Abstract
Primary biliary cirrhosis (PBC), a classic autoimmune liver disease, is characterised by a progressive T cell predominant lymphocytic cholangitis, and a serologic pattern of reactivity in the form of specific anti-mitochondrial antibodies (AMA). CD4+ T cells are particularly implicated by PBC's cytokine signature, the presence of CD4+ T cells specific to mitochondrial auto-antigens, the expression of MHC II on injured biliary epithelial cells, and PBC's coincidence with other similar T cell mediated autoimmune conditions. CD4+ T cells are also central to current animal models of PBC, and their transfer typically also transfers disease. The importance of genetic risk to developing PBC is evidenced by a much higher concordance rate in monozygotic than dizygotic twins, increased AMA rates in asymptomatic relatives, and disproportionate rates of disease in siblings of PBC patients, PBC family members and certain genetically defined populations. Recently, high-throughput genetic studies have greatly expanded our understanding of the gene variants underpinning risk for PBC development, so linking genetics and immunology. Here we summarize genetic association data that has emerged from large scale genome-wide association studies and discuss the evidence for the potential functional significance of the individual genes and pathways identified; we particularly highlight associations in the IL-12-STAT4-Th1 pathway. HLA associations and epigenetic effects are specifically considered and individual variants are linked to clinical phenotypes where data exist. We also consider why there is a gap between calculated genetic risk and clinical data: so-called missing heritability, and how immunogenetic observations are being translated to novel therapies. Ultimately whilst genetic risk factors will only account for a proportion of disease risk, ongoing efforts to refine associations and understand biologic links to disease pathways are hoped to drive more rational therapy for patients.
Keywords: Genome-wide association study, Immunochip, HLA, CD4+ T cell, Animal models, Regulatory T cell
Highlights
-
•
Primary biliary cirrhosis is a typical autoimmune disease with a strong genetic component.
-
•
Genome-wide studies have greatly expanded understanding of genetic risk.
-
•
This review highlights individual associations including HLA types and the IL-12-STAT4 pathway.
-
•
Animal models, epigenetics and novel immunological therapies are also considered.
1. Introduction
Primary biliary cirrhosis (PBC) is an idiopathic autoimmune chronic liver disease characterised by the progressive loss of small intrahepatic bile ducts with resultant cholestasis and progressive fibrosis [1]. One in 1000 women over the age of 40 liver have PBC [2], and there remains only one licensed therapy – ursodeoxycholic acid. Failure to respond to this treatment puts patients at risk of progressive ductopenia and fibrosis, which ultimately requires liver transplantation to avoid death from liver failure. Current disease models envisage an immune-driven biliary injury, resulting in secondary cholestasis, and which arises on the background of combined genetic and environmental risks. Further mechanistic insights should illuminate better therapeutic options for patients. Herein we consider the immunogenetic basis for PBC and the potential for this new knowledge to translate into improved disease management.
1.1. PBC is a typical autoimmune disease with a T-cell signature
The phenotype of PBC is typical for autoimmune disease, characterized by strong female predisposition, with a high proportion (∼53%) of patients having at least one coincident autoimmune condition [3], [4] (Table 1), and most affected individuals manifesting detectable autoantibodies against the E2 component of the pyruvate dehydrogenase enzyme found on the inner mitochondrial membrane [5]. These ‘anti-mitochondrial antibodies’ (AMA) are both sensitive and specific for diagnosis and prediction of the disease and are usually present at high titer [6]. Other autoantibodies are also frequent among PBC patients, including antibodies with highly specific anti-nuclear antibody reactivity [1].
Table 1.
Coincidence of other autoimmune disease with PBC.
| Probable or definite co-incident condition | Number (%); n = 160 |
|---|---|
| Sjögren syndrome | 40 (25) |
| Autoimmune thyroid disease | 37 (23) |
| Rheumatoid arthritis | 27 (17) |
| Scleroderma | 12 (8) |
| Raynaud's phenomenon | 38 (24) |
| Systemic lupus erythematosus | 2 (1) |
| Autoimmune thrombocytopenic purpura | 2 (1) |
| Pernicious anemia | 6 (4) |
| All conditions | 84 (53) |
Adapted from Watt et al. [3].
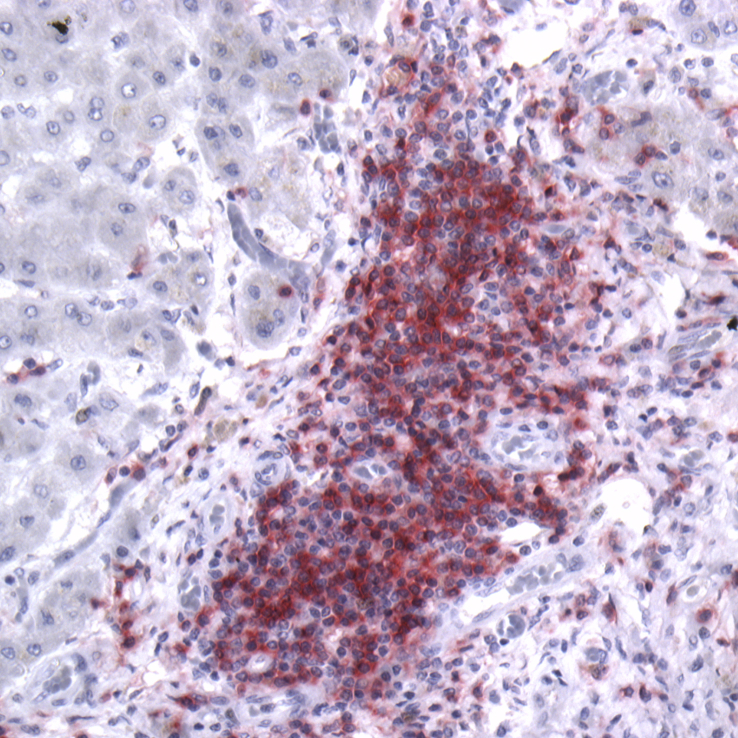
Pathologically, PBC is characterised by a progressive lymphocytic cholangitis centered on smaller intrahepatic bile ducts, often associated with the presence of granulomata in the liver. Autoantibodies against the components of mitochondria are densely localized to the apical surface of biliary epithelial cells (BEC) [7] and are associated with apoptosis [8]. A similar staining pattern may be seen on salivary epithelium in PBC patients with coincident sicca syndrome [9]. As is consistent with involvement of the adaptive immune system, the immune infiltrate is predominantly comprised of CD4+ T cells, with lesser increases in cytotoxic (CD8+) T cells [10] (Fig. 1). Numbers of CD4+ T cells are also increased in the hilar lymph nodes and the liver. Importantly, CD4+ [11] and CD8+ [12] T cells specific to mitochondrial auto-antigens have been demonstrated in the peripheral blood, livers and liver-draining lymph nodes of affected patients, while not detected in either healthy controls or patients with other liver diseases. Both MHC class I and II proteins are also expressed on BECs of PBC patients and thought to present antigen to cytotoxic CD8+ and helper CD4+ T cells, respectively [13], [14], [15].
Fig. 1.
CD4+ T cells dominate the inflammatory infiltrate of PBC. Explanted PBC liver specimen stained with rabbit anti-CD4 (clone ab133616, Abcam, UK) and revealed with alkaline phosphatase red kit (Vector laboratories, UK); hematoxylin counterstain; ×20 magnification.
The cytokine signature associated with PBC is also indicative of immune system activation with a Th1/Th17 bias. Analysis of RNA expression in explanted PBC liver samples has consistently revealed skewing of the cytokine profile with reduced IL-10 (a predominantly Th2 cytokine) and increased interferon gamma (IFNγ; a Th1 cytokine) in comparison to chronic hepatitis C explants [16], [17]. Levels of serum IL-18 – which acts to release IL-12 and activate the Th1 pathway – and IFNγ are also elevated in PBC patients relative to levels detected in healthy controls and chronic viral hepatitis patients [18], [19]. Immunohistochemical studies support these observations, with PBC liver samples showing strong staining for IFNγ and IL12RB2 with a shift to increased IL-23 and Th-17 staining in later disease [20]. Ratios of circulating Th17:regulatory T cells (Treg) [21] as well as levels of serum IgM and numbers of IgM-producing plasma cells in the liver are also frequently increased in PBC patients [22].
1.2. Immunogenetic observations from animal models of autoimmune cholangitis
There is no animal model that completely reproduces the human PBC phenotype, a situation that may relate to limitations in deriving animal models and/or the highly complex combination of environmental and genetic factors and pathogenic pathways associated with biliary injury. Among the mouse strains now used as models for PBC are a number of strains with deficiencies in Treg. One example is the Scurfy mouse, in which a mutation in the master transcription factor of Treg, FOXP3, results in the complete absence of Treg. These mice manifest peri-biliary lymphocytic infiltrate, liver damage and AMA production on a background of multi-system autoimmunity [23]. Similar disease phenotypes arise in mice expressing a dominant negative TGFβII under the control of the CD4 promoter [24] and in IL2Rα−/− mice, both of which have significant deficits in Treg function [25]. In one other murine model of PBC, a mutation that impairs function of the biliary epithelial cell and lymphocyte anion exchanger AE2 is associated with reductions in the numbers of Treg and variable periportal infiltrates with AMA [26].
