Abstract
Bacteriophages are viruses that infect bacteria. In this perspective, we discuss several aspects of a characteristic feature of bacteriophages, their host range. Each phage has its own particular host range, the range of bacteria that it can infect. While some phages can only infect one or a few bacterial strains, other phages can infect many species or even bacteria from different genera. Different methods for determining host range may give different results, reflecting the multiple mechanisms bacteria have to resist phage infection and reflecting the different steps of infection each method depends on. This makes defining host range difficult. Another difficulty in describing host range arises from the inconsistent use of the words “narrow” and especially “broad” when describing the breadth of the host range. Nearly all bacteriophages have been isolated using a single host strain of bacteria. While this procedure is fairly standard, it may more likely produce narrow rather than broad host range phage. Our results and those of others suggest that using multiple host strains during isolation can more reliably produce broader host range phages. This challenges the common belief that most bacteriophages have a narrow host range. We highlight the implications of this for several areas that are affected by host range including horizontal gene transfer and phage therapy.
Keywords: bacteriophage, host range, isolation protocol, phage therapy, bacteriophage ecology, bacteriophage evolution, phage–host interaction
Introduction
Bacteriophages (phages) are viruses that infect bacteria. Phages are estimated to be the most abundant organisms on Earth with likely more than 1031 total individual phage on the planet (Chibani-Chennoufi et al., 2004; Abedon, 2008). The types (strains or species) of bacteria that a bacteriophage is able to infect is considered the host range of the phage in question (Hyman and Abedon, 2010). It is generally believed that most bacteriophages are only capable of infecting a narrow range of bacteria that are closely related (Ackermann and DuBow, 1987; Weinbauer, 2004). This is due to a combination of factors including specificity of phages’ host binding proteins, biochemical interactions during infection, presence of related prophages or particular plasmids (especially for phages adsorbing to pili) and bacterial phage-resistance mechanisms (Adams, 1959; Hyman and Abedon, 2010; Diaz-Munoz and Koskella, 2014).
Because many bacteriophages kill the bacterial cells they infect, phages provide a possible alternative to antibiotics. This use of bacteriophages, called phage therapy, was the first use proposed for phage soon after their discovery (Summers, 2012). While displaced by antibiotics in much of the world, the increasing frequency of antibiotic-resistant bacteria has led to renewed interest in phage therapy (Brussow, 2014; Kutter et al., 2015). Host range is a key property for phage therapy as well as the biology of bacteriophages in general. One of the advantages of phage therapy is that the specificity of phage–host range spares non-pathogenic microbes from being killed during treatment. Conversely, this same specificity limits the ability of a particular phage’s use to a small set of potential pathogens requiring more specific diagnosis (Nilsson, 2014; Mapes et al., 2016).
Host Range Determination
Determining the host range of a specific phage can be somewhat difficult because measured host ranges depend on the technique used (Hyman and Abedon, 2010). Adams (1959, p. 440) states that “Host range is often, but not always, determined by success or failure of adsorption” but for phage therapy, host cell killing tends to be the key determination. Hyman and Abedon (2010) describe seven different host range types including adsorptive, penetrative (transductive), bactericidal, productive, plaquing, spotting, and lysogenic. Each host range type is dependent on phage successfully completing different steps of the infection process or determined by different methods of measuring host range. For example, a plaquing host range is found by determining whether a phage is able to form plaques on a particular species or strain of host bacteria. This is a common way to determine whether the phage can productively infect the bacteria. But not every host will allow plaquing even if the host does allow for a productive infection if infection only yields a limited number of progeny. Spot testing, in which a small volume of phage is placed on a growing lawn of bacteria, is perhaps the most common way of determining whether the phage is able to infect. While simple and rapid, this technique can sometimes cause false positives because of lysis of bacterial cells without phage infection. This is typically thought to occur when bacteria are lysed by a large number of phage adsorbing to the cell and lysing it (although it is unclear if this is a widespread mechanism) or by lysis due to residual endolysin or bacteriocins in the phage stock or other mechanisms (Abedon, 2011). Because of this, spotting host ranges tend to be an overestimate of what the true host range in terms of producing progeny is. See Khan and Nilsson (2015) for an experimental exploration of this topic. Unless explicitly indicated, for the remainder of this perspective we will use the term host range to refer to range of hosts that can produce progeny phage, the productive host range. For practical purposes, this is usually equivalent to the plaquing host range.
