Abstract
Background
The androgen receptor splice variant-7 (AR-V7) has been implicated in the development of castration-resistant prostate cancer (CRPC) and resistance to abiraterone and enzalutamide.
Objective
To develop a validated assay for detection of AR-V7 protein in tumour tissue and determine its expression and clinical significance as patients progress from hormone-sensitive prostate cancer (HSPC) to CRPC.
Design, setting, and participants
Following monoclonal antibody generation and validation, we retrospectively identified patients who had HSPC and CRPC tissue available for AR-V7 immunohistochemical (IHC) analysis.
Outcome measurements and statistical analysis
Nuclear AR-V7 expression was determined using IHC H score (HS) data. The change in nuclear AR-V7 expression from HSPC to CRPC and the association between nuclear AR-V7 expression and overall survival (OS) was determined.
Results and limitations
Nuclear AR-V7 expression was significantly lower in HSPC (median HS 50, interquartile range [IQR] 17.5–90) compared to CRPC (HS 135, IQR 80–157.5; p < 0.0001), and in biopsy tissue taken before (HS 80, IQR 30–136.3) compared to after (HS 140, IQR 105–167.5; p = 0.007) abiraterone or enzalutamide treatment. Lower nuclear AR-V7 expression at CRPC biopsy was associated with longer OS (hazard ratio 1.012, 95% confidence interval 1.004–1.020; p = 0.003). While this monoclonal antibody primarily binds to AR-V7 in PC biopsy tissue, it may also bind to other proteins.
Conclusions
We provide the first evidence that nuclear AR-V7 expression increases with emerging CRPC and is prognostic for OS, unlike antibody staining for the AR N-terminal domain. These data indicate that AR-V7 is important in CRPC disease biology; agents targeting AR splice variants are needed to test this hypothesis and further improve patient outcome from CRPC.
Patient summary
In this study we found that levels of the protein AR-V7 were higher in patients with advanced prostate cancer. A higher level of AR-V7 identifies a group of patients who respond less well to certain prostate cancer treatments and live for a shorter period of time.
Keywords: Androgen receptor, Androgen receptor variant-7, Castration-resistant prostate cancer, Metastatic biopsy, Treatment resistance, Predictor of outcome
Take Home Message
Androgen receptor splice variant-7 (AR-V7) expression increases with castration resistance and further increases following abiraterone or enzalutamide treatment, and is associated with worse outcome for metastatic castration-resistant prostate cancer, unlike total AR expression, suggesting a role in treatment resistance. Agents targeting AR-V7 are needed to test this hypothesis.
1. Introduction
Patients with advanced prostate cancer initially respond to androgen deprivation therapy (ADT), but inevitably relapse with fatal castration-resistant prostate cancer (CRPC); there is increasing evidence of ongoing androgen receptor (AR) signalling with rising prostate specific antigen (PSA), increased steroidogenesis, and occasionally activating point mutations within the AR ligand-binding domain [1], [2], [3], [4], [5]. This led to the development of abiraterone acetate (AA) and enzalutamide (EZ), which effectively target AR signalling in CRPC and improve patient outcome [6], [7], [8], [9]. However, resistance to both agents is common and may be due in part to constitutively active AR splice variants (AR-Vs), of which AR variant-7 (AR-V7) is the most well studied. Consistent with this, a recent study demonstrated limited responses to AA and EZ in patients with detectable levels of AR-V7 RNA in circulating tumour cells [3].
AR-V7 is truncated after canonical AR exon 3, with inclusion of a cryptic exon 3b (CE3b) derived from an intron in the expressed protein [10]. AR-V7 expression is associated with a rearranged AR gene and can also be produced by aberrant pre-mRNA splicing due to androgen deprivation induced by castration, AA, EZ, or ARN-509 [4], [11], [12]. The ability of the intrinsically disordered AR N-terminus to continue AR signalling in the absence of ligand binding has been proven by deletion constructs [13]. Castration-resistant cell lines including 22Rv1 and the EZ-resistant LNCaP95 harbour AR-Vs. Specific inhibition of AR-V7 with siRNA to CE3b inhibits tumour growth [1], [12], [14]. Treatment with AA or EZ results in increased expression of AR-Vs, with AR-V7 being the most highly expressed [10], [15], [16]. Cell constructs in which AR-V7 is expressed are resistant to drugs targeting the AR ligand-binding domain [14]. AR-V7 expression may have utility as a predictive biomarker and is an important therapeutic target.
Originally, studies indicated that AR-V7 heterodimerises with full-length AR (AR-FL) [14], [17]. This suggested that AR-FL blockage would inhibit AR-V7 activity. However, AR-V7 also homodimerises to itself and heterodimerises with other AR-Vs, binding to androgen response elements to generate a signal independent of AR-FL [18], [19]. Analysis of this potentially key resistance mechanism in clinical samples has been challenging because of low levels of AR-V7 mRNA and the lack of a reproducible tumour tissue assay. We established a validated assay and show for the first time in matched tumour samples from the same patients how AR-V7 expression changes from hormone-sensitive prostate cancer (HSPC) to CRPC, and evaluate its clinical significance. The data we report here are important in the interpretation of an ongoing randomised trial using this antibody to detect AR-V7 in circulating tumour cells as a putative predictive biomarker in patients whose cancer has progressed on EZ or AA (NCT02485691).