Other murine models of PBC also highlight the importance of T cells in disease pathogenesis. Interruption of selected chromosomal regions on chromosomes 3 and 4 of the non-obese diabetic mouse, for example, yields mice that develop intra- and extrahepatic inflammation and dilation along with variable AMA for which biliary disease can be transferred by the mutant T cells or prevented by T cell depletion [27]. Similarly, CD4+ T cells from mice in which AMA production is triggered by immunization with the bacterium Novosphingobium aromaticivorans, can also transfer the disease phenotype to other mice [28].
A further insight from animal models is the complexity of the processes which lead to emergence of autoimmune disease. In one mouse model, for example, in which the xenobiotic 2-octynoic acid is used to induce production of anti-mitochondrial antibodies, development of the phenotype requires both the highly immunogenic complete Freud's adjuvant and an autoimmune-prone NOD.1101 background [29].
Manipulation of immunologic pathways has also been of help in refining the relative importance of different signaling cascades to PBC-like phenotypes. For example, deletion of IL-12p40 from IL2Rα-deficient mice worsens cholangitis and fibrosis without evidence of shift to Th2 polarized responses that are thought to be associated with fibrosis [30]. Similarly, dominant negative TGFβ mice develop increased fibrosis with deletion of 12p35 [31], but, conversely, cholangitis in these mice is reduced by co-deficiency of IL-12p40 [32].
1.3. Clinical support for a strong genetic component to PBC
A significant etiologic role for genetic factors in PBC is supported by epidemiological evidence. PBC has the highest concordance in monozygotic twins of any autoimmune disease: 63% in one small series as compared to a risk in dizygotic twins close to the population level of <0.5% with no concordance between the 8 pairs studied [33]. Zygosity was carefully confirmed in each pair. Intriguingly, ages of presentation and presenting symptoms were similar among concordant twins and two of the discordant twins had other major autoimmune conditions.
The increased risk of other autoimmune conditions in PBC patients and their family members also points towards a genetic basis for disease that overlaps with the genetic factors underpinning other autoimmune diseases (Table 1). Risk for expressing AMA and for developing disease is also increased in first-degree relatives [34], [35]. and a disproportionate number of PBC patients have relatives with the disease at the point of diagnosis [2], [36]. Sibling relative risk of the disease was calculated at 10.5 [37] in a UK series and 10.7 in the USA [38]. Numbers of individuals affected by PBC are also abnormally high in selected ethnically defined sub-populations [39], [40] and in selected families within well-characterised comprehensive healthcare systems [35], [41].
2. Immunogenetic observations in PBC
High throughput genetic studies have transformed our understanding of the genes conferring risk for autoimmune diseases including PBC. Such studies involve the comparison of large cohorts of individuals with and without the disease trait. Each group is genotyped for a very large number of individual single nucleotide variants (SNV) which are selected from those sufficiently prevalent in the general population to power subsequent statistical association analyses. In the case of genome wide association studies (GWAS), these traits are chosen from sequencing efforts to dissect the genome, while in more focused studies, such as the Immunochip analyses [42], a targeted selection of SNVs is used for the study. In the case of the Immunochip, for example, SNVs implicated in immune pathways and other autoimmune diseases as well as rarer SNVs are used to allow finer-resolution analysis of loci of interest.
The analyses of large scale GWAS-derived datasets enables identification of individual variants disproportionately associated with a disease or disease trait and, by extension, risk loci and candidate genes within or near such loci [43]. The power and lack of bias given the hypothesis-free nature of such studies has revolutionized the approach to disease gene discovery and enabled a move away from candidate gene studies and primary focus on either gene array technology or whole exome genome sequencing strategies.
A key confounder in GWAS is the potential for false positives because of the multiple comparisons inherent to the analysis. Thus, careful adjustments for such comparisons is mandatory as well as the replication of associations in separate validation cohorts. Further, GWAS-identified risk variants are, by definition, more frequent in the population than is the disease per se [44]. With these caveats in mind, PBC research has benefited from several high quality GWAS [45], [46], [47], [48], meta-analyses of these datasets [49], [50], and two more focused Immunochip studies [51], [52]. The majority of these studies have focused on populations of Caucasian origin with two associations (POU2AF1 and TNFSF15) unique to the Japanese population. In Caucasians many variants associated with PBC at or below genome-wide significance levels (p < 5 × 10−8) have been identified and these are summarized in Fig. 1 and Table 2. This number will increase with larger, and higher resolution, studies. Notably, there is a high degree of overlap between the known PBC risk variants and those associated with other autoimmune conditions.
Table 2.
Risk loci associated with PBC identified by high-throughput genetic studies.
| Locus | GWAS of PBC |
Canadian/Italian/US iCHIP [51] |
UK iCHIP [52] |
Candidate gene(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | SNV/RA | OR | p-value | Peak SNV/RA | OR | p-value | SNV/RA | OR | p-value | ||
| 1p36 | [50] | rs3748816/C | 1.33 | 3.15E-08 | rs10910108/G | 1.15 | 1.81E-03 | rs10797431/A | 1.15 | 1.44E-05 | MMEL1 |
| 1p31 | [45] | rs3790567/A | 1.51 | 2.76E-11 | rs72678531/C | 1.68 | 2.66E-23 | rs72678531/G | 1.61 | 2.47E-38 | IL12RB2 |
| 1q31 | [47] | rs12134279/T | 1.34 | 2.06E-14 | rs1539414/A | 1.26 | 3.46E-06 | rs2488393/A | 1.28 | 4.29E-12 | DENND1B |
| 2q12 | [49] | rs12712133/A | 1.14 | 5.19E-9 | IL1R1, IL1RL2 | ||||||
| 2q32 | [47] | rs10931468/A | 1.5 | 2.35E-19 | rs3024921/A | 1.75 | 4.45E-11 | rs3024921/A | 1.62 | 2.59E-18 | STAT4, STAT1 |
| 2q36 | [49] | rs4973341/C | 0.82 | 2.34E-10 | CCL20 | ||||||
| 3p24 | [47] | rs1372072/A | 1.2 | 2.28E-08 | rs1025818/G | 1.2 | 3.53E-05 | rs1025818/G | 1.13 | 1.89E-04 | PLCL2 |
| 3q13 | [47] | rs2293370/G | 1.35 | 2.53E-11 | rs1131265/G | 1.42 | 4.49E-09 | rs2293370/G | 1.39 | 6.84E-16 | CD80 |
| 3q25 | [45] | rs6441286/G | 1.54 | 2.42E-14 | rs9877910/T | 1.47 | 1.02E-17 | rs2366643/A | 1.35 | 3.92E-22 | IL12A |
| 4p16 | [49] | rs11724804/A | 1.22 | 9.01E-12 | DGKQ | ||||||
| 4q24 | [47] | rs7665090/G | 1.26 | 4.06E-12 | rs7665090/G | 1.18 | 1.00E-04 | rs7665090/C | 1.26 | 8.48E-14 | NFKB1 |
| 5p13 | [47] | rs860413/A | 1.3 | 1.02E-11 | rs700172/A | 1.27 | 1.63E-06 | rs6871748/A | 1.3 | 2.26E-13 | IL7R |
| 5q21 | [49] | rs526231 | 0.87 | 1.14E-08 | C5orf30 | ||||||
| 5q33 | [49] | rs2546890 | 0.87 | 1.06E-10 | IL12B | ||||||
| 6p21 | [45] | rs2856683/C | 1.75 | 1.78E-19 | rs7775055/C | 3.71 | 1.11E-33 | rs7774434/C | 1.57 | 1.30E-48 | HLA region |
| 6q23.3 | [49] | rs6933404 | 1.18 | 1.27E-10 | TNFAIP3, OLIG3 | ||||||
| 7p14 | [47] | rs6974491/A | 1.25 | 4.44E-08 | rs17259795/T | 1.24 | 1.19E-05 | rs73112661/G | 1.22 | 1.69E-07 | ELMO1 |
| 7q32 | [50] | rs10488631/C | 1.57 | 8.66E-13 | rs10488631/C | 1.56 | 2.52E-12 | rs35188261/A | 1.52 | 6.52E-22 | IRF5 |
| 9p32 | [48] | rs4979462/T | 1.57 | 1.85E-14 | |||||||
| 11q13 | [47] | rs538147/G | 1.23 | 2.06E-10 | rs694739/A | 1.18 | 2.85E-04 | rs694739/A | 1.18 | 1.96E-07 | RPS6KA4 |
| 11q23 | [47] | rs6421571/C | 1.37 | 2.69E-12 | rs7117261/C | 1.46 | 3.18E-10 | rs80065107/A | 1.39 | 7.20E-16 | CXCR5, DDX6 |
| 11q23 | [48] | rs4938534/A | 1.38 | 3.27E-08 | POU2AF1 | ||||||
| 12p13 | [47] | rs1800693/G | 1.22 | 1.80E-09 | rs1860545/T | 1.2 | 5.27E-05 | rs1800693/G | 1.27 | 1.18E-14 | TNFRSF1A,LTBR |
| 12q24 | rs11065979/A | 1.2 | 2.87E-09 | SH2B3 | |||||||
| 13q14 | rs3862738/G | 1.33 | 2.18E-08 | TNFSF11 | |||||||
| 14q24 | [47] | rs911263/T | 1.29 | 1.76E-11 | rs911263/A | 1.25 | 3.43E-06 | rs911263/T | 1.26 | 9.95E-11 | RAD51B |
| 14q32 | [47] | rs8017161/A | 1.22 | 2.61E-13 | TNFAIP2 | ||||||
| 16p13 | [47] | rs12924729/G | 1.29 | 2.95E-12 | rs413024/T | 1.31 | 2.29E-08 | rs12708715/G | 1.29 | 2.19E-13 | SOCS1, CLEC16A |
| 16q24 | [47] | rs11117432/G | 1.31 | 4.66E-11 | rs35703946/G | 1.27 | 6.10E-04 | rs11117433/G | 1.26 | 1.41E-09 | IRF8 |
| 17q12 | [50] | rs11557467/G | 0.72 | 3.50E-13 | rs907091/C | 1.29 | 3.43E-09 | rs17564829/G | 1.26 | 6.05E-14 | ORMDL3, IKZF3 |
| 17q21 | rs17564829/G | 1.25 | 2.15E-09 | MAPT | |||||||
| 19p12 | rs34536443/G | 1.91 | 1.23E-12 | TYK2 | |||||||
| 19q13 | [111] | rs3745516/A | 1.46 | 7.97E-11 | SPIB | ||||||
| 22q13 | [47] | rs968451/T | 1.27 | 1.08E-09 | rs715505/C | 1.41 | 9.58E-12 | rs2267407/A | 1.29 | 1.29E-13 | SYNGR1 |
Risk loci for PBC that have achieved genome-wide level of significance (p < 5 × 10−8) in at least one study. GWAS, genome-wide association study; iCHIP, Illumina immunoarray association study; OR, odds ratio; RA, risk allele; SNV, single nucleotide variant.
