Abstract
Environmental microbes have been associated with both protective and adverse health effects in children and adults. Epidemiological studies often rely on broad biomarkers of microbial exposure (i.e. endotoxin, 1→3, Beta-D glucan), but fail to identify the taxonomic composition of the microbial community. Our aim was to characterize the bacterial and fungal microbiome in different types of environmental samples collected in studies of human health effects. We determined the composition of microbial communities present in home, school and outdoor air samples by amplifying and sequencing regions of rRNA genes from bacteria (16S) and fungi (18S and ITS). Samples for this pilot study included indoor settled dust (from both a Boston area birth cohort study on Home Allergens and Asthma (HAA)(n=12) and a study of school exposures and asthma symptoms (SICAS) (n=1)), as well as fine and coarse concentrated outdoor ambient particulate (CAP) samples (n=9). Sequencing of amplified 16S, 18S, and ITS regions was performed on the Roche-454 Life Sciences Titanium pyrosequencing platform. Indoor dust samples were dominated by gram-positive bacteria (Firmicutes and Actinobacteria); the most abundant bacterial genera were those related to human flora (Streptococcus, Staphylococcus, Corynebacterium and Lactobacillus). Outdoor CAPs were dominated by gram-negative Proteobacteria from water and soil sources, in particular the genera Acidovorax, and Brevundimonas (which were present at very low levels or entirely absent in indoor dust). Phylum-level fungal distributions identified by 18S or ITS regions showed very similar findings: a predominance of Ascomycota in indoor dust and Basidiomycota in outdoor CAPs. ITS sequencing of fungal genera in indoor dust showed significant proportions of Aureobasidium and Leptosphaerulina along with some contribution from Cryptococcus, Epicoccum, Aspergillus and the human commensal Malassezia. ITS sequencing detected an additional 70 fungal genera in indoor dust not observed by culture. Microbiome sequencing is feasible for different types of archived environmental samples (indoor dust, and low biomass air particulate samples), and offers the potential to study how whole communities of microbes (including unculturable taxa) influence human health.
INTRODUCTION
Environmental exposure to microbes and their components has long been linked to numerous non-infectious health effects, both beneficial and detrimental, in children and adults. These relationships are often varied and complex. For instance, increased burden of gram-negative bacteria in the environment (as assessed by endotoxin), has been associated with an elevated risk of wheeze in early life(1,2), but with protection against later childhood asthma and allergic disease. (2-6) In occupational settings, where microbial exposures are much higher, individuals may experience airway obstruction (often termed “Monday Asthma”) in response to endotoxin from gram-negative bacteria.(7) Increased levels of global biomarkers of mold (total fungal counts, ergosterol and (1→3), Beta-D glucan), have been related to both protective(8) and adverse respiratory health outcomes.(9-11) Exposure to yeasts in early life may protect against wheeze and allergic sensitization,(12) while other taxa, including Cladosporium and Alternaria, may increase wheeze risk and later development of allergic rhinitis and asthma.(12-14) Health effects of environmental microbes extend beyond respiratory outcomes. For instance, findings from a recent controlled human chamber exposure study on the health effects of concentrated ambient particles (CAPs) suggested that the endotoxin content of the particles accounted for a significant proportion of the association of CAP exposure with increases in the peripheral white cell count(15). In the same study, short term exposure to endotoxin and fungal (1→3) Beta-D-Glucan in CAPs was also associated with increased blood pressure.(16) The hematologic, immune and physiologic responses to bacteria or fungi may depend on stage of life, dose, and host factors as well as on the form and taxonomy of the organism.
In studies where global biomarkers of microbes are utilized for exposure assessment, the taxonomic composition of the sample is typically unknown. Information on taxonomy may tell us more about health effects of specific groups of microorganisms, and may help us understand heterogeneity of effects of microbial components. The activity of pathogen associated molecular patterns (PAMPs)(17), airway irritants(18), and allergens(19) is known to vary widely across taxa. Furthermore, taxonomic identification will help us pose important questions about how microbial communities, rather than individual microbes, influence health. Culture-based methods yield taxonomic information, but are time consuming, inherently biased (due to selective conditions of culture media) and must be performed immediately after environmental sample collection. Independent of the potential for culture, recent advances in metagenomics allow us to characterize the microbiome and examine its taxonomic composition across different types of exposure samples collected in environmental health studies. In addition to elucidating taxonomy through sequencing, the potential function of microbes will also be critical to uncover, as metabolic pathway genes may select for particular microbes in our environment (thereby shaping the communities of microbes we are exposed to). Characterizing the genetic potential of microbes through microbiome sequencing may also yield information about microbial metabolic and biosynthesis pathways with relevance to human health.
Our overall aim for this pilot was to sequence the microbiome in archived environmental samples from studies that have previously shown important relationships between global biomarkers of microbial exposure and health. Specifically, we wished to 1) determine whether indoor dust samples archived long-term (up to 20 years) would be suitable for microbiome analysis 2) compare fungal taxa identified by culture to fungi detected by sequencing in indoor dust samples 3) determine the feasibility of sequencing the environmental microbiome in air samples collected from a human chamber exposure study on the health effects of air pollution. (To our knowledge, this is the first study to conduct microbiome assessment of particle exposures in a human chamber exposure study.) By amplifying and sequencing rRNA genes from bacteria (16S) and fungi (18S and ITS), we determined the composition of microbial communities present in samples from three distinct studies: home floor dust from The Epidemiology of Home Allergens and Asthma Study(HAA), school classroom floor dust from the School Inner-City Asthma Study (SICAS-1); and pre-exposure calibration samples of outdoor concentrated ambient particles (CAPs) from the Cardiovascular Toxicity of Concentrated Ambient Fine, Ultrafine and Coarse Particles in Controlled Human Exposures study. For all sample types, 16S sequencing results were used to impute the functional metagenome, to yield an indirect assessment of metabolic pathway gene abundance within the bacterial communities sequenced in indoor dust and outdoor air particulates.