A second challenge to discussing host range is the use of the term “broad host range.” This term is used to describe a bacteriophage that can infect multiple species of bacteria (Greene and Goldberg, 1985; Uchiyama et al., 2008; Khan and Nilsson, 2015; Yu et al., 2016). But it is also used to describe a bacteriophage that can infect multiple strains of the same species of bacteria (Vinod et al., 2006; Gupta and Prasad, 2010; Anand et al., 2015; Xu et al., 2016). As well, the term polyvalent (or polyvalence) is sometime used equivalently to broad host range although polyvalent was previously more specifically reserved for “phages active on different bacterial genera” (Ackermann and DuBow, 1987). Whichever term is used, there is no standard as to how many or what percentage of strains/species tested must be infected for a phage to have a broad host range. For example, bacteriophage Mu is able to infect species of Escherichia coli, Citrobacter freundii, Shigella sonnei, Enterobacter, and Erwinia (Paolozzi and Ghelardini, 2006); staphylococcal phage ϕ812 infects 95% of 782 strains of Staphylococcus aureus and 43% of other Staphylococcus species tested (Pantucek et al., 1998); and bacteriophage P-27/HP infects 60% of 28 S. aureus isolates (Gupta and Prasad, 2010). All are described as having broad host range or being polyvalent. In addition to variation in numbers of bacteria species/strains tested, with a very few exceptions there are no standard collections of bacterial species. Instead, most researchers develop their own collections of bacteria usually including both previously described and newly isolated strains or species.
Bacteriophage Isolation Protocols
Isolating phage can be a fairly simple process and for the most part researchers accomplish it in a very similar manner to that used by the earliest phage biologists (d’Hérelle and Smith, 1926; De Groat, 1927). The basic method is to obtain an environmental sample that is likely to contain or have been in contact with the targeted host bacteria. This can be raw fecal matter (Markel and Eklund, 1974; Jensen et al., 2015; Xu et al., 2016), sewage samples (Lin et al., 2010; Khan and Nilsson, 2015; Yu et al., 2016), water samples (Uchiyama et al., 2008; Lin et al., 2011), soil samples (Campos et al., 1978; Greene and Goldberg, 1985; Anand et al., 2015), samples taken from infected or healthy humans (Ackermann et al., 1975; Ronda et al., 1981), etc. Broth media or buffer is added to the samples and they are then filtered to remove the bacteria and other solid material. This filtrate is then added to a fresh culture of host bacteria and incubated overnight. In some cases, the sample is added directly to a growing bacterial culture without filtering. The next day, the culture is centrifuged to remove cell and other debris, and the supernatant is filtered to remove any remaining bacteria. This filtrate is then tested for the presence of phage by either spot or plaque testing. This procedure is so common as to be the basis for the standard method of screening for coliphage in water samples (International Organization for Standardization, 2000).