Adapted from Mells and Hirschfield [112].
3. The HLA gene variants associated with PBC
The HLA region, on the short arm of chromosome 6 contains many genes related to the adaptive immune response. Key amongst these are the HLA-A, HLA-B and HLA-C genes, which are associated with the production of MHC class I and the HLA-D genes, which are associated with MHC class II. Class I proteins are primarily involved with the presentation of shorter processed fragments of intracellular antigen to cytotoxic CD8+ T cells, while MHC class II proteins present extracellular antigen to CD4+ T helper cells. Variations in the large and highly polymorphic HLA locus on chromosome 6 have been long associated with PBC [53]. Initial studies linked the DRB*08:01 allele group with disease risk, but the populations explored in these studies were relatively small [e.g. Ref. [54]].
More recently, analysis of a much larger sample of PBC in the Italian population confirmed an association of risk for PBC with the HLA-DR alleles B1*08 and B1*02 and an apparently protective effect of B1*11 and B1*13 [55]. These observations have been clarified by subsequent GWAS and Immunochip work, and a set of robust HLA associations now exists (Table 3), albeit primarily related to European Caucasian populations concerned and also the choice of variants examined on the Immunochip [42].
Table 3.
HLA associations in PBC.
| Risk conferring | ||
| HLA-DQA1*04:01-HLA-DQB1*04:02-HLA-DRB1*08:01-HLA-B*39:05 |
[51], [52] N. Am, UK, Ital |
* |
| HLA-DRB1*04:04-HLA-DQB1*03:02 |
[51], [52] N. Am, UK, Ital |
* |
| HLA-DRB1*14-HLA-DPB1*03:01 |
[113] Ital |
* |
| HLA-DRB1*08:03-HLA-DQB1*06:01 | [95], [114] Jap, Chin | |
| HLA-DRB1*04:05-HLA-DQB1*04:01 | [95] Jap | |
| Protective | ||
| HLA-DQB1*06:02-HLA-DRB1*15:01-HLA-DQA1*01:02-HLA-B*07:02 |
[51], [52] N. Am, UK, Ital |
* |
| HLA-DQB1*03:01-HLA-DRB1*11:01-HLA-DQA1*05:01-HLA-DRB1*11:04 |
[51], [52], [114] N. Am, UK, Ital, Chin |
* |
| HLA-DRB1*13:02- HLA-DQB1*06:04 | [95] Jap | |
| HLA-DRB1*11:01-HLA-DQB1*03:01 | [95] Jap | |
* Dense SNV analysis and subsequent conditional analysis of this HLA haplotype in the Italian cohort has suggested that risk-conferring/protective effects are predominantly due to variants in HLA-DRB1 with associated linkage disequilibrium [113].
Data from a relatively small study of Chinese support observations in that population, but require confirmation [114].
In terms of understanding PBC pathogenesis, it is possible that specific HLA variants either confer an overall predisposition to autoimmune disease or result in altered immune responses to specific environmental antigens. Although these possibilities require further investigation, recent exciting work has shown that T cells with the risk-conferring HLA DRB1*08:01 genotype shown high-affinity responses to specific pyruvate dehydrogenase E2 subunit peptides, response that did not develop in the presence of the protective DRB1*11:01 allele [56].
4. Non-HLA gene associations
4.1. T cell activation and the IL-12 pathway
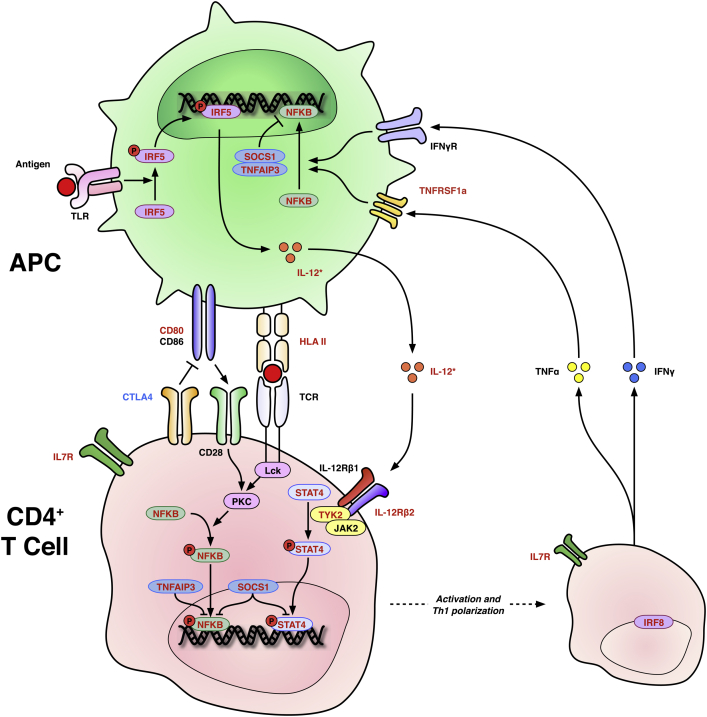
Among the HLA genes associated with risk for PBC are a number of genes associated with CD4+ T cell activation, especially with IL-12-JAK-STAT4 signaling, a pathway promoting Th1 T cell polarisation (Summarized as Fig. 3). IL-12 is a heterodimeric molecule made up of the two subunits, p35 and p40, encoded by the IL12A and IL12B genes respectively. The latter protein also heterodimerizes with IL-23p19 to form IL-23, a key signaling component in the Th-17 pathway. The IL-12 receptor is also encoded by two genes, IL12RB1, which is constitutively expressed, and IL12RB2 which is upregulated by interferon-γ (IFNγ) to act as a positive feedback loop in antigenic stimulation. The tyrosine kinase 2 (TYK2) protein is key to both IL-12 and IL-23 receptor signaling. Variants in these genes are also associated with other autoimmune diseases and in systemic lupus erythematosus appear to influence IFNγ production [57]. STAT4 deficient mice show impaired Th1 polarization and a defect in effector cytokine production that can block the development of autoimmune diabetes [58], [59]. Another gene of interest isTNFAIP3, which encodes a zinc finger protein with the ability to modify ubiquitination states and inhibit the NFκB and STAT pathways as well as the functioning of some TNF receptors [60]. Deficiency confers excess TNF sensitivity in mice and early cachexia and death. The SOCS1 protein also implicated by genetics in PBC, has a regulatory effect on both NFκB and JAK-STAT pathways including STAT4 [61].
Fig. 3.