METHODS
Sample Collection
“The Epidemiology of Home Allergens and Asthma” birth cohort study (n=12)
House dust samples, from children’s bedrooms or from the living room, were collected using a Eureka Mighty-Mite vacuum cleaner modified to hold 19 by 90-mm cellulose extraction thimble to collect house dust. For this pilot study, dust samples from the Home Allergens and Asthma cohort were chosen from 2 sampling time points (age 1 and age 12), with an over-representation of age 1 samples (as these had corresponding fungal culture data). All samples had at least 50mg dust remaining. For bedroom samples collected at one year of age, the area (2 m2) of the bedroom floor surrounding the baby’s crib was vacuumed for 5 min (n=3). Family room dust samples, from homes when subjects were 1 year of age (n=7) and at age 12 (n=2), were also collected using a Eureka Mighty-Mite vacuum. For family room dust samples, we vacuumed both a 1-m2 area of the family room floor for 2 min and an upholstered chair commonly used by the parent while holding the infant (at age 1) or by index child themselves (at age 12) for 3 min. All of the bedroom floor and living room house dust samples collected at age 1 had corresponding fungal culture data (n=10). Culture methods are described in detail elsewhere.(12)
SICAS-1 (n=1)
In the classroom, vacuuming of the settled dust sample was performed for a total of 6 minutes per sample, 3 minutes on the floor and 3 minutes on other surfaces, such as desks and chairs, as previously described.(20)
“Cardiovascular Toxicity of Concentrated Ambient Fine, Ultrafine and Coarse Particles in Controlled Human Exposures” Study (n=9)
This study included 55 healthy non-smokers, aged 18-60 years, who had no history of cardiovascular disease.(16) Using a cross-over study design, participants blinded to the study condition received up to 6 separate 130-minute exposures (coarse CAPs (2.5 to 10μm)(~200 μg/m3), fine CAPs (0.1 to 2.5 μm)(~250μg/m3), quasi-ultrafine CAPs(<0.3 μm)(~200,000 particles/cm3), filtered air and medical air) at least two weeks apart, with a physician in attendance. Controlled exposures were generated using high-flow (5,000 L/min) Harvard Ambient Particle Concentrators to draw ambient particles from a 1.8 m high PM10 inlet located, 10 m from a busy 4-lane downtown Toronto street with ~2,500 vehicles passing during the 130-minute exposure. Ambient particle exposures were concentrated and adjusted through a dilution control system to deliver target concentrations of CAPs. During the calibration phase (prior to exposing subjects), CAPs were collected onto 47mm, 2 μm pore size Teflon filters Teflo (R2PJ047, Pall Corporation, Ann Arbor, MI) with the pump set to a flow rate of 2 liters/minute while connected to the CAP airstream with the concentrator running (coarse or fine), to simulate the exposure conditions. The exposed surface on the filters was 39mm. Filters collected during the calibration phase (including both coarse and fine particle fractions) were extracted for DNA and sequenced for the bacterial and fungal microbiome. Calibration filters were chosen at random for microbiome sequencing
Microbiome Sequencing and Bio-informatics
DNA Extraction and PCR Amplification
Indoor dust samples and filters containing CAPs were extracted for metagenomic DNA in a clean hood using MP Biomedicals’ FastDNATM-96 Fungal/Bacterial DNA Kit. In each case 50mg of dust or half of the filter was included in the extraction. The extraction protocol includes both physical and chemical disruption of the microbial matter in order to extract all available DNA with a minimum of bias. The V1 through V3 hyper-variable regions (V1-V3) of 16S rRNA genes were amplified from the metagenomic DNA using primers 27F and 534R (27F: 5'- AGAGTTTGATCCTGGCTCAG-3' and 534R: 5'-ATTACCGCGGCTGCTGG-3').(21) The oligonucleotides containing the 16S primer sequences also contained an adaptor sequence for the Roche-454 Life Sciences Titanium pyrosequencing platform as well as one of 96 tag sequences unique to each sample. Analogous primers were designed for amplifying the 18S and ITS regions of small eukaryotes. (18S: F’ – GACTCAACACGGGGAAACT, R’ – ATTCCTCGTTGAAGAGCA) and (ITS: F’ (18S-F) – GTAAAAGTCGTAACAAGGTTTC, R’ (5.8S-1R) – GTTCAAAGAYTCGATGATTCAC). Negative controls were included for the extraction and PCR amplification procedures. In each case there was no indication of contaminants intrinsic to the protocols.
Sequence Processing and Analysis
The DNA sequences for each targeted amplicon were assigned to the appropriate sample based on the unique sequence tag used in the targeted amplification and using Mothur’s trim.seqs protocol.(22) Chimeric sequences were identified and removed with the Mothur implementation of UCHIME.(23) For 16S sequences, we directly aligned each sequence within the processed and cleaned (primers and barcodes removed) 16S fasta files to the Ribosomal Database Project (RDP) database using the RDP classifier version 2.8 and the 16S rRNA training set 9 (http://rdp.cme.msu.edu), using a confidence cutoff of 0.5. We then attempted to further resolve the sequences that were unclassified at the genus level by utilizing NCBI’s BLAST algorithm and their non-redundant nucleotide database (NCBI NRdb downloaded 06/22/2015). For 18S amplicons, we directly aligned each sequence within the processed and cleaned (primers and barcodes removed) 18S fasta files to a database maintained by SILVA (www.arb-silva.de), the SILVA 115 NR99 database. This reference database was chosen because it contains non-redundant, aligned 18S rRNA sequences. Sequences less than 200bp were removed from the analysis, to minimize the possibility of misclassification. Sequence classification was done using the least-common-ancestor methodology to identify a taxonomic classification for each sequence. For ITS amplicons, we directly aligned each sequence within the processed and cleaned (primers and barcodes removed) ITS fasta files to a database maintained by the Fungal Metagenomics Project (http://www.borealfungi.uaf.edu), the Fungal Internal Transcribed Spacers (ITS) rDNA sequence database fungal_its.fa.2014-12-13.
Imputation of the Functional Metagenome for Bacteria
After the 16S data was filtered for base quality, and the primers and barcodes removed, Mothur v.1.33.3 (http://www.mothur.org/wiki/Main_Page)(22)was employed to compare the DNA sequences to each other and to cluster the groups where each sequence was 97% similar to all other sequences in its group thereby creating operationally defined taxonomic units(24), OTUs. The bacterial taxonomy was then determined using the references in Greengenes 16S rRNA database version 13.5.99.(25) Then, PICRUSt version 1.0.0(26) normalized the OTUs by 16S rRNA copy number and predicted metagenomes functional content from the Kyoto Encyclopedia of Genes and Genomes (KEGG) catalogue. Finally, HUMAnN version 0.99(27) determined coverage and abundance of microbial pathways from the KEGG Ortholog results.(28) Microbial metabolic genes catalogued within the KEGG database are annotated to an entire range of microbial metabolic functions, from carbohydrate, amino acid, and lipid metabolism to xenobiotic/chemical degradation pathways (breakdown of synthetic compounds (caprolactam), pesticides (DDT, atrazine), and plant-derived compounds/fragrances (limonene/pinene)).
Principal Component Analysis and Statistical Comparisons
We performed principal component analysis on microbiome sequencing data using STAMP (Statistical Analysis of Metagenomic Profiles) software. For individual samples, we plotted scores for the first,second and third principal components to visualize similarity of community composition (samples of similar community composition will cluster together in the plot (particularly for PC1 vs. PC2 and PC1 vs. PC3), while those with markedly different composition are plotted far apart). For comparing the imputed functional metagenome by sample type (indoor dust vs. outdoor CAPs), R was used to calculate pathways that were differentially abundant (P < 0.001 for statistical significance) using a t-test.