2. Materials and methods
2.1. Antibody generation and characterisation
2.1.1. Antibody generation
Several polyclonal antibodies were generated in four rabbits immunised with a synthetic AR-V7 peptide containing the 16 amino acids of CE3b (aa 630–645). Sera collected from immunised rabbits were purified and screened by immunoprecipitation and western blotting of AR-V7–transfected M12 cells. The polyclonal antibody H6253 was selected because it demonstrated reactivity with a single band consistent with AR-V7. A hybridoma was generated by fusing splenocytes with the fusion partner cell line 240E-W2. The rabbit monoclonal antibody EP343 was selected from the final hybridoma cell line and was further characterised.
2.1.2. Cell lines
LNCaP95 cells were provided by Drs. Alan K Meeker and Jun Luo (Johns Hopkins University, Baltimore, MD, USA) and cultured in RPMI 1640 medium supplemented with 10% charcoal-stripped foetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA). M12 cells were provided by Dr. Joy Ware (Virginia Commonwealth University, Richmond, VA, USA) and cultured in RPMI 1640 supplemented with 5% FBS [20]. DU145, 22Rv1s, and PC3 cells were obtained from ATCC (Manassas, VA, USA) and grown in their recommended culture medium containing 10% FBS at 37 °C in 5% CO2. LuCap xenografts were provided by Drs. Eva Corey and Colm Morrisey (University of Washington, Seattle, WA, USA). M12 cells expressing cumate-inducible 3×FLAG-wtAR, 3×FLAG-ARv567es, and 3×FLAG-AR-V7 lentivirus were prepared using the SparQcumate switch lentivector system (Systems Biosciences, Palo Alto, CA, USA). pCDH-EF1-CymR-T2A-Puro vectors were packaged into lentiviral particles using pPACK packaging systems (System Biosciences). To prepare stable cell lines, M12 cells were infected with 1×107 virus particles per 1 × 106 cells and then selected with 1 μg/ml puromycin (Invitrogen) for 10 d. Stably transduced M12 lines were maintained in RPMI 1640 supplemented with 5% FBS, 0.01 μM dexamethasone (Sigma Aldrich, St. Louis, MO, USA), 10 ng/ml epidermal growth factor (Invitrogen), 10 ml/l insulin-transferrin-selenium (Cellgro, Tewksbury, MA, USA), 100 IU/ml penicillin, and 100 μg/μl streptomycin at 37 °C with 5% CO2.
2.1.3. siRNA and quantitative RT-PCR
siRNA studies were performed with cells reaching ∼70% confluence. Cells were transfected with 50 nM siRNA (Supplementary Table 1) for 48 h using Lipofectamine RNA iMax (Life Technologies, Carlsbad, CA, USA). Quantitative RT-PCR was performed on RNA isolated using Trizol reagent according to the manufacturer's instructions (Invitrogen). cDNA was reverse-transcribed from total RNA (1 μg) using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed using iTaq Universal Probe PCR Master Mix (Bio-Rad) on a ViiA 7 real-time PCR system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The probes used are listed in Supplementary Table 2.
2.1.4. Western blotting and co-immunoprecipitation
Western blotting was performed on whole cell extracts prepared in lysis buffer (50 mM HEPES, 150 mM NaCl, 1.5 mM EGTA, 1% Triton X-100) with a complete protease inhibitor cocktail (Roche, Basel, Switzerland). Tissue extract (from 200-μm biopsies embedded in optimal cutting temperature compound) were prepared in lysis buffer and then sonicated (two rounds of 10 s at 10 W). The extracted protein concentration was determined by bicinchoninic acid assay (Thermo Scientific, Waltham, MA, USA). Protein extracts (20 μg) were separated on 4–12% NuPAGE Bis-Tris gel (Invitrogen) by electrophoresis and subsequently electrotransferred onto Immobilon-P membranes (Millipore, Billerica, MA, USA). Membranes were blocked and antibodies were diluted in 5% milk. The antibody for the AR N-terminal domain (AR-NTD) was used at 1:5000 dilution (Dako, Glostrup, Denmark) and AR-V7 at 1:1000. Immunodetection was performed according to the manufacturer's protocol using enhanced chemiluminescence reagent (GE Health Care, Little Chalfont, UK). In co-immunoprecipitation experiments, cells were lysed in cold Pierce IP Lysis buffer (Thermo Scientific). Precleared cell lysate was incubated with anti-FLAG M2 (Sigma Aldrich), anti-AR-V7, or anti-AR-NTD (Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies overnight. Immune complexes were collected using protein A/G Plus agarose beads and analysed by immunoblotting as previously described [21].