Schematic representation of CD4+ T cell activation by antigen presenting cells and the IL-12/STAT4 pathway. Antigen activates APC through TLR, which in turn produce IL-12 after phosphorylation of IRF5. Antigen is presented to CD4+ T cells by HLA II with co-stimulation via CD80 and 86 to CD28. There is competitive inhibition of this co-stimulation by CTLA4. IL-12 activates a cascade of signaling factors including NFKB and STAT4 to promote the production of Th1-type cytokines including TNFα and IFNγ; the transcription factor IRF8 is involved. IL7R supports lymphocyte development. There is positive feedback from Th1 cytokines to APCs. Red text denotes confirmed risk associations with PBC; blue text putative associations. Arrows denote positive effects; barred lines denote negative effects. APC = antigen-presenting cell; CD = cluster of differentiation; CTLA4 = Cytotoxic T lymphocyte antigen 4; HLA II = human leucocyte antigen class II; IFN-γ = interferon-γ; IFNγR = interferon-γ receptor; IL-12 = interleukin-12; IL-12Rβ1/2 and IL-12 receptor β subunits 1 and 2; IL7R = interleukin-7 receptor; IRF5 and IRF8 = interferon response factors 5 and 8; JAK2 = Janus kinase 2; Lck = lymphocyte-specific protein tyrosine kinase; NFKB = nuclear factor kappa-light-chain-enchancer of activated B cells; PKC = protein kinase C; SOCS1 = suppressor of cytokine signaling 1; STAT4 = signal transducer and activator of transcription 4; TCR = T-cell receptor; TLR = Toll-like receptor; TNFAIP3 = tumor necrosis factor alpha-induced protein 3; TNFRSF1a = Tumor necrosis factor receptor superfamily 1a; TNFα = tumour necrosis factor alpha; TYK2 = Tyrosine kinase 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.2. CD80
CD80 (also known as B7.1) is an inducible co-stimulatory molecule on antigen-presenting cells. CD80 and the constitutively expressed CD86, together B7, provide a signal to the T cell CD28 receptor which amplifies T-cell antigen receptor signaling. By contrast, another T cell receptor, CTLA4, acts as a higher affinity competitor to CD28 to reduce T cell immune responses. Deficiency of CD80 results in a reduced plasma cell mediated humoral response to immunization [62].
4.3. IL7Rα/CD127
IL-7 is necessary for both T and B lymphocyte development and also for maintenance of T cell populations in the periphery. Mice deficient in IL-7R have markedly reduced thymic and splenic lymphoid cellularity [63]. IL-7R mutations represent one of a heterogeneous group of genetic lesions that cause Omenn syndrome, a condition characterized by reduced variation in the T cell repertoire with immune dysregulation, autoimmunity and a graft-versus host disease phenotype [64]. IL-7R is induced upon T cell positive selection and controls thymic CD8+ lineage specification and peripheral naive T-cell homeostasis [23] while also having a role in myeloid cell differentiation [24]. IL7R expression is generally reduced in Treg cells compared to other T cells [65].
4.4. Other T cell associated genes
IKZF3 encodes Ikaros family zinc finger protein 3, also known as Aiolos. The gene is one of a family of hematopoietic transcription factors and is involved in lymphocyte development and proliferation, especially in B cells [66]. A link to autoimmunity is implied by the lupus-like syndrome that develops in IKZF3 knock-out mice [67]. Subsequent work has also linked this protein to Th17 development through an interaction with the IL2 receptor, disruption of which underlies PBC in one mouse model of disease ([68]; see above).
SH2B3 encodes a member of the SH2B adaptor proteins known as SH2B3 or Lnk, and maps to a widely shared autoimmune disease locus. Lnk is involved in multiple growth factor and cytokine signaling pathways, is a negative regulator of T cell activation, tumor necrosis factor and Janus kinase 2 and 3 (JAK2/3) signaling and is required for normal hematopoiesis. Mice deficient in SH2B3 have greater levels of activated T cells and a tendency to autoimmunity [69].
4.5. B cell development, signaling and migration
In addition to genes encoding proteins such as IL7R and IRFs, expressed in T as well as B cells, results of genetic studies have identified a number of PBC risk loci containing genes that imply a role for B cells in PBC. CD80, for example, is key in the germinal center focused humoral response to immunization and the chemokine receptor, CXCR5, is involved in the migration of both T and B cells to sites of antibody production along gradients of CXCL13. CXCR5 is constitutively expressed on mature B cells and induced on T follicular helper cells in response to antigen [70] and its deficiency is associated with impaired germinal center responses.
POU2AF1 also known as Oct binding factor 1 (OBF1), is a transcription factor involved in the transcription of a number of B cell specific proteins. Mice deficient for this protein have a reduced B cell repertoire, striking reductions in class-switched immunoglobulins and disordered germinal center formation [71].
4.6. TNF ligands and receptors
TNFRSF1A encodes a member of the tumor necrosis factor family of receptors. It is predominantly expressed on antigen-presenting cells and represents a major receptor for tumor necrosis factor alpha (TNFα). Activation of this receptor can cause apoptosis through activation of NFκB and mutations leading to its constitutive activation are associated with periodic fever syndrome [72].
Loci containing two TNF receptor ligands have also been associated with risk for PBC. These proteins include TNFSF15 (or TNF-like ligand TL1A) which encodes a vascular endothelial growth inhibitor primarily expressed on endothelia with little expression in the liver (http://proteinatlas.org). Ligation with its cognate receptor DR3, which is chiefly expressed on lymphocytes, may induce apoptosis, but is also associated with costimulation, mucosal hyperplasia and autoimmune inflammation [73]. TNFSF11, or receptor activator of nuclear factor kappa-B ligand (RANKL) or TNF-related activation-induced cytokine (TRANCE) is also a TNFR ligand. This protein plays a major role in NFκB mediated control of osteoclast activity, but also has activity as a dendritic cell survival factor through the control of apoptosis. Deficiency results in impaired lymphocyte differentiation [74].
4.7. Other signaling molecules
The transcription factor NFκB plays important roles in both the innate and adaptive immune system [61], [75], influencing both B and T cell activation downstream of the antigen receptors and its promoting inflammatory responses. NFκB1 encodes the p50 subunit of NFκB by way of a precursor p105 and NFκB-deficient mice show many immune defects including susceptibility to various bacteria and impaired induction of lymphocyte antibody responses and proliferation [76].
IRF5 and IRF8 encode two members of the interferon response factor family: a set of transcription factors central to the control of type 1 interferon production and response [77]. IRF5 is ubiquitously expressed, upregulated by type 1 interferons and is activated by Toll-like receptor ligation so as to promote downstream transcription of IL6, IL12 and TNFα. IRF8 is key to generating Th1 type responses and is induced by IFNγ. Its deficiency in mice confers susceptibility to intracellular infection [78].
CCL20 is a chemoattractant for lymphocytes, and to a lesser degree, neutrophils and is strongly expressed in the liver [79]. In human alcoholic hepatitis, its expression has been linked to disease severity, and in mouse models of hepatitis, its silencing ameliorates inflammation and production of pro-fibrotic mediators [80]. CCR6 – the receptor for CCL20 – has been reported as being important for the positioning of pathogenic Th17 T cells in the inflamed liver, particularly in PBC liver, in which CCR6 concentrates around bile ducts [81].
4.8. Other genes
A number of other variants within genes involved in key immunological pathways have been suggested by GWAS data, but not attained genome-wide significance. An example is CTLA4 which encodes a protein expressed on T cells and which competes with CD28 for binding to CD80 and CD86, thus reducing pro-effector signaling through CD28 (Fig. 1) [82], [83]. Also of interest, but not yet of proven significance, is ICOSL, the cognate ligand of inducible costimulator or ICOS (Fig. 2). ICOS is a member of the CD28 family that is minimally expressed on unactivated T cells, but is rapidly upregulated after stimulation through the T-cell receptor and CD28. Excessive ICOS expression is associated with multi-system autoimmunity [84].
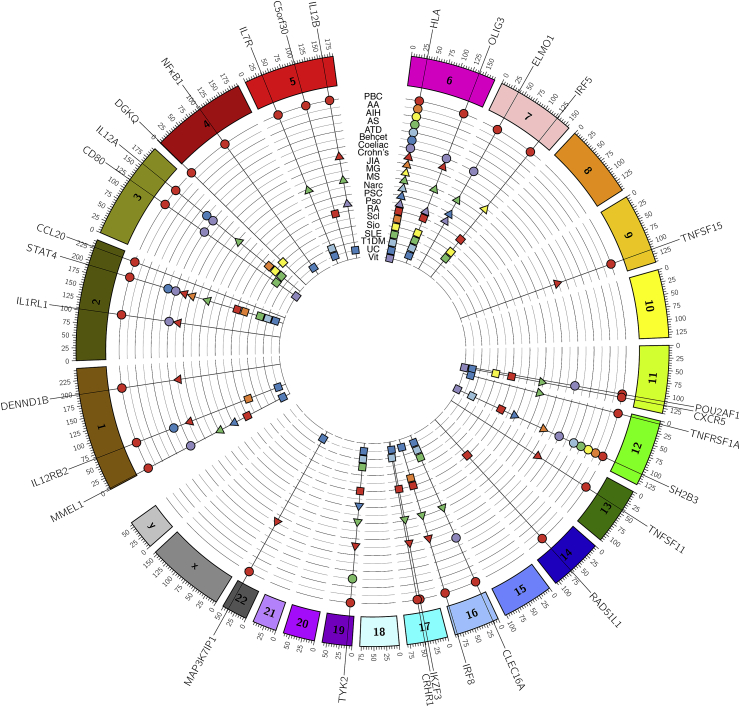
Fig. 2.
