Abstract
Higher plants have been shown to experience a juvenile vegetative phase, an adult vegetative phase, and a reproductive phase during its postembryonic development and distinct lateral organ morphologies have been observed at the different development stages. Populus euphratica, commonly known as a desert poplar, has developed heteromorphic leaves during its development. The TCP family genes encode a group of plant-specific transcription factors involved in several aspects of plant development. In particular, TCPs have been shown to influence leaf size and shape in many herbaceous plants. However, whether these functions are conserved in woody plants remains unknown. In the present study, we carried out genome-wide identification of TCP genes in P. euphratica and P. trichocarpa, and 33 and 36 genes encoding putative TCP proteins were found, respectively. Phylogenetic analysis of the poplar TCPs together with Arabidopsis TCPs indicated a biased expansion of the TCP gene family via segmental duplications. In addition, our results have also shown a correlation between different expression patterns of several P. euphratica TCP genes and leaf shape variations, indicating their involvement in the regulation of leaf shape development.
Higher plants usually experience three postembryonic developmental stages, i.e. a juvenile vegetative phase, an adult vegetative phase, and a reproductive phase1,2. These phases are defined by the morphology of lateral organs3, which is marked by a number of either continuous or discrete features. As a major lateral organ, leaf shows both continuous and discrete morphological changes in phase transition: leaf size shows a continuous variation, while several distinct shapes are seen in different plant developmental phases4. Phase transition has been shown to be regulated by both environmental factors such as photoperiod5,6 and intrinsic regulators including plant hormones such as gibberellins6, trans-acting small interfering RNAs (tasiRNAs)7,8, and most importantly, microRNAs9. MiR156, a conserved microRNA among both herbaceous and woody plants which shows response to sugar concentration10, as well as its downstream regulatory module components such as SQUAMOSA promoter-binding protein-like (SPL) genes and MiR172, has been shown as responsible for development phase transition, and thus controlling leaf morphology changes in Arabidopsis, maize, and several woody species including Eucalyptus globulus, and Populus x Canadensis11,12,13,14,15. However, more details of the downstream regulation cascade for controlling leaf shape remain to be elucidated.
The process of leaf development is composed of primordia initiation, establishment of polarity in three axes, lamina expansion, and formation of leaf margin16,17,18, and involves coordinated regulation among transcription factors, small RNAs and hormones19. Genetic approaches have demonstrated several key genes involved in leaf development. For example, suppression of the expression of Class I KNOTTED1-LIKE HOMEOBOX (KNOX) genes has been shown as critical for leaf primordia initiation20,21. Class III HD-ZIP, KANADI and YABBY gene families are involved in the establishment of dorsoventral polarity22,23,24,25,26,27. ANGUSTIFOLIA (AN) and ROTUNDIFOLIA3 (ROT3), and ANGUSTIFOLIA3 (AN3) and ROTUNDIFOLIA4 (ROT4) affected lamina expansion by regulating the shape and number of cells in the lamina, respectively17. In addition, PIN and CUC genes play crucial role in leaf margin patterning by controlling auxin-maxima formation28,29.
The TCP transcription factor family, named after its initial members teosinte branched1 (tb1) from maize (Zea mays)30, CYCLOIDEA (CYC) from snapdragon (Antirrhinum majus)31, and the PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) from rice (Oryza sativa)32, was first described in 1999, as a small group of plant specific genes encoding proteins sharing the so-called TCP domain, which is a 59-amino acid non-canonical basic helix-loop-helix (bHLH) motif that allows DNA binding, protein-protein interactions and nuclear localization32,33,34. TCPs have been identified in various plants including Arabidopsis35, rice36,37,38, tomato39 and poplar34,37. These genes have been categorized by phylogenetic analysis into two classes based mainly on differences within the TCP domain: class I (known as PCF class or TCP-P class) and class II (known as TCP-C class)33,37,40. Class I is formed by a group of relatively closely related proteins exemplified by rice PCF1 and PCF2, whereas class II is further subdivided into two clades according to differences within the TCP domain: the CIN clade exemplified by CINCINNATA (CIN) of Antirrhinum41 and the CYC/TB1 clade (or ECE clade) including CYC and tb142.
Many members of the TCP family from both Class I and Class II have been demonstrated to participate in leaf development control in Antirrhinum41, Arabidopsis43,44,45,46,47,48,49, and tomato50. They have been shown to physically interact with several proteins involved in leaf development regulation, including ASYMMETRIC LEAVES1 (AS1), AS2 and NGATHA (NGA)46,51,52. In particular, SPL9, a target of MiR156, has been reported to interact with TCP4 and this complex promoted CUC-controlled acquisition of leaf complexity in Arabidopsis53, indicating that TCPs may play important roles in the regulatory cascade of leaf shape control centered by the MiR156 module. However, it remains unknown whether TCPs are also involved in leaf morphology regulation in woody plants.
