Abstract
Malignant mesothelioma (MM) is a highly-aggressive heterogeneous malignancy, typically diagnosed at advanced stage. An important area of mesothelioma biology and progression is understanding intercellular communication and the contribution of the secretome. Exosomes are secreted extracellular vesicles shown to shuttle cellular cargo and direct intercellular communication in the tumour microenvironment, facilitate immunoregulation and metastasis. In this study, quantitative proteomics was used to investigate MM-derived exosomes from distinct human models and identify select cargo protein networks associated with angiogenesis, metastasis, and immunoregulation. Utilising bioinformatics pathway/network analyses, and correlation with previous studies on tumour exosomes, we defined a select mesothelioma exosomal signature (mEXOS, 570 proteins) enriched in tumour antigens and various cancer-specific signalling (HPGD/ENO1/OSMR) and secreted modulators (FN1/ITLN1/MAMDC2/PDGFD/GBP1). Notably, such circulating cargo offers unique insights into mesothelioma progression and tumour microenvironment reprogramming. Functionally, we demonstrate that oncogenic exosomes facilitate the migratory capacity of fibroblast/endothelial cells, supporting the systematic model of MM progression associated with vascular remodelling and angiogenesis. We provide biophysical and proteomic characterisation of exosomes, define a unique oncogenic signature (mEXOS), and demonstrate the regulatory capacity of exosomes in cell migration/tube formation assays. These findings contribute to understanding tumour-stromal crosstalk in the context of MM, and potential new diagnostic and therapeutic extracellular targets.
Malignant mesothelioma (MM) is an incurable malignancy involving serosal tissues, especially the pleura. MM has a median survival from initial diagnosis of 7–9 months1. Contributing factors such as the absence of biomarkers and different pathologic subtypes increase the difficulty of treatment, and as a result, individuals with MM generally have a median survival ranging from 11 months with chemotherapy to 7 months with supportive care2,3. In the next 25 years it is estimated that the diagnosis of MM will increase ~5–10% annually in most industrialized countries at a cost of ~$300 billion worldwide4. No single-modality MM therapy including chemotherapy, radiation therapy, immunotherapy, cyto-reductive surgery or surgery has reliably demonstrated superiority to supportive care5. Importantly, diagnosis of MM is often difficult and most patients present at an advanced stage. Many blood-based biomarkers for diagnosis of MM have been described, with soluble members of the mesothelin family being the predominant focus6,7. However, their limited specificity has meant that new tumour-specific markers are being actively sorted8,9,10. Recently, several candidate protein, glycoprotein, antibody, and miRNA markers have been reported11,12,13,14,15 but still require independent validation. Improved surveillance and early detection of MM using specific markers of initiation and progression are required to improve clinical intervention, and patient survival16.
A number of studies in animal models and human patients have demonstrated that inhalation or injection of asbestos fibres results in a chronic inflammatory response characterized primarily by recruitment of cancer-associated fibroblasts (CAFs)17 to promote production of chemokines and cytokines in the lung17 and pleura18. Exposure of human MM cells to asbestos has been shown to facilitate autocrine production and transcriptional regulation of cytokines19,20. Such findings support a malignant secretory network that can regulate the MM tumour microenvironment and fundamental to understanding the progression of various malignancies, including mesothelioma. Importantly, MM has a highly secretory cell type, and the factors released by cells may act in an autocrine or paracrine fashion on tumour and stroma, where they may modulate the extracellular environment and indeed provide a resource for putative cancer biomarkers15. Malignant pleural effusions have been demonstrated to accumulate secreted tumour-derived extracellular vesicles (EVs), specifically exosomes, bearing tumour antigens and antigen-presenting molecules, capable of facilitating anti-tumour immune responses21,22. Importantly, exosomes from different tumour cells have shown immune activity against not only syngeneic but also allogeneic tumour growth, indicating that tumour-derived exosomes may harbor common tumour antigens capable of inducing antigen-specific immune responses23. Therefore, tumour-derived exosomes are a natural and novel source of tumour antigens which could provide alternative diagnostic circulating markers for mesothelioma and its progression but also may represent attractive tumour-specific therapeutic targets21,23,24,25.
Exosomes are small (30–150 nm) nano-extracellular vesicles derived from the endosomal pathway by inward budding luminal membranes of multivesicular bodies (MVBs) to form intraluminal vesicles (ILVs); MVBs then traffic to and fuse with the plasma membrane whereupon they release their ILV contents into extracellular space (as exosomes)26,27. Exosomes have diverse roles in intercellular communication which can be conferred by mediators that are presented on their surface or contained within the lumen. Exosomes contain a specific composition of proteins, lipids, mRNA, regulatory RNA and DNA cargo components28. Increasing evidence suggests that exosomes can influence physiological processes such as cell transformation28, immunoregulation25,29, and importantly cancer progression30,31,32,33,34,35,36,37,38, vaccination against infectious disease39, and vaccines for possible cancer treatments40,41,42. These studies have led to several clinical and pre-clinical investigations of exosome/EV-based therapies43,44,45,46. In the context of therapeutic applications, exosomes of selected cell types have been used as therapeutic agents in immune therapy, vaccination trials, regenerative medicine, and drug delivery47. Exosomes also provide an as yet largely untapped source of diagnostic, prognostic, and predictive biomarkers27,29,42. Recently, various innovative therapies involving the ex vivo manipulation and subsequent reintroduction of exosome-based therapeutics into humans are being developed and validated, although no exosome-based therapeutics have yet to be brought into the clinic44,47,48.
As a first step towards characterizing the contribution of exosomes in MM, we report in this study an exosomal MM protein signature using four different human MM cell models. Using quantitative proteomic profiling of purified exosomes from each model, we reveal insights into their select oncogenic cargo, demonstrating an enrichment of components associated with immune response, angiogenesis, and cell transformation during cancer progression. Importantly, we demonstrate select proteins known to be associated with cellular migration, invasion, and metastasis enriched in mesothelioma-derived exosomes49,50,51,52. Further, we highlight the functional relevance of MM-derived exosomes by pathway analysis associated with immunoregulation. Given that MM progression is associated with vascular remodelling and angiogenesis53,54, and accumulation of tumour-associated fibroblasts17, we investigated the functional effects of oncogenic exosomes from MM on recipient cells in the tumour microenvironment, including fibroblast and endothelial cells. Although studies have shown that many populations of MM contain tumour-associated fibroblasts55, little is known about interactions and how tumour-associated fibroblasts are regulated by MM cells. These data support the assertion that selective exosomal cargo may contribute to the progressive changes of mesothelioma by tumour angiogenesis and malignancy. This definitive characterisation and comprehensive informatics analysis of the protein cargo in exosomes derived from different human MM tumours contributes to our understanding of the extracellular environment during MM and identifies potential new diagnostic targets from the extracellular and secreted factors derived from human MM cells.
Results and Discussion
In this study we have investigated exosomes derived from different human malignant mesothelioma cancer cell models by quantitative protein profiling and functional characterisation to gain insights into secreted modulators of tumourigenicity, immunoregulation, and metastasis.
Exosomes are released from malignant mesothelioma cells
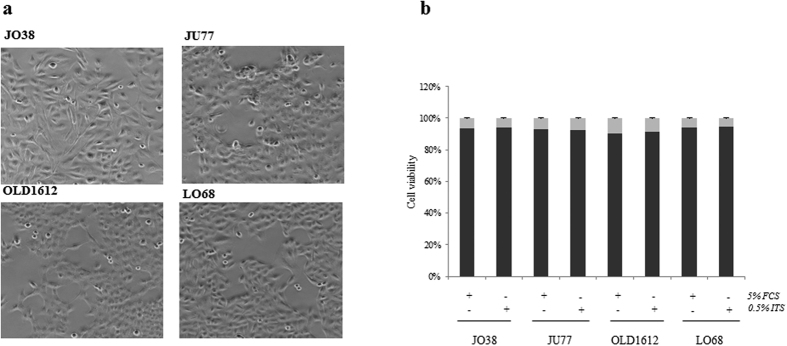
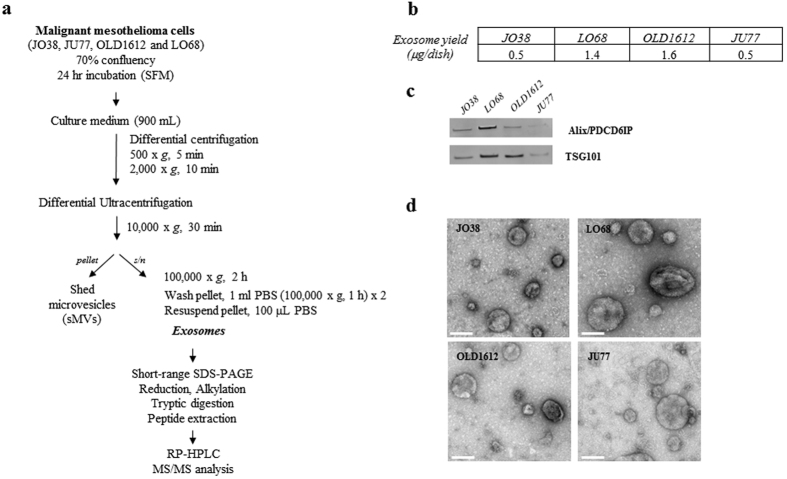
Human MM cells (JO38, JU77, OLD1612, LO68) displayed a fibroblast-like mesenchymal phenotype in culture (Fig. 1a). Exosomes were isolated from cell conditioned medium in serum-free conditions for 24 hr (>92% cell viability) (Fig. 1b). Exosome isolation and enrichment strategy is shown (Fig. 2a). Exosome yield was in range 0.5–1.6 μg protein/dish (mean ~90 μg/~8 × 108 cells) (Fig. 2b). Western blot analysis confirmed expression of exosome markers Alix/PDCD6IP and TSG101 (Fig. 2c), and transmission electron microscopy revealed a relatively homogenous population of spherical membranous vesicles 30–150 nm in diameter (Fig. 2d), which is in accordance with the typical size reported for exosomes56,57.
Figure 1. Human malignant mesothelioma cell characterisation.
(a) Phase contrast images of human MM cells JO38, JU77, OLD1612, and LO68 reveal elongated fibroblast-like morphology. (b) Cell viability as determined by the Trypan Blue dye exclusion assay after culture in presence of 5% FCS (foetal calf serum) or 0.5% ITS (insulin-transferrin-selenium) conditions for 24 hr, demonstrating that all cell lines remain >92% viable. Data representative of mean ± SEM of three independent experiments performed in triplicate.
Figure 2. Isolation and characterisation of MM-derived exosomes.
(a) Workflow showing isolation and purification of exosomes from mesothelioma cell lines using serum-free media (SFM) conditions. Exosomes (10 μg) were solubilised in SDS, separated by 1D-SDS-PAGE and fractions (n = 2) subjected to in-gel reduction, alkylation, and tryptic digestion. Extracted peptides were fractionated and identified using mass spectrometry analysis, data processing database searching, informatics and protein annotation. (b) Protein yield (μg/cell dish) for JO38, JU77, OLD1612, and LO68 exosomes is shown (average n = 3). (c) For Western blotting, exosome preparations (10 μg) were separated by 1D-SDS-PAGE, electrotransferred, and probed with exosome markers Alix/PDCD6IP and TSG101. Data representative of three independent experiments. (d) For transmission electron microscopy, exosomes (2 μg) were negatively stained using uranyl acetate and viewed by transmission EM, revealing a relatively homogenous population of round membranous vesicles 30–150 nm in size for all cell types. Scale bar, 100 nm. Representative image from n = 3 and 5 independent fields of view.