PBC shares risk loci with multiple other autoimmune diseases. Circos plot [110] of gene variants associated with PBC and other selected autoimmune conditions in high-throughput genetics studies and their meta-analyses. Note that significant pleiotropy exists with some loci implicated in multiple conditions and variable HLA associations a universal feature. PBC = primary biliary cirrhosis; AA = alopecia areata; AIH = autoimmune hepatitis; AS = ankylosing spondylitis; ATD = autoimmune thyroid disease; JIA = juvenile idiopathic arthritis; MG = myasthenia gravis; MS = multiple sclerosis; Narc = narcolepsy; PSC = primary sclerosing cholangitis; Pso = psoriasis; RA = rheumatoid arthritis; Scl = scleroderma; Sjö = Sjögren's syndrome; SLE = systemic lupus erythematosus; T1DM = type 1 diabetes mellitus; UC = ulcerative colitis; Vit = vitiligo. Only validated associations at p < 5 × 10−8 are included; supporting citations available at request.
It is likely that an increasing number of risk-affecting variants will be identified as the numbers of participants in genetic studies increases and additional meta-analyses are performed.
5. Epistasis, missing heritability and selection pressure
In the majority of instances the odds ratios for any individual risk variant associated with PBC are modest and close to or lower than 1.5 (Table 2). Calculations of the risk for disease conferred by all the known PBC susceptibility alleles together suggest these alleles account for only 5.3% of the heritable risk for this disease [85]. This low number may relate, at least in part to limitations to GWAS studies such as frequency of the studied variants and sample size [44]. It is also possible that shared environmental traits and gene-environmental interactions inflate perceived heritability or that the gap between calculated and observed heritability may be partially explained by epistasis. In epistatic interactions, the effects of one variant may be dependent on the presence and effects of one or more other variants. One example of such interactions in PBC, is the risk-conferring epistatic interaction between the 1p31 (IL12RB2) and 7q32 (IRF5) loci [51]. A potential risk-amplifying interaction between CTLA4 and TNFα variants has also been reported [83].
6. Epigenetics
A necessary corollary to the discordance seen in some monozygotic twins is the influence of epigenetic factors. Several environmental factors have been proposed and these are well reviewed elsewhere [86]. Specific epigenetic observations may however partly explain a deficiency in our understanding of the genetic basis of PBC to date: the lack of an explanation for female predominance, including the lack of identified risk loci on the X chromosome. A major observation was that rates of X chromosome monosomy in peripheral leukocytes – i.e. the presence of a single X chromosome in a usually diploid cell – are higher in PBC patients than hepatitis C or healthy controls after correction for age, with which monosomy correlates [87]. Subsequent similar observations have been made in other autoimmune diseases and the observation that autoimmunity is significantly more common in patients with constitutive monosomy X: Turner's syndrome [88], [89]. Intriguingly, there is also an increased rate of Y chromosome loss in men with PBC [90].
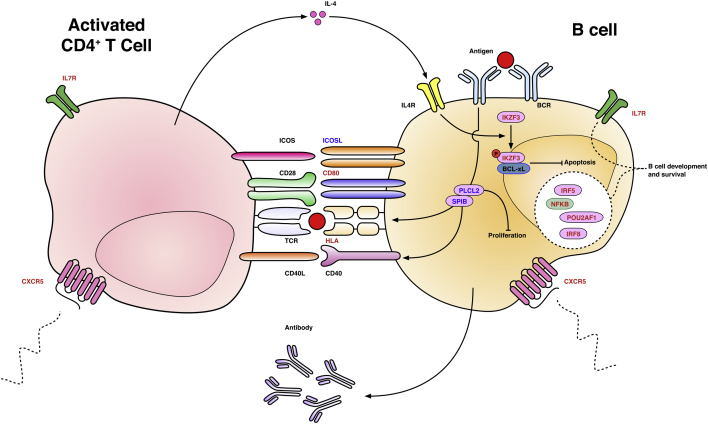
In healthy women, one X chromosome homologue is inactivated by heterochromatin packaging, DNA methylation, reduced histone acetylation and other mechanisms resulting in gene silencing. In PBC, it has been demonstrated that while X inactivation appears to be random, there is preferential loss in cells with X monosomy in contrast to that seen in health [91]. In related work, the methylation states of genes that escape modification in X chromosome inactivation to variable degrees were examined in monozygotic PBC twin pairs. In the small number of samples that were available to process, no significant differences in methylation status were observed. A further epigenetic observation of particular interest given the role of CD40-CD40L in T:B cell interactions (Fig. 4) is that there is reduced methylation of CD40L promoter regions amongst PBC patients compared with controls and that this correlates with serum IgM [92].
Fig. 4.
Schematic representation of T-cell:B-cell interaction demonstrating genes with variants associated with PBC Antigen binds B-cell receptor triggering multiple events including the suppression of apoptosis by BCL-xL in a mechanism involving the phosphorylation of IKZF3. B-cell receptor signaling is also partly mediated by the transcription factor SPIB and PLCL2; the former is involved with differentiation and the latter controls subsequent proliferation. B cells present antigen to T cells on MHC class II. Multiple co-stimulatory molecules promote both B cell activation and continued T cell activation: ICOS-ICOSL, CD28-CD80/CD86, CD40L-CD40. IL7R is involved in the development of both T and B cells. A number of other factors key to B cell development and survival are associated with PBC risk (IRF5, IRF8, NFKB and POU2AF1). CXCR5 guides both B and T cell positioning along CXCL13 chemokine gradients and facilitates migration to germinal centres. Red text denotes confirmed risk associations with PBC; blue text putative associations. Arrows denote positive effects; barred lines denote negative effects. BCR = B-cell receptor; IL-4 = interleukin-4; IL7R = Interleukin-7 receptor; ICOS(L) = inducible co-stimulator (ligand); PLCL2 = phospholipase C-like 2 protein; TCR = T-cell receptor; IRF5 & 8 = interferon regulatory factors 5 & 8; SPIB = Spi-B; NFKB = nuclear factor kappa-light-chain-enhancer of activated B cells; POU2AF1 = POU class 2 associating factor 1; IKZF3 = Ikaros family zinc finger protein 3; CXCR5 = chemokine (C-X-C motif) receptor 5.
Two studies have reported microRNA expression in PBC. Building on their work describing the anion exchanger 2 (AE2) deficient mouse [26], one group have described upregulated microRNA-506 (miR-506) – which downregulates AE2 – in PBC BECs by in situ hybridization and also by BEC culture with anti-miR506 increasing AE2 activity [93]. A second group work from PBC and control explant liver samples demonstrated down-regulation of miR-122a and miR-26a with upregulation of miR-328 and miR-299-5p [94]. All of these microRNAs are implicated in key mechanisms in PBC pathogenesis including apoptosis and the response to oxidative stress and are therefore worthy of further investigation.
7. Relating immunogenetic observations to clinical phenotypes
As summarized above, multiple HLA haplotypes have been associated with risk for PBC and also with specific phenotypes. A study of Japanese patients, for example, has reported an association of the HLA DRB1*04:05 and DRB1*08:03, alleles with presence of anti-gp210 and anti-centromere antibodies, respectively, which are in turn associated with rapidly progressing disease and with concurrent systemic sclerosis and portal hypertension, respectively. Presence of the rs9277535 variant at the HLA-DPB1 locus in PBC patients has also been associated with expression of anti-sp100 antibodies [50] and in Japanese PBC patients, the DRB1*09:01-DQB1*03:03 haplotype has been associated with increased tendency for progression to need for transplantation [95], an association not yet confirmed in European populations.
7.1. Non-HLA associations
Development of biliary cirrhosis weeks after initiating IFNγ therapy has been reported in one patient with IL-12 deficiency-related tuberculosis [96] and in a second patient with tuberculosis and autoantibodies to IFNγ, unspecified non-specific cholangitis developed together with multi-system autoimmunity following IFNγ treatment [97]. Unfortunately, the patients' autoantibody profiles or hepatic histology were not reported for these patients and while IL-12 deficiency in the first case may be relevant to the phenotypes, another and more likely possibility is that exogenous interferon induced the autoimmunity [98]. Variants at the IL12A locus do appear, however, to have some effect on the risk of PBC recurrence after transplantation, although the immunosuppression regime chosen is also relevant to this development [99].