RESULTS
Bacteria (16S Sequencing)
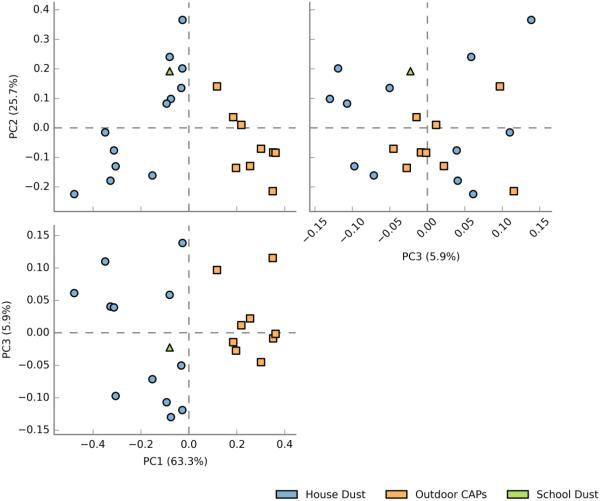
Sample characteristics, including sample type, location of indoor dust sampling, size of outdoor air particulates (coarse vs. fine), and season of sample collection are shown in Table 1. Ninety percent of the samples sequenced contained at least 3000 reads for 16S rDNA sequencing. All samples analyzed contained at least 1000 reads; one sample contained only 419 reads and was excluded from the analysis. On average, 27% of sequences (range 9 to 46%) remained unclassified after alignment to the RDP database. After conducting a BLAST search against NCBI’s NR nucleotide database, an average of 6 % of these unclassified sequences (range 0.2% to 16%) were identified. Overall, indoor dust samples showed remarkably different bacterial taxonomic profiles (PC1 vs. PC2 and PC1 vs. PC3) as compared to samples from outdoor air. Clustering of bacterial communities by sample type is visualized by a plot of principal component scores that represent underlying taxonomic distributions (Figure 1). Plant groups, namely Streptophyta, were also identified in the amplified 16S rRNA gene sequences.
Table 1.
Characteristics of Environmental Samples
| Microbiome Sequencing |
|||||||
|---|---|---|---|---|---|---|---|
| Sample ID | Sample Type | Sample Details | Season | 16S (Bacteria) |
ITS (Fungi) |
18S (Fungi) |
Culture for Fungi |
| HD_01 | House Dust | Living room floor | summer | X | X | X | X |
| HD_02 | House Dust | Child’s bedroom floor | Fall | X | X | X | X |
| HD_03 | House Dust | Living room floor | Fall | X | X | X | X |
| HD_04 | House Dust | Child’s bedroom floor | Fall | X | X | X | X |
| HD_05 | House Dust | Child’s bedroom floor | summer | X | X | X | X |
| HD_06 | House Dust | Living room floor | summer | X | X | X | X |
| HD_07 | House Dust | Living room floor | Fall | X | X | X | X |
| HD_08 | House Dust | Living room floor | Fall | X | X | X | X |
| HD_09 | House Dust | Living room floor | Winter | X | X | X | X |
| HD_10 | House Dust | Living room floor | Fall | X | X | X | X |
| HD_11 | House Dust | Living room floor | Spring | X | X | X | |
| HD_12 | House Dust | Living room floor | Spring | X | X | X | |
| SD_01 | School Dust | Classroom | X | X | X | ||
| PM_01 | Outdoor CAPs | Coarse particles (PM2.5-10) | Fall | X | X | ||
| PM_02 | Outdoor CAPs | Coarse particles (PM2.5-10) | Spring | X | X | ||
| PM_03 | Outdoor CAPs | Fine particles (PM0.1-2.5) | Winter | X | X | ||
| PM_04 | Outdoor CAPs | Fine particles (PM0.1-2.5) | Spring | X | X | X | |
| PM_05 | Outdoor CAPs | Coarse particles (PM2.5-10) | Fall | X | X | X | |
| PM_06 | Outdoor CAPs | Fine particles (PM0.1-2.5) | Fall | X | X | ||
| PM_07 | Outdoor CAPs | Coarse particles (PM2.5-10) | summer | X | X | X | |
| PM_08 | Outdoor CAPs | Coarse particles (PM2.5-10) | Fall | X | X | X | |
| PM_09 | Outdoor CAPs | Coarse particles (PM2.5-10) | Fall | X | X | X | |
Figure 1.
Principal component plot of bacterial (16S) community composition by sample type
( = house dust,
= house dust,  = outdoor CAPs,
= outdoor CAPs,  = classroom dust)
= classroom dust)
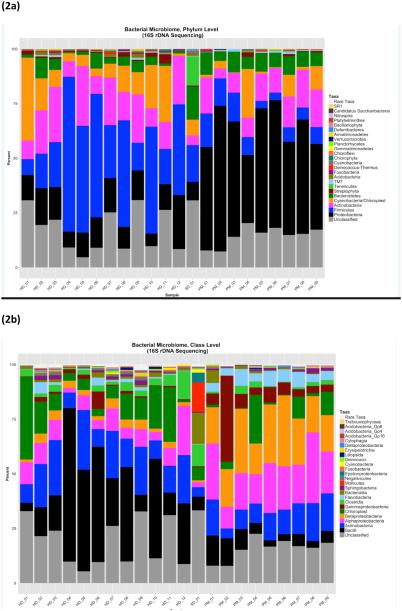
Bacteria in indoor dust appeared to be dominated by gram-positive microbes, as represented by the phylum Firmicutes (the majority of which are gram-positive), as well as Actinobacteria (comprised entirely of gram-positive microbes) (Figure 2a). Taxonomic identification at the class and genus levels (Figures 1b and 1c, respectively) revealed that most of the Firmicutes dominating indoor dust samples originate from the taxonomic class Bacilli (with variable amounts of the genera Staphylococcus, Streptococcus, Lactobacillus). Two of the indoor dust samples were unique, in that the Bacilli detected were dominated by either Solibacillus or Brocothrix. Actinobacteria in indoor dust was comprised of genera related to common skin flora: Corynebacterium and Propionibacterium. All of the indoor dust samples contained some level of Cyanobacteria. Gram-negative bacteria (including members of the phyla Bacteroidetes and Proteobacteria) are also represented in indoor dust, albeit at much lower percentages than gram-positives (Figure 2a).For the gram-negative phylum Proteobacteria, microbes from the Alpha-proteobacteria and Beta-proteobacteria classes (specifically the Massilia and Paracoccus genera) were observed (Figure 2b and 2c). Interestingly, the school indoor dust sample contained Ureaplasma, a type of Mollicute (phylum Tenericutes), which was not detected in any of the house dust samples (Figure 2a). Mollicutes are a unique class of bacteria distinguished by the absence of a cell wall. In contrast to indoor dust, bacterial taxonomic composition of outdoor CAPs was dominated by gram negative bacteria, with large proportions of the phylum Proteobacteria, as well as some contribution from Bacteroidetes. Composition of the mainly gram-positive Firmicutes was much lower in outdoor CAPs vs. indoor dust. (Comparison by sample type at the phylum level is shown in Supplemental Figure 1a-c). Within Proteobacteria, members of the taxonomic classes alpha, beta and gamma-proteobacteria were all detected in outdoor CAPs. The genera Brevundimonas (alpha-proteobacteria), Acidovorax and Delftia (beta-proteobacteria) were present in the majority of CAP samples. While proportions of gamma-proteobacteria in outdoor CAPs were generally quite low, one sample showed a large percentage of Escherichia. Flavobacterium, a genera within Bacteroidetes, was present in the majority of CAP samples. Gram-positive bacteria detectable in CAPs are represented by the phyla Firmicutes and Actinobacteria. While present in much lower amounts, the gram-positive micro-organisms encountered in CAPs included many of the taxa also detected in indoor dust: Staphylococcus, Streptococcus, Lactobacillus, Propionibacterium, Corynebacterium, and Nocardiodes.