Proteome analysis of human MM-derived exosomes
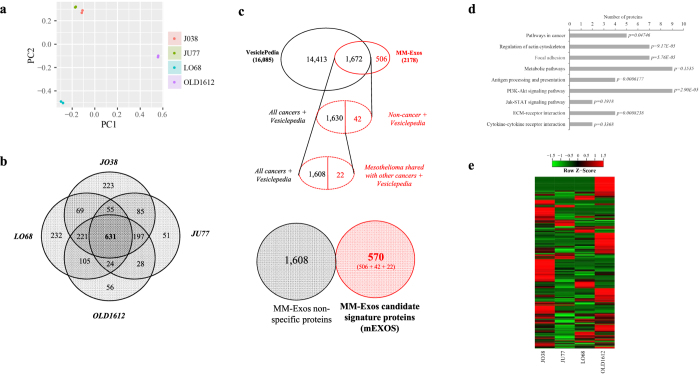
Quantitative proteomic profiling identified a total of 2,178 proteins in exosomes from different human-derived MM cells (biological duplicate) (Supplementary Dataset). We evaluated the spatial separation for these proteins in the studied groups using principal component analysis (PCA). The result of PCA demonstrated good separation between MM models (Fig. 3a). Distribution of identified proteins between each MM-derived exosome type is shown, revealing a common subset of 631 exosomal proteins identified between all models (Fig. 3b). The 631 common exosomal proteins included proteins involved in exosome biogenesis (including TSG101, VPS26A, VPS29, VPS35, and Alix58) and intracellular vesicle trafficking (including tetraspanins CD63 and CD9, CD81/CD82, and small Rab GTPases59,60,61) (Table S1). The majority of the common mesothelioma exosome proteins (508/631 (80%)) have previously been observed in exosomes released from diverse cell types (see EV database compendium Vesiclepedia containing 93,980 protein/mRNA entries (16,085 human proteins/mRNAs)62). Similarly, several key proteins associated with exosome biogenesis (ESCRT-associated, tetraspanins), sorting/trafficking (including Rab GTPases, ADP-ribosylation factors, clathrin and coatomer subunits, lipid raft flotillins), and vesicle release (including synaptotagmin 1/2, dynamin 1/2, synaptogyrin, VAMP3 and VAT1) (Table S2) were identified, though were not common to all cells. Nearly a quarter of the total number of exosomal proteins identified from JO38, JU77, OLD1612, and LO68 cells (506/2,178 (23%)) have not been previously reported in the EV database, Vesiclepedia (Supplementary Dataset). Besides common proteins associated with vesicle biogenesis and trafficking and the fact that only a single other proteomic study associated with mesothelioma and EVs is reported in Vesiclepedia63, this indicates the selectivity of unique oncogenic cargo within MM-derived exosomes, in addition to improvements in proteomics technologies and mass spectrometry.
Figure 3. Characterisation of mesothelioma-derived exosomes reveals an exosomal-specific signature (mEXOS).
(a) Principal component analysis (PCA). Each symbol represents a biological replicate, and the colour represents the group (model). (b) A four-way Venn diagram of proteins distributed between each MM-derived exosome type is shown, revealing 631 proteins common to each dataset. (c) To determine the classification of proteins in mEXOS we applied a stringent filtering criteria. The total MM-derived exosomal proteins (2,178) were compared with the Vesiclepedia database (comprising 16,085 human entries) and literature searching, of which 506 were unique to this study and not previously reported in the context of extracellular vesicles. Among the 1,672 co-identified proteins, 42 were non-cancer proteins reported in Vesiclepedia. with a further 22 proteins reported shared with mesothelioma and other cancers in Vesiclepedia. Therefore, to determine the unique MM exosome protein signature (mEXOS), we summated the 506, 42, and 22 proteins from these categories to reveal 570 proteins as select exosomal and MM-derived components (Table S3). (d) KEGG pathway analysis of mEXOS, with p-values indicated. (e) Correlation matrix of mEXOS, representing differential abundance based on normalised spectral count (SpC) values between each of the MM models investigated, showing that each individual sample represents clear distribution and similarity with other MM models.
In comparison to exosomes derived from human mesothelioma models, Hegmans et al.63 reported the proteomic characterisation of exosomes derived from two different human mesothelioma cells (PMR-MM7 and PMRMM8) by MALDI-TOF mass spectrometry. The study identified 19 proteins, with components associated with antigen presentation, including MHC class I molecules and Hsp90, although no mesothelioma-associated antigens were identified. In comparison, we report all these proteins in mesothelioma-derived exosomes, and importantly 15 proteins were in the common 631 exosomal protein list (Table S3). Further, Clayton et al.64 detected Her2-neu, mesothelin, CD9, CD81, LAMP-1 and MHC-like MICA by Western blotting in tumour-derived exosomes from advanced pleural MM. In comparison, this study identified mesothelin, CD9, and CD81 in exosomes, with CD9 and CD81 being common to all mesothelioma-derived exosomes. Clayton et al.65 further demonstrated a direct interaction between the activating receptor natural killer group 2D (NKG2D) receptor (MHC class-I related) on exosomes from various cancer cell models/primary cells and natural killer (NK) cells or CD8+ T cells. This interaction was shown to implicate NKG2D as a target for exosome-mediated tumour immune evasion. Primary mesothelioma cell-derived exosomes were shown to harbour typical exosome markers including MHC class I and TSG101. Expression of MHC-like molecules MICA/MICB were also demonstrated. Interestingly, we report such proteins in MM-derived exosomes (OLD1612 cells). More recently, Manfredi et al.15 have investigated the secretome using proteomic profiling from different human MM models to reveal proteins commonly expressed between these studies (i.e., MM98: 78/208 common, REN: 46/112). Of note, we report 2,073 exosomal proteins unique to our study. Therefore, this current study on proteomic profiling of MM-derived exosomes provides an extensive insight into the protein cargo of cancer exosomes, direct comparison with previous reports on MM-derived exosomes and the secretome, and significantly increased coverage of exosomal-associated proteins not previously reported in Vesiclepedia or identified from human MM models.
Mesothelioma-enriched exosome protein cancer signature (mEXOS)
To address the contribution of exosome cargo proteins to mesothelioma in comparison to other cancer types (i.e., mesothelioma-enriched exosome protein cancer signature, mEXOS) we used a stringent filtering criteria utilising a combination of Vesiclepedia, and literature searching of exosomal studies investigating mesothelioma cells/pleural effusion (Fig. 3c) 21,23,63,64,65. Of the 2,178 proteins identified in this study, 1,672 were common to various cancer-types reported in Vesiclepedia (i.e., bladder, breast, colorectal, melanoma, ovarian, prostate, stomach cancers), while the remaining 506 proteins were categorised as unique to MM only. These unique exosomal proteins represent a select and novel dataset for the comprehensive analysis of MM and exosomal proteins. Among the 1,672 proteins, 42 were non-cancer proteins reported in Vesiclepedia, with a further 22 proteins shared by mesothelioma and other cancers in Vesiclepedia (Fig. 3c). In total, we report 570 proteins in defining a unique MM exosome protein signature (mEXOS) containing new candidate biomarkers for MM (Fig. 3c) (Table S4). Interestingly, 142/570 proteins were identified in all MM-derived exosomes. For proteins classified into mEXOS, KEGG pathway analysis and Gene Ontology were used to determine pathways enriched, and protein information on subcellular location, biological function, and cellular component (Figure S1). Of note we observe KEGG enrichment (Fig. 3d) associated with “pathways in cancer” (ko05200), “cytokine receptor interaction” (ko04060), “regulation of actin cytoskeleton” (ko04810), “focal adhesion” (ko04510), “metabolic pathways” (ko01100), “antigen processing and presentation” (ko04612), “extracellular matrix-receptor interaction” (ko04512), “PI3K-Akt signalling pathway” (ko04151) and “Jak-STAT signalling pathway” (ko04630).
Several select protein networks were identified in mEXOS, demonstrating the variability in protein expression between distinct human MM models (Fig. 3a,e). This cluster and normalised heat map analysis revealed that MM-derived exosomes from JU77 and JO38 were most similar in expression profiles, while similarities were identified between LO68 and OLD1612 models. These protein expression cluster analyses align the cell models with the amount of exosomes released from malignant mesothelioma cells, with JU77 and JO38 (low yield producing) and LO68 and OLD1612 (high yield producing) (Figs 2b and 3a). There were several exosomal cargo proteins whose expression and abundance profile were similar across all models, including villin 2 (VIL2), transferrin receptor (TFRC), thrombospondin 1 fragment (THBS1), and lactadherin (MFGE8). Interestingly, such proteins have been shown to be involved in cell-to-cell and cell-to-matrix interactions66,67,68,69,70,71, and as for MFGE8, an important role in the maintenance of intestinal epithelial homeostasis, promotion of mucosal healing, and neovascularization72,73. A salient finding has been the recent implication of MFGE8 in modulating the tumour microenvironment and promote tumourigenicity in primary lung cancer cells74. Such MM-specific information provide an integrative analysis of the selective extracellular cargo components associated with different human-derived MM models in vitro and importantly new candidate extracellular biomarkers for MM.
Exploration of candidate exosomal biomarkers for MM
We report several select MM specific and abundant markers in mEXOS which have direct clinical relevance and correlation with previously identified expression profiling studies investigating MM (Table S4). We report 5 known tubulin isotypes in mEXOS, including proteins TUBB4A (470 SpC), Q8IWP6 (class IVb beta tubulin) (544 SpC), and B3KPS3 (455 SpC). Emerging evidence suggests that tubulins and the tubulin-microtuble network are critically involved in cell stress responses involved in cancer75. Tubulins are now considered important targets for chemotherapeutic drugs in many solid tumour malignancies including mesothelioma76,77. Further, we report galectin-3-binding protein (LG3BP/B4DVE1) (465 SpC) in mEXOS, and importantly, the first report of LG3BP in MM-derived exosomes. As an important mediator of integrin cell adhesion, expression of LG3BP/B4DVE1 has been shown to be significantly up-regulated in malignant pleural mesothelioma78 and it has been reported in other tumour-derived exosomes including colorectal, breast, and bladder79.
Further potential markers within mEXOS include alpha-enolase (ENO1), annexin A1 (ANXA1), and glucose-6-phosphate 1-dehydrogenase (A8K8D9/G6PD). ENO1 (307 SpC) is a multifunctional enzyme that, as well as its role in glycolysis, plays a part in various processes such as cell control, hypoxia, and the innate immune response80. Further, ENO1 is suggested to function in the intravascular and pericellular fibrinolytic system due to its ability to serve as a receptor and activator of plasminogen on the cell surface of several cell-types such as leukocytes and neurons, and further stimulates immunoglobulin production81. Of note, ENO1 has previously been identified in mesothelioma exosomes63. ANX1 (102 SpC) is known to contribute to the adaptive immune response by enhancing signalling cascades that are triggered by T-cell activation, regulates differentiation and proliferation of activated T-cells, inflammation and wound healing, cell polarization, and cell migration82. ANX1 has previously been shown to be important in the response to oxidative stress in mesothelioma cells83. Moreover, G6PD (190 SpC) is necessary for oxidative ribose production, controlling the pentose phosphate pathway84. Importantly, G6PD has been revealed to be involved in apoptosis, angiogenesis, and the efficacy to anti-cancer therapy, making it a promising target in directing and monitoring cancer therapy85. High levels of G6PD are observed in some cancers and its expression can transform fibroblasts to induce tumour formation in vivo86. These proteins were identified across all MM exosomes, and provide potential common markers that define a MM exosome-related signature. The fact that these proteins can be identified in exosomes is an exciting indication that identification and production of MM factors may be used as biomarkers of tumour development or as indicators of patient responses to therapeutic regimens or surgical resection of MM.
In the context of previous studies investigating novel tumour-derived biomarkers for MM, Suraokar et al.87 reported gene expression microarray profiling (global transcriptomic microarray analysis) of 53 surgically resected MMs tumours along with paired normal tissue. In comparison to mEXOS, we report 9 of their gene products (BCAT1, CALB2, CCDC68, CFB, HPGD, LAMA1, MAMDC2, MAP2 and SULF1), with 5 genes upregulated in expression correlating with MMs tumours (BCAT1, CALB2, CFB, LAMA1, and SULF1). Crispi et al.88 performed Affymetrix HGU133A plus 2.0 microarray analysis to molecularly dissect mesothelioma tumour pathways and identify new tumour biomarkers that could be used as early diagnostic markers and possibly as specific molecular therapeutic targets. In comparison, we report 3 proteins (ANXA6, LAMA1 and NRP2) in mEXOS. Annexin A6 (ANXA6) has been identified in exosomes derived from human mesothelioma cells63 and shown to regulate membrane-cytoskeleton dynamics and membrane-fusion events between intracellular compartments, and further play a role in the inward vesiculation process89. Neuropilins (NRPs) are multifunctional non–tyrosine kinase receptors, expressed at the surface of endothelial cells, with NRP2 expressed in the lymphatic system90. The expression of NRP2 contributes to tumour-angiogenesis and lymphangiogenesis91, and importantly contribute to cell communication processes, influencing cell positioning and behaviour, and tissue morphogenesis92. In comparison to additional known factors identified in mEXOS and associated with MM, we report cyclooxygenase-2 (COX-2) and calretinin (CALB2). COX2 is implicated in many events in the tumorigenic process, producing highly reactive products that can affect cell growth, immune response, apoptosis, and angioneogenesis93. Importantly, high COX-2 expression is a marker of poor prognosis in MM94. CALB2 is a vitamin D–dependent calcium-binding protein involved in calcium signalling, and recently shown to be an important sensitive and specific diagnostic marker for MM in serous effusions95. CALB2 is an established immunohistochemical marker used in the diagnosis of mesothelioma96 and frequently used for the diagnosis of MM (as apposed to lung adenocarcinoma)97. It has further been reported as a blood-based marker of mesothelioma98. Interestingly, CALB2 has previously been found in exosomes from malignant pleural effusion99.