As mentioned above, the association of CXCR5 variants with PBC risk points to the involvement of both B and T cells destined to migrate to germinal centers. Consistently, numbers of peripheral T-follicular helper (Tfh) cells have been reported in PBC as compared with control and autoimmune hepatitis subjects. Such elevations in Tfh were also correlated with lack of biochemical response to UDCA [100], [101]. Of related interest is the observation that expression of the CXCR5 ligand, CXCL13, is upregulated in the spleens of PBC patients, but not in spleens of patients with chronic viral hepatitis [102].
A variety of variants have been associated with particular disease sub-phenotypes in PBC, but most of these have not been validated. In one study, for example, PBC susceptibility and progression was associated with specific variants in TNFα, CTLA4 and AE2 genes [103], but these genes have been identified by subsequent GWAS or Immunochip data.
In follow-up studies of GWAS datasets, associations of STAT4 variation with ANA status and CTLA4 variants with anti-centromere and gp210 have been identified in Japanese PBC patients [104] and TNFSF15 polymorphisms have been linked to functional changes in the gene product. Differences in mRNA and protein expression were also seen in both PBC and healthy subjects carrying this variant, although a link to outcome or phenotype remains unclear [105].
8. Translating immunogenetic observations to novel therapies
To date, a single proof-of-concept study has examined the effect of the anti-human IL-12 and IL-23 agent ustekinumab in PBC [106] and revealed no significant change in the primary outcome measure of serum alkaline phosphatase. By contrast, ustekinumab is efficacious in the T-cell mediated disease psoriasis, which also has an IL-12 signature, but IL-23 antagonism alone is also highly effective in that disease [107].
Building on genetic observations and those seen in murine studies [108], a trial of the CTLA4 fusion protein abatacept is now underway in PBC patients (NCT02078882). Inhibition of CD40-CD40L interactions is also being studied as a PBC treatment option, these proteins not being specifically highlighted by genetic association data, but represent key players in T cell:B cell interactions (Fig. 4; NCT02193360). As noted above, differences in CD40L promoter methylation are reported in PBC [92].
9. Conclusion
PBC is a complex classical autoimmune disease with a clear heritable component intertwined with strong environmental influences. There is also now very strong evidence linking PBC to numerous immune pathways, especially those centering on antigen-presentation to T cells.
PBC also remains a therapeutically challenging disease because our increased understanding of its immune basis has not yet translated to marked improvements in patient care [109]. Variants in key immunologic pathways highlight genes involved primarily in the adaptive immune system including antigen-presenting cells, B cells and especially T cells. Of particular note are the genes related to the IL-12-STAT4 Th1-polarizing pathway. These data provide the framework for further animal work, prospective analysis of patient populations and, ultimately the discovery and/or application of novel or repurposed therapeutic agents. Select animal studies suggest that the gene targets identified may have hepato-protective as well as risk effects, but further work is necessary to connect genetic associations to clinical presentation and outcomes. The immunogenetic future lies in larger, finer resolution, high-throughput genetic studies coupled to careful assessment of patient characteristics and trials informed by biological studies.
Acknowledgements
GW is a Medical Research Council Clinical Research Training Fellow. GH is a co-investigator for UK-PBC (www.uk-pbc.com) supported by a Stratified Medicine Award from the UK Medical Research Council, and Principal Investigator for UK-PSC, a NIHR Rare Disease Translational Collaboration.
The views expressed are those of the authors(s) and not necessarily those of the National Health System, the National Institute for Health Research, or the Department of Health.
References
- 1.Hirschfield G.M., Gershwin M.E. The immunobiology and pathophysiology of primary biliary cirrhosis. Annu. Rev. Pathol. Mech. Dis. 2013;8(1):303–330. doi: 10.1146/annurev-pathol-020712-164014. [DOI] [PubMed] [Google Scholar]
- 2.Boonstra K., Beuers U., Ponsioen C.Y. Epidemiology of primary sclerosing cholangitis and primary biliary cirrhosis: A systematic review. J. Hepatol. 2012;56(5):1181–1188. doi: 10.1016/j.jhep.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 3.Watt F.E., James O.F.W., Jones D.E.J. Patterns of autoimmunity in primary biliary cirrhosis patients and their families: a population-based cohort study. QJM. 2004;97(7):397–406. doi: 10.1093/qjmed/hch078. [DOI] [PubMed] [Google Scholar]
- 4.Somers E.C. Are individuals with an autoimmune disease at higher risk of a second autoimmune disorder? Am. J. Epidemiol. 2009;169(6):749–755. doi: 10.1093/aje/kwn408. [DOI] [PubMed] [Google Scholar]
- 5.Gershwin M.E. Identification and specificity of a cDNA encoding the 70 kd mitochondrial antigen recognized in primary biliary cirrhosis. J. Immunol. 1987;138(10):3525–3531. [PubMed] [Google Scholar]
- 6.Bogdanos D.P., Komorowski L. Disease-specific autoantibodies in primary biliary cirrhosis. Clin. Chim. Acta. 2011;412(7–8):502–512. doi: 10.1016/j.cca.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Migliaccio C. Monoclonal antibodies to mitochondrial E2 components define autoepitopes in primary biliary cirrhosis. J. Immunol. Baltim. Md. 1950. 1998;161(10):5157–5163. [PubMed] [Google Scholar]
- 8.Matsumura S. Contribution to antimitochondrial antibody production: cleavage of pyruvate dehydrogenase complex-E2 by apoptosis-related proteases. Hepatology. 2002;35(1):14–22. doi: 10.1053/jhep.2002.30280. [DOI] [PubMed] [Google Scholar]
- 9.Tsuneyama K. Human combinatorial autoantibodies and mouse monoclonal antibodies to PDC-E2 produce abnormal apical staining of salivary glands in patients with coexistent primary biliary cirrhosis and Sjogren's syndrome. Hepatology. 1994;20(4 Pt 1):893–898. doi: 10.1002/hep.1840200418. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto E. Immunohistochemical characterization of hepatic lymphocytes in primary biliary cirrhosis in comparison with primary sclerosing cholangitis and autoimmune chronic active hepatitis. Mayo Clin. Proc. 1993;68(11):1049–1055. doi: 10.1016/s0025-6196(12)60897-0. [DOI] [PubMed] [Google Scholar]
- 11.Shimoda S. Identification and precursor frequency analysis of a common T cell epitope motif in mitochondrial autoantigens in primary biliary cirrhosis. J. Clin. Invest. 1998;102(10):1831–1840. doi: 10.1172/JCI4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kita H. Quantitative and functional analysis of PDC-E2–specific autoreactive cytotoxic T lymphocytes in primary biliary cirrhosis. J. Clin. Invest. 2002;109(9):1231–1240. doi: 10.1172/JCI14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baba N. Gene expression profiling in biliary epithelial cells of primary biliary cirrhosis using laser capture microdissection and cDNA microarray. Transl. Res. 2006;148(3):103–113. doi: 10.1016/j.trsl.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Van den Oord J.J., Sciot R., Desmet V.J. Expression of MHC products by normal and abnormal bile duct epithelium. J. Hepatol. 1986;3(3):310–317. doi: 10.1016/s0168-8278(86)80483-4. [DOI] [PubMed] [Google Scholar]
- 15.Barbatis C. Immunocytochemical analysis of HLA class II (DR) antigens in liver disease in man. J. Clin. Pathol. 1987;40(8):879–884. doi: 10.1136/jcp.40.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagano T. Cytokine profile in the liver of primary biliary cirrhosis. J. Clin. Immunol. 1999;19(6):422–427. doi: 10.1023/a:1020511002025. [DOI] [PubMed] [Google Scholar]
- 17.Harada K. In situ nucleic acid hybridization of cytokines in primary biliary cirrhosis: predominance of the Th1 subset. Hepatology. 1997;25(4):791–796. doi: 10.1002/hep.510250402. [DOI] [PubMed] [Google Scholar]
- 18.Fracchia M. Serum interferon gamma in primary biliary cirrhosis: effect of ursodeoxycholic acid and prednisone therapy alone and in combination. Eur. J. Gastroenterol. Hepatol. 2000;12(4):463–468. doi: 10.1097/00042737-200012040-00016. [DOI] [PubMed] [Google Scholar]
- 19.Yamano T. Serum interferon-gamma-inducing factor/IL-18 levels in primary biliary cirrhosis. Clin. Exp. Immunol. 2000;122(2):227–231. doi: 10.1046/j.1365-2249.2000.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C.-Y. IL-12/Th1 and IL-23/Th17 biliary microenvironment in primary biliary cirrhosis: implications for therapy. Hepatology. 2014;59:1944–1953. doi: 10.1002/hep.26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rong G. Imbalance between T helper type 17 and T regulatory cells in patients with primary biliary cirrhosis: the serum cytokine profile and peripheral cell population. Clin. Exp. Immunol. 2009;156(2):217–225. doi: 10.1111/j.1365-2249.2009.03898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels J.A. Immunostaining of plasma cells in primary biliary cirrhosis. Am. J. Clin. Pathol. 2009;131(2):243–249. doi: 10.1309/AJCP8WHR0IEVUUOJ. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W. Deficiency in regulatory T cells results in development of antimitochondrial antibodies and autoimmune cholangitis. Hepatology. 2008;49(2):545–552. doi: 10.1002/hep.22651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oertelt S. Anti-mitochondrial antibodies and primary biliary cirrhosis in TGF-beta receptor II dominant-negative mice. J. Immunol. Baltim. Md. 1950. 2006;177(3):1655–1660. doi: 10.4049/jimmunol.177.3.1655. [DOI] [PubMed] [Google Scholar]