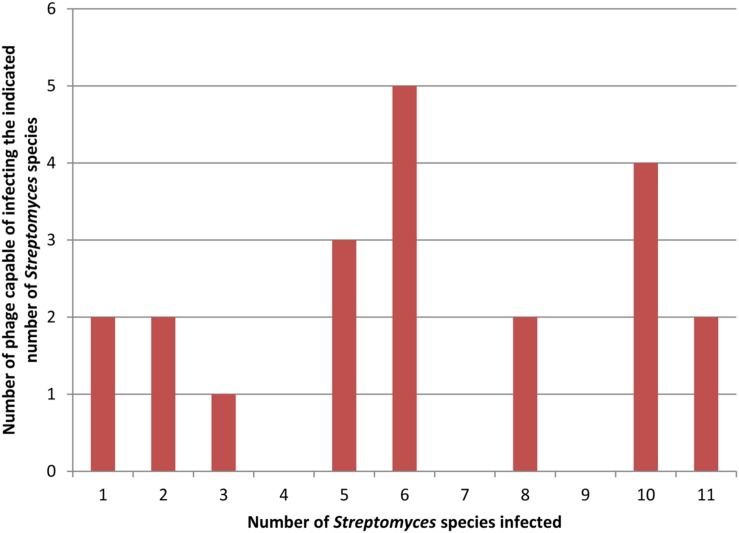
Most often, this procedure is performed using a single bacterial strain for phage isolation. The isolated phages are then tested against a collection of other bacterial strains and species to test host range breadth if host range is being measured. In many cases, this procedure produces phages with a narrow host range (Campos et al., 1978; Auad et al., 1997; Lu et al., 2003; Lin et al., 2011). The same procedure, however, can produce broader host range phages as well (Greene and Goldberg, 1985; Vinod et al., 2006; Uchiyama et al., 2008; Anand et al., 2015; Khan and Nilsson, 2015). When used to isolate many phages, both types of phages may be found together. For example, Greene and Goldberg (1985) screened about 700 soil samples for phages infecting Streptomyces avermitilis and found 57 phages that appeared unique by restriction analysis. They determined the host range of 21 of these that produced turbid plaques on S. avermitilis, suggesting a temperate life cycle. To do so, they used 10 additional Streptomyces species and found a diversity of host ranges based on plaque testing and as shown in Figure 1. Two phages were capable of infecting only a single host strain and two phages were capable of infecting all eleven host species with no obvious distribution of the phages infecting intermediate numbers of hosts. Jensen et al. (2015) found a similar mix of host range breadths by spot testing 12 novel phages against seven strains of S. aureus. Figure 1 also shows the difficulty in determining the definitional borderline between narrow and broad host range as discussed above.
FIGURE 1.
Distribution of host range breadths of Streptomyces temperate phages. Data for this figure is taken from Table 3 of Greene and Goldberg (1985). Twenty-one distinct phages were tested on 11 different host strains. In this figure both clear and turbid plaques (as indicated in the original results) are combined to indicate host susceptibility to a phage.
In order to isolate broader host range phage in a more controlled manner, we and other groups have modified the basic isolation procedure to use multiple hosts during phage isolation. Jensen et al. (1998) compared phages isolated on single hosts to phages isolated on pairs of hosts using E. coli, Pseudomonas aeruginosa and Sphaerotilus natans (all Gram-negative Proteobacteria). They found phages that could productively infect two or all three of the hosts in both the single- and double-host isolated phages. Mapes et al. (2016) developed a host range expansion (HRE) protocol to broaden the host range of P. aeruginosa-specific phages. They cultured a mixture of four phages with a mix of 16 different host strains and isolated individual phage strains by plaque isolation after multiple passages of phage mix onto fresh host mix. Over the course of 30 cycles host range was expanded as indicated by spot testing on both the 16 host strains and an additional 10 P. aeruginosa strains. We have isolated phages against Enterococcus faecalis using either one or a mixture of two host strains during isolation. As shown in Table 1, the two phages (AUEF4 and AUEF5) isolated using two strains had broader host ranges than the phage isolated using a single host (AUEF3) or another phage (VD13) previously isolated on the same host (Ackermann et al., 1975). The host ranges of AUEF4 and AUEF5 also included a strain of E. faecium.
Table 1.
Host ranges of Enterococcus faecalis bacteriophages on E. faecalis and E. faecium strains.