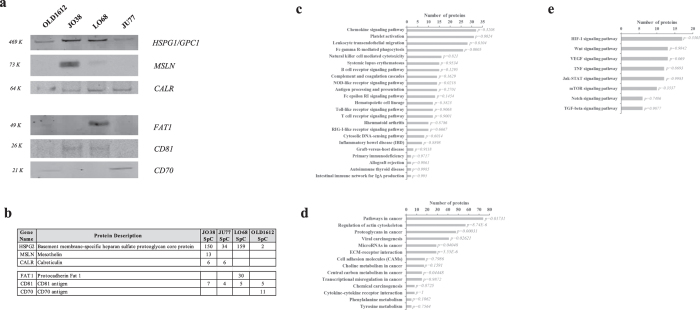
Additionally, we report several key proteins in MM-derived exosome cargo (although not specific to mEXOS) known to be expressed in mesothelioma, including mesothelin (MSLN), calreticulin (CALR), RuvB-like proteins (RUVBL1/2), proteins signal transducer and activator of transcription 1 (STAT1), vimentin (VIM), and superoxide dismutase (SOD) 1/2 (Table 1) (Fig. 4a,b)100. MSLN is an established marker in mesothelioma diagnosis6,96. MSLN has been identified enriched in tumour-derived exosomes21,23,101 and exosomes from malignant pleural effusion99. The RUVBL1/2 proteins are involved in chromatin remodelling102, DNA interaction and repair103, and promote interaction of the modified histones with other proteins which positively regulate transcription104. These proteins may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene-mediated growth induction, and DNA repair. There has been extensive work on several DNA-inducible genes as human MM biomarkers of exposure to these agents, including p53 induction of DNA strand breaks, p53 expression, and apoptosis in cell lines, particularly in cultured mesothelial cells105. Such in vivo findings highlight the importance of oxidative damage in asbestos-induced carcinogenesis, however were unable to define the specific differentially expressed components and molecular basis of asbestos-induced disease and MM progression. Further, STAT1 which is known to have an oncogenic function and is constitutively activated in many human cancer cells including human MM106,107, has not previously been observed in mesothelioma, or lung cancer exosomes. The proteins identified in this profiling study, and specifically the oncogenic signature (mEXOS), represents an extensive and important catalogue of proteins attributed in exosomes specifically in the context of human MM and may represent new selective extracellular and circulating targets in MM progression, diagnosis, and monitoring.
Table 1. Mesothelioma-associated protein cargo in exosomes.
| Gene Namea | Protein Accessiona | Protein Descriptiona | SpC Combined (4 cell lines)b | Reported in VesiclePediac |
|---|---|---|---|---|
| MSLN | Q13421 | Mesothelin (CAK1 antigen) | 13 | Y |
| TNFAIP2 | Q03169 | Tumor necrosis factor alpha-induced protein 2 | 64 | Y |
| CALB2 | P22676 | Calretinin | 7 | Y |
| CALR | P27797 | Calreticulin (CRP55) | 12 | Y |
| TNFRSF11B | O00300 | Tumor necrosis factor receptor superfamily member 11B | 9 | Y |
| GOLT1B | Q9Y3E0 | Vesicle transport protein GOT1B | 7 | Y |
| Q59FU8_HUMAN | Q59FU8 | Tumor necrosis factor receptor superfamily, member 6 isoform 1 variant | 4 | |
| LGALS1 | P09382 | Galectin-1 | 60 | Y |
| SOD1 | P00441 | Superoxide dismutase 1 | 9 | Y |
| SOD2 | B3KUK2 | Superoxide dismutase 2 | 7 | Y |
| STAT1 | P42224 | Signal transducer and activator of transcription 1-alpha/beta | 139 | Y |
| STAT2 | P52630 | Signal transducer and activator of transcription 2 (p113) | 6 | Y |
| STAT3 | K7ENL3 | Signal transducer and activator of transcription | 25 | Y |
| TNFRSF10A | O00220 | Tumor necrosis factor receptor superfamily member 10A | 8 | Y |
| TPT1 | Q5W0H4 | Translationally-controlled tumor protein | 7 | Y |
| VIM | P08670 | Vimentin | 180 | Y |
aProtein accession and description derived from Gene Ontology (UniProt).
bCombined spectral counts (SpC) observed for mesothelioma exosome datasets. Contribution of individual SpC for each cell lines reported in Supplementary Dataset.
cComparison with extracellular vesicle database, Vesiclepedia62. Y indicates reported in the database.
Figure 4. Validation and pathway analysis of mesothelioma-derived exosome cargo.
(a) To validate the relative abundance of proteins identified using GeLC-MS/MS, Western blot analysis was used to compare expression for selected proteins; glypican-1 (HSPG1/GPC1), mesothelin (MSLN), calreticulin (CALR), and other exosome components protocadherin Fat 1 (FAT1), CD81, and CD70. For Western blotting, exosomes (10 μg) were separated by 1D-SDS-PAGE, electrotransferred, and probed with markers as indicated (n = 2). (b) Relative quantitation of label-free spectral counting (SpC) are shown for selected proteins validated using Western immunoblotting (n = 3). (c) Immune system and immune disease related pathways in MM-derived exosomes, with p-values indicated. (d) Cancer cell biology related pathways in MM-derived exosomes, with p-values indicated. (e) Signal transduction related pathways in MM-derived exosomes, with p-values indicated.
Mesothelioma-derived exosomes contain immunoregulatory components
Tumour-derived exosomes have been shown to be immunomodulatory during cancer progression25. In this study, KEGG pathway annotation for the 2,178 proteins revealed pathways related to immune system and immune diseases (Fig. 4c). For these data, we identified a total of 111 proteins associated with immunoregulation in MM-derived exosomes (Table S5), of which 26 were identified in mEXOS, including oncostatin-M receptor (OSMR), multidrug resistance-associated protein 1 (ABCC1), and the SUMO-1 activating receptor, SAE1. OSMR is a multifunctional cell surface cytokine receptor, which induces several pro-malignant effects, including a pro-angiogenic phenotype and increased cell migration and invasiveness108. Recently, OSMR mRNA has been shown to be significantly elevated in malignant pleural mesothelioma, compared with benign asbestos-related pleural effusion109. Further, OSMR which is a multifunctional cell surface cytokine, previously found to be over-expressed in mesothelioma, has been suggested to be a candidate for antibody-mediated targeted inhibition110. Expression of the ATP-binding cassette superfamily member ABCC1 has been previously demonstrated in non-small-cell lung carcinoma111 and is an important immunoregulatory component of chemotherapy resistance112. The CD70 antigen is expressed by limited subsets of normal lymphocytes and dendritic cells (DCs), but aberrantly expressed by a broad range of hematologic malignancies and some solid tumours113,114,115. CD70 is associated with MHC class II where it acts as a co-stimulatory molecule, with identification of CD70 previously associated with exosomes in contributing to their immunostimulatory capacity116,117.
Of the immune-associated components, 30 different proteins were identified associated with MHC I antigen processing and presentation (e.g., TCP-1-beta, protein-methionine sulfoxide oxidase MICAL1, and plastin-3), and 21 proteins identified associated with MHC II antigen processing and presentation (e.g., ACLY variant protein, copine-1, and guanine nucleotide-binding protein subunit beta-2-like 1). Furthermore, various proteins were identified associated with the B-cell response (e.g., vitronectin, and toll interacting protein), NK-cell response (e.g., SUMO-1 activating enzyme, and long-chain-fatty-acid-CoA ligase 4), T-cell response (e.g., tensin-3, and TELO2-interacting protein 1), and other related components associated with the immune response (e.g., STAT1, MX1, and MX2). Further, 28 proteins (24 associated with B-cell response, 4 associated with T-cell response) have been identified in previous studies associated with EVs117,118,119.
Interestingly, we observed exosomal protein components co-identified in a previous immune signature of malignant pleural mesothelioma based on clustering of relevant enriched genes to mesothelioma (tissue, cell lines) compared to non-malignant mesothelial cells120. In comparison, this current study identified 9 components in exosomes derived from MM, including the interferon-induced proteins IFIT1, MX1/2, and STAT1. EVs have previously been shown to mediate the transfer and specific activation of STAT1 and pro-inflammatory cytokine signalling in target cells121. This current study represents a significant subset of proteins associated with immunoregulation, not previously associated with exosomes or EVs, or attributed to MM (Table S5). Collectively, we highlight the importance of MM-derived exosomes revealed by pathway analysis and bioinformatics analysis associated with immunoregulation Given the chronic inflammatory response characteristic of MM progression, it remains to be determined whether such components are functionally active in tumour-derived exosomes, or in target recipient cells associated with the tumour microenvironment.
Exosomes as carriers of tumour-derived antigens
Immune responses have been shown to be beneficial in MM patients122,123, therefore, focusing immunotherapeutic strategies on promoting these immune responses is an attractive approach. Towards this aim, tumour- and immune cell-derived exosomes have been shown to carry tumour antigens and modulate the immune response, leading to eradication of established tumours by CD8+ T cells and CD4+ T cells, as well as directly suppressing tumour growth and resistance to malignant tumour development24,25,124. Therefore, in this study, to investigate the presence of tumour-specific antigens within exosomes in the context of MM, we utilised a combination of bioinformatics and gene ontology analyses to reveal 16 select tumour-derived antigens as cargo components in MM-derived exosomes, including CD70 antigen, cleavage and polyadenylation specificity factor subunit 1 and 3 (CPSF1/3), melanoma-associated antigen D2 (11B6), glypican-1, BJ-HCC-24 tumour antigen, and mesothelin (CAK1 antigen) (Table 2). Several candidates were validated using immunoblotting (Fig. 4a) and correlated with mass spectrometry expression (Fig. 4b). Tumour proteins (including mesothelin) derived from patients with mesothelioma have been shown to induce a spontaneous humoral immune response125,126. Such responses against tumour-derived antigens include antibodies against these antigens, and may be useful as diagnostic tumour markers and targets for immune-based therapies127,128,129. Previously, exosomes are known to transfer tumour antigens, including mesothelin, to DCs for antigen presentation and cross-presentation21,23,130. The tumour-associated antigen MAGE proteins have attracted considerable interest in vaccine-based cancer immunotherapies131,132. MAGE-D2 has recently been shown to suppress expression of the apoptotic death receptor 2 (TRAIL-R2), promoting and protecting melanoma cells from apoptosis133. Several annexins (ANXA1-6, 11) were further identified across various MM-derived exosomes. Annexins have been implicated in several functions including membrane trafficking, cell signalling, ion transport, inflammation, apoptosis and haemostasis. Previously, ANXA2 has been reported to be a specific and selective antigen overexpressed in lung cancer tissues with high asbestos exposure and capable of inducing humoral immunity in MM patients134. Recently, ANXA2 has been identified as an antigenic target for pancreatic cancer immunotherapy135.