- 25.Wakabayashi K. IL-2 receptor α−/− mice and the development of primary biliary cirrhosis. Hepatology. 2006;44(5):1240–1249. doi: 10.1002/hep.21385. [DOI] [PubMed] [Google Scholar]
- 26.Salas J.T. Ae2a,b-deficient mice develop antimitochondrial antibodies and other features resembling primary biliary cirrhosis. Gastroenterology. 2008;134(5):1482–1493. doi: 10.1053/j.gastro.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Irie J. NOD.c3c4 congenic mice develop autoimmune biliary disease that serologically and pathogenetically models human primary biliary cirrhosis. J. Exp. Med. 2006;203(5):1209–1219. doi: 10.1084/jem.20051911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattner J. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe. 2008;3(5):304–315. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakabayashi K. Induction of autoimmune cholangitis in non-obese diabetic (NOD).1101 mice following a chemical xenobiotic immunization. Clin. Exp. Immunol. 2009;155(3):577–586. doi: 10.1111/j.1365-2249.2008.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y. Distinct from its canonical effects, deletion of IL-12p40 induces cholangitis and fibrosis in interleukin-2Rα−/− mice. J. Autoimmun. 2014;51(c):99–108. doi: 10.1016/j.jaut.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda M. Deletion of interleukin (IL)-12p35 induces liver fibrosis in dominant-negative TGFbeta receptor type II mice. Hepatology. 2013;57(2):806–816. doi: 10.1002/hep.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida K. Deletion of interleukin-12p40 suppresses autoimmune cholangitis in dominant negative transforming growth factor beta receptor type II mice. Hepatology. 2009;50(5):1494–1500. doi: 10.1002/hep.23132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selmi C. Primary biliary cirrhosis in monozygotic and dizygotic twins: genetics, epigenetics, and environment. YGAST. 2004;127(2):485–492. doi: 10.1053/j.gastro.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Lazaridis K.N. Increased prevalence of antimitochondrial antibodies in first-degree relatives of patients with primary biliary cirrhosis. Hepatology. 2007;46(3):785–792. doi: 10.1002/hep.21749. [DOI] [PubMed] [Google Scholar]
- 35.Mantaka A. Primary biliary cirrhosis in a genetically homogeneous population: disease associations and familial occurrence rates. BMC Gastroenterol. 2012;12(1) doi: 10.1186/1471-230X-12-110. 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selmi C. Genes and (auto)immunity in primary biliary cirrhosis. Genes. Immun. 2005;6(7):543–556. doi: 10.1038/sj.gene.6364248. [DOI] [PubMed] [Google Scholar]
- 37.Jones D.E. Familial primary biliary cirrhosis reassessed: a geographically-based population study. J. Hepatol. 1999;30(3):402–407. doi: 10.1016/s0168-8278(99)80097-x. [DOI] [PubMed] [Google Scholar]
- 38.Gershwin M.E. Risk factors and comorbidities in primary biliary cirrhosis: A controlled interview-based study of 1032 patients. Hepatology. 2005;42(5):1194–1202. doi: 10.1002/hep.20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arbour L. Characteristics of primary biliary cirrhosis in British Columbia's first nations population. Can. J. Gastroenterol. 2005;19(5):305–310. doi: 10.1155/2005/203028. [DOI] [PubMed] [Google Scholar]
- 40.Delgado J. The epidemiology of primary biliary cirrhosis in southern Israel. Israel Med. Assoc. J. IMAJ. 2005;7(11):717–721. [PubMed] [Google Scholar]
- 41.Tsuji K. Familial primary biliary cirrhosis in Hiroshima. J. Autoimmun. 1999;13(1):171–178. doi: 10.1006/jaut.1999.0299. [DOI] [PubMed] [Google Scholar]
- 42.Parkes M. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 2013;14(9):661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 43.Lessard C.J. The genomics of autoimmune disease in the era of genome-wide association studies and beyond. Autoimmun. Rev. 2012;11(4):267–275. doi: 10.1016/j.autrev.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manolio T.A. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirschfield G.M. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 Variants. N. Engl. J. Med. 2009;360(24):2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. 2010;42(8):658–660. doi: 10.1038/ng.627. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mells G.F. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2011;43(4):329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura M. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet. 2012;91(4):721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cordell H.J. International genome-wide meta-analysis in primary biliary cirrhosis discovers six new risk loci and highlights the role of JAK-STAT/IL12/IL27 signaling and cytokine-cytokine pathways in disease etiology. Nat. Commun. 2015:1–13. (in press) [Google Scholar]
- 50.Hirschfield G.M. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. 2010;42(8):655–657. doi: 10.1038/ng.631. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juran B.D. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum. Mol. Genet. 2012;21(23):5209–5221. doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J.Z. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. 2012;44(10):1137–1141. doi: 10.1038/ng.2395. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ercilla G. Primary biliary cirrhosis associated with HLA-DRw3. Tissue Antigens. 1979;14(5):449–452. doi: 10.1111/j.1399-0039.1979.tb00874.x. [DOI] [PubMed] [Google Scholar]
- 54.Manns M.P. HLA DRw8 and complement C4 deficiency as risk factors in primary biliary cirrhosis. Gastroenterology. 1991;101(5):1367–1373. doi: 10.1016/0016-5085(91)90090-8. [DOI] [PubMed] [Google Scholar]
- 55.Invernizzi P. Human leukocyte antigen polymorphisms in italian primary biliary cirrhosis: a multicenter study of 664 patients and 1992 healthy controls. Hepatology. 2008;48(6):1906–1912. doi: 10.1002/hep.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chow I.T. Differential binding of pyruvate dehydrogenase complex-E2 epitopes by DRB1*08:01 and DRB1*11:01 is predicted by their structural motifs and correlates with disease risk. J. Immunol. 2013;190(9):4516–4524. doi: 10.4049/jimmunol.1202445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishizaki M. Involvement of tyrosine kinase-2 in both the IL-12/Th1 and IL-23/Th17 axes in vivo. J. Immunol. 2011;187(1):181–189. doi: 10.4049/jimmunol.1003244. [DOI] [PubMed] [Google Scholar]
- 58.Chang H.-C. Impaired development of human Th1 cells in patients with deficient expression of STAT4. Blood. 2009;113(23):5887–5890. doi: 10.1182/blood-2008-09-179820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Z. Autoimmune diabetes is blocked in Stat4-deficient mice. J. Autoimmun. 2004;22(3):191–200. doi: 10.1016/j.jaut.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Vereecke L., Beyaert R., van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30(8):383–391. doi: 10.1016/j.it.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 61.Strebovsky J. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 2011;25(3):863–874. doi: 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- 62.Good-Jacobson K.L. CD80 expression on B cells regulates murine T follicular helper development, germinal center B cell survival, and plasma cell generation. J. Immunol. 2012;188(9):4217–4225. doi: 10.4049/jimmunol.1102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peschon J.J. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 1994;180(5):1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liston A., Enders A., Siggs O.M. Unravelling the association of partial T-cell immunodeficiency and immune dysregulation. Nat. Rev. Immunol. 2008;8(7):545–558. doi: 10.1038/nri2336. [DOI] [PubMed] [Google Scholar]
- 65.Liu W. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J. Exp. Med. 2006;203(7):1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma S. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol. Cell. Biol. 2010;30(17):4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun J. Lack of the transcriptional coactivator OBF-1 prevents the development of systemic lupus erythematosus-like phenotypes in Aiolos mutant mice. J. Immunol. 2003;170(4):1699–1706. doi: 10.4049/jimmunol.170.4.1699. [DOI] [PubMed] [Google Scholar]
- 68.Quintana F.J. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 2012;13(8):770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katayama H. Lnk prevents inflammatory CD8(+) T-cell proliferation and contributes to intestinal homeostasis. Eur. J. Immunol. 2014;44(6):1622–1632. doi: 10.1002/eji.201343883. [DOI] [PubMed] [Google Scholar]