| Isolated on 8413 | Isolated on 8413 + OG1RF | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| VD13 | AUEF3 | AUEF4 | AUEF5 | ||||||
| E. faecalis strain | Reference | Spot testing | Plaque | Spot testing | Plaque | Spot testing | Plaque | Spot testing | Plaque |
| 8413 (ATCC 29200) | Ackermann et al., 1975 | +++ | + | +++ | + | +++ | + | +++ | + |
| COM1 | McBride et al., 2007 | - | NT | - | NT | - | NT | - | NT |
| DS5 | McBride et al., 2007 | + | - | - | NT | + | - | + | - |
| DS16 | McBride et al., 2007 | + | +∗ | +++ | + | +++ | + | +++ | + |
| JH2 | McBride et al., 2007 | - | NT | - | NT | - | NT | - | NT |
| OG1RF | McBride et al., 2007 | + | - | + | - | - | NT | - | NT |
| Pan7 | McBride et al., 2007 | + | - | - | NT | + | - | + | - |
| V583 | McBride et al., 2007 | - | NT | - | NT | - | NT | - | NT |
| EF-1 | Yoong et al., 2004 | - | NT | - | NT | - | NT | - | NT |
| EF-17 | Yoong et al., 2004 | - | NT | + | - | + | - | + | - |
| EF-24 | Yoong et al., 2004 | - | NT | - | NT | - | NT | - | NT |
| EF-25 | Yoong et al., 2004 | - | NT | - | NT | + | + | + | + |
| E. faecium strains | |||||||||
| EFSK2 | Yoong et al., 2004 | - | NT | - | NT | - | + | + | + |
| EFSK16 | Yoong et al., 2004 | - | NT | - | NT | - | NT | - | NT |
| EFSK33 | Yoong et al., 2004 | - | NT | - | NT | - | NT | - | NT |
∗Plaques seen but much smaller than plaques on 8413.
For spot testing +++indicates complete lysis, +indicates turbid lysis, -indicates no lysis. For plaque testing +indicates plaques seen, -indicates no plaques observed. NT indicates phage/host combinations not tested as lack of any apparent lysis in spot testing suggests no interaction of that pair. For all phage, efficiency of plating was reduced on all hosts relative to 8413.
Yu et al. (2016) have tested two sequential multiple host isolation methods after failing to find sufficiently broad host range phages using the simultaneous multi-host protocol described above. In method A, a mixed phage stock and host 1 is plated and plaques are allowed to form. All of these plaques are then collected, added to host 2, and then plated. This process can be continued with a third, fourth or more hosts and, in theory, phages from the last set of plaques collected should be able to infect all of the previous hosts. Similarly in method B, a phage stock is added to host 1 and the phages are allowed to infect and adsorb to the bacterial cells. The free phages and adsorbed phages are separated by centrifugation and the adsorbed phages are collected and allowed to grow on the first host for a few hours. The phages from this enrichment culture are then added to host 2. At least some of these phages will adsorb to and infect the bacterial cells. The free and adsorbed phages are again separated and the adsorbed phages are collected and allowed limited growth before transfer to another host. As with method A, this process can be continued and the last set of enriched phage should be able to infect all of the previous hosts. In both methods, single phage strains were then isolated using plaque purification. Yu et al. (2016) tested each method with several strains of E. coli and P. aeruginosa as well as one strain each of P. putida and P. syringae. Both methods produced phages able to infect all their Pseudomonas strains as well as some that could also infected E. coli.
Implications and Conclusion
While there is some agreement as to the number of bacteria and viruses in various environments (Wommack and Colwell, 2000; Chibani-Chennoufi et al., 2004; Suttle, 2007), one of the unresolved questions of phage ecology is how frequently a particular bacteriophage encounters a susceptible host outside of the laboratory (Koskella and Meaden, 2013). The relative ease of finding phages capable of infecting multiple strains or species of bacteria, as in the studies cited here, suggests that this may be less of an issue as many phages may be capable of infecting multiple types of hosts. It may also be an error to consider host range as a stable property with a particular host always in or out of a phage’s host range (Hyman and Abedon, 2010; Koskella and Meaden, 2013) as hosts may evolve phage resistance and phages can evolve to overcome this resistance (Stern and Sorek, 2011; Samson et al., 2013; Chaturongakul and Ounjai, 2014). This has many implications for phage biology as well as for practical applications of phages.