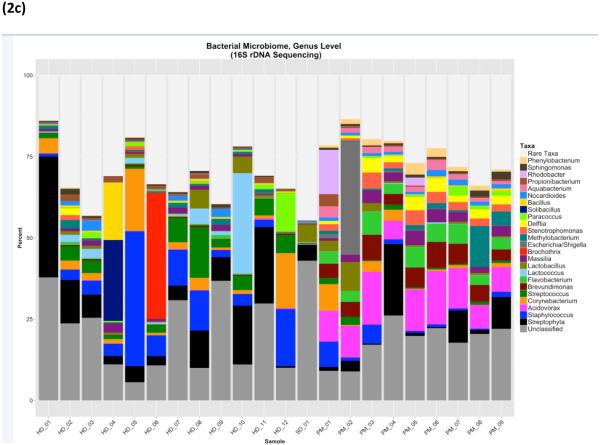
Figure 2.
Bacterial microbiome by 16S shown in panels a-c at three different taxonomic levels: (a) Phylum (b) Class (c) Genus
For indoor dust, the top 5 most abundant genera, as calculated by ranking median abundance across samples, were Streptococcus, Staphylococcus, Corynebacterium, Lactobacillus and Massilia. Abundance of these taxa was highly variable within indoor dust samples The top 5 most abundant bacterial genera in outdoor CAPs were Acidovorax, Brevundimonas, Massilia, Delftia (Proteobacteria) and Flavobacterium (Bacteroidetes).
Fungi (ITS and 18S Sequencing)
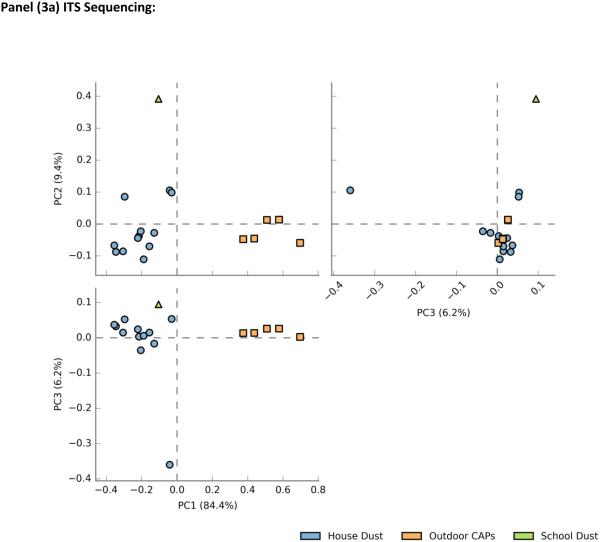
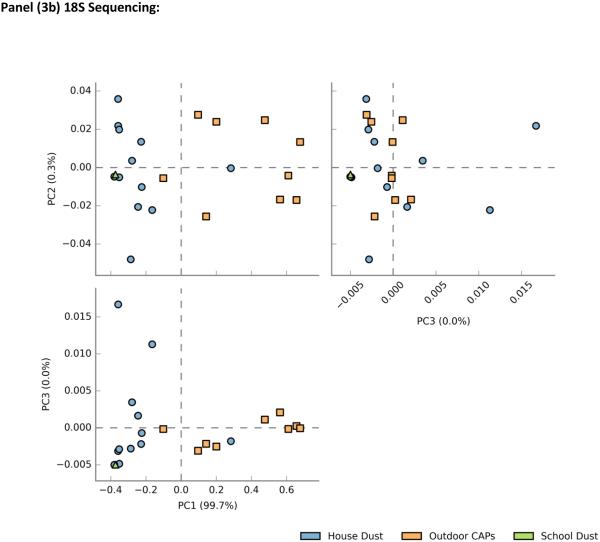
In sequencing runs for 18S, all analyzed samples had greater than 1,000 reads; one sample had 70 reads and was excluded from the analysis. For ITS, five of the outdoor CAP samples had too few reads (<1000) and were excluded from the analysis. Plots of principal component scores for overall fungal microbiome composition revealed separate clusters for indoor dust and outdoor air (Figure3a-b). In addition to showing the differentiation between indoor dust and outdoor air fungal communities, the ITS principal component plot also demonstrated distinct separation between the two types of indoor samples (school dust and house dust) (Figure 3a). In general, fungal community composition by ITS showed clearer differentiation by sample type as compared to 18S (Figure 3b). A slight overlap between indoor dust and outdoor CAP composition was observed, and the school dust sample clustered with house dust on the principal component plot.
Figure 3.