2.2. Patient cohort and tissue analysis
2.2.1. Patients and tissue samples
Patients were identified from a population of men with metastatic CRPC treated at the Royal Marsden NHS Foundation Trust. Patients with a diagnosis of prostate adenocarcinoma with sufficient formalin-fixed, paraffin-embedded (FFPE) matched archival and CRPC tissue for AR-V7 immunohistochemistry (IHC) were selected. One patient who had received a drug targeting the AR N-terminus (EZN-4176) and two patients with histology consistent with neuroendocrine differentiation were excluded [22]. Archival tissue was obtained from prostate needle biopsy, transurethral resection of the prostate (TURP), or prostatectomy procedures. CRPC tissue was obtained from metastases within bone, lymph node, soft tissue, or visceral organs. All tissue blocks were freshly sectioned and only considered for IHC analyses if adequate material was present (≥50 tumour cells; reviewed by D.N.R.). All patients had given written informed consent and were enrolled in institutional protocols approved by the Royal Marsden NHS Foundation Trust Hospital (London, UK) ethics review committee (reference no. 04/Q0801/60). A total of 37 patients with CRPC had archival (diagnostic) and CRPC tissue sufficient for testing; four patients had archival tissue from CRPC TURP samples and therefore 33 patients had matched HSPC and CRPC tissue samples. Of the 37 patients who had CRPC biopsies, 35 had treatment with AA or EZ and 12 had their CRPC biopsy performed before AA treatment and had fully evaluable PSA response data. Demographic and clinical data for each patient were retrospectively collected from the hospital electronic patient record system.
2.2.2. Tissue analysis
IHC was performed using the rabbit AR-V7 monoclonal antibody clone EP343 (Epitomics). Antigen retrieval was achieved by microwaving slides in citrate buffer (pH 6.0) for 18 min at 800 W. Endogenous peroxidase was blocked using 3% H2O2 solution. Blocking was performed using the protein block solution from a Novolink polymer detection system (Leica, Wetzlar, Germany). EP343 was diluted 1:200 and tissue was incubated for 1 h. The reaction was visualised using the Novolink polymer and DAB chromogen. Cases were scored by a pathologist (D.N.R) blinded to clinical data using the modified H score (HS) method, a semi-quantitative assessment of staining intensity that reflects antigen concentration [23]. HS was determined according to the formula: [(% of weak staining) × 1] + [(% of moderate staining) × 2] + [(% of strong staining) × 3], yielding a range from 0 to 300 [23]. We used a mouse monoclonal antibody clone (AR441, Dako) to test AR-NTD expression at 1:5000 dilution. Antigen retrieval was performed using pH 8.1 Tris/EDTA solution heated in a water bath. Cell pellets from 22RV1 and LNCaP95 cells were used as positive controls for AR-V7 IHC and cell pellets from DU145 and PC3 cells were selected as negative controls. For AR-NTD IHC, positive controls included cell pellets from 22RV1 and LNCAP cells and benign prostate tissue, and negative controls included cell pellets from DU145 and PC3 cells.
RNA in situ hybridisation was performed on freshly cut 4-μm sections derived from FFPE blocks using an RNAscope 2.5 platform (Advanced Cell Diagnostics [ACD], Hayward, CA, USA). Three ACD target probes were used: AR probe, AR-V7, and PPIB (cyclophilin B) as a positive control. For all probes, xenograft sections were subjected to mild pretreatment conditions, while the standard pretreatment protocol was used for patient sections. The slides were processed for standard signal amplification steps according to the manufacturer's instructions.
2.2.3. Statistical analysis
HS data are reported as median values with interquartile range (IQR). Wilcoxon signed-rank and Mann-Whitney tests were used to compare differences in expression levels. Time to CRPC was defined as the time from diagnosis to biochemical and/or radiological progression on luteinising hormone-releasing hormone (LHRH) agonist alone or with anti-androgen if started before/or with LHRH agonist with castrate serum level of testosterone. Median time to CRPC was estimated using the Kaplan-Meier method; Cox regression modelling was used to determine association with HS. Overall survival (OS) was measured from the date of CRPC biopsy to the date of last contact (1 patient on regular follow up; censored at last contact) or date of death from any cause (36 patients). The Kaplan-Meier product-limit method was used to estimate median OS for patients grouped by tertiles (to ensure approximately 10 events per group) for nuclear AR-V7 expression, nuclear AR-NTD expression, and the nuclear AR-V7/AR-NTD expression ratio. The association between protein expression and OS was determined using univariate Cox regression models; linear trend tests were evaluated by determining orthogonal polynomials. The association between the magnitude of PSA decline at 12 wk and nuclear AR-V7 expression was evaluated using univariate linear regression models. Biochemical response to AA was defined as ≥50% decline in PSA from baseline at 12 wk. Statistical analyses were performed using Prism v.6 (GraphPad, La Jolla, CA, USA) and SPSS v.21 (IBM, Armonk, NY, USA).
