-tree pruning and regrafting enabled with 100 bootstrap replicates. In addition to the maximum-likelihood analysis, a Bayesian inference analysis was also conducted using MrBayes v3.2.4 x64 (Ronquist et al., 2012). A mixed model with gamma estimation and invariable sites was used. The mixed model settled on a GTR submodel with only one parameter difference from the default GTR model with a posterior probability > 0.8. The standard GTR model accounted for the remainder of the model probability. The analysis used two cold chains with three heated chains each and ran for one million generations. After the run finished, convergence was assessed using average standard deviation of split frequencies of the cold chains, potential scale reduction factors of parameters, and minimum effective sample sizes of parameters. All criteria indicated the runs had converged.
Average nucleotide identity (ANI) was calculated using JSpecies 1.2.1 (Richter and Rosselló-Móra, 2009). The calculations were made using the MUMmer aligner with its default options. Contig files were generated for this analysis using the seqret function of the EMBOSS package (Rice et al., 2000). The reciprocal comparisons were averaged for reporting. Estimates of in silico DDH were made using the Genome-to-genome distance calculator 2.1 (Meier-Kolthoff et al., 2013) using the BLAST+ alignment method and the formula 2 algorithm outputs.
Select genomes were compared using the BLAST Ring Generator (BRIG) v1.0 (Alikhan et al., 2011). Default BLAST options were used. A whole genome alignment was generated using the Mauve program v2.3.1 (Darling et al., 2010). The progressiveMauve algorithm was used with default options.
Homoserine Lactone Detection
Homoserine lactone (HSL) production was detected using a well-diffusion assay with the HSL-sensing bacterium Agrobacterium tumefaciens NTL4 pZLR4 (Cha et al., 1998) as previously described (Ravn et al., 2001; Collins et al., 2015). In brief, A. tumefaciens NTL4 was grown in 3 mL of LB with 30 μg/mL gentamicin for 24 h at 30°C. This culture was used to inoculate 50 mL of AB minimal media with 0.5% casamino acids and 0.5% glucose (Chilton et al., 1974), and allowed to grow for another 24 h at 30°C. This culture was used to inoculate 100 mL of AB minimal media to which 1.2% agar had been added and a final concentration of 0.5% casamino acids, 0.5% glucose, and 75 μg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) was added after autoclaving. The inoculated molten agar was allowed to solidify in Petri dishes and wells were cut into the media using a sterile borer.
Leisingera sp. JC1 and Leisingera sp. ANG1 were grown overnight at 30°C in 3 mL of SWT broth with 30 μM FeCl3, 0.5% glucose, and 0.5% casamino acids to induce HSL production. Cells were pelleted and the supernatant was collected and filtered through a 0.22 μm filter (Thermo Scientific, Agawam, MA, USA). Cell-free supernatant (60 μl) was added to the wells in the A. tumefaciens plates. The N-3-oxohexanoyl homoserine lactone standard was serially diluted and added to wells of an A. tumefaciens plate as a control and for semi-quantitative comparison. All plates were incubated at 28°C for 24 h before imaging.
Siderophore Detection
To qualitatively detect siderophore production, Leisingera sp. JC1 was plated in triplicate on chrome azurol S (CAS) indicator agar, modified for marine bacteria as previously described (Whistler and Ruby, 2003), and incubated at 28°C for 24 h before imaging. Sequestration of iron from CAS causes a color change from blue to orange, indicating siderophore production.
Detection of Indigoidine Biosynthesis Genes in JC1 Genomic DNA
To confirm the presence of indigoidine biosynthesis genes in JC1, genomic DNA was extracted and quantified as described for genomic sequencing above. Primers were designed (Supplemental Table S1) to amplify the igiCDR genes based on the draft genome assembly and using Primer3 software (Untergasser et al., 2012). PCR amplification was performed using the standard GoTaq Green Master Mix (Promega, Madison, WI, USA) protocol with 30 cycles and 55°C annealing temperature.
Leisingera sp. JC1 Large Scale Culture
Leisingera sp. JC1 was cultured for extraction using SWT media (as described above except without addition of glycerol, delineated hereafter as SWTng). A three step culturing process was employed to produce sufficient scale for secondary metabolite extraction, while ensuring that the bacterium was in late stationary phase for optimal production of secondary metabolites (Ruiz et al., 2010). First, small scale cultures were prepared by inoculating a JC1 colony into 5 mL of media in a 24 deep well plate, which was incubated for 3 days at room temperature while shaking at 200 rpm. Then, medium scale cultures were prepared by transferring 1.5 mL of the small scale cultures into 125 mL baffled flasks with 50 mL media, which were incubated for 3 days at room temperature while shaking at 125 rpm. Lastly, large scale cultures were prepared by transferring 15 mL of medium scale cultures into 1 L baffled flasks with 500 mL of media, which were incubated for 3 days at room temperature while shaking at 125 rpm.
Extraction of Leisingera sp. JC1
All extraction solvents were ACS grade and purchased from Sigma Aldrich (St. Louis, MO, USA).
Normal Extraction
Diaion HP20 resin (Supelco, Bellefonte, PA, USA) was pre-washed by sequentially rinsing resin with methanol and Millipore water (EMD Millipore, Billerica, MA, USA). Large scale JC1 cultures were sonicated to lyse cells prior to addition of pre-washed Diaion HP20 resin (50 g, 10% w/v), followed by incubation for 24 h at room temperature while shaking at 125 rpm. Bacterial culture and resin were then filtered using a coarse glass frit filter and washed with Millipore water to remove aqueous media components. The resin and bacterial culture were then sequentially extracted with methanol, dichloromethane, and acetone (2 × 150 mL). Organic portions were combined, extracted with ethyl acetate to remove residual aqueous material, and concentrated.
Indigoidine Enriched Extraction
Because indigoidine is poorly soluble in water and most organic solvents, a second extraction protocol was utilized to prepare an indigoidine enriched extract following modified literature procedures (Yu et al., 2013). Briefly, large scale cultures were sonicated to lyse cells and transferred to centrifuge tubes. Cells were then separated from supernatant by low-speed centrifugation (850 g × 5 min; Beckman Coulter Avanti J-E Centrifuge, Brea, CA, USA). Supernatant was transferred to new tubes and subjected to high-speed centrifugation (21,000 g × 10 min) to obtain an indigoidine enriched pellet. The pellet was washed with methanol, transferred to a microcentrifuge tube, dried under N2 gas, and dissolved in dimethyl sulfoxide (DMSO).
Detection of Indigoidine Production by Leisingera sp. JC1 via LC–MS
All HPLC grade solvents and reagents were purchased from Sigma-Aldrich. LC–MS data were collected on an Agilent ESI single quadrupole mass spectrometer coupled to an Agilent 1260 HPLC system with a G1311 quaternary pump, G1322 degasser, and a G1315 diode array detector (Agilent Technologies, Santa Clara, CA). A gradient elution was used from 10% methanol in H2O to 90% methanol in H2O over 25 min using an Agilent Eclipse XDB-C18 RP-HPLC column (4.6 mm × 150 mm, 5 μm) and a flow rate of 1 mL/min. Indigoidine enriched extracts were prepared at 5 mg/mL in DMSO. Indigoidine eluted at retention time (tR) 10.7 min in agreement with literature (Yu et al., 2013).
Zone of Inhibition Assays
To observe inhibition of vibrio strains and ANG isolate strains by Leisingera sp. JC1 (Supplementary Table S3), a zone of inhibition (ZOI) assay was used. The vibrio strains V. anguillarum 775, V. parahaemolyticus KNH1, V. fischeri ES114, V. harveyi B392, and Photobacterium leiognathi KNH6 were grown for 2.5 h (to stationary phase) at 30°C in YTSS (4 g/L tryptone, 2.5 g/L yeast extract, 15 g/L Instant Ocean sea salts) broth and then serially diluted from 107 to 104 CFU/mL in YTSS broth to observe density dependent inhibition. Each dilution was plated in triplicate on YTSS agar using a sterile swab to form a lawn. All ANG isolates tested were grown overnight (~4 × 108 CFU/mL) in SWT broth at 30°C and plated on SWT agar using a sterile swab to form a lawn. Leisingera sp. JC1 was grown overnight to a density of ~1 × 108 CFU/mL in SWT when testing with ANG isolates and in YTSS when testing with vibrio strains. This overnight broth of Leisingera sp. JC1 was spotted (10 μL) on the surface of each lawn in quadruplicate. All plates were incubated at 28°C for 24 h before imaging and ZOI measurements around the Leisingera sp. JC1 colonies. SWT or YTSS broth (10 μL) were spotted on each lawn as media controls, and 10 μL of the overnight culture of Leisingera sp. JC1 was spotted in quadruplicate on SWT or YTSS agar without any bacterial lawns as a growth control.