Principal component plot for Phylum-level Fungal community composition by sample type a) ITS sequencing b) 18S sequencing type
( = house dust,
= house dust,  = outdoor CAPs,
= outdoor CAPs,  = classroom dust)
= classroom dust)
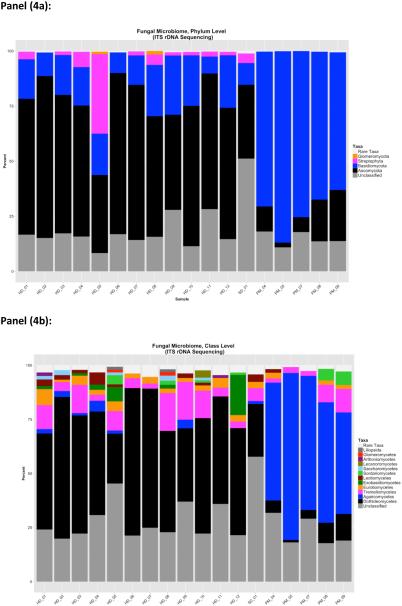
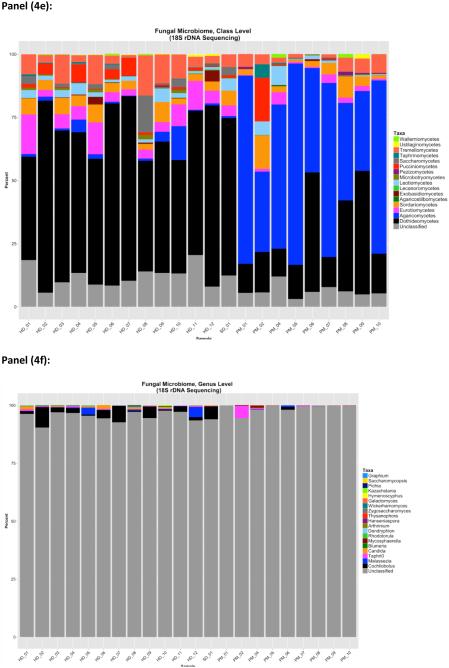
Trends for overall phylum and class-level distributions were similar by ITS and 18S sequencing (Figure 4a-b, Figure 4d-e). In indoor dust samples, the fungal microbiome was dominated by Ascomycota, with a consistent presence (at a much lower percent composition) of Basidomycota. Two of the indoor dust samples contained fungi from the phylum Glomeromycota (ITS only). Class level distributions demonstrated a predominance of Dothidiomycetes and Eurotiomycetes (Ascomycota) in indoor dust samples, along with smaller fractions of Agaricomycetes and Tremellomycetes (Basidiomycota). Outdoor air CAPs contained large fractions of Basidiomycota, with lower levels of Ascomycota. Agaricomycetes was the taxonomic class contributing most to the predominance of Basidomycota in outdoor CAPs. The Ascomycota in outdoor CAPs were comprised of Dothidomycetes (which constituted medium to large percentages of total CAP fungal composition) and Eurotiomycetes which were generally present in very low amounts.
Figure 4.
Fungal microbiome by ITS (panels a-c) at three taxonomic levels (a) phylum (b)class (c) and genus. Fungal microbiome by 18S (panels d-f) at (d)Phylum (e) Class (f) and genus levels
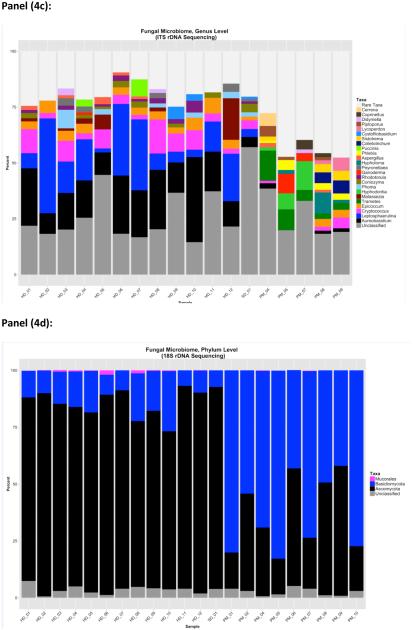
Genus level resolution was possible for fungal classification by ITS (Figure 4c), but not 18S (Figure 4f). Indoor dust samples contained fairly large percentages of the genera Aureobasidium and Leptosphaerulina, along with some contribution from Cryptococcus, Epicoccum, Aspergillus and Malassezia to fungal community composition. Outdoor CAPs included the fungal genera Trametes, Epicoccum, Aspergillus, Cryptococcus, Hyphodontia and Hypholoma. For a subset of house dust samples (n=10) a comparison of the fungi detected by culture and those observed by ITS sequencing is shown in Table 2. Aureobasidium and Alternaria were consistently detected in all samples by both culture and by ITS sequencing. Aspergillus and Epicoccum were detected more often by ITS sequencing than by culture.. Two relatively rare fungi, Paecilomyces and Trichoderma, were detected by culture but not by ITS sequencing. Although culture conditions were suitable for growing Botrytis, it was detected in two samples by ITS sequencing but not by culture. In all, ITS sequencing detected more than 70 (at ≥0.1% composition) additional fungal genera in house dust samples that were not detected by culture (either because culture conditions were not suitable for growth of these fungi, or because they were non-viable at the time of culture). A table listing these taxa, along with the percentage of house dust samples with detectable levels of these fungi, is shown in the supplementary file (Supplemental table 1). This supplemental file also lists plant groupings detected within the ITS amplicons, including Alnus and Nothofagus.
Table 2.
Fungal Genera Cultured from House Dust Samples and Detection by ITS Sequencing
| Alternaria | Aspergillus | Aureobasidium | Botrytis | Cladosporium | Curvularia | Epicoccum | Paecilomyces | Penicillium | Trichoderma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS | Cul | ITS |
| HD_01 | • | • | • | • | • | • | • | • | • | • | • | |||||||||
| HD_02 | • | • | • | • | • | • | • | • | • | |||||||||||
| HD_03 | • | • | • | • | • | • | • | • | • | • | • | • | ||||||||
| HD_04 | • | • | • | • | • | • | • | • | • | • | • | |||||||||
| HD_05 | • | • | • | • | • | • | • | • | • | • | • | • | ||||||||
| HD_06 | • | • | • | • | • | • | • | • | • | • | • | • | ||||||||
| HD_07 | • | • | • | • | • | • | • | • | • | • | • | • | • | |||||||
| HD_08 | • | • | • | • | • | • | • | • | • | • | • | |||||||||
| HD_09 | • | • | • | • | • | • | • | • | • | • | • | • | ||||||||
| HD_10 | • | • | • | • | • | • | • | • | ||||||||||||
Cul= Detectable by culture; ITS=Detectable by ITS sequencing at 0.1% relative abundance or greater.
(•) indicates if a fungal genera was detected in a given sample
Overall, identification of fungi was more complete at the higher taxonomic levels by 18S sequencing (3 to 10% unclassified sequences, for phylum and class level identification, respectively), as opposed to profiling by ITS (18 % to 28% unclassified sequences at phylum and class level) (Figure 3). However, ITS outperformed 18S sequencing for taxonomic identification at the genus level (~25% unclassified sequences for ITS vs. 97% for 18S).