The unresolved question of how often a phage finds a susceptible host has implications for the role of host range in horizontal gene transfer (HGT) of the bacterial hosts. Bacteriophages can move their own genome between host genomes by forming lysogens in the hosts. Lysogenic conversion, the expression of phage genes from the prophage, plays a role in pathogenesis of several bacterial species (Brüssow et al., 2004; Hyman and Abedon, 2008; Fortier and Sekulovic, 2013). For example, E. coli strains such as E. coli O157:H7 that produce Shiga-toxin are lysogens for a lambdoid prophage that contains the genes for the toxin (Unkmeir and Schmidt, 2000; Hyman and Abedon, 2008). Likewise, transduction, the movement of bacterial genome fragments via a phage capsid, is an important mechanism of HGT (Brüssow, 2008; Hendrix and Casjens, 2008; Kelly et al., 2009). Transductive host range describes which bacteria are susceptible to HGT by a particular phage and differs from productive host range in that the phage need not be able to replicate on the host but only needs to insert DNA into the recipient cell (Hyman and Abedon, 2010). As phages may utilize HGT themselves to evolve by exchanging segments between infecting phage and prophages, host range in general may similarly affect phage evolution (Campbell, 1988; Labrie and Moineau, 2007).
Broad host range phages are seen as more useful as well in some applications of bacteriophages. For phage therapy, a broad host range phage that kills multiple species of bacteria would be the equivalent of a broad spectrum antibiotic. Currently, multiple phage species are often mixed into a cocktail in order to treat several different bacteria that may be the cause of infections (Gill and Hyman, 2010; Kutateladze and Adamia, 2010; Lu and Koeris, 2011; Chan and Abedon, 2012). A smaller number of broad host range phages could be more useful than a larger number of narrow host range phages. On the other hand, the findings of studies such as that shown in Figure 1 may mean that even phages isolated on a single host may unexpectedly infect other hosts. This might partly obviate one of the commonly cited advantages of phage therapy, host specificity that spares normal flora bacteria (Loc-Carrillo and Abedon, 2011; Nilsson, 2014). Other potential uses of phages that depend on host specificity such as using phage to deliver therapeutic or other genes to bacteria (Westwater et al., 2003; Clark et al., 2012) and the use of bacteriophage and bacteriophage proteins as biosensors for pathogenic bacteria (Zourob and Ripp, 2010; Singh et al., 2013; Peltomaa et al., 2016) might be similarly affected.
In conclusion, bacteriophage host range is not a fixed property of each species of bacteriophage. Rather, it is one that can evolve over time and can show unexpected plasticity. Modifying procedures and growth conditions can favor the isolation of novel phages with broader host ranges. Narrow host range cannot be assumed but must be tested just as life cycle, transduction potential and carriage of toxin genes are screened for when phage are isolated for other applications. We strongly encourage the use of plaque testing for determining host range when testing for phage therapy and related applications rather than spot testing as plaque formation shows that phages are capable of productive infection. Unless one is certain of being able to use a sufficient dose of phage to attack all bacteria in a single treatment, phage progeny production is needed to kill a bacterial population. Furthermore, for uses such as phage therapy, testing on clinical isolates is better than testing on laboratory host strains. Lastly, we encourage more judicious use of the term broad host range and recommend reserving its use for phages that have been shown to infect multiple bacterial species at a minimum and preferably, multiple genera.
Author Contributions
All three authors substantially contributed to this work and all have read and approved it. SW isolated AUEF 4 and AUEF5, determined their host ranges and completed the primary analysis of all four phages. AR measured host ranges, did the literature review and primary preparation of the manuscript. PH conceived of and designed the host range project as well as helping draft and editing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We want to thank Stephen T. Abedon for his helpful suggestions in developing this manuscript. We also want to thank the reviewers for raising interesting issues that improved this Perspective.
Footnotes
Funding. The research described in this paper was supported by funding from the AU Biology/Toxicology department. AR was also supported by funds from the Choose Ohio First scholarship program.