To quantify inhibition, an average of three ZOI diameters were measured and an average of three diameters of the JC1 colonies were measured using ImageJ (Schneider et al., 2012). Due to slight variations in JC1 colony size across trials, the measurements were normalized by subtracting the average JC1 colony diameter from the average ZOI diameter. To determine if differences in ZOIs across lawn densities per organism were statistically significant, one-way ANOVAs were performed. If the results of the one-way ANOVA indicated statistically significant differences, multiple comparisons post hoc Tukey tests were performed to determine which lawn densities were significantly different.
96-Well Liquid Assays
Leisingera sp. JC1 extracts were tested for antibacterial activity against V. fischeri ES114, V. anguillarum 775, and V. parahaemolyticus KNH1. High throughput assays with these bacterial strains were developed based on similar assays with natural product extracts and human pathogens (Zgoda and Porter, 2001), including obtaining CFU counts and growth curves for each of the vibrio strains as well as determining proper incubation times and temperatures and finding appropriate controls. These assays were performed in 96-well plates (Corning Costar, Corning, NY, USA) with SWT media and incubated at 28°C while shaking at 200 rpm. The bacterial inocula were prepared by adding select colonies into 5 mL of media and adjusted to OD600 0.1 (approximately 1–2 × 108 CFU/mL as per Clinical and Laboratory Standards Institute, 2012). Colony forming unit (CFU) counts were manually confirmed to ensure accurate approximation for each vibrio strain.
Extracts were screened as previously described (Zgoda and Porter, 2001) with the following modifications. Briefly, master mix was prepared by addition of 1.6 mL adjusted vibrio inoculum, 7.84 mL sterile water, and 6.4 mL of SWT media. To each well, 198 μL of master mix was added with 2 μL of either positive control (chloramphenicol, final testing concentration 2.5 μg/mL), negative control (DMSO), or extract prepared in DMSO (screened at final concentration of 500 μg/mL; MIC performed using serial dilutions). Sterility control wells consisted of 98 μL sterile water, 100 μL of SWT media, and 2 μL of DMSO. All controls and samples were tested in technical triplicates with experiments repeated a minimum of three times to confirm results. Plates were read at 600 nm every 2 h from 0 to 10 h with a final reading at 24 h using a Synergy H1 Hybrid Reader (Biotek, Winooski, VT, USA). Results are given as percent control activity (PCA) calculated in comparison with DMSO, the negative control.
Localization of Indigoidine Production by Leisingera sp. JC1 Using DART-MS
Direct analysis in real time-mass spectrometry (DART-MS) analysis was performed using a JEOL AccuTOF with DART ion source (IonSense, Inc., Saugus, MA, USA). High purity helium 5.0–6.0 grade (greater than 99.999% purity) was heated to 300°C and used for ionization. Five locations were selected on JC1 colonies in the presence or absence of V. fischeri, including (A) center of colony, (B) midpoint between center and edge of colony, (C) edge of colony, (D) ZOI (in the absence of V. fischeri sample was obtained from a point equidistant from colony edge), and (E) outside ZOI. At each location a sterile single use syringe needle (BD Medical, Franklin Lakes, NJ, USA) was placed in the sample and then placed between the DART ion source and the MS inlet. Positive ion MS data were obtained over a m/z range of 60–700 and relative percent abundance was obtained for the indigoidine ion. Standards were run after sampling each colony and mass spectral data were monitored in real time to ensure no residual indigoidine remained after each sample. DART-MS is only semi-quantitative due to the potential for differential ionization, suppression of ions, and/or changes in sample concentration in the DART ion source (Sanchez et al., 2011). Therefore, relative indigoidine ion abundance was used to generate heatmaps representing a gradient from less abundance (black) to more abundance (red).
Measurement of Indigoidine Production by Leisingera sp. JC1 in Co-culture
JC1 bacterial inoculum was prepared by adding JC1 colonies into 5 mL of SWTng media in a 24 deep well plate, incubated for 24 h at room temperature while shaking at 200 rpm. Bacterial inocula for the vibrios were prepared by adding bacterial colonies of each species separately into 5 mL of SWTng media in 24 deep well plates, incubated for 2 h at 28°C while shaking at 200 rpm. All bacterial inocula (JC1, V. fischeri, V. anguillarum, V. parahaemolyticus) were adjusted to OD600 0.1 prior to use.
Co-cultures of JC1 with individual vibrios were prepared by adding 1 mL of adjusted JC1 inoculum to 10 mL SWTng media in 125 mL baffled flasks, incubated for 24 h at room temperature while shaking at 125 rpm, followed by addition of 200 μL of V. fischeri, V. anguillarum, or V. parahaemolyticus. After addition of the vibrio strain, co-cultures were incubated for an additional 24 h at room temperature while shaking at 125 rpm. Monocultures of JC1, V. fischeri, V. anguillarum, and V. parahaemolyticus were prepared by adding 1 mL of adjusted inoculum to 10 mL SWTng media in 125 mL baffled flasks, incubated for 48 h while shaking at 125 rpm.
All co-cultures and monocultures were extracted using the indigoidine enriched protocol described above. LC–MS data was obtained on the Agilent LC–MS system described above, using an isocratic method to ensure minimal baseline variation (10% acetonitrile in H2O with 0.1% formic acid over 15 min at a flow rate of 1 mL/min with 20 μL injection volume). Extracts were prepared at 5 mg/mL in DMSO. Indigoidine was detected and quantitated via measurement of area under the curve at UV absorbance 299 nm and confirmed by MS.
Results and Discussion
Genome Characteristics and General Metabolism
Taxonomic Placement of JC1
Leisingera sp. JC1 has a draft genome size of 5.19 Mb and GC content of 62.3% (Table 1), which is average for members of the roseobacter clade and similar to other squid-associated isolates (Collins et al., 2015). This larger genome size reflects the generalist lifestyle and ability to use diverse energy sources common of roseobacters (Newton et al., 2010). The repABC genes for plasmid replication are present as well as tra genes necessary for conjugative plasmid transfer, indicating the potential presence of extrachromosomal DNA. Further sequencing is necessary to confirm the number, size, and content of these putative plasmids.
Table 1.
Genome statistics of Leisingera sp. JC1.
| Genome size (Mb) | Number of contigs | N50 (bp) | G + C content (%) | Number of genes | Missing genes∗ (% of total) | Fold coverage |
|---|---|---|---|---|---|---|
| 5.19 | 168 | 123,213 | 62.3 | 5,074 | 54 (1.1) | 37 |
∗As predicted by the RAST server (Aziz et al., 2008).
Phylogenetic reconstruction methods (Bayesian and maximum-likelihood) used with the 33-gene concatenation returned identical topologies with overall strong statistical supports (Figure 1), placing JC1 close to the Leisingera taxa previously isolated from the ANG. Average nucleotide identity (ANI) and in silico DNA–DNA hybridization estimates (isDDH) support this placement. JC1 had higher ANI (90.5–91.7%) (Supplementary Table S2) and isDDH (38.8–44.6%) values with the ANG isolates than with any other Leisingera sp. These results also show that JC1 does not group with either Leisingera sp. ANG-M7 or the ANG1 group, but is still related to both (Figure 1; Supplementary Table S3), which is not unusual since other ANG isolates also fall outside the main ANG1 clade (Collins et al., 2015).
FIGURE 1.