Metabolic pathway prediction based on 16S rRNA survey results
The metabolic pathways that were most abundant in the imputed functional metagenome of indoor dust samples were pathways related to amino acid metabolism (ko00290: Valine, leucine and isoleucine biosynthesis), carbohydrate metabolism (ko00660: C5-Branched dibasic acid metabolism) and lipid metabolism (ko00061: Fatty acid biosynthesis). The top three most abundant metabolic pathways detected in outdoor CAPs were related to metabolism of co-factors and vitamins (ko00760: Nicotinate and nicotinamide metabolism), energy metabolism (ko00190: Oxidative phosphorylation) and carbohydrate metabolism (ko00010: Glycolysis / Gluconeogenesis). In terms of xenobiotic degradation, microbiota sequenced in indoor dust showed the capability of metabolizing environmental chemicals known to influence human health including pesticides (DDT and atrazine) and dioxin. A complete list of all metabolic pathways identified through imputation of the functional metagenome, along with their relative abundances in all samples, is shown in supplemental file 2.
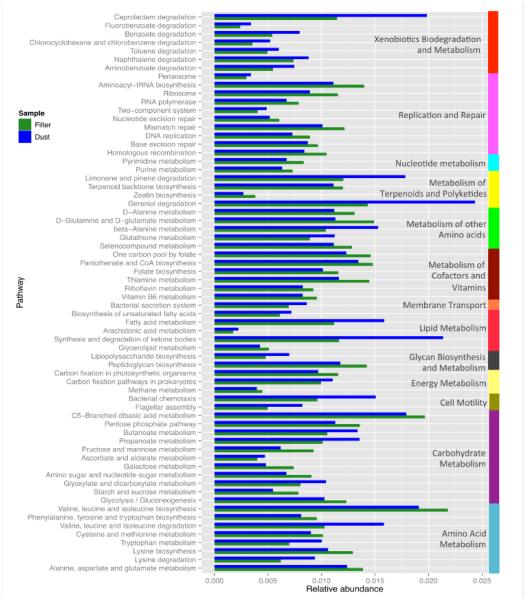
We chose to compare indoor dust and filter (CAPs) samples, however the findings from this comparison should be treated with caution, as samples were collected from different regions and at different time points. We found 66 pathways that were significantly differentially abundant between the filter and the dust samples (Figure 5). The seven pathways classified as xenobiotics biodegradation and metabolism were all overrepresented in the dust samples. For instance, the caprolactam degradation pathway (ko00930) was almost two times more abundant in the dust samples than in the filter samples. Interestingly, caprolactam is used mainly for the manufacture of synthetic fibers, resins, synthetic leather and plasticizers [products that are prevalent in home environments].
Figure 5.
Differentially Abundant Functional Gene/Metabolic Pathways in indoor dust vs outdoor CAPs (filter) samples (p<0.0001 for all comparisons)
The pathways associated with the metabolism of terpenoids and polyketides, limonene and pinene degradation (ko00903) and geraniol degradation (ko00281) were also found at higher abundances in dust samples. Limonene and pinene are aromatic compounds found in a variety of trees and herbs. Meanwhile, geraniol is produced by many flowering plants such as geraniums and citrus fruits. Those three components are often used as scents or as flavor additives. The geraniol degradation pathway ends with the production of 3-methylcrotonyl-CoA. This metabolite is part of the valine, leucine and isoleucine degradation pathway (ko00280), which was also found overrepresented in the dust samples.
Discussion
In this pilot study, we demonstrated the feasibility of microbiome assessment of archived samples from epidemiologic and controlled human exposure studies. We sequenced marker genes of the bacterial and fungal microbiome in house dust samples archived for over 20 years from a birth cohort study of home exposures and asthma incidence. We also successfully characterized the environmental microbiome in concentrated outdoor air particulate samples containing relatively low biomass.
Indoor dust samples were dominated by gram-positive micro-organisms, including a subset of bacterial genera known to be associated with human sources (Staphylococcus, Streptococcus, Lactobacillus, Corynebacterium and Priopionibacterium). Presence of these bacterial genera, which are common in human skin and gut flora, has been documented in other studies of the home microbiome .(29-31) The human contribution to bacteria within the home is further confirmed in a definitive report by Lax et al(32), demonstrating that abundance patterns of bacterial OTUs (operational taxonomic units) in samples from the home closely mirror the microbial profiles of its human residents. Humans are also potential sources of fungi indoors. Although we detected some known human-associated fungi, in particular the commensal yeast Malassezia, the majority of fungal taxa sequenced in our indoor samples were associated with soil (Aureobasidium,Penicillium), water (Aureobasidium) or plant (Leptosphaerulina, Epicoccum, Cladosporium, Alternaria) sources. This finding is consistent with work by Adams et al(30), in which indoor samples from a students’ dormitory complex showed a consistent, but relatively low, abundance of the human-derived taxon Malassezia, with much higher percentages of the fungal classes Dothidiomycetes (Leptosphaerulina, Cladoporium, Alternaria) and Eurotiomycetes (Penicillium, Aspergillus). As in the Adams et al study, we also consistently detected a moderate percent abundance of the soil-derived genus Cryptococcus; in some of our indoor dust samples Cryptococcus was the third most abundant taxon at the genus level. (It is important to note that the majority of Cryptococci are non-pathogenic to humans, with the exception of a few well-known pathogenic species).In addition to humans, soil, plant and water-based sources, pets are another important microbial reservoir that may influence the composition of the home microbiome. The indoor microbiome of dog owners’ homes shows greater microbial diversity(33), and may contain higher levels of Lactobacillus Johnsonii, a taxon that may mitigate allergic disease response.(34)Microbes found indoors may also be influenced by the characteristics of the home itself. Dannemiller and colleagues found that water leaks, longer AC use, and suburban (vs.urban) homes were associated with greater diversity in house dust samples.(31) Although we did not have the power to examine these relationships, sequencing of additional samples within the two prospective cohorts described in the present work (HAA and SICAS) may help us to identify characteristics in homes and schools that are associated with particular microbial communities.