3. Results
3.1. Antibody production and specificity
A polyclonal antibody (H6253) to CE3b of AR-V7 was initially developed. Antibody specificity was determined by immunoprecipitation and western blotting of AR-V7–transfected M12 cell lysate (Supplementary Fig. 1A,B). Following confirmation, fusions were performed and a rabbit monoclonal antibody that detected AR-V7, but not AR-FL or AR-V567es, was generated (EP343; Fig. 1A,B). Next, endogenous expression of AR-V7 in prostate cancer cell lines was examined (Fig. 1C). Of note, nonspecific bands were observed for all cell lines. RT-PCR of PC3 mRNA showed that no AR-V7–specific RNA was present (Supplementary Fig. 1C). To further confirm this negative result, RNA in situ hybridisation for AR and AR-V7 was carried out. As expected, VCAP and 22Rv1 were positive for AR and AR-V7 RNA, while PC3 was negative for both (Supplementary Fig. 2). To further demonstrate the ability of the antibody to detect AR-V7, LNCaP95 cells were transfected with siRNAs to ARexon1, ARexon7, AR-V7 CE3b, or siRNA scrambled. Suppression of AR-V7 protein was observed with siRNAs to exon 1 and CE3b, but not the scrambled siRNA or exon 7, an exon absent from AR-V7 (Fig. 1D). Overall, these data indicate that the antibody binds to AR-V7 but may bind to other proteins.
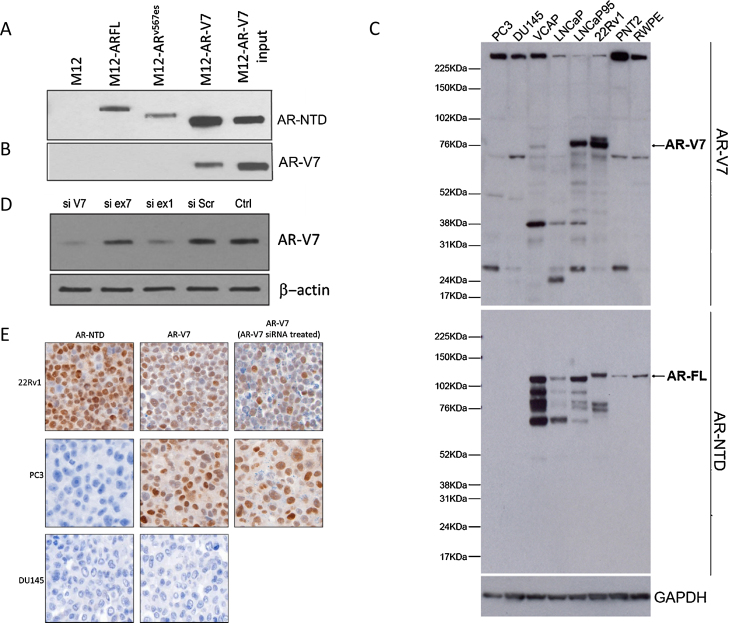
Fig. 1.
EP343 identifies the AR-V7 splice variant. Immunoprecipitation (IP) of M12 cells stably transfected with PCDNA3 plasmid expressing 3×FLAG-tagged cDNAs encoding either AR-FL, ARv567es, or AR-V7. Following IP of cell lysates with a FLAG antibody, blots were probed for (A) AR-NTD or (B) AR-V7 to determine the specificity of the EP343 antibody. (C) Immunoblot staining of various prostate cancer cell lines, both positive and negative for AR-V7 and AR-FL, shows AR-V7 staining (top arrow) along with a number of nonspecific bands and AR-FL (bottom arrow). (D) LNCaP95s were transfected with siRNAs targeting either cryptic exon 3B (si V7), exon 7 (si ex7), exon 1 (si ex1), scrambled SiRNA (si SCR), or an input control. Note that siRNAs directed at components of AR-V7 (si V7 and si ex 1) reduced EP343 staining, but siRNA directed at exon 7 had no effect. (E) IHC on 22Rv1 (positive for AR-NTD and AR-V7), DU145 (negative for AR-NTD and AR-V7) and PC3s which are negative for AR-NTD and should be negative for AR-V7 but have obvious EP343 staining. Both PC3 and 22Rv1 were transfected with AR-V7 (cryptic exon 3B) siRNA and then pelleted for EP343 IHC staining. The targeted siRNA reduced AR-V7 in 22Rv1 IHC but not in PC3.
3.2. IHC
The AR-V7 antibody was next optimised for IHC. 22Rv1 was positive for AR-V7 (HS 150) while DU145 showed no reactivity (HS 0; Fig. 1E). To confirm that this was AR-V7, 22Rv1 was treated with CE3b siRNA and a decrease in staining was observed (HS 70; Fig. 1E). However, PC3 was negative for AR-NTD but showed AR-V7 positivity that was not decreased by CE3b siRNA (Fig. 1E). These data are consistent with the western blot finding that this antibody can react with nonspecific protein targets as well as AR-V7. We next tested LuCaP xenografts for AR-V7 and AR-NTD expression. In neuroendocrine lines, very low or undetectable levels of AR-FL and AR-V7 were observed (Supplementary Fig. 3C). By contrast, xenografts with detectable AR-V7 mRNA yielded positive IHC results (Supplementary Fig. 3A,B), further confirming that EP343 detects AR-V7 in tissue by IHC analysis. Similar results were acquired by both laboratories (LNCaP95, 22Rv1, PC3, and DU145 cell pellets), demonstrating assay reproducibility.