MLSA analysis places Leisingera sp. JC1 in a sister group to other previously isolated ANG bacteria. Based on a comparison of 33 single-copy housekeeping genes, the egg jelly coat (JC) isolate JC1 is placed in the previously described roseobacter clade, “Clade 1” (Newton et al., 2010), along with seven previously isolated ANG bacteria (Collins et al., 2015). The phylogenetic tree reflects both Bayesian inference and maximum-likelihood methods with both posterior probability and bootstrap supports displayed.
There are indications that JC1 may be more similar to Leisingera sp. ANG-M7 than to the Leisingera sp. ANG1 group. Both the ANI and isDDH values between JC1 and ANG-M7 are elevated in comparison to their values with the ANG1 group. There are no support statistics for ANI so it is uncertain if the 1.2% (JC1-M7 ANI versus JC1 compared to the ANG group) and 1.6% (M7-JC1 ANI versus M7 compared to the ANG group) higher values are significantly different. However, isDDH values are supported by 95% confidence intervals. The lower interval for JC1-M7 does not overlap with the upper interval for any comparison with a member of the ANG group, suggesting the isDDH values are significantly different. Additionally, the Bayesian inference found a small fraction of topologies in which the placements of JC1 and ANG-M7 were reversed, while the maximum-likelihood analysis found this occurrence in 33 of 100 bootstrap replicates. Overall, these analyses suggest that Leisingera sp. JC1 is distinct from, but related to the current ANG isolates.
Isolates having similar pigmentation to Leisingera sp. JC1 were cultured from other egg clutches, an ANG, and ovary from different females (data not shown). Among these, colonies with a similar dark blue morphology were isolated from the JCs of 1 and 23 day old eggs laid by different females. Similar colonies were isolated from the ANG of one of these females and the ovary of another female. Preliminary 16S sequencing placed two of these isolates in the genus Leisingera (data not shown), and further sequencing will reveal if these are the same strain as JC1. In addition, the production of the pigment indigoidine was confirmed by these additional strains (see below). These data suggest that Leisingera sp. JC1 and/or other indigoidine-producing strains may be selected for in the ANG/JC symbiosis.
Primary Metabolism
Leisingera sp. JC1 has a complete Entner-Duodoroff pathway and tricarboxylic acid cycle for metabolism of glucose. JC1 lacks any orthologs of phosphofructokinase, a major enzyme of glycolysis, but does contain a glucokinase and two distinct glucose-6-phosphate-1-dehydrogenases (GAPDHs). A glucose-6-phosphate-1-dehydrogenase (GPDH) is present, which catalyzes the first step of the alternative pathways for glucose metabolism, indicating that the Entner-Duodoroff pathway is probably used instead of glycolysis. Leisingera sp. JC1 only has the first two enzymes of the oxidative pentose phosphate pathway, but any 6-phosphate-gluconate produced can be further dehydrated by the Entner-Duodoroff pathway. Glycolate is a dissolved organic carbon often excreted by phytoplankton, and can be a carbon source for marine heterotrophic bacteria (Edenborn and Litchfield, 1985). Leisingera sp. JC1 is predicted to oxidize glycolate to glyoxylate by a glycolate oxidase. JC1 has one system for glycerol uptake, the Ugp system, which can transport glycerol-3-phosphate against the concentration gradient. Sulfur oxidation genes are present, as well as a complete denitrification pathway with a copper-containing nitrite reductase. An assimilatory nitrate reductase is also present, which can convert nitrate to nitrite. An ammonia assimilation pathway is present with a ferrodoxin-dependent GOGAT, but no adenylyltransferase gene (GlnE) is present.
Transport
The high-affinity inorganic phosphate transport genes pstABCS and their regulatory genes phoBUR are present in JC1. The siderophore biosynthesis genes asbAB and siderX456, which encode high-affinity iron chelators, and the ferric iron ABC transporter, pitADC, are also present. JC1 has ABC transporters for dipeptides, oligopeptides, branched-chain amino acids, alkylphosphonate, and tungstate. The tripartite ATP-independent periplasmic (TRAP) transporter genes dctMPQ are present for unknown substrates, as well as the twin-arginine translocation (TAT) system genes, tatABC.
Leisingera sp. JC1 contains all 13 genes that encode the structural proteins essential for the Type VI Secretion System (T6SS) to function (Cianfanelli et al., 2016). The T6SS is a one-step mechanism for delivery of effectors across the Gram-negative outer membrane and membrane of the target cell, be it bacterial or eukaryotic. Widespread amongst the Proteobacteria, some T6SSs have been implicated in eukaryotic virulence (Pukatzki et al., 2006; Sana et al., 2012), but the majority are believed to play a role in bacterial competition (Hood et al., 2010; Schwarz et al., 2010). While it is possible for one T6SS system to affect both bacterial and eukaryotic targets (Jiang et al., 2014) it is believed that the system evolved for interactions with other bacteria, even in the case of intraspecific competition (Unterweger et al., 2014). Little work has been done, however, to investigate the role of T6SSs in beneficial host-symbiont relationships. Eleven of the 12 previously described ANG isolates also possess a T6SS (Collins et al., 2015), and it is possible that this system plays a role in interactions with other ANG or JC bacteria and/or the squid host. In the ANG, bacteria are partitioned into densely packed, epithelium-lined tubules, where each tubule is dominated by a particular taxon (Collins et al., 2012). These ANG/JC isolates may utilize the T6SS to outcompete other bacteria to establish colonization of a single tubule. While Leisingera sp. JC1 groups closely with other ANG isolates that also possess a T6SS (Figure 1, Collins et al., 2015), intraspecific effectors may facilitate competition between these strains, since ANG tubules are often highly pigmented with a single color (e.g., all dark blue matching the pigmentation of JC1 or all red-orange matching the pigmentation of several ANG isolates). Future studies will investigate the nature of JC1’s T6SS effector proteins in the ANG symbiosis. There are numerous classes of evolved effector VgrG proteins, each with their own enzymatic function (reviewed in Durand et al., 2014). Understanding the number and type of effectors that JC1 can produce and deliver may help elucidate any role in the symbiosis.
Secondary Metabolite Biosynthesis
Analysis with the antibiotic and Secondary Metabolite Analysis Shell (antiSMASH, Weber et al., 2015) predicted several potential secondary metabolite biosynthesis gene clusters (Supplementary Table S4). These results included three separate siderophore clusters, one bacteriocin, one HSL, one type 1 polyketide synthase (T1 PKS), one other PKS (not type 1,2,3, or trans-AT), and two clusters classified as “other.” Of these two “other” clusters, one contains the biosynthesis cluster for the known antimicrobial metabolite, indigoidine (Cude et al., 2012), while the other contains a previously described putative hybrid polyketide synthase/non-ribosomal peptide synthetase (PKS/NRPS) gene cluster known to be conserved amongst roseobacters (Martens et al., 2007). This PKS/NRPS gene cluster encodes a polyketide synthase, glycosyl transferase, non-ribosomal peptide synthetase, and phosphopantetheinyl transferase, but the product of this cluster has not yet been identified. The top homologous gene cluster of the T1 PKS is 45% similar to a cluster in the ANG isolate, Leisingera sp. ANG-M7. While some roseobacters are capable of producing the novel secondary metabolite TDA (Bruhn et al., 2006, 2007; Geng et al., 2008), genes for synthesis of this molecule were not found nor was the molecule detected via LC–MS (data not shown).