- 70.Haynes N.M. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 2007;179(8):5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 71.Schubart D.B. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383(6600):538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 72.Kumpfel T., Hohlfeld R. Multiple sclerosis. TNFRSF1A, TRAPS and multiple sclerosis. Nat. Rev. Neurol. 2009;5(10):528–529. doi: 10.1038/nrneurol.2009.154. [DOI] [PubMed] [Google Scholar]
- 73.Meylan F., Richard A.C., Siegel R.M. TL1A and DR3, a TNF family ligand-receptor pair that promotes lymphocyte costimulation, mucosal hyperplasia, and autoimmune inflammation. Immunol. Rev. 2011;244(1):188–196. doi: 10.1111/j.1600-065X.2011.01068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green E.A., Flavell R.A. TRANCE-RANK, a new signal pathway involved in lymphocyte development and T cell activation. J. Exp. Med. 1999;189(7):1017–1020. doi: 10.1084/jem.189.7.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayden M.S., Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes. Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sha W.C. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80(2):321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 77.Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 78.Turcotte K. Icsbp1/IRF-8 is required for innate and adaptive immune responses against intracellular pathogens. J. Immunol. 2007;179(4):2467–2476. doi: 10.4049/jimmunol.179.4.2467. [DOI] [PubMed] [Google Scholar]
- 79.Hieshima K. Molecular cloning of a novel human CC chemokine liver and activation-regulated chemokine (LARC) expressed in liver. Chemotactic activity for lymphocytes and gene localization on chromosome 2. J. Biol. Chem. 1997;272(9):5846–5853. doi: 10.1074/jbc.272.9.5846. [DOI] [PubMed] [Google Scholar]
- 80.Affo S. CCL20 mediates lipopolysaccharide induced liver injury and is a potential driver of inflammation and fibrosis in alcoholic hepatitis. Gut. 2014;63(11):1782–1792. doi: 10.1136/gutjnl-2013-306098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oo Y.H. CXCR3-dependent recruitment and CCR6-mediated positioning of Th-17 cells in the inflamed liver. J. Hepatol. 2012;57(5):1044–1051. doi: 10.1016/j.jhep.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agarwal K. CTLA-4 gene polymorphism confers susceptibility to primary biliary cirrhosis. J. Hepatol. 2000;32(4):538–541. doi: 10.1016/s0168-8278(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 83.Juran B.D. Carriage of a tumor necrosis factor polymorphism amplifies the cytotoxic T-lymphocyte antigen 4 attributed risk of primary biliary cirrhosis: evidence for a gene-gene interaction. Hepatology. 2010;52(1):223–229. doi: 10.1002/hep.23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simpson T.R., Quezada S.A., Allison J.P. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS) Curr. Opin. Immunol. 2010;22(3):326–332. doi: 10.1016/j.coi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 85.Tang, R., et al., The cumulative effects of known susceptibility variants to predict primary biliary cirrhosis risk. (2015) 1–6. [DOI] [PubMed]
- 86.Juran B.D., Lazaridis K.N. Environmental factors in primary biliary cirrhosis. Semin. Liver Dis. 2014;34(3):265–272. doi: 10.1055/s-0034-1383726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Invernizzi P. Frequency of monosomy X in women with primary biliary cirrhosis. Lancet. 2004;363(9408):533–535. doi: 10.1016/S0140-6736(04)15541-4. [DOI] [PubMed] [Google Scholar]
- 88.Invernizzi P. X chromosome monosomy: a common mechanism for autoimmune diseases. J. Immunol. 2005;175(1):575–578. doi: 10.4049/jimmunol.175.1.575. [DOI] [PubMed] [Google Scholar]
- 89.Jorgensen K.T. Autoimmune diseases in women with Turner's syndrome. Arthritis Rheum. 2010;62(3):658–666. doi: 10.1002/art.27270. [DOI] [PubMed] [Google Scholar]
- 90.Lleo A. Y chromosome loss in male patients with primary biliary cirrhosis. J. Autoimmun. 2013;41:87–91. doi: 10.1016/j.jaut.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 91.Miozzo M. Preferential X chromosome loss but random inactivation characterize primary biliary cirrhosis. Hepatology. 2007;46(2):456–462. doi: 10.1002/hep.21696. [DOI] [PubMed] [Google Scholar]
- 92.Lleo A. Immunoglobulin M levels inversely correlate with CD40 ligand promoter methylation in patients with primary biliary cirrhosis. Hepatology. 2012;55(1):153–160. doi: 10.1002/hep.24630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Banales J.M. Up-regulation of microRNA 506 leads to decreased Cl−/HCO3− anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology. 2012;56(2):687–697. doi: 10.1002/hep.25691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Padgett K.A. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J. Autoimmun. 2009;32(3–4):246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Umemura T. Human leukocyte antigen class II molecules confer both susceptibility and progression in Japanese patients with primary biliary cirrhosis. Hepatology. 2012;55(2):506–511. doi: 10.1002/hep.24705. [DOI] [PubMed] [Google Scholar]
- 96.Pulickal A.S. Biliary cirrhosis in a child with inherited interleukin-12 deficiency. J. Trop. Pediatr. 2008;54(4):269–271. doi: 10.1093/tropej/fmm119. [DOI] [PubMed] [Google Scholar]
- 97.Döffinger R. Autoantibodies to interferon-gamma in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2004;38(1):e10-4. doi: 10.1086/380453. [DOI] [PubMed] [Google Scholar]
- 98.Dumoulin F.L. Autoimmunity induced by interferon-alpha therapy for chronic viral hepatitis. Biomed. Pharmacother. 1999;53(5–6):242–254. doi: 10.1016/S0753-3322(99)80095-X. [DOI] [PubMed] [Google Scholar]
- 99.Carbone M. Calcineurin inhibitors and the IL12A locus influence risk of recurrent primary biliary cirrhosis after liver transplantation. Am. J. Transpl. 2013;13(4):1110–1111. doi: 10.1111/ajt.12132. [DOI] [PubMed] [Google Scholar]
- 100.Wang L. CXCR5 +CD4 +T follicular helper cells participate in the pathogenesis of primary biliary cirrhosis. Hepatology. 2015 Feb;61(2):627–638. doi: 10.1002/hep.27306. Epub 2015 Jan 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Webb G.J., Hirschfield G.M. Follicles, germinal centers, and immune mechanisms in primary biliary cirrhosis. Hepatology. 2015;61(2):424–427. doi: 10.1002/hep.27552. [DOI] [PubMed] [Google Scholar]
- 102.Kikuchi K. Splenic lymph follicles generate immunoglobulin M-producing B cells in primary biliary cirrhosis. Hepatol Res. 2014 Oct;44(10):E253–E256. doi: 10.1111/hepr.12231. [DOI] [PubMed] [Google Scholar]
- 103.Poupon R. Genetic factors of susceptibility and of severity in primary biliary cirrhosis. J. Hepatol. 2008;49(6):1038–1045. doi: 10.1016/j.jhep.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 104.Aiba Y. Genetic polymorphisms in CTLA4 and SLC4A2 are differentially associated with the pathogenesis of primary biliary cirrhosis in Japanese patients. J. Gastroenterol. 2011;46(10):1203–1212. doi: 10.1007/s00535-011-0417-7. [DOI] [PubMed] [Google Scholar]
- 105.Hitomi Y. Human primary biliary cirrhosis-susceptible allele of rs4979462 enhances TNFSF15 expression by binding NF-1. Hum Genet. 2015 Jul;134(7):737–747. doi: 10.1007/s00439-015-1556-3. [DOI] [PubMed] [Google Scholar]
- 106.Hirschfield G.M. P367 phase 2 study evaluating the efficacy and safety of ustekinumab in patients with primary biliary cirrhosis WHO had an inadequate response to whom it may concern: ursodeoxycholic acid. J. Hepatol. 2014;60(1):S189–S190. [Google Scholar]
- 107.Kopp T. Clinical improvement in psoriasis with specific targeting of interleukin-23. Nature. 2015;521(7551):222–226. doi: 10.1038/nature14175. [DOI] [PubMed] [Google Scholar]
- 108.Dhirapong A. Therapeutic effect of cytotoxic T lymphocyte antigen 4/immunoglobulin on a murine model of primary biliary cirrhosis. Hepatology. 2013;57(2):708–715. doi: 10.1002/hep.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dyson J.K. Unmet clinical need in autoimmune liver diseases. J. Hepatol. 2014;62(1):208–218. doi: 10.1016/j.jhep.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 110.Krzywinski M.I. Circos: an information aesthetic for comparative genomics. Genome Res. 2009 Sep;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu X. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. 2010;42(8):658–660. doi: 10.1038/ng.627. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mells G.F., Hirschfield G.M. Genetics of primary biliary cirrhosis. eLS. 2013 [Google Scholar]
- 113.Invernizzi P. Classical HLA-DRB1 and DPB1 alleles account for HLA associations with primary biliary cirrhosis. 2012;13(6):461–468. doi: 10.1038/gene.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao D.T. Human leucocyte antigen alleles and haplotypes and their associations with antinuclear antibodies features in Chinese patients with primary biliary cirrhosis. Liver Int. 2014;34(2):220–226. doi: 10.1111/liv.12236. [DOI] [PubMed] [Google Scholar]