Bacteria detected in outdoor samples were predominately gram-negative Proteobacteria; in particular the taxa Acidovorax (associated with plant matter), Brevundimonas, and Flavobacterium (a genera within Bacteriodetes often detected in environmental water and soil samples). These three gram-negative genera showed little to no abundance in the indoor samples analyzed. However, the gram-positive taxa identified in these outdoor samples showed considerable overlap with genera detectable in indoor samples (including Staphylococcus, Streptococcus and Corynebacterium). Fungal distributions contained a predominance of the phylum Basidiomycota, which was mainly comprised of Agaricomycetes at the class level. Interestingly, characterization of fungal microbiota in a much different locale (Berkeley, California) also showed a high proportion of Agaricomycetes in outdoor air, with significant representations of Dothidiomycetes and Tremellomycetes as secondary taxa.(35) Season and particle size (fine vs. coarse CAPs) were not associated with different types of microbial (bacterial or fungal) communities (data not shown); however, the relatively small sample size in this study may have obscured our ability to detect community differences between these particle size fractions. A study by Cao et al of the environmental microbiome in outdoor air particulates during a Beijing smog event reported a higher percentage of eukaryotes in larger particulates (PM2.5-10μm )as opposed to particles < 2.5 μm in diameter.(36) The microbial signatures found in air particulates are unlikely to be from whole microbial cells (which are often larger than 2 or 10μm), but rather fragments of these micro-organisms. For both indoor and outdoor samples, 18S sequencing yielded a more complete taxonomic profile at the Phylum and Class level, but identified only a small percentage of the fungal genera. Since the 18S rRNA gene encodes for a transcribed (as well as functional) portion of rRNA, it shows greater conservation across fungi overall but still contains sufficient variability to differentiate between fungi at higher taxonomic levels. Conversely, ITS is an intronic region of rDNA, and is less conserved across fungal taxa. Hypervariable regions within the ITS rDNA locus serve as “barcodes” for specific fungal genera, with little homogeneity at higher at taxonomic levels.(37)
Unfortunately, no bioinformatic technique exists for combining sequencing information from these two different rDNA regions into an overall relative abundance profile when utilizing current targeted sequencing technologies. The choice of which rDNA region to use will depend upon the level of taxonomic classification desired by the investigator.Expansion of the existing fungal databases, such as SILVA, UNITE and the Fungal Metagenomics Project database, will eventually increase ITS coverage, so that the majority of fungal taxa, from phylum to species level, will be identifiable in environmental samples. In a subset of our indoor dust samples, we were able to compare ITS sequence data with culturable fungi. While Aureobasidium was consistently detected in all samples by both methods, sensitivity of detection varied for other genera.Amplification and sequencing by ITS may capture dead or unculturable fungi (potentially explaining detection by ITS but not culture), and culture techniques may selectively grow relatively rare fungi that are missed due to inadequate sequencing read depth or representation in an ITS database. In all, ITS sequencing yielded a much broader survey of fungi present within the environmental samples studied (with an excess of 80 additional taxa detectable by ITS, as in contrast to the 9 detectable by culture). It is important to note that the culture methods used here for comparison with ITS sequencing (performed 20 years ago, prior to the advent of metagenomic sequencing) were originally designed to characterize indoor mold exposure in an epidemiological study, and were not optimized for a direct culture vs. metagenomic sequencing comparisons.
In addition to characterizing the microbiome with respect to taxonomic composition, we were also able to impute the functional metagenome of bacterial communities. The role of microbial metabolic function/xenobiotic degradation may be of particular importance indoors, where human use of household chemicals may alter the microbiome. Thus far, limited information exists on the relationship between household chemical use and indoor bacterial and fungal levels; however, the ability of bacteria and fungi to degrade synthetic substrates has been clearly demonstrated in environmental microbiology.(38,39) Indoor chemicals (pesticides, phthalates, triclosan, fragrances and synthetic polymers) may exert selective pressure on microbes, with the potential to drastically alter microbial community composition (taxa with the ability to degrade or utilize these chemicals as substrates may proliferate, while others that are susceptible to their anti-microbial activity may be reduced). Furthermore, xenobiotic degradation by microbes may either increase or decrease toxicity of a chemical compound with respect to human health, depending upon the activity of the parent compound vs. its metabolites. In this pilot study, we were able to identify microbial metabolic pathway genes in indoor dust that degrade known toxic compounds (pesticides, bisphenol, dioxin), synthetic materials found indoors (caprolactam) and fragrance additives used in household products (limonene, pinene). Future work is needed to uncover whether the potential to metabolize these compounds selects for particular microbes indoors and to determine whether microbial metabolism of these compounds alters their toxicity.
In summary, characterization of the bacterial and fungal microbiome in the indoor dust and outdoor air samples presented in this work show both the potential and limitations for this type of exposure assessment in studies of human health effects. Methods for taxonomic identification of microbial communities through metagenomics approaches to DNA sequencing are more developed for bacteria than for fungi, where reference databases are more incomplete, and, up until recently, developed for purposes other than the relation of the environmental microbiome to human physiologic or health outcomes. While tremendous gains are made in identifying additional taxa that are not amenable to culture techniques, a fairly large percentage of sequences (for both environmental bacteria and fungi) cannot yet be assigned to a highly resolved (i.e. genus or species) taxonomic level. This limitation will be less of an issue as databases expand to include a greater number of micro-organisms. In all, microbiome sequencing provides an additional assessment of microbes in our environment that offers the option to identify both culturable and non-culturable as well as live and dead organisms that exist in the environment. Thus, one has the potential to study a more complete view of microbial communities, or even previously unstudied individual taxa, that may influence human health.
Supplementary Material
Characterization of the microbiome in both indoor and outdoor samples, as demonstrated in this pilot, can be applied to numerous types of environmental health studies. These include studies on home microbial communities in early life and childhood allergic disease development, as well as the assessment of the microbiome in particulate air pollution and its contribution to respiratory, inflammatory and hematologic health outcomes. Furthermore, sequencing of the microbiome in outdoor air particulates may aid our understanding of how this exposure has fluctuated with the changing temperature and weather patterns associated with global warming.