Quorum Sensing
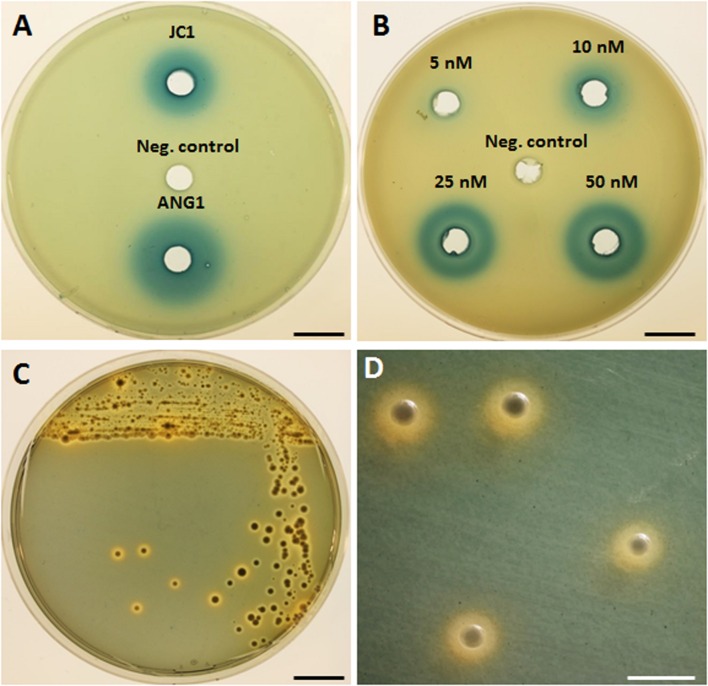
AntiSMASH predicted one luxIR homolog in Leisingera sp. JC1, flanked by an acyltransferase, crotonyl-CoA reductase, helicase, and oxidoreductase, similar to the previously published gene arrangement in bacterial isolates from the ANG (Collins et al., 2015). Production of HSLs by JC1 was confirmed in the A. tumefaciens NTL4 reporter assay, in which cell-free supernatant of a JC1 culture did induce β-galactosidase activity, indicating the presence of HSLs (Figures 2A,B). When compared to a dilution series of the N-3-oxohexanoyl HSL, JC1 produced a halo similar to that seen by 25 nM of HSL standard. The HSL production of JC1 was also slightly less than that of a closely related ANG isolate, Leisingera sp. ANG1.
FIGURE 2.
Detection of homoserine lactone production and siderophore production by Leisingera sp. JC1. (A) Homoserine lactones were detected by β-galactosidase activity in cell-free supernatant of Leisingera sp. JC1 and compared with previously tested Leisingera sp. ANG1 (Collins et al., 2015); SWT broth was used as a negative control. (B) Dilution of N-3-oxohexanoyl homoserine lactone used as a positive control for the HSL assay; DMSO was the HSL standard solvent and was the negative control. (C) Leisingera sp. JC1 plated on CAS agar. Sequestration of iron changes the media from blue to orange, indicating siderophore production. (D) Magnified view of JC1 colonies from the plate in (C), showing the orange halos in the media indicative of siderophore production. Scale bars, 1.5 cm (A–C), 4 mm (D).
Understanding the gene regulation by quorum sensing will be an important avenue of research for Leisingera sp. JC1 and the other E. scolopes ANG isolates due to the different habitats these bacteria experience. It is hypothesized that cephalopod ANGs are colonized via horizontal transmission from the environment (Kaufman et al., 1998), and potential symbionts must switch from living at very low cell densities in the seawater to very high cell densities in the ANG tubules (Collins et al., 2012). When ANG bacteria are deposited into the JC layers of eggs, these bacteria again experience a switch from the very high densities of the ANG to a lower density in the eggs. Due to this change in environments and cell densities, quorum sensing may play a role in gene regulation for ANG/egg JC bacteria.
Quorum sensing is also important in host-microbe interactions involving other roseobacters. For example, quorum sensing regulates motility and biofilm formation during host colonization in the sponge symbiont Ruegeria sp. KLH11 (Zan et al., 2012) and is necessary for colonization of the alga, Ulva australis by Phaeobacter gallaeciensis 2.10 (Rao et al., 2007). In other roseobacters, quorum sensing regulates secondary metabolite production, such as TDA in Phaeobacter gallaeciensis (Berger et al., 2011). In the indigoidine producing roseobacter, Leisingera sp. Y4I, there are two quorum sensing systems that regulate indigoidine production, pgaIR and phaIR (Cude et al., 2015). The JC1 luxI homolog has a 72% amino acid similarity to pgaI (RBY4I_1689) in Y4I, and the JC1 luxR homolog has an 81% amino acid similarity to pgaR (RBY4I_3631) in Y4I. The second set of luxIR homologs in Y4I, phaIR (RBY4I_3464 and RBY4I_1027), is not present in JC1. PgaI synthesizes the C8-HSL, produced by several proteobacteria, while PhaI synthesizes the 3OHC12:1-HSL, which may be species specific. JC1 lacking the phaIR system may reflect its divergence from Leisingera sp. Y4I. Further analyses will be needed to understand if indigoidine production in Leisingera sp. JC1 is regulated by quorum sensing.
Siderophore Production
Three separate siderophore biosynthesis gene clusters were detected in the genome, as described above, and production of iron chelators was confirmed by plating on CAS agar (Figures 2C,D). Appearance of an orange halo around colonies indicates that iron was sequestered from the chrome-azurol S dye in the media. Siderophores are high-affinity iron chelators, and can provide a growth advantage to cells in iron-limited environments, such as in seawater and in colonization of hosts. Although, the presence of siderophore biosynthesis genes in the genomes of currently sequenced roseobacter clade members is rare, 10 of the 12 previously sequenced E. scolopes ANG roseobacter symbionts did have the genes and/or demonstrate production of siderophores (Collins et al., 2015). Similar to the majority of the squid-associated roseobacter clade, the Leisingera sp. JC1 genome contains siderophore biosynthesis genes, indicating that siderophore production may play a role in the ANG symbiosis.
Indigoidine Biosynthesis Genes
The indigoidine biosynthesis gene cluster in Leisingera sp. JC1 contains all six biosynthesis genes previously described for Leisingera sp. Y4I (Cude et al., 2012) and shares a similar genome arrangement (Figure 3). To confirm the presence of individual members of the indigoidine biosynthesis gene cluster, primers were designed to three different components of the pathway, the non-ribosomal peptide synthetase (igiD), the transcriptional regulator (igiR), and one of the three indigoidine modification genes (igiC). The presence of these genes in JC1 genomic DNA was confirmed by PCR (Supplementary Figure S1).
FIGURE 3.
Leisingera sp. JC1 indigoidine biosynthesis operon. Operon organization obtained from the published genome sequence (OBY26161.1, OBY26149.1, OBY26162.1, OBY26150.1, OBY26151.1, OBY26152.1). Arrows drawn to scale.
Indigoidine biosynthesis genes have been detected in a diverse group of bacteria, including the Actinobacteria (Streptomyces), and Alpha-, Beta-, and Gamma-proteobacteria. The JC1 indigoidine biosynthesis operon shares the closest homology to the operon in Leisingera sp. Y4I, with 90–95% amino acid similarity for all gene products (Table 2). Other indigoidine biosynthesis operons share the non-ribosomal peptide synthetase, igiD, but many lack the same accessory genes required to modify indigoidine. When compared to other indigoidine producing strains, the igiD of JC1 is functionally homologous to other NRPS genes, sharing 49–53% amino acid similarity with Vogesella indigofera, Streptomyces lavendulae, and Dickeya dadantii 3937 (Table 2). A comparison with the genome of Leisingera sp. Y4I also confirmed that the indigoidine gene cluster is shared between these strains although absent from related ANG isolate Leisingera sp. M7 (Supplementary Figures S4 and S5).
Table 2.
Comparison of indigoidine biosynthesis operon in Leisingera sp. JC1 to other indigoidine producing strains.
| Gene | Annotation | % Amino acid identity to Leisingera sp. Y4I operon | % Amino acid identity to Vogesella indigofera operon | % Amino acid identity to Streptomyces lavendulae operon | % Amino acid identity to Dickeya dadantii 3937 operon |
|---|---|---|---|---|---|
| igiE | Cyanate transport protein, CynX | 95 | 58 | NA | NA |
| igiF | 4-oxalocrotonate tautomerase | 91 | NA∗ | NA | NA |
| igiD | Peptide synthetase | 91 | 53 | 50 | 49 |
| igiC | Arylmalonate decarboxylase | 95 | 56 | NA | NA |
| igiB | Hydroxyisobutyrate dehydrogenase | 93 | 51 | NA | NA |
| igiR | Transcriptional regulator, TetR | 90 | 42 | NA | NA |
∗NA, not applicable, no homolog of the gene present in that organism.