Table 2. Tumour-associated antigens identified in mesothelioma-derived exosomes.
| Gene Namea | Protein Accessiona | Protein Descriptiona | SpC Combined (4 cell lines)b | Reported in VesiclePediac | |
|---|---|---|---|---|---|
| Tumour antigens | MCAM | A8K6A6 | Melanoma cell adhesion molecule | 31 | Y |
| CD70 | P32970 | CD70 antigen (CD27 ligand) | 11 | Y | |
| MAGED2 | Q9UNF1 | Melanoma-associated antigen D2 | 10 | Y | |
| GPC1 | H7C410 | Glypican-1 | 44 | Y | |
| CPSF1 | Q10570 | Cleavage and polyadenylation specificity factor subunit 1 | 9 | Y | |
| CPSF3 | Q05BZ5 | CPSF3 protein | 2 | Y | |
| Q9NXW1_HUMAN | Q9NXW1 | BJ-HCC-24 tumor antigen | 6 | Y | |
| ARMC9 | Q7Z3E5 | LisH domain-containing protein ARMC9 (Melanoma/melanocyte-specific tumor antigen) | 10 | Y | |
| MSLN | Q13421 | Mesothelin (CAK1 antigen) | 13 | Y | |
| ANXA1 | P04083 | Annexin A1 | 268 | Y | |
| ANXA2 | P07355 | Annexin A2 | 643 | Y | |
| ANXA3 | P12429 | Annexin A3 | 95 | Y | |
| ANXA4 | P09525 | Annexin A4 | 167 | Y | |
| ANXA5 | P08758 | Annexin A5 | 351 | Y | |
| ANXA6 | A6NN80 | Annexin A6 | 205 | Y | |
| ANXA11 | P50995 | Annexin A11 | 102 | Y | |
| Overexpressed/accumulated in tumours | MFI2 | P08582 | Melanotransferrin (Melanoma-associated antigen p97) | 87 | Y |
| CDK1 | P06493 | Cyclin-dependent kinase 1 | 48 | Y | |
| MUC5AC | P98088 | Mucin-5AC | 176 | Y | |
| MMP2 | P08253 | 72 kDa type IV collagenase | 12 | Y | |
| FGFBP1 | Q14512 | Fibroblast growth factor-binding protein 1 | 11 | Y | |
| PXDN | Q92626 | Peroxidasin homolog (Melanoma-associated antigen MG50) | 60 | Y | |
| MUC5B | E9PBJ0 | Mucin-5B | 55 | Y | |
| FAT1 | Q14517 | Protocadherin Fat 1 | 30 | Y | |
| MUC13 | Q9H3R2 | Mucin-13 | 4 | Y | |
| TGFBI | Q15582 | Transforming growth factor-beta-induced protein ig-h3 | 112 | Y | |
| MIF | P14174 | Macrophage migration inhibitory factor | 18 | Y |
aProtein accession and description derived from Gene Ontology (UniProt).
bCombined spectral counts (SpC) observed for mesothelioma exosome datasets. Contribution of individual SpC for each cell lines reported in Supplementary Dataset.
cComparison with extracellular vesicle database, VesiclePedia62. Y indicates reported in the database.
In addition to tumour-associated antigens, we further report several proteins attributed to overexpression or accumulation in human tumours and biofluids, including melanotransferrin (melanoma-associated antigen p97), protocadherin Fat 1 (FAT1), fibroblast growth factor-binding protein 1, and mucins 5AC/5B/13136,137,138 (Table 2). Several of these proteins were validated using immunoblotting (Fig. 4a) and correlated with mass spectrometry results (Fig. 4b). Interestingly, antibodies against tumour antigens on mucins are widely used clinically as diagnostic tools (serum assays) for different cancer types, including colorectal, breast, ovarian and pancreatic adenocarcinoma138. Exosomes have been shown to mediate the transfer of modified tumour antigens, including mucins, to generate an immune response against tumours, highlighting the role of such vesicles as carriers of exogenous tumour antigens and in immunoregulation130.
Cancer signalling networks reflected in mesothelioma-derived exosome protein cargo
To identify pathways associated with cancer signalling in MM exosomes, we performed KEGG pathway annotation for the 2,178 proteins to reveal pathways related to cancer cell biology (cell motility, ECM-receptor interaction) (Fig. 4d), and signal transduction (including Jak-STAT signalling, TGF-β, TNF, mTOR, Wnt, VEGF, and Notch pathways (Fig. 4e). With relevance to Jak-STAT signalling, exosomes have been attributed to IFN-α-induced cell-to-cell transfer of antiviral components from LNPCs to HBV-infected hepatocytes both in vitro and in vivo139. The correlation of activation of Jak-STAT pathway with TGF-β expression in mesothelioma-derived exosomes has been previously associated with anti-proliferation64 and implicated in progression of MM107,140. TGF-β signalling has been attributed to transcription regulation of connective tissue growth factor (CTGF), enhancing expression of CTGF and directly controlling pro-oncogenic effects (cell proliferation, extracellular matrix remodelling) in MM140. Expression of STAT1/3/5 have been shown dysregulated in human MM (up-regulation of STAT1/5, down-regulation of STAT3), associated with downstream EGFR signalling, and inversely correlate with patient survival107. Further, selective MEK or PI3K kinase inhibitors are equally effective in down-regulating the pro-metastatic phenotype, suggesting that MEK or PI3K are appropriate targets for development of molecular therapeutics for MM141. Tumour-derived exosomes from pleural effusions of mesothelioma patients have been shown to partially modulate recipient immune cell function, through TGF-β signalling65. Associated with cancer cell signalling and various pathways related to cancer cell biology, we report heparan sulfate proteoglycan glypican-1 (HSPG1/GPC1) in exosomes derived from all MM models in this study. Previously, exosomal GPC1 has been shown to modulate the angiogenic and metastatic potential of human and mouse cancer cells142. GPC1 has been attributed to melanoma development and progression143, and increased expression in human glioma tumours and glioma-derived cell lines144. Further, GPC1 has recently been identified to be specifically enriched on cancer-cell-derived exosomes37. Tumour-derived GPC1-positive exosomes were capable of specifically distinguishing healthy subjects and patients with a benign pancreatic disease from patients with early- and late-stage pancreatic cancer, and correlate with tumour burden and survival of pre- and post-surgical patients with pancreatic cancer (carcinoma in situ, stage I and stages II–IV)37, supporting its utility as a biomarker for all stages of pancreatic cancer and its potential for early detection. As discussed, we highlight the importance of MM-derived exosomal cargo revealed by pathway and network analysis and bioinformatics analysis associated with immunoregulation. These findings may suggest that MICA (or other such related molecules), as cargo components of tumour exosomes, may facilitate targeting tumour exosomes to defined immune cells including CD8+ T cells and NK cells, to deliver ligands in a cell-type selective manner. Interestingly, we report both the identification of MICA-L1 and –L3, and significant elevated expression of components associated with TGF-β signalling in mesothelioma-derived exosomes. It remains unknown of the mechanisms of how tumour-derived exosomes selectively target and transfer ligands to specific immune cells and recipient cells associated with the tumour microenvironment to modulate the immunogenic response.
Mesothelioma exosomes regulate recipient cells of the tumour microenvironment
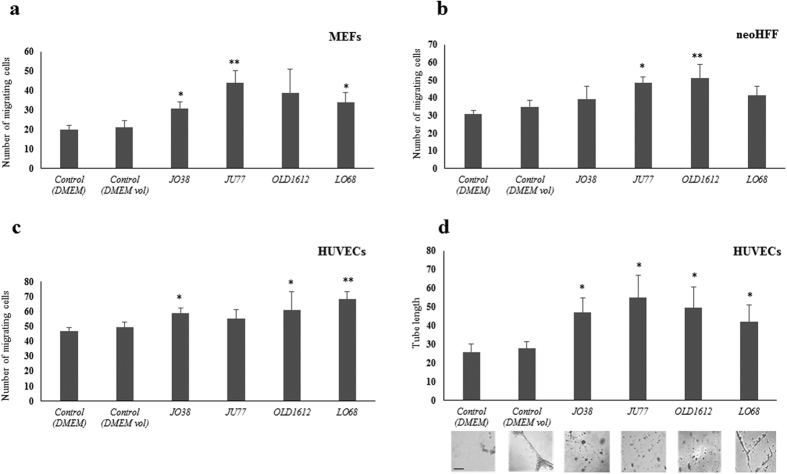
To demonstrate that MM-derived exosomes transport functionally active cargo, exosomes were investigated for their ability to regulate fibroblast and endothelial cells within the tumour microenvironment. Various mouse and human fibroblast models (MEFs/neoHFFs) and human endothelial cells (HUVECs) were cultured in DMEM-supplemented with MM exosomes (30 μg/mL), and over a 24 hr period, displayed significantly higher migration rates compared with vehicle controls alone (Fig. 5a–c). Recipient fibroblast cell migration was monitored using the transwell assay, which demonstrated that MM exosomes (30 μg/mL) significantly increased MEF cell migration for exosomes derived from JO38, OLD1612, and LO68 cells compared to vehicle treated cells (Fig. 5a), and exosomes derived from JU77 and OLD1612 significantly increased neoHFF cell migration (Fig. 5b). Further, exosomes from JO38, OLD1612, and LO68 cells resulted in significantly increased endothelial cell migration of HUVECs (Fig. 5c). Further, supplementation with MM-derived exosomes resulted in significantly increased HUVEC tube length formation compared to vehicle (control) treated cells (Fig. 5d). Together, these data demonstrate that exosomes derived from MM cells can differentially promote migratory capacity in recipient fibroblast and endothelial cells and HUVEC angiogenesis.
Figure 5. Mesothelioma exosomes regulate recipient cells of the tumour microenvironment.
To explore the potential exosome-mediated regulation of cells in the tumour microenvironment, fibroblasts (MEFs, neoHFFs) and endothelial cells (HUVECs) were exposed to MM cell-derived exosomes and recipient cell function analysed. For recipient cell migration, MEF (a), neoHFF (b) and HUVEC (c) cells were investigated using transwell assays over 24 hr in response to exosomes (30 μg/mL) derived from JO38, JU77, OLD1612, and LO68 cells. Vehicle control (DMEM), in addition to exosome-free control (exosome-depleted) were used. Transversing cells were stained with DAPI, imaged, and counted (n = 3; average ± sem; *p < 0.05, **p < 0.01). Tube formation assays using HUVEC cells (7 × 104), supplemented with MM-derived exosomes (30 μg/mL) and controls and seeded onto Matrigel (1 mg/mL). After 24 hr, tube formation was analysed, imaged, and quantified. Scale bar = 50 μm. (representative images from n = 3, average ± sem; *p < 0.05).
Previously, tumour exosomes (30 μg/mL) derived from mammary epithelial BT-474 cells have been shown to significantly increase cell proliferation of parental BT-474 cells145. Further, gastric cancer SGC7901-cell-derived exosomes have been shown to promote activation of PI3K/Akt and proliferation of SGC7901 and BGC823 cells146. Interestingly, proteomic analysis has revealed that abundant cell migratory and angiogenic factors are present in MM-derived exosomes147. Exosome uptake was shown to induce upregulation of angiogenesis-related genes and result in enhanced endothelial cell proliferation, migration, and sprouting148. Therefore, MM- and tumour-derived exosomes can directly facilitate reprogramming of recipient cells of the tumour microenvironment to facilitate cell migration and angiogenesis.
Mesothelioma-derived exosomes contain factors associated with metastasis
Tumour-derived exosomes have been shown to modulate the metastatic niche30,149,150,151. Importantly, various changes which facilitate metastasis have been described in the lung. These pre-formed lung niches encompass cells recruited from the bone marrow and resident cells such as club/Clara cells152 and alveolar macrophages153. Factors produced by resident cells in metastatic niches can enhance the survival and outgrowth of disseminated tumour cells. To gain insights into specific components from mesothelioma-derived exosomes that may reprogram the lung microenvironment and promote metastatic formation, we compared our data with several key studies investigating factors associated with the metastatic niche, including the lung30,150,151,154,155,156,157,158,159. Several mediators of tissue invasion, intravasation and metastasis were identified including growth factor receptors, oncoproteins, proteases, and chemoattractants (Table 3). Macrophage migration inhibitory factor (MIF) was expressed in all mesothelioma-derived exosomes, and interestingly has been reported critical in pancreatic exosomes in regulating liver pre-metastatic niche formation and metastasis30. Further, compared with patients whose pancreatic tumours did not progress, MIF expression in exosomes was markedly higher from stage I pancreatic cancer patients who later developed liver metastasis30. Recent studies indicate that exosomes contain fibronectin on their external surface which facilitate interaction with target cells through heparin sulfate160. As the accumulation of fibronectin (FN1) in the metastatic niche is one of the earliest stages of metastasis formation, exosomes are now considered an early and fundamental driver of microenvironment reprogramming. In fact, in this study we observed FN1 in mEXOS and for MM models JU77, LO68, and OLD1612 (Table S4). Interestingly, we note the significant abundance of peptides identified for this protein between these models, indicating the significant expression of FN1 cargo in these exosomes.