With its genome well sequenced and annotated54, P. trichocarpa (black cottonwood) has been widely employed as a model tree for genomic and genetic studies. P. trichocarpa possesses almost identical leaf shapes during development phase transitions. In contrast, the desert poplar P. euphratica employs linear-laceolate, lanceolate, oval and wide-oral leaves sequentially along the process of development with a dramatic increase in leaf area, thickness, dry weight, as well as specific leaf area55,56,57. Recently, the genome sequence of P. euphratica has also been sequenced and assembled58,59, making it a suitable model for studying the roles of TCP transcription factors in leaf morphology establishment at genome-wide scale. Therefore, in this study, we firstly carried out genome-wide identification of TCP transcription factors in P. trichocarpa and P. euphratica, and then we conducted expression analysis of TCP genes in various leaf types of P. euphratica to establish a correlation between their expression and various leaf morphologies.
Results
Identification of TCP genes in P. trichocarpa and P. euphratica
We first performed genome-wide search of putative TCP genes in both P. trichocarpa and. P. euphratica. At least 60 putative TCP proteins were identified in the P. trichocarpa genome, all of which were annotated as TCP family protein in the Plant Transcription Factor Database 3.060, confirming the reliability of our initial search. Among them, 23 were identified as redundant sequences resulting from alternative splicing and thus discarded. Multiple-alignment was carried out for the remaining protein sequences together with the Arabidposis TCPs. Manual inspection on the alignment revealed one peptide sequence without a TCP domain which was thus discarded. The 36 putative TCPs genes were named PtrTCP1 to PtrTCP36 in the order of their access number in the Phytozome database61 (Supplementary Table S1). Similar HMMER search and manual inspection were conducted against the P. euphratica genome and 33 non-redundant putative genes were identified and named as PeuTCP1 to PeuTCP33 according to their accession number (Supplementary Table S2). These poplar TCPs have a peptide length ranging from 41 to 623 amino acids, a molecular weight between 4787.43 and 68064.55, and an isoelectric point (pI) value between 5.84 and 9.58 (Supplementary Fig. S1, Supplementary Tables S1 and S2). The molecular weight and pI values of these poplar TCPs showed similar distribution as those of Arabidopsis TCPs (Supplementary Fig. S1).
Phylogenetic analysis of TCP proteins
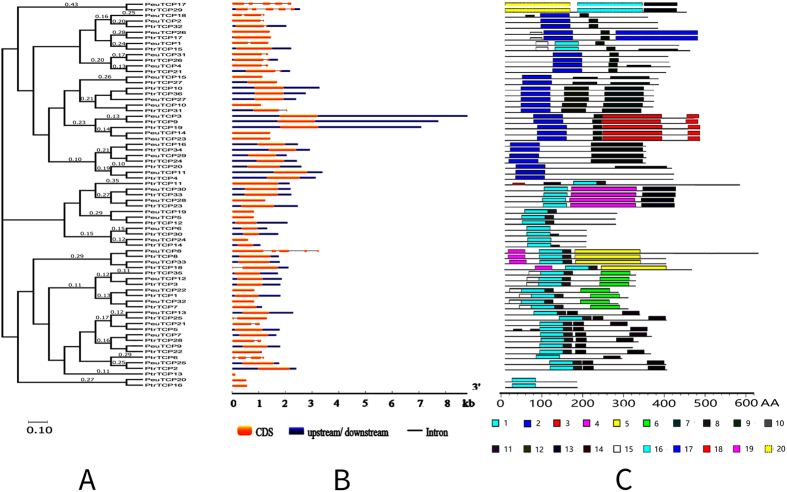
In order to elucidate phylogenetic relations among the poplar TCPs, a maximum likelihood (ML) phylogenetic tree was built based on multiple-alignment of the TCP domain sequences of the poplar TCPs and their Arabidopsis homologues. As shown in Fig. 1, the 93 TCPs were classified into two classes, Class I (red) and Class II, where Class II was further divided into two clades, CYC (orange) and CIN (yellow). All Arabidopsis TCPs fell in the same Class or clade as previously reported34, confirming the reliability of our phylogenetic tree. Similar phylogenetic trees were obtained by the minimal evolution, maximal parsimony, neighbour joining methods with minor differences at some branches (Supplementary Figs S2–S4). According to the phylogenetic tree, both poplar species possessed an expanded TCP family with approximate 1.5-fold size compared with Arabidopsis. Interestingly, the expansion in both poplar species was biased, which occurred mainly in Class I and the CYC clade, while the CIN clade remained largely the same size as in Arabidopsis (Table 1).
Figure 1. Phylogenetic tree of Arabidopsis and Populus TCPs.