Detection of Indigoidine Production by Leisingera sp. JC1
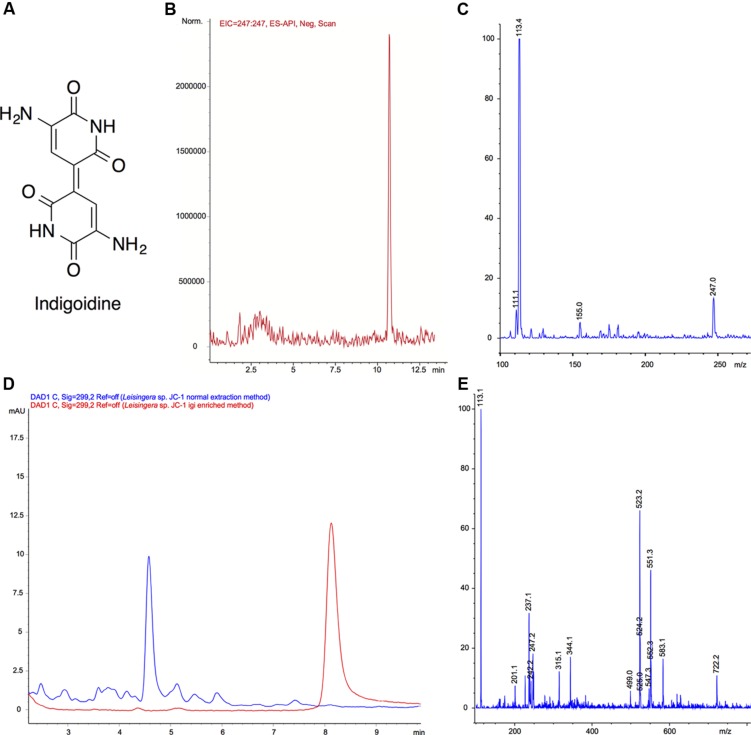
Because of the distinctive morphology and the genetic evidence for indigoidine biosynthesis, Leisingera sp. JC1 was cultured and extracted to obtain chemical evidence of indigoidine production. Using a three-step culture process, a deep blue liquid culture was obtained. However, upon extraction using a typical resin-based organic extraction protocol, most of the blue color was insoluble in organic solvents and little evidence of indigoidine production was observed via liquid chromatography-mass spectrometry (LC–MS, see Figures 4D,E), integrating to only 0.8% of the JC1 normal extract and indicative of negligible indigoidine extraction using this method. Therefore, an indigoidine enriched extraction protocol was utilized to pellet the insoluble indigoidine away from other media and cellular components followed by dissolving the sample in DMSO (Yu et al., 2013), resulting in an indigoidine enriched extract with 91.1% indigoidine. Analysis via LC–MS confirmed the presence of indigoidine (Figure 4A) in the indigoidine enriched extract (Figures 4B,C) with a peak eluting at 10.7 min with an [M-H]- of 247.0, consistent with the molecular weight and fragmentation pattern of indigoidine (248.2 g/mol) and in agreement with literature precedent (Yu et al., 2013). In addition, indigoidine was detected in two other JC and ANG isolates that exhibited a similar dark blue coloration in culture (data not shown).
FIGURE 4.
Mass spectral confirmation of indigoidine production by Leisingera sp. JC1 and comparison of normal and igi enriched extracts. (A) Structure of indigoidine. (B) Extracted ion chromatogram (EIC) of indigoidine to confirm presence in extract ([M-H]- 247.0) using a previously reported LC–MS solvent gradient (Yu et al., 2013). (C) Negative ionization mass spectrum of peak at 10.7 min. The [M-H]- peak of 247.0 is consistent with a molecular formula of C10H7N4O4, confirming the presence of indigoidine (248.19 g/mol). (D) LC–MS UV chromatogram overlays (299 nm) of normal extract (blue) with indigoidine (igi) enriched extract (red). The peak eluting at tR 8.2 min in the igi enriched extract was confirmed to be indigoidine with a purity of 91.1% (tR different due to use of isocratic solvent system to minimize baseline variation). Although, a small peak was present at tR 8.2 min in the normal extract, this integrates to only 0.8% indicating that the Leisingera sp. JC1 normal extract contains negligible amounts of indigoidine. (E) Negative ionization mass spectrum of peak at tR 8.2 min for normal extraction method. Although there was an ion corresponding to [M-H]- of 247.2, this was not the most dominant ion and the more quantitative UV analysis in D shows only a very small peak, indicative of negligible indigoidine in the JC1 normal extract.
Antibacterial Activity of Leisingera sp. JC1
Zone of Inhibition
Zone of inhibition assays were performed to test the ability of Leisingera sp. JC1 to inhibit other marine bacteria, both free-living and symbiotic (Supplementary Table S3). JC1 was tested against the E. scolopes light organ symbiont, V. fischeri ES114; another bioluminescent member of the Vibrionaceae, P. leiognathi KNH6, isolated from Hawaiian seawater; V. harveyi B392; V. parahaemolyticus KNH1 and V. anguillarum 775. These bacteria were plated at lawn densities from 104 to 107 CFU/mL to test the efficacy of possible inhibition at varying densities which more closely reflect biologically relevant concentrations. Overall, Leisingera sp. JC1 differentially inhibited the five vibrios tested (Figure 5). For two of the strains tested, V. fischeri (F3,76 = 12.63, P < 0.0001) and P. leiognathi (F3,60 = 137.5, P < 0.0001), JC1 showed significantly greater inhibition at lower lawn densities (Figure 5A; Supplementary Figure S2). When measured ZOIs were normalized for variations in JC1 colony diameter, there was an average 4.2 mm ZOI at 104 CFU/mL of V. fischeri, while at the 107 CFU/mL density, there was a 2.3 mm ZOI (Supplementary Figures S2I–L). A multiple comparisons post hoc Tukey test determined that the ZOI for the 104–106 CFU/mL lawn densities of V. fischeri were significantly greater than the ZOI at the 107 CFU/mL density. The change in ZOI with test strain lawn density was most apparent for P. leiognathi, where the average ZOI at the 104–105 CFU/mL lawn densities ranged from 2.4 to 2.9 mm, and then dropped to 0 mm at the 106–107 CFU/mL densities (Figures 5A,D; Supplementary Figures S2A–D). A multiple comparisons post hoc Tukey test showed that the ZOI at 104 CFU/mL of P. leiognathi was significantly different from the ZOI at 105 CFU/mL, and that both ZOIs at 104 and 105 CFU/mL were significantly different from the 106–107 CFU/mL results. Leisingera sp. JC1 showed a trend toward inhibition of V. anguillarum with ZOIs ranging from 2.7 to 3.3 mm (Figures 5A,E; Supplementary Figures S2E–H) although, a one-way ANOVA determined that the ZOIs were not statistically different (F3,60 = 2.553, P = 0.0639). No inhibition was observed when JC1 was tested against V. parahaemolyticus or V. harveyi at any lawn density (Figures 5A,F,G; Supplementary Figures S2M–P).
FIGURE 5.
Leisingera sp. JC1 differentially inhibits pathogenic and non-pathogenic vibrio species. (A) Zones of inhibition around JC1 colonies were measured with increasing lawn densities of each vibrio species. Leisingera sp. JC1 differentially inhibited five vibrio strains. Significantly greater inhibition of Vibrio fischeri was seen between the 104–106 CFU/ml and the 107 CFU/ml lawn density. Significantly greater inhibition of Photobacterium leiognathi was seen between the 104 and 105 CFU/ml lawn densities, as well as between the 104–105 and the 106–107 CFU/ml densities. JC1 inhibited Vibrio anguillarum uniformly across all lawn densities and did not inhibit Vibrio parahaemolyticus or Vibrio harveyi. Letters indicate significantly different groups based on results from post hoc Tukey tests (see Results and Discussion). Representative 24 h images of (B) JC1 plated alone on YTSS agar and with 106 CFU/ml of (C) V. fischeri, (D) P. leiognathi, (E) V. anguillarum, (F) V. parahaemolyticus, and (G) V. harveyi. Scale bars: 5 mm (B–G).
Leisingera sp. JC1 was also tested in a ZOI assay against the 12 previously described ANG isolates (Collins et al., 2015) and one additional ANG isolate, Muricauda sp. ANG21. All ANG isolates were only tested at a lawn density of approximately 108 CFU/mL. Inhibition was observed against Ruegeria sp. ANG-S4, with an average ZOI of 6.3 mm (±0.7) and against Muricauda sp. ANG21, with an average ZOI of 5.9 mm (±0.6; Supplementary Figure S3). Leisingera sp. JC1 was not able to inhibit any of the other Leisingera spp. previously isolated from ANGs. Since partitioning between bacterial taxa is observed in the ANG tubules some activity against other ANG isolates may contribute to competition between strains during colonization (Collins et al., 2012).