References
- Abedon S. T. (2008). “Phages, ecology, evolution,” in Bacteriophage Ecology, ed. Abedon S. T. (Cambridge: Cambridge University Press; ), 1–28. [Google Scholar]
- Abedon S. T. (2011). Lysis from without. Bacteriophage 1 46–49. 10.4161/bact.1.1.13980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann H. W., Caprioli T., Kasatiya S. S. (1975). A large new Streptococcus bacteriophage. Can. J. Microbiol. 21 571–574. 10.1139/m75-080 [DOI] [PubMed] [Google Scholar]
- Ackermann H.-W., DuBow M. S. (1987). “Phage multiplication,” in Viruses of Prokaryotes: General Properties of Bacteriophages Vol. 1 eds Ackermann H.-W., DuBow M. S. (Boca Raton, Florida: CRC Press, Inc.), 49–85. [Google Scholar]
- Adams M. H. (1959). Bacteriophages. New York, NY: InterScience. [Google Scholar]
- Anand T., Vaid R. K., Bera B. C., Barua S., Riyesh T., Virmani N., et al. (2015). Isolation and characterization of a bacteriophage with broad host range, displaying potential in preventing bovine diarrhoea. Virus Genes 51 315–321. 10.1007/s11262-015-1222-9 [DOI] [PubMed] [Google Scholar]
- Auad L., de Ruiz holgado A. A. P., Forsman P., Alatossava T., Raya R. R. (1997). Isolation and characterization of a new Lactobacillus delbrueckii ssp.bulgaricus temperate bacteriophage. J. Dairy Sci. 80 2706–2712. 10.3168/jds.S0022-0302(97)76231-3 [DOI] [Google Scholar]
- Brüssow H. (2008). “Phage-bacterium co-evolution and its implication for bacterial pathogenesis,” in Horizontal Gene Transfer in the Evolution of Pathogenesis, eds Hensel M., Schmidt H. (Cambridge: Cambridge University Press; ), 49–77. [Google Scholar]
- Brussow H. (2014). Phage therapy: quo vadis? Clin. Infect. Dis. 58 535–536. 10.1093/cid/cit776 [DOI] [PubMed] [Google Scholar]
- Brüssow H., Canchaya C., Hardt W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68 560–602. 10.1128/MMBR.68.3.560-602.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. (1988). “Phage evolution and speciation,” in The Bacteriophages Vol. 1 ed. Calendar R. (New York, NY: Plenum Press; ), 1–14. [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. (1978). Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J. Mol. Biol. 119 167–178. 10.1016/0022-2836(78)90431-X [DOI] [PubMed] [Google Scholar]
- Chan B. K., Abedon S. T. (2012). Phage therapy pharmacology: phage cocktails. Adv. Appl. Microbiol. 78 1–23. 10.1016/B978-0-12-394805-2.00001-4 [DOI] [PubMed] [Google Scholar]
- Chaturongakul S., Ounjai P. (2014). Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens. Front. Microbiol. 5:442 10.3389/fmicb.2014.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S., Bruttin A., Dillmann M. L., Brüssow H. (2004). Phage-host interaction: an ecological perspective. J. Bacteriol. 186 3677–3686. 10.1128/JB.186.12.3677-3686.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J., Abedon S. T., Hyman P. (2012). “Phages as therapeutic delivery vehicles,” in Bacteriophages in Health and Disease, eds Hyman P., Abedon S. T. (Wallingford: CABI Press; ), 86–100. [Google Scholar]
- De Groat A. F. (1927). The bacteriophage: a method of isolation. J. Immunol. 14 175–179. [Google Scholar]
- d’Hérelle F., Smith G. H. (1926). The Bacteriophage and Its Behavior [English translation]. Baltimore: The Williams &Wilkins Co. [Google Scholar]
- Diaz-Munoz S. L., Koskella B. (2014). Bacteria-phage interactions in natural environments. Adv. Appl. Microbiol. 89 135–183. 10.1016/B978-0-12-800259-9.00004-4 [DOI] [PubMed] [Google Scholar]
- Fortier L. C., Sekulovic O. (2013). Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4 354–365. 10.4161/viru.24498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J. J., Hyman P. (2010). Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 11 2–14. 10.2174/138920110790725311 [DOI] [PubMed] [Google Scholar]
- Greene J., Goldberg R. B. (1985). Isolation and preliminary characterization of lytic and lysogenic phages with wide host range within the streptomycetes. J. Gen. Microbiol. 131 2459–2465. 10.1099/00221287-131-9-2459 [DOI] [PubMed] [Google Scholar]
- Gupta R., Prasad Y. (2010). Efficacy of polyvalent bacteriophage P-27/HP to control multidrug resistant Staphylococcus aureus associated with human infections. Curr. Microbiol. 62 255–260. 10.1007/s00284-010-9699-x [DOI] [PubMed] [Google Scholar]