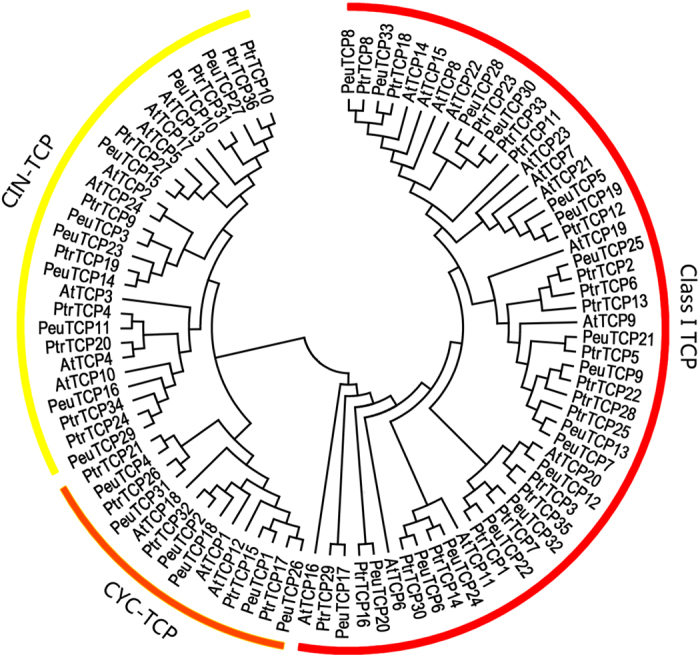
The phylogenetic tree was built based on multiple alignment of the TCP domain in the TCP proteins using the Neighbor-Joining method with 1000 bootstrap replicates.
Table 1. TCP genes in Arabidopsis and the two Populus species.
| Species | Class I TCPs | CYC TCPs | CIN TCPs |
|---|---|---|---|
| Arabidopsis thaliana | 13 | 3 | 8 |
| Populus trichocarpa | 21 | 5 | 10 |
| Populus euphratica | 18 | 6 | 9 |
Chromosomal location analysis of PtrTCP genes
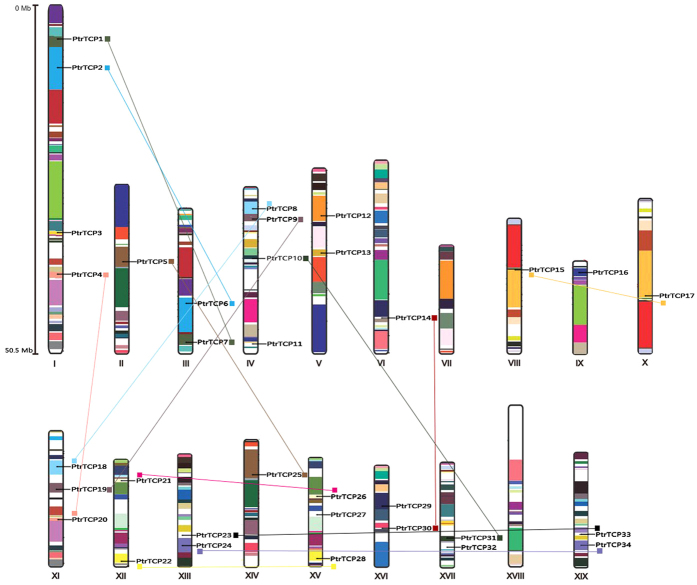
Among the 36 P. trichocarpa TCPs, 34 members were located at the 19 chromosomes assembled in the P. trichocarpa genome v 3.0, and the other two were located at two unmapped scaffolds, viz. Scaffold 41 and Scaffold 457, respectively. TCP genes are distributed on 17 out of the 19 P. trichocarpa chromosomes in an uneven manner, with the number of TCP genes per chromosome ranging from 0 to 4. Chromosomes 1 and 4 contain four genes, while no TCP gene is found on Chromosomes 7 and 18 (Fig. 2). Additionally, as shown in Fig. 2, 13 pairs of duplicated genes were identified, accounting for about 72% of the P. trichocarpa TCP family, complying with the ~1.5-fold expansion of the poplar genome as compared with Arabidopsis54. In fact, as the two genes located to unmapped scaffolds also show high identity to other genes, there could be even more duplication events. Further, all the paralogous gene pairs we identified located on different chromosomes, suggesting that they result from segment duplications rather than tandem duplications.
Figure 2. Physical locations of TCP genes on P. trichocarpa chromosomes.
The TCP genes are located according to the JGI P. trichocarpa genome V. 3.0 annotations, and possible gene duplication events are indicated by colored lines.
Genomic structure of the TCP genes
As shown in Fig. 3B, most (53 out of 69) of the poplar TCP genes contain only one exon and 11 members contain one intron and two exons. Only PeuTCP18 compromises two introns and three exons and PtrTCP6 possesses three introns and four exons, whereas three genes (PeuTCP17, PtrTCP29 and PeuTCP8) consist of four introns and five exons. Moreover, similar exon/intron structures were found in poplar genes within the same phylogenetic subfamily, further confirming the reliability of our phylogenetic analysis.
Figure 3. Genomic structure and motif composition of poplar TCPs.
(A) Phylogenetic tree of P. trichocarpa and P. euphratica TCP proteins. (B) Genomic structure of poplar TCP genes. Exons, introns, and UTRs are indicated with yellow boxes, blue boxes and black lines, respectively. (C) Motif composition of poplar TCP proteins. Conserved motifs in the poplar TCP proteins are indicated by colored boxes.