Table 3. Metastatic-niche-associated proteins in mesothelioma exosomes.
| Gene Namea | Protein Descriptiona | SpC Combined (4 cell lines)b | Reported in VesiclePediac |
|---|---|---|---|
| ADAM10 | Disintegrin and metalloproteinase domain-containing protein 10 | 20 | Y |
| ADAMTS1 | A disintegrin and metalloproteinase with thrombospondin motifs 1 | 15 | Y |
| ANXA1 | Annexin A1 (Annexin I) | 268 | Y |
| ANXA11 | Annexin A11 (56 kDa autoantigen) | 102 | Y |
| ARHGDIA | Rho GDP-dissociation inhibitor 1 | 29 | Y |
| CD44 | CD44 antigen | 97 | Y |
| GRB2 | Growth factor receptor-bound protein 2 | 6 | Y |
| ITGA2 | Integrin alpha-2 (CD49 antigen-like family member B) | 53 | Y |
| ITGB4 | Integrin beta | 64 | Y |
| MET | Hepatocyte growth factor receptor (HGF receptor) | 21 | Y |
| MIF | Macrophage migration inhibitory factor | 18 | Y |
| MMP2 | 72 kDa type IV collagenase | 12 | Y |
| RPLP2 | 60S acidic ribosomal protein P2 (Renal carcinoma antigen NY-REN-44) | 39 | Y |
| S100A10 | S100 calcium binding protein A10 | 42 | Y |
| S100A11 | Protein S100-A11 (Calgizzarin) | 41 | Y |
| S100A6 | Protein S100-A6 | 18 | Y |
| SRC | Proto-oncogene tyrosine-protein kinase Src | 52 | Y |
| STUB1 | E3 ubiquitin-protein ligase CHIP | 9 | Y |
| TNC | TNC variant protein | 16 | Y |
aProtein accession and description derived from Gene Ontology (UniProt).
bCombined spectral counts (SpC) observed for mesothelioma exosome datasets. Contribution of individual SpC for each cell lines reported in Supplementary Dataset.
Tenascin C (TNC) is associated with extracellular matrix remodelling and deposition and formation of the metastatic niche. TNC has recently been shown from primary breast cancer cells to colonize lung metastatic niche formation155,161. TNC is associated with development and progression of pulmonary micrometastases161, and suggested to promote tumour cell dissemination and survival during metastasis by growth factor binding, and interactions with extracellular matrix components including fibronectin, proteoglycans, fibrinogen, matrix metalloproteinases, and cell surface receptors, EGFR and integrins162. Melanoma-derived exosomes have been demonstrated, through a MET signalling-dependent pathway, to promote the metastatic process in vivo32. Upon activation, various cytoplasmic effector molecules including growth factor receptor-bound protein 2 (GRB2), and SRC are further recruited to the MET receptor163. In this study, we report the identification of MET, GRB2 and SRC in mesothelioma-derived exosomes. Previously, in comparison of human metastatic and primary colorectal cancer cell-derived exosomes, we reported the identification and significant up-regulation of MET, GRB2, and SRC49. The selective enrichment of metastatic factors in MM-derived exosomes contributes to our understanding of the crosstalk between tumour and stromal cells in the tumour microenvironment in the context of local disease progression. Understanding the functional role of the secretome15 – specifically exosome components - in this lung-specific environment remains to be further investigated in the context of MM.
In addition, we report several proteases implicated in the metastatic niche, including matrix metalloproteinases MMP-2, MMP-14, a disintegrin and metalloproteinase 10 (ADAM10), and ADAM with thrombospondin motif 1 (ADAMTS1). Secretion of matrix-degrading enzymes, including MMP-2 and -14 play an essential role in oncogenic cell transformation and subsequent tumour migration/invasion, serving to mediate the breakdown of basement membrane barriers164. Primary tumour-derived exosomes have also been demonstrated to mediate changes in expression of MMP-2, and MMP-9 in the metastatic niche149,159. Further, the chemoattractant S100 calcium binding proteins S100A6/A10/A11 were identified in all mesothelioma cell-derived exosomes. S100 proteins are commonly up-regulated in tumours and typically associated with tumour progression, including metastasis165. Of interest, S100A10 has been attributed to tumour growth and invasion, and a key component in metastatic evolution through recruitment of tumour-associated macrophages to the tumour microenvironment, mediating inflammation, angiogenesis, suppressing antitumor immunity, and matrix remodelling166. Further, S100A11 is associated with tumour lymph node metastasis in metastatic non-small cell lung cancer and metastatic hepatocellular carcinoma167, and pancreatic adenocarcinoma165. Recently, secreted S100A11 in normal human keratinocytes was shown to promote cell proliferation, survival, and invasion via activation of EGF family members including EGF168. Collectively, although it is evident that specific growth factor receptors, oncoproteins, proteases, and chemoattractants are key regulators of the lung and other sites of tumour metastasis, changes in the lung tumour microenvironment to facilitate local progression of MM by EVs and other secreted networks17 remains to be investigated in the context of MM and its progression.
Summary
Our findings highlight that exosomes derived from MM tumour cells contain select cargo proteins known to be associated with angiogenesis, cell migration, metastasis, and immunoregulation. Using quantitative proteomics, pathway and network analyses, EV database resources, and bioinformatics analyses, we report a mesothelioma-enriched exosome protein cancer signature (mEXOS), associated with tumour antigens and various cancer-specific signalling (HPGD, ENO1, EDIL3, OSMR) and secreted modulators (FN1, ITLN1, MAMDC2, PDGFD, GBP1). To our knowledge, this is the first demonstration of selective enrichment in exosomes of immunomodulatory components (T- and B-cell immune responses and MHC I/II-peptide antigen processing and presentation), signal transduction molecules (ALCAM, HSP90AA1, LGALS1, TNIK), and metastatic factors (MET, MIF, S100A10, S100A11, TNC, MMP2, ADAM10, ADAMTS1) in different human mesothelioma models. We also demonstrate the functional importance of tumour exosomes, revealing for the first time, that MM-derived exosomes from distinct human models, stimulate fibroblast and endothelial cell migration, and promote endothelial cell angiogenesis.
Presently, there appear to be more questions than answers in terms of what mechanisms, functions, and role of these distinct exosomes associated with MM progression in various cell types, including stromal and tumour cells, in addition to immune cells. Chronic pulmonary inflammation has long been a hallmark of asbestos deposition and is thought to contribute to asbestos-related carcinogenesis. Measures of inflammation such as high neutrophil/lymphocyte ratio have been attributed with angiogenesis, cellular proliferation and prognosis in MM patients169. Taken together, comprehensive quantitative proteomic analysis of exosomes secreted by MM tumour cells has revealed a large number of candidate and clinically-relevant extracellular molecules in the regulation of tumour progression, migration, cell transformation, metastasis and in immunoregulation. This information could represent potential specific diagnostic targets for factors of MM origin. The biological significance of our findings are highlighted by the oncogenic effect of exosomes on directly influencing cells in the tumour microenvironment (fibroblasts/endothelial cells). Further functional insights and clinical correlation of these enriched exosomal cargo components in biofluids will extend our understanding of the development of mesothelioma, and mechanisms of how the extracellular environment can contribute to and regulate its progression. Tumour-derived exosomes and their cargo therefore represent exciting and potentially early targets for circulating markers of MM (including tumour-derived antigens), and in the design of targeted immunomodulatory therapeutics.
Methods
Cell culture
Human mesothelioma JO38, JU77, OLD1612, and LO68 cells were established from different patients presenting with malignant pleural mesothelioma from the National Centre for Asbestos Related Diseases (NCARD) as described170. Cells were cultured and maintained in RPMI 1640 (Invitrogen-GIBCO, Carlsbad, USA) supplemented with 2 mM L-Glutamine, 25 mM HEPES, and 5% foetal calf serum (FCS) (Invitrogen-GIBCO), at 37 °C with 10% CO2. Mouse embryonic fibroblasts (MEFs) and human endothelial HUVECs were obtained from American Type Culture Collection (Manassas, VA, USA). MEFs and HUVECs were cultured and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% FCS at 37 °C with 10% CO2.
Exosome isolation
JO38, JU77, LO68, and OLD1612 cells (150-mm culture dish, total of 90 dishes, ~8 × 108) were cultured to 70% confluence in RPMI-1640 + 5% FCS, washed three times with RPMI 1640 supplemented with 0.5% insulin-transferrin-selenium (ITS), and 100 μg/mL streptomycin, and cultured in this medium for 24 hr. These serum-free cell culture conditions were initially optimized with assessment for cell morphology and viability. Cell viability of MM cells were measured using the Trypan blue assay following 24 hr culture in RPMI 1640 containing 5% FCS. Viability was expressed as percentage of viable cells from total cells and presented as mean ± SEM. Conditioned media (CM) was collected (~900 mL) and centrifuged (500× g for 5 min, 2000× g for 10 min) as described49,171. Supernatants were centrifuged at 10,000× g for 30 min at 4 °C, and the resulting supernatants at 100,000× g for 1 hr to isolate exosomes50,56,57,171. Exosomes were washed in PBS, and ultracentrifuged at 100,000× g for 1 hr, as previously described172. Pellets were resuspended in 50 μl of PBS for downstream analysis56.
Protein quantification and immunoblotting
Protein content was estimated by 1D-SDS-PAGE/SYPRO® Ruby protein staining densitometry, as previously described173. For immunoblotting (10 μg), membranes were probed with primary antibodies [mouse anti-TSG101 (BD Transduction Laboratories; 1:500), mouse anti-Alix (Cell Signaling Technology; 1:1000), rabbit anti-CD70 (Abcam; 1:1000), mouse anti-HSPG1 (Abcam; 1:200), rabbit anti-MSLN (Cell Signaling Technology; 1:1000), rabbit anti-FAT1 (GeneTex; 1:2000), mouse anti-CD81 (Santa Cruz Biotechnology; 1:1000), rabbit anti-CALR (Abcam; 1:1000) for 3 hr at room temperature (RT) in 50 mM Tris, 150 mM NaCl, 0.05% (v/v) Tween 20 (TTBS) followed by incubation with either IRDye 800 goat anti-mouse or goat IgG or IRDye 680 goat anti-rabbit IgG (1:15000, LI-COR Biosciences) for 1 hr at RT in TTBS. Immunoblots were visualized using the Odyssey Infrared Imaging System and Image Studio™ Software (v3.0, LI-COR Biosciences, Nebraska USA).
Transmission electron microscopy
Exosome samples (2 μg in 10 μl PBS) were applied to 400 mesh carbon-coated copper grids and negatively stained with 10 μL of a 2% uranyl acetate solution for 10 min (ProSciTech, Queensland, Australia). Grids were air dried and viewed using a JEOL JEM-2010 transmission electron microscope operated at 80 kV as previously described173.
Proteomic analysis
Proteomic experiments were performed in biological duplicate (n = 2) as previously described, with MIAPE-compliance171,174. Briefly, exosomes from each cell line (10 μg) were lysed in SDS sample buffer (4% (w/v) SDS, 20% (v/v) glycerol, 0.01% (v/v) bromophenol blue, 0.125 M Tris-HCl, pH 6.8), and proteins separated by short-range SDS-PAGE (10 × 6 mm), and visualized by Imperial Protein Stain (Invitrogen). Individual samples were excised into equal fractions (n = 2), destained (50 mM ammonium bicarbonate/acetonitrile), reduced (10 mM DTT (Calbiochem) for 30 min), alkylated (50 mM iodoacetic acid (Fluka) for 30 min) and trypsinized (0.2 μg trypsin (Promega Sequencing Grade) for 16 hr at 37 °C). A nanoflow UPLC instrument (Ultimate 3000 RSLCnano, Thermo Fisher Scientific) was coupled on-line to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) with a nanoelectrospray ion source (Thermo Fisher Scientific). Peptides were loaded (Acclaim PepMap100, 5 mm × 300 μm i.d., μ-Precolumn packed with 5 μm C18 beads, Thermo Fisher Scientific) and separated (Acquity UPLC M-Class Peptide BEH130, C18, 1.7 μm, 75 μm × 250 mm, Waters). Data was acquired using Xcalibur software v2.1 (Thermo Fisher Scientific). Details of the operation of the mass spectrometer are described previously174.
Database searching and protein identification
Raw data were processed using Proteome Discoverer (v2.1, Thermo Fisher Scientific). MS2 spectra were searched with Mascot (v2.4, Matrix Science), and Sequest HT (v2.1, Thermo Fisher Scientific) against a database of 133,798 ORFs (UniProtHuman, Apr 2016). Peptide lists were generated from a tryptic digestion with up to two missed cleavages, carbamidomethylation of cysteines as fixed modifications, and oxidation of methionines and protein N-terminal acetylation as variable modifications. Precursor mass tolerance was 10 ppm, product ions were searched at 0.06 Da tolerances, minimum peptide length defined at 6, maximum peptide length 144, and max delta CN 0.05. Peptide spectral matches (PSM) were validated using Percolator based on q-values at a 1% false discovery rate (FDR)175,176. With Proteome Discoverer, peptide identifications were grouped into proteins according to the law of parsimony and filtered to 1% FDR177. Scaffold Q+S (v4.5.3, Proteome Software Inc) was employed to validate MS/MS-based peptide and protein identifications from database searching. Initial peptide identifications were accepted if they could be established at greater than 95% probability (PEP 5%) as specified by the Peptide Prophet algorithm178. Protein probabilities were assigned by the Protein Prophet algorithm177. Protein identifications were accepted, if they reached greater than 99% probability and contained at least 2 identified unique peptides. These identification criteria typically established <1% false discovery rate based on a decoy database search strategy at the protein level. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone, were grouped to satisfy the principles of parsimony. Contaminants, and reverse identification were excluded from further data analysis. UniProt was used for protein annotation. Raw mass spectrometry data is deposited in the PeptideAtlas and can be accessed at http://www.peptideatlas.org/PASS/PASS00812.