- Hendrix R. W., Casjens S. R. (2008). “The role of bacteriophages in the generation and spread of bacterial pathogens,” in Horizontal Gene Transfer in the Evolution of Pathogenesis, eds Hensel M., Schmidt H. (Cambridge: Cambridge University Press; ), 79–112. [Google Scholar]
- Hyman P., Abedon S. T. (2008). “Phage ecology of bacterial pathogenesis,” in Bacteriophage Ecology, ed. Abedon S. T. (Cambridge: Cambridge University Press; ), 353–385. [Google Scholar]
- Hyman P., Abedon S. T. (2010). Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70 217–248. 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- International Organization for Standardization (2000). Water Quality – Detection and Enumeration of Bacteriophages, Part 2: Enumeration of Somatic Coliphages. ISO 10705-2:2000(E) Geneva: International Organization for Standardization. [Google Scholar]
- Jensen E. C., Schrader H. S., Rieland B., Thompson T. L., Lee K. W., Nickerson K. W., et al. (1998). Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. C., Hair B. B., Wienclaw T. M., Murdock M. H., Hatch J. B., Trent A. T., et al. (2015). Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS ONE 10:e0131714 10.1371/journal.pone.0131714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B. G., Vespermann A., Bolton D. J. (2009). The role of horizontal gene transfer in the evolution of selected foodborne bacterial pathogens. Food Chem. Toxicol. 47 951–968. 10.1016/j.fct.2008.02.006 [DOI] [PubMed] [Google Scholar]
- Khan M. M., Nilsson A. S. (2015). Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10:e0118557 10.1371/journal.pone.0118557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B., Meaden S. (2013). Understanding bacteriophage specificity in natural microbial communities. Viruses 5 806–823. 10.3390/v5030806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze M., Adamia R. (2010). Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28 591–595. 10.1016/j.tibtech.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Kutter E. M., Kuhl S. J., Abedon S. T. (2015). Re-establishing a place for phage therapy in western medicine. Future Microbiol. 10 685–688. 10.2217/fmb.15.28 [DOI] [PubMed] [Google Scholar]
- Labrie S. J., Moineau S. (2007). Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J. Bacteriol. 189 1482–1487. 10.1128/JB.00728-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Han J., Ji X., Hong W., Huang L., Wei Y. (2011). Isolation and characterization of a new bacteriophage MMP17 from Meiothermus. Extremophiles 15 253–258. 10.1007/s00792-010-0354-z [DOI] [PubMed] [Google Scholar]
- Lin N. T., Chiou P. Y., Chang K. C., Chen L. K., Lai M. J. (2010). Isolation and characterization of ϕAB2: a novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 161 308–314. 10.1016/j.resmic.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Loc-Carrillo C., Abedon S. T. (2011). Pros and cons of phage therapy. Bacteriophage 1 111–114. 10.4161/bact.1.2.14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T. K., Koeris M. S. (2011). The next generation of bacteriophage therapy. Curr. Opin. Mirobiol. 14 524–531. 10.1016/j.mib.2011.07.028 [DOI] [PubMed] [Google Scholar]
- Lu Z., Breidt F. J., Fleming H. P., Altermann E., Klaenhammer T. R. (2003). Isolation and characterization of a Lactobacillus plantarum bacteriophage, ϕJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 84 225–235. 10.1016/S0168-1605(03)00111-9 [DOI] [PubMed] [Google Scholar]
- Mapes A. C., Trautner B. W., Liao K. S., Ramig R. F. (2016). Development of expanded host range phage active on biofilms of multi-drug resistant Pseudomonas aeruginosa. Bacteriophage 6:e1096995 10.1080/21597081.2015.1096995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel D. E., Eklund C. (1974). Isolation, characterization, and classification of three bacteriophage isolates for the genus Levinea. Int. J. Syst. Bacteriol. 24 230–234. 10.1099/00207713-24-2-230 [DOI] [Google Scholar]