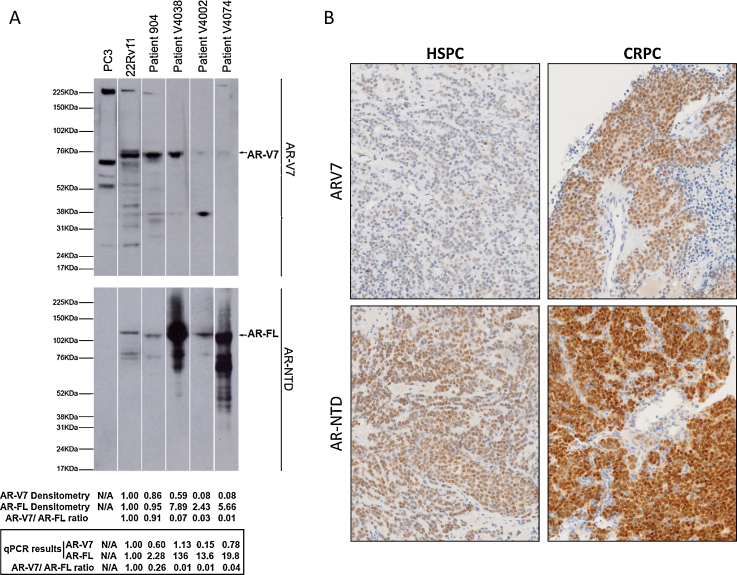
In CRPC liver metastases, AR-V7 nuclear positivity was observed in bile ducts that were AR-NTD negative (Supplementary Fig. 4A). Similarly, AR-V7 positivity was observed for colorectal cancer and basal cells of benign respiratory epithelium (Supplementary Figs. 4C and 5A). Critically, no AR-V7 RNA was observed by RNA in situ hybridisation (Supplementary Figs. 4B,D and 5B). These data indicate that the antibody may also detect a non-CE3b target. Critically, to ensure that the antibody was detecting AR-V7 in our patient cohort, we carried out protein and RNA isolation followed by western blotting and qPCR analysis on fresh tissue from a selection of CRPC patients. Expression of AR-V7 at the correct size was the major protein detected in western blots (Fig. 2A).
Fig. 2.
AR-V7 and AR-NTD protein and mRNA in prostate cancer cell lines and patient samples. (A) Protein and mRNA expression results for cell lines and patient castration resistant prostate cancer (CRPC) biopsy lysates. Patient protein samples show that EP343 binds to AR-V7 and some nonspecific band/s. Densitometry and qPCR results are normalised against 22Rv1 results. (B) AR-V7 and AR-NTD staining of patient hormone-sensitive prostate cancer (HSPC) and CRPC tissues, highlighting increases in both AR-V7 and AR-NTD.
3.3. Nuclear AR-V7 and AR-NTD expression increases as prostate cancer becomes castration resistant, and increases further with AA and/or EZ resistance
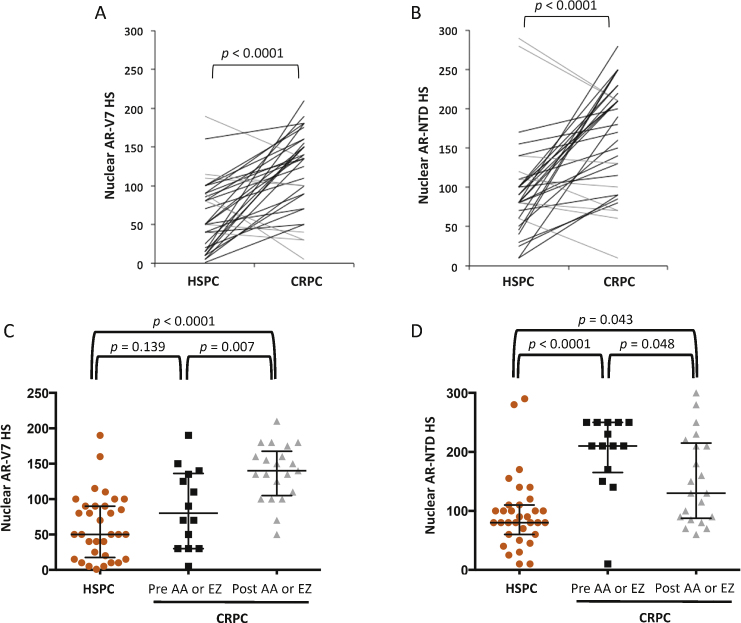
Matched HSPC and CRPC tissue was available from 33 patients to evaluate nuclear expression of AR-V7 and AR-NTD (matched cohort; Supplementary Table 3). To determine changes in nuclear AR-V7 and AR-NTD expression during progression from HSPC to CRPC, HS data for expression were calculated from IHC (Fig. 2B). Nuclear AR-V7 expression levels increased significantly (p < 0.0001) as patients progressed from HSPC (median HS 50, IQR 17.5–90) to CRPC (HS 135, IQR 80–157.5; Fig. 3A). Although nuclear AR-V7 increased in 26 (79%) patients, seven (21%) patients had tumours with higher nuclear AR-V7 expression in HSPC (HS 90, IQR 50–115) compared to CRPC (HS 40, IQR 30–100). Similar to AR-V7, median nuclear AR-NTD expression in patients with HSPC (HS 80, IQR 60–110) significantly increased (p < 0.0001) as tumours progressed to CRPC (HS 180, IQR 95–215; Fig. 3B).