Label-free spectral counting
Significant spectral count (SpC) and fold change ratios (Rsc) were determined as previously described49,50,56,171,172,173. The relative abundance of a protein within a sample was estimated using normalized SpC, where for each individual protein, significant peptide MS/MS spectra (i.e., ion score greater than identity score) were summated, and normalized by the total number of significant MS/MS spectra identified in the sample. For each protein the Fisher’s exact test was applied to significant assigned spectra. The resulting p-values were corrected for multiple testing using the Benjamini-Hochberg procedure179 and statistics performed as previously described50. For pathway analyses, KEGG (http://www.genome.jp/kegg/pathway.html) and DAVID (http://david.abcc.ncifcrf.gov/) resources were utilised. Clustering of samples was performed by principal component analysis (PCA) and visualised using ggplot2180 and ggfortify (https://cran.r-project.org/web/packages/ggfortify/index.html). The heat map of proteins was performed using gplots (https://cran.r-project.org/web/packages/gplots/index.html).
Cell migration assay
Cell transwell migration assay was performed as described181. Briefly, MEF, neoHFF, and HUVEC cells (5 × 104) in suspension were pelleted at 500× g for 5 min, and resuspended in 100 μL of DMEM. Cells were overlaid onto Transwell® polycarbonate membrane cell culture inserts (8.0 μm pore size, Corning), and inserts placed into 24-well companion plates. The bottom chamber contained DMEM (0% FCS) and was supplemented with vehicle control (DMEM), and volume control (DMEM), or MM-derived exosomes (30 μg/μL). Cell migration through the transwell was performed at 37 °C for 24 hr. Inserts were removed, and cells fixed (4% (v/v) formaldehyde, 10 min) and nuclei stained with DAPI. Non-migrating cells were removed from the upper side of the inserts using cotton swabs. Migrating cells were imaged using an inverted Nikon Eclipse TE300 microscope equipped with an attached 12.6 mp digital camera (Nikon DXM1200C) (n = 3; average ± SEM, *p < 0.05, **p < 0.01).
Endothelial cell tube formation assay
Endothelial cell tube formation assays were performed as previously described174. Briefly, HUVECs (7 × 104 cells/well) were re-suspended in DMEM + 5% FBS, and seeded onto growth factor-reduced BD matrigel (1 mg/mL) for 1 hr. Cells were supplemented for 2 hr with vehicle control (DMEM), volume control (DMEM), or MM-derived exosomes (30 μg/μL). Tube formation was analysed (24 hr culture at 37 °C) and imaged using inverted Nikon Eclipse TE300 microscope equipped with an attached 12.6 mp digital camera (Nikon (n = 3; average ± SEM, *p < 0.05)).
Experimental design and statistical rationale
All methods were carried out in accordance with the approved guidelines of La Trobe Institute for Molecular Science. Functional cell assays were conducted by a minimum of 3 independent biological experiments. For all assays, statistical analysis was performed using Student’s t-tests using GraphPad Prism (v5.0), with *p < 0.05 and **p < 0.01 considered statistically significant. Mass spectrometry analysis of exosome proteins was performed in biological replicates, and only proteins identified in both biological replicates used for label-free quantification. Statistical testing of proteomic data was performed using a Poisson distribution with EdgeR software (v3.2), with *p < 0.05 considered statistically significant. Furthermore, selected proteomic findings were validated using orthogonal approaches including western immuno-blotting performed in biological replicates.
Additional Information
How to cite this article: Greening, D. W. et al. Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo. Sci. Rep. 6, 32643; doi: 10.1038/srep32643 (2016).
Supplementary Material
Acknowledgments
Authors are supported, in part, by the National Health and Medical Research Council of Australia program grant APP487922 (R.J.S.), National Health and Medical Research Council Centres of Research Excellence 628727 (R.J.S., B.W.S.R., J.C.). D.W.G. is supported by the LIMS Molecular Biology Stone Fellowship, La Trobe University Research Focus Area Leadership Grant, and La Trobe University Start-up Fund. M.C. is supported by a La Trobe University Postgraduate Scholarship. Proteomics in this study was supported using the Mass Spectrometry and Proteomics facility, La Trobe University.
Footnotes
Author Contributions D.W.G. and R.J.S. conceived and designed the experiments performed by D.W.G. and H.J., D.W.G., M.C., B.W.S.R., I.M.D., J.C. and R.J.S. were involved in data interpretation, and all authors provided input to D.W.G. who wrote the manuscript.
References
- Roe O. D. & Stella G. M. Malignant pleural mesothelioma: history, controversy and future of a manmade epidemic. European Respiratory Review 24, 115–131, 10.1183/09059180.00007014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. Advances in the diagnosis, treatment and prognosis of malignant pleural mesothelioma. Ann Transl Med 3, 182, 10.3978/j.issn.2305-5839.2015.07.03 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faig J. et al. Changing pattern in malignant mesothelioma survival. Transl Oncol 8, 35–39, 10.1016/j.tranon.2014.12.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M. et al. Malignant mesothelioma: facts, myths, and hypotheses. J Cell Physiol 227, 44–58, 10.1002/jcp.22724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott F. E. Mesothelioma: a review. Ochsner J 12, 70–9 (2012). [PMC free article] [PubMed] [Google Scholar]
- Robinson B. W. et al. Mesothelin-family proteins and diagnosis of mesothelioma. Lancet 362, 1612–6, 10.1016/S0140-6736(03)14794-0 (2003). [DOI] [PubMed] [Google Scholar]
- Pass H. I. et al. Soluble mesothelin-related peptide level elevation in mesothelioma serum and pleural effusions. The Annals of thoracic surgery 85, 265–72; discussion 272, 10.1016/j.athoracsur.2007.07.042 (2008). [DOI] [PubMed] [Google Scholar]
- Creaney J. & Robinson B. W. Serum and pleural fluid biomarkers for mesothelioma. Current opinion in pulmonary medicine 15, 366–370, 10.1097/MCP.0b013e32832b98eb (2009). [DOI] [PubMed] [Google Scholar]
- Hollevoet K. et al. Serum mesothelin for diagnosing malignant pleural mesothelioma: an individual patient data meta-analysis. Journal of clinical oncology 30, 1541–1549, 10.1200/JCO.2011.39.6671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos I., Boura P., Xanthos T. & Syrigos K. Effectiveness of mesothelin family proteins and osteopontin for malignant mesothelioma. The European respiratory journal 41, 706–715, 10.1183/09031936.00226111 (2013). [DOI] [PubMed] [Google Scholar]
- Posadas E. M. et al. Proteomic analysis for the early detection and rational treatment of cancer–realistic hope? Annals of oncology 16, 16–22, 10.1093/annonc/mdi004 (2005). [DOI] [PubMed] [Google Scholar]
- Cerciello F. et al. Identification of a seven glycopeptide signature for malignant pleural mesothelioma in human serum by selected reaction monitoring. Clinical proteomics 10, 16, 10.1186/1559-0275-10-16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff R. M. et al. Early detection of malignant pleural mesothelioma in asbestos-exposed individuals with a noninvasive proteomics-based surveillance tool. PloS one 7, e46091, 10.1371/journal.pone.0046091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L. et al. Association of MiR-126 with soluble mesothelin-related peptides, a marker for malignant mesothelioma. PloS one 6, e18232, 10.1371/journal.pone.0018232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi M. et al. The secretome signature of malignant mesothelioma cell lines. J Proteomics, 10.1016/j.jprot.2016.02.021 (2016). [DOI] [PubMed] [Google Scholar]
- van der Bij S. et al. Markers for the non-invasive diagnosis of mesothelioma: a systematic review. Br J Cancer 104, 1325–1333, 10.1038/bjc.2011.104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Pleural mesothelioma instigates tumor-associated fibroblasts to promote progression via a malignant cytokine network. Am J Pathol 179, 1483–1493, 10.1016/j.ajpath.2011.05.060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill G. D., Mangum J. B., Moss O. R. & Everitt J. I. Soluble ICAM-1, MCP-1, and MIP-2 protein secretion by rat pleural mesothelial cells following exposure to amosite asbestos. Exp Lung Res 29, 277–290 (2003). [DOI] [PubMed] [Google Scholar]
- Hillegass J. M. et al. Utilization of gene profiling and proteomics to determine mineral pathogenicity in a human mesothelial cell line (LP9/TERT-1). J Toxicol Environ Health A 73, 423–436, 10.1080/15287390903486568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A. et al. Alterations in gene expression in human mesothelial cells correlate with mineral pathogenicity. Am J Respir Cell Mol Biol 41, 114–123, 10.1165/rcmb.2008-0146OC (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre F. et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360, 295–305, 10.1016/S0140-6736(02)09552-1 (2002). [DOI] [PubMed] [Google Scholar]
- Bard M. P. et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. American journal of respiratory cell and molecular biology 31, 114–121, 10.1165/rcmb.2003-0238OC (2004). [DOI] [PubMed] [Google Scholar]
- Wolfers J. et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nature Med. 7, 297–303, 10.1038/85438 (2001). [DOI] [PubMed] [Google Scholar]
- Robbins P. D. & Morelli A. E. Regulation of immune responses by extracellular vesicles. Nature reviews. Immunology 14, 195–208, 10.1038/nri3622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening D. W. et al. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol 40, 72–81, 10.1016/j.semcdb.2015.02.009 (2015). [DOI] [PubMed] [Google Scholar]
- Xu R. et al. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest 126, 1152–62, 10.1172/JCI81129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz M. et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol 11, 688–701, 10.1038/nrurol.2014.301 (2014). [DOI] [PubMed] [Google Scholar]
- Greening D. W. et al. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol 40, 60–71, 10.1016/j.semcdb.2015.02.008 (2015). [DOI] [PubMed] [Google Scholar]
- Nawaz M. et al. Extracellular vesicles in ovarian cancer: applications to tumor biology, immunotherapy and biomarker discovery. Expert Rev Proteomics 13, 395–409, 10.1586/14789450.2016.1165613 (2016). [DOI] [PubMed] [Google Scholar]
- Costa-Silva B. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 17, 816–826, 10.1038/ncb3169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A. et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335, 10.1038/nature15756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Med. 18, 883–891, 10.1038/nm.2753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luga V. et al. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 151, 1542–1556, 10.1016/j.cell.2012.11.024 (2012). [DOI] [PubMed] [Google Scholar]
- Vaksman O., Trope C., Davidson B. & Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 35, 2113–2120, 10.1093/carcin/bgu130 (2014). [DOI] [PubMed] [Google Scholar]
- Webber J. P. et al. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene, 10.1038/onc.2013.560 (2014). [DOI] [PubMed] [Google Scholar]
- Ono M. et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Science signaling 7, ra63, 10.1126/scisignal.2005231 (2014). [DOI] [PubMed] [Google Scholar]
- Melo S. et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 26, 707–721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atay S. et al. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proceedings of the National Academy of Sciences of the United States of America 111, 711–716, 10.1073/pnas.1310501111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S. et al. Dendritic cell-derived exosomes for cancer immunotherapy: what’s next? Cancer Res 70, 1281–1285, 10.1158/0008-5472.CAN-09-3276 (2010). [DOI] [PubMed] [Google Scholar]
- Gehrmann U. et al. Synergistic induction of adaptive antitumor immunity by codelivery of antigen with alpha-galactosylceramide on exosomes. Cancer Res 73, 3865–3876, 10.1158/0008-5472.CAN-12-3918 (2013). [DOI] [PubMed] [Google Scholar]
- Mignot G. et al. Prospects for exosomes in immunotherapy of cancer. J Cell Mol Med 10, 376–88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak J. Extracellular vesicles - biomarkers and effectors of the cellular interactome in cancer. Front Pharmacol 4, 21, 10.3389/fphar.2013.00021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzas E. I. et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol 10, 356–364, 10.1038/nrrheum.2014.19 (2014). [DOI] [PubMed] [Google Scholar]