Prediction of conserved motifs in TCP proteins
Conserved motifs in the poplar TCP proteins were analyzed with the MEME program62 and shown in Fig. 3C. The MEME analysis discovered 20 putative motifs in total, namely motif 1 to motif 20 (Supplementary Fig. S5). Motif 1 and Motif 2 were both identified as the conserved TCP domain, while no matches were found for the other motifs. Either motif 1 or motif 2 is present in every poplar TCP protein we have identified, providing further support for the reliability of our identification. Additionally, although the functions of the rest 18 motifs are unknown, similar motif composition was observed in TCP proteins of the same subfamily, while significant differences were observed between different subfamilies, indicating possible intra-subfamily functional redundancy and inter-subfamily function divergence.
Global expression profiling in P. trichocarpa
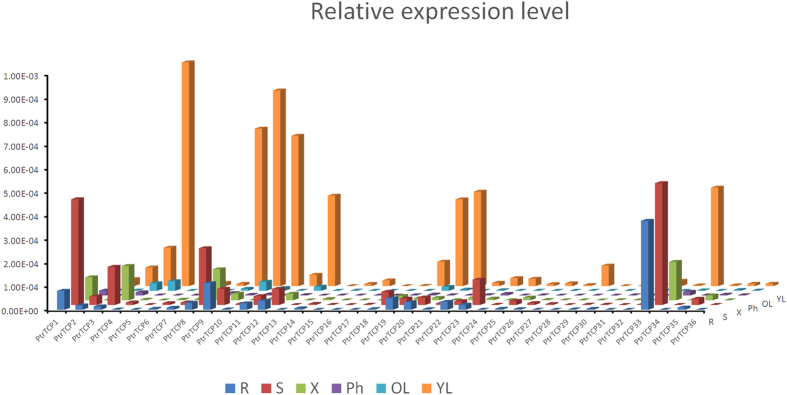
Relative transcript abundance of the 36 PtrTCPs in six P. trichocarpa tissues are shown in Fig. 4. Generally, 18 out of the 36 genes showed high expression levels in the young leaf, four genes (PtrTCP1, PtrTCP23, PtrTCP33 and PtrTCP35) with high expression levels in the stem, and one gene (PtrTCP33) with high level of expression in the root. Moreover, poplar TCP genes with close phylogenetic relationship showed both similar and divergent expression patterns. For example, the paralogous pair PtrTCP9 and PtrTCP19 were both expressed highly in the young leaf, at a moderate level in the root, and at a low level in the stem, and the expression level of PtrTCP9 was higher than that of PtrTCP19 in each tissue we tested. In contrast, another paralogous pair, PtrTCP1 and PtrTCP7, divergent expression patterns were found. PtrTCP7 was almost undetectable in all tissues we examined, while PtrTCP1 was expressed highly in the stem, and moderately in the root and xylem.
Figure 4. Transcriptional profiling of P. trichocarpa TCP genes in root, stem, xylem, phloem, old leaf and young leaf.
Each bar represents three biological replicates.
Correlation of TCP expression levels and leaf morphogenesis in P. euphratica
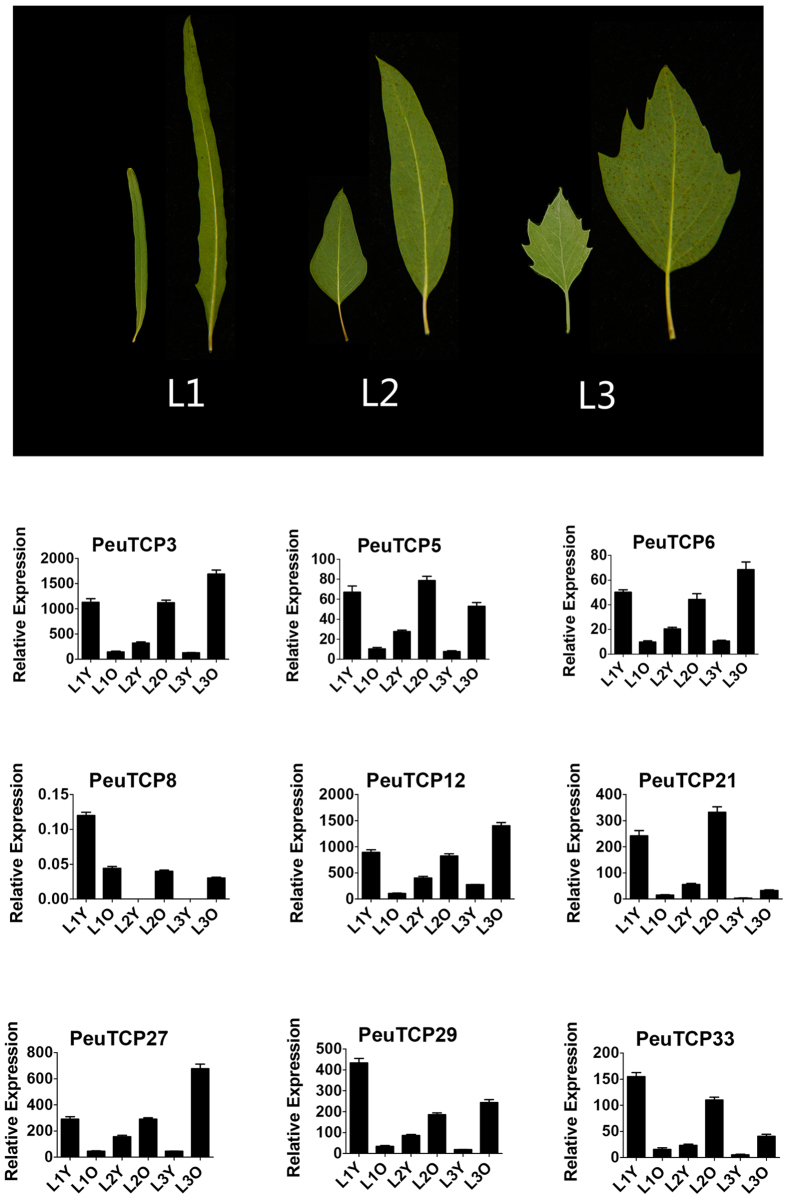
In P. euphratica, linear-laceolate leaves are found in juvenile (first year) shoots, and lanceolate and oval leaves are sequentially seen later in adult phase shoots (Fig. 5, top panel). Previous studies have shown that A. thaliana TCPs from Class I and the CIN clade, but not from the CYC clade, i.e. AtTCP2, AtTCP3, AtTCP4, AtTCP10, AtTCP24, AtTCP5, AtTCP13, AtTCP7, AtTCP21, AtTCP23, AtTCP9, AtTCP20, AtTCP11, AtTCP14 and AtTCP1543,44,45,46,47,48,49 are involved in leaf morphology regulation. Based on our phylogenetic analysis as well as transcriptome sequencing data previously reported58, we selected 9 representative TCP genes that are both closely related to the Arabidopsis TCPs motioned above and highly expressed according to the transcriptome analysis, i.e. PeuTCP3 (homologous to AtTCP2 and AtTCP24), PeTCP5 (homologous to AtTCP7, AtTCP21 and AtTCP23), PeuTCP6 (homologous to AtTCP11), PeuTCP8 (homologous to AtTCP14 and AtTCP15), PeuTCP12 (homologous to AtTCP20), PeuTCP21 (homologous