Acknowledgments
Funding sources
This publication was made possible by the following grants: NIH grants R01AI073964, R01AI035786, P03ES000002, USEPA RD-83241601, Health Canada, Environment Canada, and AllerGen NCE. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the USEPA. Further, USEPA does not endorse the purchase of any commercial products or services that might be mentioned in the publication.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5em00639b
References
- 1.Park JH, Gold DR, Spiegelman DL, Burge HA, Milton DK. House dust endotoxin and wheeze in the first year of life. Am J Respir Crit Care Med. 2001 Feb;163(2):322–328. doi: 10.1164/ajrccm.163.2.2002088. [DOI] [PubMed] [Google Scholar]
- 2.Perzanowski MS, Miller RL, Thorne PS, Barr RG, Divjan A, Sheares BJ, et al. Endotoxin in inner-city homes: associations with wheeze and eczema in early childhood. J Allergy Clin Immunol. 2006 May;117(5):1082–1089. doi: 10.1016/j.jaci.2005.12.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tischer C, Gehring U, Chen CM, Kerkhof M, Koppelman G, Sausenthaler S, et al. Respiratory health in children, and indoor exposure to (1,3)-beta-D-glucan, EPS mould components and endotoxin. Eur Respir J. 2011 May;37(5):1050–1059. doi: 10.1183/09031936.00091210. [DOI] [PubMed] [Google Scholar]
- 4.Gehring U, Strikwold M, Schram-Bijkerk D, Weinmayr G, Genuneit J, Nagel G, et al. Asthma and allergic symptoms in relation to house dust endotoxin: Phase Two of the International Study on Asthma and Allergies in Childhood (ISAAC II) Clin Exp Allergy. 2008 Dec;38(12):1911–1920. doi: 10.1111/j.1365-2222.2008.03087.x. [DOI] [PubMed] [Google Scholar]
- 5.Sordillo JE, Hoffman EB, Celedon JC, Litonjua AA, Milton DK, Gold DR. Multiple microbial exposures in the home may protect against asthma or allergy in childhood. Clin Exp Allergy. 2010 Jun;40(6):902–910. doi: 10.1111/j.1365-2222.2010.03509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002 Sep 19;347(12):869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 7.Castellan RM, Olenchock SA, Kinsley KB, Hankinson JL. Inhaled endotoxin and decreased spirometric values. An exposure-response relation for cotton dust. N Engl J Med. 1987 Sep 3;317(10):605–610. doi: 10.1056/NEJM198709033171005. [DOI] [PubMed] [Google Scholar]
- 8.Schram-Bijkerk D, Doekes G, Douwes J, Boeve M, Riedler J, Ublagger E, et al. Bacterial and fungal agents in house dust and wheeze in children: the PARSIFAL study. Clin Exp Allergy. 2005 Oct;35(10):1272–1278. doi: 10.1111/j.1365-2222.2005.02339.x. [DOI] [PubMed] [Google Scholar]
- 9.Blatter J, Forno E, Brehm J, Acosta-Perez E, Alvarez M, Colon-Semidey A, et al. Fungal exposure, atopy, and asthma exacerbations in Puerto Rican children. Ann Am Thorac Soc. 2014 Jul;11(6):925–932. doi: 10.1513/AnnalsATS.201402-077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maheswaran D, Zeng Y, Chan-Yeung M, Scott J, Osornio-Vargas A, Becker AB, et al. Exposure to Beta-(1,3)-D-glucan in house dust at age 7-10 is associated with airway hyperresponsiveness and atopic asthma by age 11-14. PLoS One. 2014 Jun 6;9(6):e98878. doi: 10.1371/journal.pone.0098878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norback D, Markowicz P, Cai GH, Hashim Z, Ali F, Zheng YW, et al. Endotoxin, ergosterol, fungal DNA and allergens in dust from schools in Johor Bahru, Malaysia- associations with asthma and respiratory infections in pupils. PLoS One. 2014 Feb 11;9(2):e88303. doi: 10.1371/journal.pone.0088303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behbod B, Sordillo JE, Hoffman EB, Datta S, Webb TE, Kwan DL, et al. Asthma and allergy development: contrasting influences of yeasts and other fungal exposures. Clin Exp Allergy. 2015 Jan;45(1):154–163. doi: 10.1111/cea.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reponen T, Lockey J, Bernstein DI, Vesper SJ, Levin L, Khurana Hershey GK, et al. Infant origins of childhood asthma associated with specific molds. J Allergy Clin Immunol. 2012 Sep;130(3):639–644. doi: 10.1016/j.jaci.2012.05.030. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark PC, Celedon JC, Chew GL, Ryan LM, Burge HA, Muilenberg ML, et al. Fungal levels in the home and allergic rhinitis by 5 years of age. Environ Health Perspect. 2005 Oct;113(10):1405–1409. doi: 10.1289/ehp.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behbod B, Urch B, Speck M, Scott JA, Liu L, Poon R, et al. Endotoxin in concentrated coarse and fine ambient particles induces acute systemic inflammation in controlled human exposures. Occup Environ Med. 2013 Nov;70(11):761–767. doi: 10.1136/oemed-2013-101498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong J, Urch B, Speck M, Coull BA, Koutrakis P, Thorne PS, et al. Endotoxin and beta-1,3-d-Glucan in Concentrated Ambient Particles Induce Rapid Increase in Blood Pressure in Controlled Human Exposures. Hypertension. 2015 Sep;66(3):509–516. doi: 10.1161/HYPERTENSIONAHA.115.05342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabanov DS, Prokhorenko IR. Structural analysis of lipopolysaccharides from Gram-negative bacteria. Biochemistry (Mosc) 2010 Apr;75(4):383–404. doi: 10.1134/s0006297910040012. [DOI] [PubMed] [Google Scholar]
- 18.Thrasher JD, Crawley S. The biocontaminants and complexity of damp indoor spaces: more than what meets the eyes. Toxicol Ind Health. 2009 Oct-Nov;25(9-10):583–615. doi: 10.1177/0748233709348386. [DOI] [PubMed] [Google Scholar]
- 19.Crameri R, Garbani M, Rhyner C, Huitema C. Fungi: the neglected allergenic sources. Allergy. 2014 Feb;69(2):176–185. doi: 10.1111/all.12325. [DOI] [PubMed] [Google Scholar]
- 20.Sheehan WJ, Hoffman EB, Fu C, Baxi SN, Bailey A, King EM, et al. Endotoxin exposure in inner-city schools and homes of children with asthma. Ann Allergy Asthma Immunol. 2012 Jun;108(6):418–422. doi: 10.1016/j.anai.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012 Jun 13;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009 Dec;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6(12):e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005 Mar;71(3):1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006 Jul;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013 Sep;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8(6):e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012 Jan;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taubel M, Rintala H, Pitkaranta M, Paulin L, Laitinen S, Pekkanen J, et al. The occupant as a source of house dust bacteria. J Allergy Clin Immunol. 2009 Oct;124(4):834–40. doi: 10.1016/j.jaci.2009.07.045. e47. [DOI] [PubMed] [Google Scholar]
- 30.Adams RI, Miletto M, Lindow SE, Taylor JW, Bruns TD. Airborne bacterial communities in residences: similarities and differences with fungi. PLoS One. 2014 Mar 6;9(3):e91283. doi: 10.1371/journal.pone.0091283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dannemiller KC, Gent JF, Leaderer BP, Peccia J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air. 2015 Mar;:31. doi: 10.1111/ina.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014 Aug 29;345(6200):1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, et al. Man's best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010 Aug;126(2):410–2. doi: 10.1016/j.jaci.2010.05.042. 412.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014 Jan 14;111(2):805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams RI, Miletto M, Taylor JW, Bruns TD. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013 Jul;7(7):1262–1273. doi: 10.1038/ismej.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao C, Jiang W, Wang B, Fang J, Lang J, Tian G, et al. Inhalable microorganisms in Beijing's PM2.5 and PM10 pollutants during a severe smog event. Environ Sci Technol. 2014 Feb 4;48(3):1499–1507. doi: 10.1021/es4048472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee S, Karlovsky P. Removal of the endocrine disrupter butyl benzyl phthalate from the environment. Appl Microbiol Biotechnol. 2010 Jun;87(1):61–73. doi: 10.1007/s00253-010-2570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R. Bioremediation approaches for organic pollutants: a critical perspective. Environ Int. 2011 Nov;37(8):1362–1375. doi: 10.1016/j.envint.2011.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.