- Gyorgy B., Hung M. E., Breakefield X. O. & Leonard J. N. Therapeutic Applications of Extracellular Vesicles: Clinical Promise and Open Questions. Annu Rev Pharmacol Toxicol, 10.1146/annurev-pharmtox-010814-124630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader P., Breakefield X. O. & Wood M. J. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol Med 20, 385–393, 10.1016/j.molmed.2014.03.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaloussi S. E. L., Mager I., Breakefield X. O. & Wood M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews. Drug discovery 12, 347–357, 10.1038/nrd3978 (2013). [DOI] [PubMed] [Google Scholar]
- Fais S. et al. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano 10, 3886–3899, 10.1021/acsnano.5b08015 (2016). [DOI] [PubMed] [Google Scholar]
- Lasser C. Exosomes in diagnostic and therapeutic applications: biomarker, vaccine and RNA interference delivery vehicle. Expert Opin Biol Ther 15, 103–117, 10.1517/14712598.2015.977250 (2015). [DOI] [PubMed] [Google Scholar]
- Ji H. et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 13, 1672–1686, 10.1002/pmic.201200562 (2013). [DOI] [PubMed] [Google Scholar]
- Tauro B. J. et al. Oncogenic H-ras reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol Cell Proteomics 12, 2148–2159, 10.1074/mcp.M112.027086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servais E. L. et al. Mesothelin overexpression promotes mesothelioma cell invasion and MMP-9 secretion in an orthotopic mouse model and in epithelioid pleural mesothelioma patients. Clin Cancer Res 18, 2478–2489, 10.1158/1078-0432.CCR-11-2614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras K. & Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res 316, 1324–1331, 10.1016/j.yexcr.2010.02.045 (2010). [DOI] [PubMed] [Google Scholar]
- Edwards J. G. et al. Angiogenesis is an independent prognostic factor in malignant mesothelioma. Br J Cancer 85, 863–868, 10.1054/bjoc.2001.1997 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Singh S. et al. Angiogenic cytokines in mesothelioma: a study of VEGF, FGF-1 and -2, and TGF beta expression. J Pathol 189, 72–78, 10.1002/(SICI)1096-9896(199909)189:1 (1999). [DOI] [PubMed] [Google Scholar]
- Harvey P. et al. Immunoreactivity for hepatocyte growth factor/scatter factor and its receptor, met, in human lung carcinomas and malignant mesotheliomas. J Pathol 180, 389–394, (1996). [DOI] [PubMed] [Google Scholar]
- Greening D. W. et al. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol 1295, 179–209, 10.1007/978-1-4939-2550-6_15 (2015). [DOI] [PubMed] [Google Scholar]
- Xu R. et al. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11–25, 10.1016/j.ymeth.2015.04.008 (2015). [DOI] [PubMed] [Google Scholar]
- Simpson R. J., Lim J. W., Moritz R. L. & Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics 6, 267–283, 10.1586/epr.09.17 (2009). [DOI] [PubMed] [Google Scholar]
- Fukuda M. Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol Life Sci 65, 2801–2813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. T., Mishra A. & Lambright D. G. Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 10, 1377–1389, 10.1111/j.1600-0854.2009.00942.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature reviews. Molecular cell biology 10, 513–525 (2009). [DOI] [PubMed] [Google Scholar]
- Kalra H. et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS biology 10, e1001450, 10.1371/journal.pbio.1001450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegmans J. P. et al. Proteomic analysis of exosomes secreted by human mesothelioma cells. The American journal of pathology 164, 1807–1815, 10.1016/S0002-9440(10)63739-X (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A. et al. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer research 67, 7458–7466, 10.1158/0008-5472.CAN-06-3456 (2007). [DOI] [PubMed] [Google Scholar]
- Clayton A. et al. Human tumor-derived exosomes down-modulate NKG2D expression. Journal of immunology 180, 7249–7258 (2008). [DOI] [PubMed] [Google Scholar]
- Chen H., Herndon M. E. & Lawler J. The cell biology of thrombospondin-1. Matrix Biol 19, 597–614 (2000). [DOI] [PubMed] [Google Scholar]
- Daniels T. R. et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta 1820, 291–317, 10.1016/j.bbagen.2011.07.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S. & George S. P. Regulation of cell structure and function by actin-binding proteins: villin’s perspective. FEBS Lett 582, 2128–39, 10.1016/j.febslet.2008.02.040 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallero M. A. et al. Thrombospondin 1 binding to calreticulin-LRP1 signals resistance to anoikis. FASEB J 22, 3968–3979, 10.1096/fj.07-104802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinale F. G. Cell-matrix signaling and thrombospondin: another link to myocardial matrix remodeling. Circ Res 95, 446–448, 10.1161/01.RES.0000142315.88477.42 (2004). [DOI] [PubMed] [Google Scholar]
- Ensslin M. A. & Shur B. D. Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell 114, 405–417 (2003). [DOI] [PubMed] [Google Scholar]
- Bu H. F. et al. Milk fat globule-EGF factor 8/lactadherin plays a crucial role in maintenance and repair of murine intestinal epithelium. J Clin Invest 117, 3673–3683, 10.1172/JCI31841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestre J. S. et al. Lactadherin promotes VEGF-dependent neovascularization. Nat Med 11, 499–506, 10.1038/nm1233 (2005). [DOI] [PubMed] [Google Scholar]
- Jinushi M. et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci USA 108, 12425–12430, 10.1073/pnas.1106645108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A. L., Kavallaris M. & McCarroll J. A. Microtubules and their role in cellular stress in cancer. Front Oncol 4, 153, 10.3389/fonc.2014.00153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani M. et al. Class III beta-tubulin (TUBB3): more than a biomarker in solid tumors? Curr Mol Med 11, 726–731 (2011). [DOI] [PubMed] [Google Scholar]
- Zimling Z. G. et al. A biomarker profile for predicting efficacy of cisplatin-vinorelbine therapy in malignant pleural mesothelioma. Cancer Chemother Pharmacol 70, 743–754, 10.1007/s00280-012-1965-0 (2012). [DOI] [PubMed] [Google Scholar]
- Singhal S. et al. Gene expression profiling of malignant mesothelioma. Clin Cancer Res 9, 3080–97 (2003). [PubMed] [Google Scholar]
- Fontana S., Saieva L., Taverna S. & Alessandro R. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: state of the art and new perspectives. Proteomics 13, 1581–1594, 10.1002/pmic.201200398 (2013). [DOI] [PubMed] [Google Scholar]
- Ji H. et al. Progress in the biological function of alpha-enolase Animal Nutrition 2, 12–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capello M., Ferri-Borgogno S., Cappello P. & Novelli F. A-enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 278(27), 1064–74 (2011). [DOI] [PubMed] [Google Scholar]
- Lim L. H. & Pervaiz S. Annexin 1: the new face of an old molecule. FASEB J. 21(4), 968–975 (2007). [DOI] [PubMed] [Google Scholar]
- Rihn B. H. et al. Differential gene expression in mesothelioma. FEBS Lett 480, 95–100 (2000). [DOI] [PubMed] [Google Scholar]
- Mathupala S. P., Heese C. & Pedersen P. L. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem 272, 22776–22780 (1997). [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang Z., Zhu Y. & Qin S. Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer Agents Med Chem 14, 280–289 (2014). [DOI] [PubMed] [Google Scholar]
- Kuo W., Lin J. & Tang T. K., Human glucose-6-phosphate dehydrogenase (G6PD) gene transforms NIH 3T3 cells and induces tumors in nude mice. Int J Cancer 85, 857–864 (2000). [DOI] [PubMed] [Google Scholar]
- Suraokar M. B. et al. Expression profiling stratifies mesothelioma tumors and signifies deregulation of spindle checkpoint pathway and microtubule network with therapeutic implications. Ann Oncol 25, 1184–1192, 10.1093/annonc/mdu127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispi S. et al. Global gene expression profiling of human pleural mesotheliomas: identification of matrix metalloproteinase 14 (MMP-14) as potential tumour target. PLoS One 4, e7016, 10.1371/journal.pone.0007016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H. et al. Role of annexin A6 in cancer. Oncol Lett 10, 1947–1952, 10.3892/ol.2015.3498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasarre P., Potiron V., Drabkin H. & Roche J. Guidance molecules in lung cancer. Cell Adh Migr 4, 130–145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L. et al. Abnormal lymphatic vessel development in neuropilin 2 mutant mice. Development 129, 4797–806 (2002). [DOI] [PubMed] [Google Scholar]
- Nasarre P., Gemmill R. M. & Drabkin H. A. The emerging role of class-3 semaphorins and their neuropilin receptors in oncology. Onco Targets Ther 7, 1663–87, 10.2147/OTT.S37744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. G. et al. Cyclooxygenase-2 expression is a novel prognostic factor in malignant mesothelioma. Clin Cancer Res 8, 1857–62 (2002). [PubMed] [Google Scholar]
- Baldi A. et al. Prognostic significance of cyclooxygenase-2 (COX-2) and expression of cell cycle inhibitors p21 and p27 in human pleural malignant mesothelioma. Thorax 59, 428–433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wang B., Long H. & Wen F. Diagnostic accuracy of calretinin for malignant mesothelioma in serous effusions: a meta-analysis. Sci Rep 5, 9507, 10.1038/srep09507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez N. G. Immunohistochemical diagnosis of epithelioid mesothelioma: an update. Arch Pathol Lab Med 129, 1407–14, 10.1043/1543-2165(2005)129[1407:IDOEMA]2.0.CO;2 (2005). [DOI] [PubMed] [Google Scholar]
- Kushitani K. et al. Immunohistochemical marker panels for distinguishing between epithelioid mesothelioma and lung adenocarcinoma. Pathol Int 57, 190–199, 10.1111/j.1440-1827.2007.02080.x (2007). [DOI] [PubMed] [Google Scholar]
- Raiko I. et al. Development of an enzyme-linked immunosorbent assay for the detection of human calretinin in plasma and serum of mesothelioma patients. BMC Cancer 10, 242, 10.1186/1471-2407-10-242 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. O. et al. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics 13, 2125–2134, 10.1002/pmic.201200323 (2013). [DOI] [PubMed] [Google Scholar]
- Tomasetti M. & Santarelli L. Biomarkers for early detection of malignant mesothelioma: diagnostic and therapeutic application. Cancers (Basel) 2, 523–548, 10.3390/cancers2020523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton J. L. et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics 9, 1324–1338, 10.1074/mcp.M000063-MCP201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Mizuguchi G., Hamiche A. & Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544, 10.1038/35020123 (2000). [DOI] [PubMed] [Google Scholar]
- Ikura T. et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000). [DOI] [PubMed] [Google Scholar]
- Jha S. & Dutta A. RVB1/RVB2: running rings around molecular biology. Mol Cell 34, 521–533, 10.1016/j.molcel.2009.05.016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski B. et al. Levels of 8-hydroxy-2′-deoxyguanosine in DNA of white blood cells from workers highly exposed to asbestos in Germany. Mutat Res 468, 195–202 (2000). [DOI] [PubMed] [Google Scholar]
- Arzt L. et al. Signal transducer and activator of transcription 1 (STAT1) acts like an oncogene in malignant pleural mesothelioma. Virchows Arch 465, 79–88, 10.1007/s00428-014-1584-8 (2014). [DOI] [PubMed] [Google Scholar]
- Kothmaier H. et al. EGFR and PDGFR differentially promote growth in malignant epithelioid mesothelioma of short and long term survivors. Thorax 63, 345–351, 10.1136/thx.2007.085241 (2008). [DOI] [PubMed] [Google Scholar]
- Winder D. M. et al. Overexpression of the oncostatin M receptor in cervical squamous cell carcinoma cells is associated with a pro-angiogenic phenotype and increased cell motility and invasiveness. J Pathol 225, 448–462, 10.1002/path.2968 (2011). [DOI] [PubMed] [Google Scholar]
- Ak G. et al. MicroRNA and mRNA features of malignant pleural mesothelioma and benign asbestos-related pleural effusion. Biomed Res Int 2015, 635748, 10.1155/2015/635748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarel M. M. & Coleman N. Oncostatin M receptor is a novel therapeutic target in cervical squamous cell carcinoma. J Pathol 232, 386–390, 10.1002/path.4305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W. et al. Multidrug resistance markers P-glycoprotein, multidrug resistance protein 1, and lung resistance protein in non-small cell lung cancer: prognostic implications. J Cancer Res Clin Oncol 131, 355–363, 10.1007/s00432-004-0653-9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunicka T. & Soucek P. Importance of ABCC1 for cancer therapy and prognosis. Drug Metab Rev 46, 325–342, 10.3109/03602532.2014.901348 (2014). [DOI] [PubMed] [Google Scholar]