Fig. 3.
Nuclear AR-V7 and AR-NTD expression increases from hormone-sensitive prostate cancer (HSPC) to castration-resistant prostate cancer (CRPC). (A) Nuclear AR-V7 immunohistochemical H score (HS) for matched HSPC and CRPC patient samples. Patients with decreased (grey line) and increased (black line) nuclear AR-V7 expression are shown, with p values for a Wilcoxon matched-pairs signed-rank test. (B) Nuclear AR-NTD HS for matched HSPC and CRPC patient samples. Patients with decreased (grey line) and increased (black line) nuclear AR-NTD expression are shown, with p values for a Wilcoxon matched-pairs signed-rank test. (C) Nuclear AR-V7 HS for HSPC biopsies (orange circles), CRPC biopsies before abiraterone acetate (AA) and enzalutamide (EZ) treatment (black squares), and CRPC biopsies after AA or EZ (grey triangles). Black crosshairs denote the median HS with interquartile range, and p values are for Mann-Whitney tests. (D) Nuclear AR-NTD HS for HSPC biopsies (orange circles), CRPC biopsies before AA or EZ (black squares), and CRPC biopsies after AA or EZ (grey triangles). Black crosshairs denote the median HS with interquartile range, and p values are for Mann-Whitney tests.
To determine the effect of AA or EZ treatment on nuclear AR-V7 and AR-NTD expression, patients with CRPC biopsies (n = 35; 31 patients from the matched cohort and 4 patients with CRPC biopsies only) who received treatment with AA or EZ were divided into two groups: patients who had biopsies before AA or EZ treatment (n = 14) and patients who had biopsies after AA or EZ treatment (n = 21). These were compared to the HSPC biopsies (33 patients from matched cohort). The median time of CRPC biopsy was 3.9 mo (IQR 0–7.7) before AA or EZ and 9.6 mo (IQR 6–23.6) after AA or EZ treatment. Mean nuclear AR-V7 expression increased from HSPC biopsies (HS 50, IQR 17.5–90) to a higher level following castration but before AA or EZ treatment (HS 80, IQR 30–136.3) to the highest level after AA or EZ treatment (HS 140, IQR 105–167.5; Fig. 3C). The increase in nuclear AR-V7 expression between HSPC and after AA or EZ treatment was highly significant (p < 0.0001), as was the increase in nuclear AR-V7 between pre and post AA or EZ (p = 0.007) treatment biopsies. However, the difference in nuclear AR-V7 expression between HSPC and biopsies before AA or EZ treatment was not significant (p = 0.139; Fig. 3C). Median nuclear AR-NTD expression increased from HSPC biopsies (HS 80, IQR 60–110) to CRPC biopsies before AA or EZ treatment (HS 210, IQR 165–250; Fig. 3D). However, median nuclear AR-NTD expression decreased in CRPC biopsies after (HS 130, IQR 87.5–215) compared to before AA or EZ treatment (Fig. 3D). Differences in AR-NTD expression levels among all the groups were statistically significant.
3.4. Nuclear AR-V7 expression in HSPC is correlated with time to castration resistance
Thirty-three patients had HSPC biopsies and all patients developed CRPC whether treated with systemic therapy or radical treatment (prostatectomy and/or radiotherapy). The median time to development of CRPC was 22.3 months (95% confidence interval [CI] 14.3–30.3). Fifteen (45.5%) patients were treated with LHRH agonist with or without anti-androgen therapy, and eighteen (54.5%) patients received radical (radiotherapy or surgery) treatment. The median time to CRPC was longer in patients that received radical therapy than among those treated with androgen deprivation alone (34.2 vs 15.3 mo; hazard ratio [HR] 0.37; 95% CI 0.17–0.81; p = 0.014). Nuclear AR-V7 expression at HSPC biopsy was associated with time to castration resistance in patients treated with systemic therapy exclusively (HR: 1.02, 95% CI 1.002–1.032; p = 0.03), but not among those who received radical treatment (HR 1, 95% CI 0.988–1.008; p = 0.634; Supplementary Fig. 6A,B).