to AtTCP9), PeuTCP27 (homologous to AtTCP5 and AtTCP13), PeuTCP29 (homologous to AtTCP3, AtTCP4 and AtTCP10) and PeuTCP33 (homologous to AtTCP14 and AtTCP15) and performed qRT-PCR in P. euphratica leaves with different shape to examine expression patterns of TCP genes during leaf development. In general, TCP members of both Class I and Class II showed a decrease of expression level in young leaves along the developmental process (Fig. 5, bottom panel). Interestingly, two different expression patterns were observed in linear-laceolate leaves and the other two-type leaves. In the linear-laceolate leaves (L1), the expression levels for all the TCPs were higher in young leaves than adult ones, while lower expression levels were observed in young lanceolate and oval leaves than corresponding adult leaves (L2 and L3).
Figure 5. Expression pattern of selected TCP genes in different P. euphratica leaves.
Each bar represents three biological replicates and error bar represents S.D.
To determine whether such expression patterns are specifically correlated with the leaves with different shape, we conducted similar qRT-PCR test for TCP genes in P. tomentosa leaves from juvenile and adult shoots, as this poplar species employs a single leaf shape during its whole life span.The expression patterns of PtoTCP genes were different from their homologues in P. euphratica: higher expression levels were observed in young leaves than adult leaves from both juvenile (one year) and adult phase (multiple year) shoots (Supplementary Fig. S6). In addition, as SPLs have been shown as important developmental markers in both Arabidopsis13 and hybrid poplar14, we also conducted qRT-PCR analysis for P. euphratica SPL genes homologous to AtSPL3 and AtSPL9, i.e. PeuSPL6 and PeuSPL23, and PeuSPL8, PeuSPL17 and PeuSPL27, respectively. These SPL genes showed similar expression patterns as the TCP genes in P. euphratica. (Supplementary Fig. S7).
Discussion
TCP family transcription factors are plant specific transcription factor that play various roles in multiple aspects of plant growth and development. In the present study, we identified and characterized genes encoding TCP proteins in P. euphratica and P. trichocarpa. The TCP family was expanded by ~1.5 fold in both poplar species compared with Arabidopsis and the expansion was due to segmental duplications rather than tandem duplications. Further, as indicated by phylogenetic analysis, such expansion was carried out in an uneven manner that Class I and the CYC clade TCPs were expanded in poplar, while the number of CIN clade TCPs remained almost the same as in Arabidopsis. Moreover, while 23 out of 33 PeuTCPs were clustered closely to their P. trichocarpa orthologues, the others suggested both putative gene duplication and gene loss events in either species. For example, PtrTCP3 and PtrTCP35, PtrTCP10 and PtrTCP36, PeuTCP7 and PeuTCP13 were shown as closest homologs to each other, indicating possible gene duplications in the respective species. Meanwhile, the orthologues of PtrTCP5, PtrTCP6, PtrTCP13, PeuTCP18 and PeuTCP18 were missing, suggesting the occurrence of gene loss events in the divergence of the two poplar species.