- McBride S. M., Fischetti V. A., Leblanc D. J., Moellering R. C., Jr., Gilmore M. S. (2007). Genetic diversity among Enterococcus faecalis. PLoS ONE 2:e582 10.1371/journal.pone.0000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A. S. (2014). Phage therapy–constraints and possibilities. Ups. J. Med. Sci. 119 192–198. 10.3109/03009734.2014.902878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantucek R., Rosypalova A., Doskar J., Kailerova J., Ruzicková V., Borecka P., et al. (1998). The polyvalent staphylococcal phage ϕ812: its host-range mutants and related phages. Virology 246 241–252. 10.1006/viro.1998.9203 [DOI] [PubMed] [Google Scholar]
- Paolozzi L., Ghelardini P. (2006). “The bacteriophage Mu,” in The Bacteriophages, eds Calendar R., Abedon S. T. (Oxford: Oxford University Press; ), 469–496. [Google Scholar]
- Peltomaa R., Lopez-Perolio I., Benito-Pena E., Barderas R., Moreno-Bondi M. C. (2016). Application of bacteriophages in sensor development. Anal. Bioanal. Chem. 408 1805–1828. 10.1007/s00216-015-9087-2 [DOI] [PubMed] [Google Scholar]
- Ronda C., Lopez R., Garcia E. (1981). Isolation and characterization of a new bacteriophage, Cp-1, infecting Streptococcus pneumoniae. J. Virol. 40 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson J. E., Magadan A. H., Sabri M., Moineau S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11 675–687. 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- Singh A., Poshtiban S., Evoy S. (2013). Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors (Basel) 13 1763–1786. 10.3390/s130201763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A., Sorek R. (2011). The phage-host arms race: shaping the evolution of microbes. Bioessays 33 43–51. 10.1002/bies.201000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers W. C. (2012). The strange history of phage therapy. Bacteriophage 2 130–133. 10.4161/bact.20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. A. (2007). Marine viruses – major players in the global ecosystem. Nat. Rev. Microbiol. 5 801–812. 10.1038/nrmicro1750 [DOI] [PubMed] [Google Scholar]
- Uchiyama J., Rashel M., Maeda Y., Takemura I., Sugihara S., Akechi K., et al. (2008). Isolation and characterization of a novel Enterococcus faecalis bacteriophage ϕEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 278 200–206. 10.1111/j.1574-6968.2007.00996.x [DOI] [PubMed] [Google Scholar]
- Unkmeir A., Schmidt H. (2000). Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68 4856–4864. 10.1128/IAI.68.9.4856-4864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinod M. G., Shivu M. M., Umesha K. R., Rajeeva B. C., Krohne G., Karunasagar I., et al. (2006). Isolation of Vibrio harveyi bacteriophage with a potential for biocontrol of luminous vibriosis in hatchery environments. Aquaculture 255 117–124. 10.1016/j.aquaculture.2005.12.003 [DOI] [Google Scholar]
- Weinbauer M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28 127–181. 10.1016/j.femsre.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Westwater C., Kasman L. M., Schofield D. A., Werner P. A., Dolan J. W., Schmidt M. G., et al. (2003). Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob. Agents Chemother. 47 1301–1307. 10.1128/AAC.47.4.1301-1307.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack K. E., Colwell R. R. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64 69–114. 10.1128/MMBR.64.1.69-114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chen M., He L., Zhang S., Ding T., Yao H., et al. (2016). Isolation and characterization of a T4-like phage with a relatively wide host range within Escherichia coli. J. Basic Microbiol. 56 405–421. 10.1002/jobm.201500440 [DOI] [PubMed] [Google Scholar]
- Yoong P., Schuch R., Nelson D., Fischetti V. A. (2004). Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 186 4808–4812. 10.1128/JB.186.14.4808-4812.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Mathieu J., Li M., Dai Z., Alvarez P. J. (2016). Isolation of polyvalent bacteriophages by sequential multiple-host approaches. Appl. Environ. Microbiol. 82 808–815. 10.1128/AEM.02382-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourob M., Ripp S. (2010). “Bacteriophage-based biosensors,” in Recognition Receptors in Biosensors, ed. Zourob M. (Berlin: Springer Science; ), 415–448. [Google Scholar]