96-Well Liquid Assay
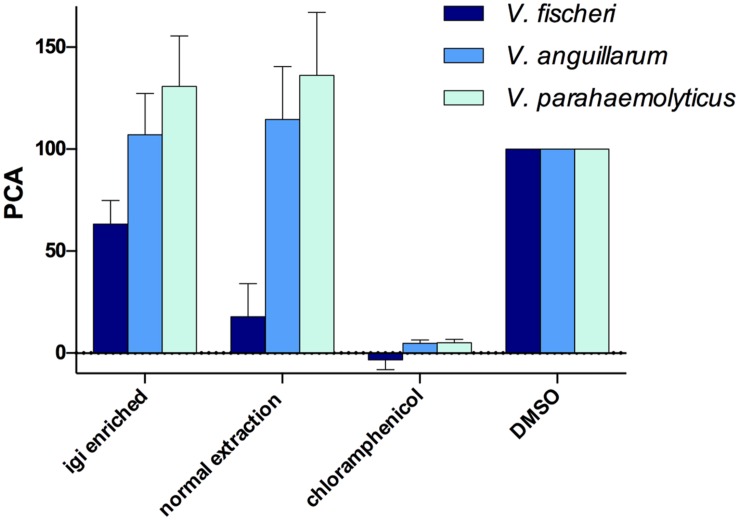
Both the normal and indigoidine enriched JC1 extracts were screened for activity using 96-well plate liquid assays with several of the vibrios tested above, including V. fischeri ES114, V. anguillarum 775, and V. parahaemolyticus KNH1 (Figure 6). Both extracts were initially tested at 500 μg/mL with minimum inhibitory concentrations (MICs) determined for active samples. The JC1 normal extract was found to strongly inhibit growth of V. fischeri with a PCA value of 17.9 ± 9.3 during screening and was determined to have a MIC of 250 μg/mL. The JC1 indigoidine enriched extract also exhibited moderate inhibition of V. fischeri with a PCA of 63.3 ± 6.7.
FIGURE 6.
Leisingera sp. JC1 extracts inhibit V. fischeri in 96-well liquid assays. JC1 cultures were grown at large scale and extracts were prepared using either an indigoidine enriched procedure (igi enriched) or a normal extraction method. All extracts were screened at 500 μg/mL against V. fischeri, V. anguillarum, and V. parahaemolyticus. The JC1 normal extract strongly inhibited growth of V. fischeri (PCA 17.9 ± 9.3) with less activity of the JC1 igi enriched extract against V. fischeri (PCA 63.3 ± 6.7). No inhibition of V. anguillarum or V. parahaemolyticus was observed. Data shown are average PCA values from three experiments (PCA, percent control activity, as compared with DMSO negative control).
In contrast to the ZOI data above, no inhibition was observed for either extract when tested against V. anguillarum, potentially due to differences between the activity of indigoidine in agar versus liquid assays, as seen with Leisingera sp. Y41 and hypothesized to result from changes in the redox state of indigoidine (Cude et al., 2012). These results may also be attributed to differences in the chemical composition between extracts and the bacteria in situ (e.g., aqueous soluble metabolites are generally excluded from the extraction protocols used in this study). Neither JC1 extract inhibited V. parahaemolyticus, in agreement with the ZOI data above.
Previous studies with a mutant of Leisingera sp. Y4I that did not produce indigoidine suggested that production of the compound is required for inhibition of V. fischeri (Cude et al., 2012). However, with the more potent inhibition of V. fischeri seen in the JC1 normal extract versus the indigoidine enriched extract in this study (Figure 6), indigoidine production does not seem to be the only mechanism of inhibition for Leisingera sp. JC1. Given that the JC1 normal extract contains only minimal amounts of indigoidine (0.8% as discussed above), the bacterium may be utilizing other secondary metabolites in conjunction with indigoidine for chemical defense. The JC1 genome includes several other secondary metabolite biosynthetic gene clusters for HSL, siderophore, bacteriocin, PKS, and PKS/NRPS production and thus Leisingera sp. JC1 likely utilizes one or more of the compounds encoded by these pathways for chemical defense, in addition to the defensive capabilities attributed to indigoidine. Creating an indigoidine mutant of Leisingera sp. JC1 will help test this hypothesis, in conjunction with identification of additional metabolite(s) responsible for JC1 antimicrobial activity.
Localization of Indigoidine Production by Leisingera sp. JC1
While performing ZOI assays, there was a dramatic change in colony pigmentation of Leisingera sp. JC1 when grown alone (Figure 5B) as compared to growth under challenge with various vibrio strains (Figures 5C–G). Deep blue pigment production was observed uniformly when JC1 was grown in monoculture and appeared to localize to the outer edges of the colonies when presented with vibrio strains. Direct analysis in real time-mass spectrometry (DART-MS) is an ambient ionization technique in which samples can be analyzed without sample preparation or extraction (Sanchez et al., 2011). DART-MS was utilized to chemically confirm the visual observations of localization of indigoidine production of JC1 in monoculture and co-culture with V. fischeri over the course of 7 days (Figure 7). Five locations were selected on each colony including center (A), midpoint (B), edge (C), ZOI (D), and outside ZOI (E). In the absence of V. fischeri, indigoidine was uniformly produced throughout JC1 colonies (locations A–C). However, in the presence of V. fischeri there was little to no indigoidine production in the center or midpoints of JC1 colonies, but intense indigoidine detected around colony edges (Figure 7, location C only). Indigoidine was only minimally detected in the ZOI or outside the ZOI for either monoculture or co-culture. Trends in the localization of indigoidine production were even more apparent upon measurement after 7 days.
FIGURE 7.
Leisingera sp. JC1 localizes indigoidine production to outer edge of colony in the presence of V. fischeri. Five locations (A–E) were analyzed for the presence of indigoidine via direct analysis in real time-mass spectrometry (DART-MS) over 7 days. (A) In the absence of V. fischeri, indigoidine production was more uniformly distributed across the colony. (B) In the presence of V. fischeri, indigoidine production was primarily localized to outer edge of colony (location C) whereas minimal indigoidine was detected in the center of the colony (location A). Very little indigoidine was detected in the ZOI or outside ZOI in the presence or absence of V. fischeri. Heatmaps represent a gradient of indigoidine ion abundance from less abundance (black, 0%) to more abundance (red, 100%).
There are several examples of pigment production being induced when in the presence of other bacteria, such as in Staphylococcus aureus when co-cultured with Pseudomonas aeruginosa (Antonic et al., 2013) or production of a red pigment by Streptomyces lividans TK23 when co-cultured with Tsukamurella pulmonis TP-B0596 (Onaka et al., 2011). Pigment production can also be induced under other stress response conditions, such as protection from UV radiation (Tong and Lighthart, 1997). Pigment production has also been tied to photosynthesis (Orf and Blankenship, 2013), however, Leisingera sp. JC1 lacks genes associated with photosynthesis or carbon fixation (data not shown).
When grown alone, Leisingera sp. JC1 exhibited a uniform blue–black pigmentation across the colony which was confirmed by mass spectrometry to be essentially uniform production of indigoidine. Secondary metabolite biosynthesis is an energy intensive endeavor and production of antimicrobial compounds would typically be thought to be reserved for defensive situations. Since indigoidine is produced throughout the colony when in monoculture, and given its relatively moderate antibacterial activity as suggested by assays with the indigoidine enriched extract, it is also possible that indigoidine serves multiple functions for Leisingera sp. JC1. However, Leisingera sp. JC1 localized indigoidine production to the outer edges of the colony when co-cultured with V. fischeri and other vibrios. If utilized as a defensive compound, indigoidine may be localized to points of direct interaction with other microorganisms. Secondary metabolite production can be localized to susceptible locations such as in plants, sponges, and other sessile terrestrial and marine organisms (Amsler et al., 2001; Furrow et al., 2003; Van Dyck et al., 2010). The role of Leisingera sp. JC1 has yet to be examined directly in the ANG symbiosis but localized production of indigoidine or other secondary metabolites may play a role in egg defense or inhibition of other bacteria from colonizing the ANG (see conclusions below).