In Arabidopsis, several TCP transcription factors from both Class I and Class II have been shown to participate in the regulation of leaf shaping. For example, dominant negative repression of AtTCP11 led to curled rosette leaves47, and AtTCP14 and AtTCP15 have been shown to influence leaf cell size and number in a redundant manner48. In addition, partial redundancy and functional divergence in the regulation of leaf shape was found among Arabidopsis TCPs with close phylogenetic relationship. For instance, members of the CIN Clade, i.e. AtTCP2, AtTCP3, AtTCP4, AtTCP10 and AtTCP24 are shown to influence leaf shape in a complementary manner4,63,64,65. Therefore, P. euphratica TCPs with close phylogenetic relationship to these AtTCPs could also play a role in the regulation of leaf shape, and functional divergence could be expected.
Moreover, we discovered a correlation between the expression pattern of TCP genes and leaf shape regulation in P. euphratica, and such altered changes were also observed for SPL genes, whose homologues had been established as developmental markers13,14, indicating their possible involvement in the developmental transition. Changes in the expression pattern of TCP genes in different P. euphratica leaves suggests the involvement of P. euphratica TCP genes from both Class I and Class II in leaf shape regulation. However, their regulatory roles could be divergent. For example, it has been shown that Arabidopsis Class II TCP AtTCP4 regulates the serration at the leaf margin by influencing auxin distribution via the miR164-CUC pathway63. PeuTCP29, the closest P. euphratica homologue of AtTCP4, could play similar role in leaf development process. We speculate that, at the juvenile stage, high level of PeuTCP29 in the young leaf represses serration at the leaf margin, and when the leaf grows older, the expression of PeuTCP29 is reduced, resulting in formation of mild serrations. While at the adult phase, the expression of PeuTCP29 is repressed in the young leaf, leading to formation of deep serrations, and in old leaf, although the repression is somehow removed, the serrations have been established. Other Class II P. euphratica TCP genes (PeuTCP3 and PeuTCP27) could play similar role as PeuTCP29. On the contrary, Class I TCP genes have been shown to influence cell growth and proliferation, and their altered expression in the leaf could result in changes in cell number and size48,66, or the disruption of the balance between the growth rate in inner regions of the leaf lamina and that at the leaf margins, leading to curled leaf shape47. Therefore, the inverted expression pattern of Class I P. euphratica TCP genes (PeuTCP5, PeuTCP6, PeuTCP8, PeuTCP12, PeuTCP27 and PeuTCP33) could be correlated to the altered length-width ratio of P. euphratica leaves. However, the detail functions of these genes in P. euphratica still need to further determine in the future.
Materials and Methods
Sequence retrieval and TCP gene identification
The Hidden Markov Model (HMM) profile of the TCP domain (PF03634) was obtained from the pfam database67 and putative TCP protein sequences were identified by HMMER68 search against the P. euphratica genome58 and the publicly available P. trichocarpa genome v 3.054 in the Phytozome database61 with the HMM profile. In cases of multiple putative proteins from the same gene locus, the longest variant was kept for further analysis.
Phylogenetic analysis
Amino acid sequences (full-length sequences or the TCP domain sequences) of the putative TCP proteins were used for multiple alignment and phylogenetic analysis with the program MEGA version 669 with default settings.
Analysis of protein features
Molecular weights and theoretical isoelectric point (pI) values of the TCP proteins were calculated with the ProtParam program at the ExPAsy bioinformatics resource portal70. The peptide sequences of the poplar TCPs were submitted to the Multiple Em for Motif Elicitation (MEME) server62 and motif discovery was done with the following settings: motif discovery mode: normal mode; site distribution: any number of repetitions; number of motifs: 20; background motif: 0-order model of sequences; motif width: minimum: 6, maximum: 200. The motifs identified by the MEME analysis were searched in the PROSITE database71.
Analysis of genomic organization
Chromosome size and chromosomal location information for the PtrTCP genes was obtained from the Phytozome database61, and the genes were mapped to each chromosome proportionally.
Genomic and CDS sequences of the poplar TCPs were submitted to the Gene Structure Display Server (GSDS) 2.072 and the output gene structures were arranged according to a phylogenetic tree of the poplar TCPs. Paralogous gene pairs were identified according to genomic sequence identity, and the following criteria were employed: the shorter sequences should cover at least 70% of the longer sequence and the identity of aligned regions should be no less than 70%73.