3.5. Nuclear AR-V7 expression and the nuclear AR-V7/AR-NTD expression ratio are associated with worse CRPC prognosis
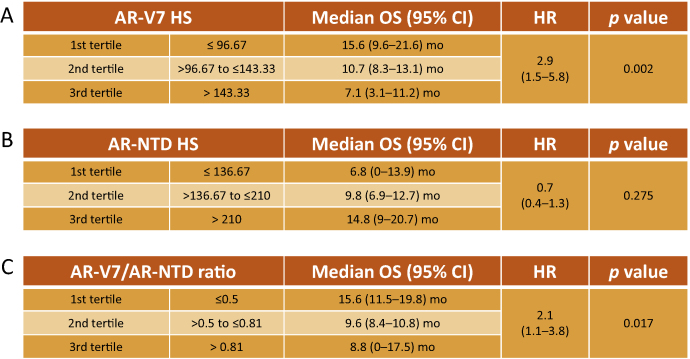
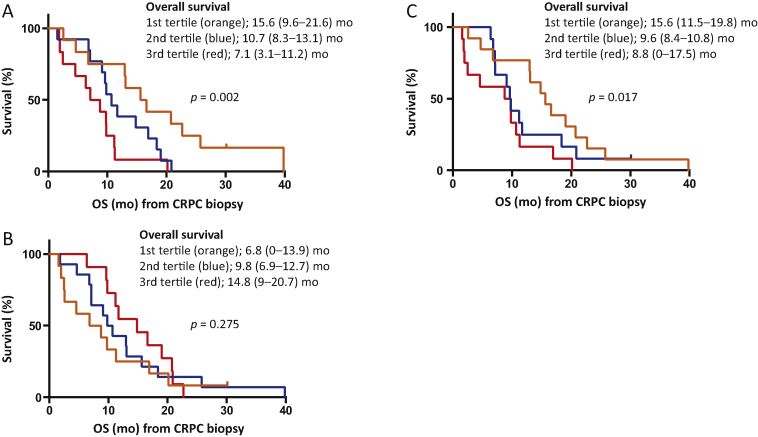
Thirty-seven patients had CRPC biopsies and were evaluable for OS from time of CRPC biopsy. As the prognostic significance of AR-V7 protein levels using EP343 has not been evaluated in patient tissue, we investigated the association of nuclear AR-V7 expression, nuclear AR-NTD expression, and the nuclear AR-V7/AR-NTD ratio in CRPC biopsies with OS from time of CRPC biopsy. Baseline characteristics for the 37 patients included in these analyses are shown in Table 1. Median OS from time of CRPC biopsy was 10.7 mo (95% CI 8.9–12.5). Nuclear AR-V7 (continuous variable) was significantly associated with OS (HR 1.012, 95% CI 1.004–1.020; p = 0.003). We divided the patient cohort into tertiles of nuclear AR-V7 expression; median OS was 15.6 mo (95% CI 9.6–21.6) for patients in the first tertile (HS ≤96.7), 10.7 mo (95% CI 8.3–13.1) for patients in the second tertile (HS >96.7–143.3), and 7.1 mo (95% CI 3.1–11.2) for patients in the third tertile (HS >143.3; Fig. 4, Fig. 5A). There was a significant linear trend in the univariate Cox regression model (p = 0.002).
Table 1.
Baseline characteristics for 37 patients with castration-resistant prostate cancer
| Parameter | Results |
|---|---|
| Median age, yr (IQR) | 67.5 (64.2–75.3) |
| ECOG PS, n (%) | |
| 0 | 4 (10.8) |
| 1 | 32 (86.5) |
| 2 | 1 (2.7) |
| Visceral metastasis (n) | |
| Present | 12 |
| Lung | 4 |
| Liver | 7 |
| Adrenal | 1 |
| Median haemoglobin, g/l (IQR) | 113.0 (105.0–124.5) |
| Median ALP, IU/l (IQR) | 142.0 (69.5–448.5) |
| Median albumin, g/l (IQR) | 34.0 (32.0–36.5) |
| Median LDH, IU/l (IQR) | 204.0 (149.3–290.3) |
| Treatment at/before biopsy, n (%) | |
| Docetaxel | 28 (75.7) |
| Cabazitaxel | 9 (24.3) |
| Abiraterone | 20 (54.1) |
| Enzalutamide | 3 (8.1) |
IQR = interquartile range; ECOG PS = Eastern Cooperative Oncology Group performance status; ALP = alkaline phosphatase; LDH = lactate dehydrogenase.
Fig. 4.
Nuclear AR-V7 and AR-V7/AR-NTD ratio but not nuclear AR-NTD expression are correlated with overall survival (OS) from castration-resistant prostate cancer biopsy. The patient cohort was divided by tertiles for (A) nuclear AR-V7, (B) nuclear AR-NTD, and (C) nuclear AR-V7/AR-NTD expression ratio, and p values for a linear trend test (orthogonal polynomial coefficients) are reported. HS = immunohistochemical H score; CI = confidence interval; HR = hazard ratio.
Fig. 5.