Regulation of Indigoidine Production by Leisingera sp. JC1
After observing localized production of indigoidine when grown on solid media with V. fischeri, additional co-culture experiments were undertaken in liquid media using several of the vibrios from the antimicrobial assays above. Leisingera sp. JC1 was grown in monoculture and in the presence of V. fischeri, V. anguillarum, and V. parahaemolyticus, followed by extraction and measurement of indigoidine production (Figure 8). Monocultures of all three vibrios were also grown and extracted as controls. Addition of V. fischeri to established cultures of Leisingera sp. JC1 resulted in a 1.38 fold increase in indigoidine. Co-cultures with V. anguillarum and V. parahaemolyticus resulted in a decrease in indigoidine production of approximately 0.5 fold for both organisms. Vibrio monocultures confirmed that these species do not produce indigoidine. Changes in indigoidine production were also visually evident with darker, more intense blue observed for extracts cultured with V. fischeri in comparison with JC1 monoculture, as well as lighter blue extracts observed for V. anguillarum and V. parahaemolyticus co-cultures (Figure 8A).
FIGURE 8.
Leisingera sp. JC1 upregulates indigoidine production in the presence of V. fischeri. JC1 was cultured alone and in the presence of V. fischeri, V. anguillarum, and V. parahaemolyticus (each vibrio was also monocultured). (A) Extracts were prepared in DMSO at 5 mg/mL for LC–MS (from left to right): JC1 monoculture, JC1 + V. fischeri, JC1 + V. anguillarum, JC1 + V. parahaemolyticus, V. fischeri monoculture, V. anguillarum monoculture, and V. parahaemolyticus monoculture. As shown, the highest pigment production was observed when JC1 was co-cultured with V. fischeri. (B) LC–MS UV chromatograms (299 nm) for all extracts. JC1 indigoidine production was upregulated when co-cultured with V. fischeri (red). Down regulation was observed when co-cultured with V. anguillarum or V. parahaemolyticus (green and pink, respectively). (C) Following extraction, indigoidine production was measured using HPLC and quantitated via measurement of the area under the curve for UV absorption at 299 nm. Fold change was calculated as compared with JC1 monoculture.
The increase in indigoidine production of JC1 with V. fischeri is consistent with the antibacterial activity observed for Leisingera sp. JC1 on both solid and liquid media (Figures 5 and 6), strengthening the hypothesis that indigoidine may play a protective role in association with E. scolopes. In addition, the downregulation of production with V. anguillarum and V. parahaemolyticus also supports the liquid culture bioassay data (Figure 6). The differential antimicrobial activity and indigoidine production between the three vibrios may be due to the purported role of the ANG and JC bacteria in the host. The ability of Leisingera sp. JC1 to inhibit V. fischeri may be related to the fact that the ANG is located directly posterior to the light organ, which harbors high densities of the sole symbiont, V. fischeri (McFall-Ngai, 2014). Each day 95% of viable V. fischeri cells in the light organ are expelled directly into the mantle cavity of the host as part of the regulatory mechanisms of that association (Boettcher et al., 1996; Nyholm and McFall-Ngai, 1998). A study from another squid, Doryteuthis pealeii (Kaufman et al., 1998) suggests that ANG bacteria are environmentally transmitted during development. Given that V. fischeri is not detected in the ANG (Collins et al., 2012), the inhibitory effect of Leisingera sp. JC1 and other ANG isolates may prevent V. fischeri and other vibrios from colonizing the ANG and thus help shape the consortium during development. Alternatively, inhibition against vibrios may play a role in egg defense since eggs are exposed to seawater for approximately three weeks and vibrios are known to be common members of the bacterioplankton.
Conclusions
Genome analyses confirm that Leisingera sp. JC1 is part of the squid-associated roseobacter clade. Both in silico and in vitro analyses confirmed the secondary metabolite potential and production of siderophores, acyl-homoserine lactones associated with quorum sensing, and the pigment indigoidine. Leisingera sp. JC1 and its extracts had inhibitory activity against a variety of marine bacteria including the light organ symbiont V. fischeri. Furthermore, JC1 challenged with V. fischeri led to increased localized production of indigoidine as well as an increased production of indigoidine when co-cultured in liquid media. Taken together these results suggest that Leisingera sp. JC1 may play a protective role in egg defense and/or in shaping the microbial community of the ANG. The importance of defensive symbioses in nature is becoming increasingly more evident (Flórez et al., 2015). A number of both terrestrial and marine organisms use novel secondary metabolites produced by bacteria toward defense from potential pathogens and fouling microorganisms. Since roseobacters have been found in the ANGs of a number of cephalopods from diverse marine environments (Kaufman et al., 1998; Grigioni et al., 2000; Barbieri et al., 2001; Pichon et al., 2005; Collins et al., 2012) there may be a conserved function of this group in this symbiosis. Further studies from this group may reveal novel compounds that are important for the biology of these associations and that exhibit antimicrobial activity.
Author Contributions
MB, SN, SG, and AS conceptualized and designed research; SG, AS, MF, and JLG conducted experiments; MB, SN, SG, AS, MF, and JPG analyzed data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Anne A. Sung for assistance with CFU counts, Alison Buchan for donation of Leisingera sp. Y4I, Allison H. Kerwin for helpful comments, and Kewalo Marine Laboratory of the University of Hawaii for assistance with animal collections.
Funding. This research was funded by NSF IOS-1557914 to SN and MB, University of Connecticut Office of the Vice President for Research to SN, and the University of Connecticut Outstanding Multicultural Scholars Program to AS.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01342
References
- Alikhan N.-F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsler C. D., McClintock J. B., Baker B. J. (2001). Secondary metabolites as mediators of trophic interactions among Antarctic marine organisms. Am. Zool. 41 17–26. [Google Scholar]
- Antonic V., Stojadinovic A., Zhang B., Izadjoo M. J., Alavi M. (2013). Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect. Drug Resist. 6 175–186. 10.2147/IDR.S49039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E., Barry K., Child A., Wainwright N. (1997). Antimicrobial activity in the microbial community of the accessory nidamental gland and egg cases of Loligo pealei (Cephalopoda: Loliginidae). Biol. Bull. 193 275–276. [DOI] [PubMed] [Google Scholar]
- Barbieri E., Paster B., Hughes D., Zurek L., Moser D., Teske A., et al. (2001). Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda:Loliginidae). Environ. Microbiol. 3 151–167. 10.1046/j.1462-2920.2001.00172.x [DOI] [PubMed] [Google Scholar]
- Berger M., Neumann A., Schulz S., Simon M., Brinkhoff T. (2011). Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193 6576–6585. 10.1128/JB.05818-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher K. J., Ruby E. G., McFall-Ngai M. J. (1996). Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol. 179 65–73. 10.1007/BF00193435 [DOI] [Google Scholar]
- Bruhn J. B., Gram L., Belas R. (2007). Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 73 442–450. 10.1128/AEM.02238-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn J. B., Haagensen J. A. J., Bagge-Ravn D., Gram L. (2006). Culture conditions of Roseobacter strain 27-4 affect its attachment and biofilm formation as quantified by real-time PCR. Appl. Environ. Microbiol. 72 3011–3015. 10.1128/AEM.72.4.3011-3015.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha C., Gao P., Chen Y. C., Shaw P. D., Farrand S. K. (1998). Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 11 1119–1129. 10.1094/MPMI.1998.11.11.1119 [DOI] [PubMed] [Google Scholar]