RNA isolation and quantitative RT-PCR
P. trichocarpa materials were collected from wild-type plants grown in the greenhouse of School of Life Science, Southwest University, Chongqing with three biological replicates. P. euphratica leaves were collected from 6-year old trees grown in the common garden, Lanzhou University, different leaves from the same tree were used for comparison and leaves series from three different trees were taken as biological replicates. Total RNA was isolated from various tissues of P. trichocarpa and P. euphratica with a BioFlux Biospin Plant Total RNA Extraction Kit (Bioer Technology) following the manufacturer’s instructions and reverse-transcription was performed with a PrimeScript RT reagent Kit with gDNA Eraser (47A) (Takara, Dalian). Quantitative real-time RT-PCR (qRT-PCR) was carried out on a Takara Thermal Cycler Dice real time system (Takara, Japan) with a GoTaq qPCR Master Mix (Promega). Primers used in in this work are listed in Supplementary Table S3.
Additional Information
How to cite this article: Ma, X. et al. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 6, 32795; doi: 10.1038/srep32795 (2016).
Supplementary Material
Acknowledgments
The present work was supported by the National Natural Science Foundation of China (31300990, 31370672, 31171620), the Natural Science Foundation Project of CQ (CSTC2013JJB8007), One hundred Talents Programme of the Chinese Academy of Sciences, the Fundamental Research Funds for the Central Universities (XDJK2014a005, XDJK2013B032).
Footnotes
Author Contributions X.M. and K.L. conceived the study, X.M. and J.M. conducted bioinformatics analysis, X.M., C.L. and Y.J. performed experiments, X.M. drafted the manuscript, D.F. and K.L. edited the manuscript.
References
- Poethig R. S. Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923 (1990). [DOI] [PubMed] [Google Scholar]
- Bäurle I. & Dean C. The timing of developmental transitions in plants. Cell 125, 655–664 (2006). [DOI] [PubMed] [Google Scholar]
- Poethig R. S. Small RNAs and developmental timing in plants. Curr Opin Genet Devel 19, 374–378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R. S. Phase change and the regulation of developmental timing in plants. Science 301, 334–336 (2003). [DOI] [PubMed] [Google Scholar]
- Chien J. C. & Sussex I. M. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111, 1321–1328 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A., Bollman K. M. & Poethig R. S. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654 (1997). [DOI] [PubMed] [Google Scholar]
- Vazquez F. et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16, 69–79 (2004). [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A. M. & Carrington J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I. & Weigel D. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16, 258–264 (2011). [DOI] [PubMed] [Google Scholar]
- Yu S., Lian H. & Wang J.-W. Plant developmental transitions: the role of microRNAs and sugars. Curr Opin Plant Biol 27, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M. & Moose S. P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 102, 9412–9417 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. & Poethig R. S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W. et al. miRNA control of vegetative phase change in trees. PLoS Genet 7, e1002012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Conway S. R. & Poethig R. S. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 138, 245–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. Leaf shape: genetic controls and environmental factors. Int J Devel Biol 49, 547–555, 10.1387/ijdb.041921ht (2005). [DOI] [PubMed] [Google Scholar]
- Tsukaya H. Mechanism of leaf-shape determination. Annu Rev Plant Biol 57, 477–496, 10.1146/annurev.arplant.57.032905.105320 (2006). [DOI] [PubMed] [Google Scholar]
- Nicotra A. B. et al. The evolution and functional significance of leaf shape in the angiosperms. Funct Plant Biol 38, 535–552, 10.1071/FP11057 (2011). [DOI] [PubMed] [Google Scholar]
- Moon J. & Hake S. How a leaf gets its shape. Curr Opin Plant Biol 14, 24–30, 10.1016/j.pbi.2010.08.012 (2011). [DOI] [PubMed] [Google Scholar]
- Smith L. G., Greene B., Veit B. & Hake S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116, 21–30 (1992). [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B. & Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413 (1994). [Google Scholar]
- Siegfried K. R. et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128 (1999). [DOI] [PubMed] [Google Scholar]
- Sawa S. et al. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes & Devel 13, 1079–1088 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Baum S. F., Perea J. V. & Bowman J. L. Establishment of polarity in lateral organs of plants. Current Biol 11, 1251–1260, 10.1016/S0960-9822(01)00392-X (2001). [DOI] [PubMed] [Google Scholar]
- Kerstetter R. A., Bollman K., Taylor R. A., Bomblies K. & Poethig R. S. KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709 (2001). [DOI] [PubMed] [Google Scholar]
- McConnell J. R. et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 (2001). [DOI] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M. J., Drews G. N. & Clark S. E. REVOLUTA regulates meristem initiation at lateral positions. Plant J 25, 223–236, 10.1111/j.1365-313X.2001.00959.x (2001). [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E. & Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet 40, 1136–1141 (2008). [DOI] [PubMed] [Google Scholar]
- Kawamura E., Horiguchi G. & Tsukaya H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J 62, 429–441, 10.1111/j.1365-313X.2010.04156.x (2010). [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A. & Hubbard L. The evolution of apical dominance in maize. Nature 386, 485–488, 10.1038/386485a0 (1997). [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L. & Coen E. Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799, 10.1038/383794a0 (1996). [DOI] [PubMed] [Google Scholar]