- Lens S. M. et al. Aberrant expression and reverse signalling of CD70 on malignant B cells. British journal of haematology 106, 491–503 (1999). [DOI] [PubMed] [Google Scholar]
- Baba M. et al. Highly enhanced expression of CD70 on human T-lymphotropic virus type 1-carrying T-cell lines and adult T-cell leukemia cells. Journal of virology 82, 3843–3852, 10.1128/JVI.02013-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker K. et al. CD70: a new tumor specific biomarker for renal cell carcinoma. The Journal of urology 173, 2150–2153, 10.1097/01.ju.0000158121.49085.ba (2005). [DOI] [PubMed] [Google Scholar]
- Szollosi J. et al. Supramolecular complexes of MHC class I, MHC class II, CD20, and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. Journal of immunology 157, 2939–2946 (1996). [PubMed] [Google Scholar]
- Buschow S. I. et al. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunology and cell biology 88, 851–856, 10.1038/icb.2010.64 (2010). [DOI] [PubMed] [Google Scholar]
- Miguet L. et al. Proteomic analysis of malignant lymphocyte membrane microparticles using double ionization coverage optimization. Proteomics 6, 153–171, 10.1002/pmic.200500133 (2006). [DOI] [PubMed] [Google Scholar]
- Wubbolts R. et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. The Journal of biological chemistry 278, 10963–10972, 10.1074/jbc.M207550200 (2003). [DOI] [PubMed] [Google Scholar]
- Mohr S. et al. Immune Signature of Malignant Pleural Mesothelioma as Assessed by Transcriptome Analysis. Cancer Gen Prot 2, 125–136 (2005). [PubMed] [Google Scholar]
- Cossetti C. et al. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell 56, 193–204, 10.1016/j.molcel.2014.08.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bograd A. J. et al. Immune responses and immunotherapeutic interventions in malignant pleural mesothelioma. Cancer Immunol Immunother 60, 1509–1527, 10.1007/s00262-011-1103-6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. et al. Chronic inflammation in tumor stroma is an independent predictor of prolonged survival in epithelioid malignant pleural mesothelioma patients. Cancer Immunol Immunother 60, 1721–8, 10.1007/s00262-011-1073-8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuschenbach M., von Knebel Doeberitz M. & Wentzensen N. A systematic review of humoral immune responses against tumor antigens. Cancer Immunol Immunother 58, 1535–1544, 10.1007/s00262-009-0733-4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M. et al. Humoral immune response to mesothelin in mesothelioma and ovarian cancer patients. Clin Cancer Res 11, 3814–3820, 10.1158/1078-0432.CCR-04-2304 (2005). [DOI] [PubMed] [Google Scholar]
- Robinson C., Robinson B. W. & Lake R. A. Sera from patients with malignant mesothelioma can contain autoantibodies. Lung Cancer 20, 175–184 (1998). [DOI] [PubMed] [Google Scholar]
- Yasuda M. et al. Tumor-infiltrating B lymphocytes as a potential source of identifying tumor antigen in human lung cancer. Cancer Res 62, 1751–6 (2002). [PubMed] [Google Scholar]
- Mizukami M. et al. Antitumor effect of antibody against a SEREX-defined antigen (UOEH-LC-1) on lung cancer xenotransplanted into severe combined immunodeficiency mice. Cancer Res 67, 8351–8357, 10.1158/0008-5472.CAN-06-3889 (2007). [DOI] [PubMed] [Google Scholar]
- Hassan R., Bera T. & Pastan I. Mesothelin: a new target for immunotherapy. Clin Cancer Res 10, 3937–3942, 10.1158/1078-0432.CCR-03-0801 (2004). [DOI] [PubMed] [Google Scholar]
- Cho J. A. et al. Exosomes: a new delivery system for tumor antigens in cancer immunotherapy. Int J Cancer 114, 613–622, 10.1002/ijc.20757 (2005). [DOI] [PubMed] [Google Scholar]
- Acres B., Beverley P. & Scholl S. Tumor immunology and the Battle of Waterloo. Mol Cancer Ther 1, 651–5 (2002). [PubMed] [Google Scholar]
- Corbiere V. et al. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer Res 71, 1253–1262, 10.1158/0008-5472.CAN-10-2693 (2011). [DOI] [PubMed] [Google Scholar]
- Tseng H. Y. et al. The melanoma-associated antigen MAGE-D2 suppresses TRAIL receptor 2 and protects against TRAIL-induced apoptosis in human melanoma cells. Carcinogenesis 33, 1871–1881, 10.1093/carcin/bgs236 (2012). [DOI] [PubMed] [Google Scholar]
- Yasuda M. et al. Identification of a tumour associated antigen in lung cancer patients with asbestos exposure. Anticancer Res 30, 2631–9 (2010). [PubMed] [Google Scholar]
- Zheng L. & Jaffee E. M. Annexin A2 is a new antigenic target for pancreatic cancer immunotherapy. Oncoimmunology 1, 112–114, 10.4161/onci.1.1.18017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. et al. Discovery of melanotransferrin as a serological marker of colorectal cancer by secretome analysis and quantitative proteomics. J Proteome Res 13, 4919–4931, 10.1021/pr500790f (2014). [DOI] [PubMed] [Google Scholar]
- Fijneman R. J. et al. Proximal fluid proteome profiling of mouse colon tumors reveals biomarkers for early diagnosis of human colorectal cancer. Clin Cancer Res 18, 2613–2624, 10.1158/1078-0432.CCR-11-1937 (2012). [DOI] [PubMed] [Google Scholar]
- Hollingsworth M. A. & Swanson B. J. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4, 45–60, 10.1038/nrc1251 (2004). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nature immunology 14, 793–803, 10.1038/ni.2647 (2013). [DOI] [PubMed] [Google Scholar]
- Fujii M. et al. TGF-beta synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. The Journal of experimental medicine 209, 479–494, 10.1084/jem.20111653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. W. Jr. et al. Suppression of pro-metastasis phenotypes expression in malignant pleural mesothelioma by the PI3K inhibitor LY294002 or the MEK inhibitor UO126. Anticancer research 26, 809–821 (2006). [PubMed] [Google Scholar]
- Aikawa T. et al. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. J Clin Invest 118, 89–99, 10.1172/JCI32412 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsura T. et al. Identification of glypican-3 as a novel tumor marker for melanoma. Clinical cancer research: an official journal of the American Association for Cancer Research 10, 6612–6621, 10.1158/1078-0432.CCR-04-0348 (2004). [DOI] [PubMed] [Google Scholar]
- Su G. et al. Glypican-1 is frequently overexpressed in human gliomas and enhances FGF-2 signaling in glioma cells. The American journal of pathology 168, 2014–2026, 10.2353/ajpath.2006.050800 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga K. et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res 25, 3703–7 (2005). [PubMed] [Google Scholar]
- Qu J. L. et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis 41, 875–880, 10.1016/j.dld.2009.04.006 (2009). [DOI] [PubMed] [Google Scholar]
- Park J. E. et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics 9, 1085–1099, 10.1074/mcp.M900381-MCP200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I. et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res 70, 1668–1678, 10.1158/0008-5472.CAN-09-2470 (2010). [DOI] [PubMed] [Google Scholar]
- Peinado H., Lavotshkin S. & Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Seminars in cancer biology 21, 139–146, 10.1016/j.semcancer.2011.01.002 (2011). [DOI] [PubMed] [Google Scholar]
- Sleeman J. P. The metastatic niche and stromal progression. Cancer metastasis reviews 31, 429–40, 10.1007/s10555-012-9373-9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana D. et al. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1-CX3CR1 axis. Cancer Res. 69, 785–793, 10.1158/0008-5472.CAN-08-1946 (2009). [DOI] [PubMed] [Google Scholar]
- Tomita T., Sakurai Y., Ishibashi S. & Maru Y. Imbalance of Clara cell-mediated homeostatic inflammation is involved in lung metastasis. Oncogene 30, 3429–3439, 10.1038/onc.2011.53 (2011). [DOI] [PubMed] [Google Scholar]
- Vadrevu S. K. et al. Complement c5a receptor facilitates cancer metastasis by altering T-cell responses in the metastatic niche. Cancer Res 74, 3454–3465, 10.1158/0008-5472.CAN-14-0157 (2014). [DOI] [PubMed] [Google Scholar]
- Nguyen D. X., Bos P. D. & Massague J. Metastasis: from dissemination to organ-specific colonization. Nature reviews. Cancer 9, 274–284, 10.1038/nrc2622 (2009). [DOI] [PubMed] [Google Scholar]
- Minn A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524, 10.1038/nature03799 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaila B. & Lyden D. The metastatic niche: adapting the foreign soil. Nature reviews. Cancer 9, 285–293, 10.1038/nrc2621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce J. A. & Pollard J. W. Microenvironmental regulation of metastasis. Nature reviews. Cancer 9, 239–252, 10.1038/nrc2618 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T. et al. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 11, 1093–1105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange C. et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 71, 5346–5356, 10.1158/0008-5472.CAN-11-0241 (2011). [DOI] [PubMed] [Google Scholar]
- Purushothaman A. et al. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J Biol Chem 291, 1652–1663, 10.1074/jbc.M115.686295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T. et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 17, 867–874, 10.1038/nm.2379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei I., Ghajar C. M. & Lyden D. A TeNaCious foundation for the metastatic niche. Cancer cell 20, 139–141, 10.1016/j.ccr.2011.08.004 (2011). [DOI] [PubMed] [Google Scholar]
- Gherardi E., Birchmeier W., Birchmeier C. & Vande Woude G. Targeting MET in cancer: rationale and progress. Nature reviews. Cancer 12, 89–103, 10.1038/nrc3205 (2012). [DOI] [PubMed] [Google Scholar]
- Wels J., Kaplan R. N., Rafii S. & Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev 22, 559–574, 10.1101/gad.1636908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xu C., Jin Q. & Liu Z. S100 protein family in human cancer. Am J Cancer Res 4, 89–115 (2014). [PMC free article] [PubMed] [Google Scholar]
- Phipps K. D., Surette A. P., O’Connell P. A. & Waisman D. M. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer research 71, 6676–6683, 10.1158/0008-5472.CAN-11-1748 (2011). [DOI] [PubMed] [Google Scholar]
- Tian T. et al. Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer science 98, 1265–1274, 10.1111/j.1349-7006.2007.00514.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M. et al. S100A11, an dual mediator for growth regulation of human keratinocytes. Mol Biol Cell 19, 78–85, 10.1091/mbc.E07-07-0682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato D. J. et al. Inflammation-based prognostic indices in malignant pleural mesothelioma. J Thorac Oncol 7, 587–594, 10.1097/JTO.0b013e31823f45c1 (2012). [DOI] [PubMed] [Google Scholar]
- Manning L. S. et al. Establishment and characterization of five human malignant mesothelioma cell lines derived from pleural effusions. Int J Cancer 47, 285–290 (1991). [DOI] [PubMed] [Google Scholar]
- Greening D. W. et al. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol Reprod, 10.1095/biolreprod.115.134890 (2016). [DOI] [PubMed] [Google Scholar]
- Tauro B. J. et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304, 10.1016/j.ymeth.2012.01.002 (2012). [DOI] [PubMed] [Google Scholar]
- Tauro B. J. et al. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteomics 12, 587–598, 10.1074/mcp.M112.021303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal S. K. et al. YBX1/YB-1 induces partial EMT and tumourigenicity through secretion of angiogenic factors into the extracellular microenvironment. Oncotarget 6, 13718–13730, 10.18632/oncotarget.3764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening D. W. et al. Colon tumour secretopeptidome: insights into endogenous proteolytic cleavage events in the colon tumour microenvironment. Biochimica et biophysica acta 1834, 2396–407, 10.1016/j.bbapap.2013.05.006 (2013). [DOI] [PubMed] [Google Scholar]
- Brosch M., Yu L., Hubbard T. & Choudhary J. Accurate and sensitive peptide identification with Mascot Percolator. J Proteome Res 8, 3176–3181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I. & Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics 4, 1419–1440 (2005). [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A. I., Kolker E. & Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry 74, 5383–5392 (2002). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer Science & Business Media, (2009). [Google Scholar]
- Chen Y. S. et al. Proteomics profiling of Madin-Darby canine kidney plasma membranes reveals Wnt-5a involvement during oncogenic H-Ras/TGF-beta-mediated epithelial-mesenchymal transition. Mol Cell Proteomics 10, M110 001131, 10.1074/mcp.M110.001131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.