- Chilton M. D., Currier T. C., Farrand S. K., Bendich A. J., Gordon M. P., Nester E. W. (1974). Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. U.S.A. 71 3672–3676. 10.1073/pnas.71.9.3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli F. R., Monlezun L., Coulthurst S. J. (2016). Aim, load, and fire: the Type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24 51–62. 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. M07A9, 9th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Collins A. J., Fullmer M. S., Gogarten J. P., Nyholm S. V. (2015). Comparative genomics of Roseobacter clade bacteria isolated from the accessory nidamental gland of Euprymna scolopes. Front. Microbiol. 6:123 10.3389/fmicb.2015.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. J., LaBarre B. A., Won B. S., Shah M. V., Heng S., Choudhury M. H., et al. (2012). Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl. Environ. Microbiol. 78 4200–4208. 10.1128/AEM.07437-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. J., Nyholm S. V. (2011). Draft genome of Phaeobacter gallaeciensis ANG1, a dominant member of the accessory nidamental gland of Euprymna scolopes. J. Bacteriol. 193 3397–3398. 10.1128/JB.05139-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cude W. N., Mooney J., Tavanaei A. A., Hadden M. K., Frank A. M., Gulvik C. A., et al. (2012). Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization by the marine Roseobacter Phaeobacter sp. strain Y4I. Appl. Environ. Microbiol. 78 4771–4780. 10.1128/AEM.00297-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cude W. N., Prevatte C. W., Hadden M. K., May A. L., Smith R. T., Swain C. L., et al. (2015). Phaeobacter sp. strain Y4I utilizes two separate cell-to-cell communication systems to regulate production of the antimicrobial indigoidine. Appl. Environ. Microbiol. 81 1417–1425. 10.1128/AEM.02551-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9 772 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E., Cambillau C., Cascales E., Journet L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22 498–507. 10.1016/j.tim.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Edenborn H. M., Litchfield C. D. (1985). Glycolate metabolism by Pseudomonas sp., strain S227, isolated from a coastal marine sediment. Mar. Bio. 88 199–205. 10.1007/BF00397167 [DOI] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flórez L. V., Biedermann P. H., Engl T., Kaltenpoth M. (2015). Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32 904–936. 10.1039/c5np00010f [DOI] [PubMed] [Google Scholar]
- Furrow F. B., Amsler C. D., McClintock J. B., Baker B. J. (2003). Surface sequestration of chemical feeding deterrents in the Antarctic sponge Latrunculia apicalis as an optimal defense against sea star spongivory. Mar. Bio. 143 443–449. 10.1007/s00227-003-1109-5 [DOI] [Google Scholar]
- Geng H., Bruhn J. B., Nielsen K. F., Gram L., Belas R. (2008). Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74 1535–1545. 10.1128/AEM.02339-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigioni S., Boucher-Rodoni R., Demarta A., Tonolla M., Peduzzi R. (2000). Phylogenetic characterisation of bacterial symbionts in the accessory nidamental glands of the sepioid Sepia officinalis (Cephalopoda: Decapoda). Mar. Biol. 136 217–222. 10.1007/s002270050679 [DOI] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R. S., et al. (2010). A Type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Waterfield N. R., Yang J., Yang G., Jin Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15 600–610. 10.1016/j.chom.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Kaufman M., Ikeda Y., Patton C., van Dykhuizen G., Epel D. (1998). Bacterial symbionts colonize the accessory nidamental gland of the squid Loligo opalescens via horizontal transmission. Biol. Bull. 194 36–43. 10.2307/1542511 [DOI] [PubMed] [Google Scholar]
- Martens T., Gram L., Grossart H. P., Kessler D., Müller R., Simon M., et al. (2007). Bacteria of the Roseobacter clase show potential for secondary metabolite production. Microb. Ecol. 54 31–42. 10.1007/s00248-006-9165-2 [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. J. (2014). The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Ann. Rev. Microbiol. 68 177–194. 10.1146/annurev-micro-091313-103654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H.-P., Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R. J., Griffin L. E., Bowles K. M., Meile C., Gifford S., Givens C. E., et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J. 4 784–798. 10.1038/ismej.2009.150 [DOI] [PubMed] [Google Scholar]
- Nyholm S. V., McFall-Ngai M. J. (1998). Sampling the light-organ microenvironment of Eupyrmna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol. Bull. 195 89–97. 10.2307/1542815 [DOI] [PubMed] [Google Scholar]
- Onaka H., Mori Y., Igarashi Y., Furumai T. (2011). Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77 400–406. 10.1128/AEM.01337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orf G. S., Blankenship R. E. (2013). Chlorosome antenna complexes from green photosynthetic bacteria. Photosynth. Res. 116 315–331. 10.1007/s11120-013-9869-3 [DOI] [PubMed] [Google Scholar]
- Pichon D., Gaia V., Norman M. D., Boucher-Rodoni R. (2005). Phylogenetic diversity of epibiotic bacteria in the accessory nidamental glands of squids (Cephalopoda: Loliginidae and Idiosepiidae). Mar. Biol. 147 1323–1332. 10.1007/s00227-005-0014-5 [DOI] [Google Scholar]
- Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103 1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D., Webb J. S., Holmström C., Case R., Low A., Steinberg P., et al. (2007). Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73 7844–7852. 10.1128/AEM.01543-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn L., Christensen A. B., Molin S., Givskov M., Gram L. (2001). Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44 239–251. 10.1016/S0167-7012(01)00217-2 [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Mark P., van der Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz B., Chávez A., Forero A., García-Huante Y., Romero A., Sánchez M., et al. (2010). Production of microbial secondary metabolites: regulation by the carbon source. Crit. Rev. Microbiol. 36 146–167. 10.3109/10408410903489576 [DOI] [PubMed] [Google Scholar]
- Sana T. G., Hachani A., Bucior I., Soscia C., Garvis S., Termine E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and fur and modulates internalization in epithelial cells. J. Biol. Chem. 287 27095–27105. 10.1074/jbc.M112.376368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L. M., Curtis M. E., Bracamonte B. E., Kurita K. L., Navarro G., Sparkman O. D., et al. (2011). Versatile method for the detection of covalently bound substrates on solid supports by DART mass spectrometry. Org. Lett. 13 3770–3773. 10.1021/ol201404v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher T., Nyholm S. (2011). Characterizing the host and symbiont proteomes in the association between the Bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS ONE 6:e25649 10.1371/journal.pone.0025649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. W., Donia M. S. (2010). Life in cellulose houses: symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr. Opin. Biotechnol. 21 827–833. 10.1016/j.copbio.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., Hood R. D., Mougous J. D. (2010). What is type VI secretion doing in all those bugs? Trends Microbiol. 18 531–537. 10.1016/j.tim.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J. (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3 331–335. 10.1038/nchem.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y. Y., Lighthart B. (1997). Solar radiation is shown to select for pigmented bacteria in the ambient outdoor atmosphere. Photochem. Photobiol. 65 103–106. 10.1111/j.1751-1097.1997.tb01884.x [DOI] [Google Scholar]
- Tritt A., Eisen J. A., Facciotti M. T., Darling A. E. (2012). An integrated pipeline for de novo assembly of microbial genomes. PLoS ONE 7:e42304 10.1371/journal.pone.0042304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40 1–12. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterweger D., Miyata S. T., Bachmann V., Brooks T. M., Mullins T., Kostiuk B., et al. (2014). The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Comm. 5 3549 10.1038/ncomms4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck S., Flammang P., Meriaux C., Bonnel D., Salzet M., Fournier I., et al. (2010). Localization of secondary metabolites in marine invertebrates: contribution of MALDI MSI for the study of saponins in Cuvierian tubules of H. forskali. PLoS ONE 5:e13923 10.1371/journal.pone.0013923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., et al. (2015). antiSMASH 3.0 - a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43 W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whistler C. A., Ruby E. G. (2003). GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 185 7202–7212. 10.1128/JB.185.24.7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Xu F., Valiente J., Wang S., Zhan J. (2013). An indigoidine biosynthetic gene cluster from Streptomyces chromofuscus ATCC 49982 contains an unusual IndB homologue. J. Ind. Microbiol. Biotechnol. 40 159–168. 10.1007/s10295-012-1207-9 [DOI] [PubMed] [Google Scholar]
- Zan J., Cicirelli E. M., Mohamed N. M., Sibhatu H., Kroll S., Choi O., et al. (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85 916–933. 10.1111/j.1365-2958.2012.08149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgoda J. R., Porter J. R. (2001). A convenient microdilution method for screening natural products against bacteria and fungi. Pharm. Biol. 39 221–225. 10.1076/phbi.39.3.221.5934 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.