- Kosugi S. & Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619, 10.1105/tpc.9.9.1607 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J. & Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18, 215–222 (1999). [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M. & Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci 15, 31–39, 10.1016/j.tplants.2009.11.003 (2010). [DOI] [PubMed] [Google Scholar]
- Riechmann J. L. et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110 (2000). [DOI] [PubMed] [Google Scholar]
- Xiong Y. et al. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59, 191–203 (2005). [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D. & Herve C. TCP transcription factors predate the emergence of land plants. J Mol Evol 65, 23–33, 10.1007/s00239-006-0174-z (2007). [DOI] [PubMed] [Google Scholar]
- Yao X., Ma H., Wang J. & Zhang D. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J Int Plant Biol 49, 885–897 (2007). [Google Scholar]
- Parapunova V. et al. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol 14, 157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S. & Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30, 337–348 (2002). [DOI] [PubMed] [Google Scholar]
- Crawford B. C., Nath U., Carpenter R. & Coen E. S. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135, 244–253 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D. G. & Donoghue M. J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA 103, 9101–9106 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R. A. & Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proceedings of the National Academy of Sciences of the United States of America 102, 12978–12983 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C. et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6, e230, 10.1371/journal.pbio.0060230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C. et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiology 149, 1462–1477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K. & Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588, 10.1105/tpc.110.075598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I. L., Uberti Manassero N. G., Ripoll R. & Gonzalez D. H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435, 143–155, 10.1042/BJ20101019 (2011). [DOI] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R. & Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68, 147–158, 10.1111/j.1365-313X.2011.04674.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez J. A. & Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4, 406, 10.3389/fpls.2013.00406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N. et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39, 787–791, 10.1038/ng2036 (2007). [DOI] [PubMed] [Google Scholar]
- Li Z., Li B., Shen W. H., Huang H. & Dong A. TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J 71, 99–107, 10.1111/j.1365-313X.2012.04973.x (2012). [DOI] [PubMed] [Google Scholar]
- Ballester P., Navarrete-Gomez M., Carbonero P., Onate-Sanchez L. & Ferrandiz C. Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol Plant 10.1111/ppl.12327 (2015). [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I. et al. Temporal Control of Leaf Complexity by miRNA-Regulated Licensing of Protein Complexes. Current Biol 24, 2714–2719, 10.1016/j.cub.2014.09.058 (2014). [DOI] [PubMed] [Google Scholar]
- Tuskan G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604, 10.1126/science.1128691 (2006). [DOI] [PubMed] [Google Scholar]
- Zeng F., Yan H. & Arndt S. K. Leaf and whole tree adaptations to mild salinity in field grown Populus euphratica. Tree Physiol 29, 1237–1246, 10.1093/treephys/tpp055 (2009). [DOI] [PubMed] [Google Scholar]
- Huang W., Li Z., Yang Z. & Bai G. The structural traits of Populus euphratica heteromorphic leaves and their correlations. Acta Ecol Sini 30, 4636–4642 (2010). [Google Scholar]
- Huang W., Li Z., Yang Z., Liang J. & Bai G. Heteromorphic leaves structural characteristics and their correlations with diameter at brest height of Populus euphratica. Chin J Ecol 29, 2347–2352 (2010). [Google Scholar]
- Qiu Q. et al. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol, tpr015 (2011). [DOI] [PubMed]
- Ma T. et al. Genomic insights into salt adaptation in a desert poplar. Nature communications 4 (2013). [DOI] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G. & Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucl Acids Res. gkt1016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucl Acids Res 40, D1178–D1186, 10.1093/nar/gkr944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. & Elkan C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc Int Conf Intell Syst Mol Biol 2, 28–36 (1994). [PubMed] [Google Scholar]
- Schommer C., Debernardi J. M., Bresso E. G., Rodriguez R. E. & Palatnik J. F. Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7, 1533–1544, 10.1093/mp/ssu084 (2014). [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M. & Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484, 10.1105/tpc.106.044792 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K. & Nath U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67, 595–607, 10.1111/j.1365-313X.2011.04616.x (2011). [DOI] [PubMed] [Google Scholar]
- Wang B. Functional analysis of Arabidopsis thaliana Class I TCP genes AtTCP7 and its roles in plant development, University of Trento (2015). [Google Scholar]
- Finn R. D. et al. Pfam: the protein families database. Nucl Acids Res 42, D222–D230, 10.1093/nar/gkt1223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. Accelerated profile HMM searches. PLoS Comput Biol 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E. et al. Protein identification and analysis tools on the ExPASy server. (Springer, 2005). [DOI] [PubMed] [Google Scholar]
- Sigrist C. J. et al. New and continuing developments at PROSITE. Nucl Acids Res. gks1067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297, 10.1093/bioinformatics/btu817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci Rep 4, 6645, 10.1038/srep06645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.