High nuclear AR-V7 and AR-V7/AR-NTD expression ratio are associated with shorter overall survival (OS) in patients with castration-resistant prostate cancer (CRPC). Kaplan-Meier curves show OS from time of CRPC biopsy for patients divided by tertiles for (A) nuclear AR-V7, (B) nuclear AR-NTD, and (C) nuclear AR-V7/AR-NTD expression ratio. Median OS and hazard ratio with 95% confidence intervals are shown, and p values are for a linear trend test (orthogonal polynomial coefficients).
In contrast to nuclear AR-V7 expression, nuclear AR-NTD expression was not significantly associated with OS from time of CRPC biopsy (HR 0.998, 95% CI 0.993–1.003; p = 0.411). Median OS was 6.8 mo (95% CI 0–13.9) for patients in the first tertile (HS ≤136.7), 9.8 mo (95% CI 6.9–12.7) for patients in the second tertile (HS >136.7–210), and 14.8 mo (95% CI 9–20.7) for patients in the third tertile (HS >210; Fig. 5). The linear trend test was not significant (p = 0.275; Fig. 4, Fig. 5B). Next we determined whether the nuclear AR-V7/AR-NTD expression ratio was associated with OS from time of CRPC biopsy. Patients with a lower AR-V7/AR-NTD ratio had significantly longer OS (HR 2.583, 95% CI 1.424–4.684; p = 0.002). Median OS was 15.6 mo (95% CI 11.5–19.8) for patients in the first tertile (ratio ≤0.5), 9.6 mo (95% CI 8.4–10.8) for patients in the second tertile (ratio >0.5–0.81), and 8.8 mo (95% CI 0–17.5) for patients in the third tertile (ratio >0.81; Fig. 4, Fig. 5C). There was a significant linear trend in the univariate Cox regression model (p = 0.017). There were no significant differences in baseline patient characteristics (age, haemoglobin, albumin, alkaline phosphatase, lactate dehydrogenase, visceral metastasis, and performance status) among tertiles for nuclear AR-V7, nuclear AR-NTD, or the nuclear AR-V7/AR-NTD expression ratio (data not shown).
3.6. Nuclear AR-V7 expression and response to AA
Twelve patients had CRPC biopsies before AA treatment and evaluable PSA response data (Supplementary Table 3). The median time for CRPC biopsy before AA initiation was 2.0 mo (IQR 0–9). There was a significant association between the magnitude of PSA decline at 12 wk and the AR-V7 HS (p = 0.043). Two (17%) patients experienced PSA responses (≥50% change from baseline) after 12 wk of treatment (Supplementary Fig. 7). Patients who responded to AA had lower median nuclear AR-V7 (n = 2; HS 30, IQR 30–30) than patients who did not respond (n = 10; HS 80, IQR 45–128.8), although the difference was not significant (p = 0.197).
4. Discussion
The primary objective of this study was analytical validation and clinical qualification of a new antibody to the cryptic exon of AR-V7. This monoclonal antibody detects AR-V7 but may also bind additional, as yet unidentified, proteins, although we have shown that the antibody primarily binds AR-V7 in patient tumour samples. Our AR-V7 IHC assay is best utilised in conjunction with an AR-NTD antibody to minimise false positives, as conducted in our studies. We then evaluated the clinical relevance of IHC staining with this antibody for the following reasons: (1) the majority of published studies have focused on AR-V7 mRNA expression in circulating tumour cells, with AR-V7 protein expression having a much longer half-life than AR-V7 mRNA [24]; (2) there are no published data available on matched HSPC and CRPC tumour samples taken from the same patient; and (3) this antibody is currently being used to detect AR-V7 protein in circulating tumour cells as a putative predictive biomarker in a randomised clinical trial (NCT02485691).
We observed a significant increase in nuclear AR-V7 protein levels from HSPC to CRPC in our patient cohort, of which nearly all patients had received docetaxel and AA. Interestingly, a small subset of patients actually showed the reverse trend, with a decrease in nuclear AR-V7. We hypothesise that while some tumours express AR-V7 in response to androgen ablation, some tumours may acquire resistance to AA or EZ via increased androgen biosynthesis that results in lower AR-V7 expression [12]. An alternative explanation for the decrease may be an increase in other AR variants that are constitutively active. Although AR-V7 is the most common constitutively active AR variant, we have recently described a number of AR-Vs detectable in metastatic biopsy specimens from patients relapsing following AA or EZ therapy [25]. Regardless of the mechanism, however, such biopsies only capture a single time point in the growth of lethal metastases, so the heterogeneity of changes in AR-V7 staining is not unexpected.
Importantly, this study highlights that AR-V7 protein expression is a biomarker of poorer prognosis and increases with emerging drug resistance. Hornberg et al [26] also showed that patients with AR-V7 mRNA expression in the highest quartile had poorer prognosis compared to patients with lower AR-V7 levels [26]. This association was confirmed in a study by Antonarakis et al [3] in which AR-V7 mRNA was determined in metastases as well as in circulating tumour cells [3]. The present study demonstrates a significant trend towards poorer outcome among patients with increased AR-V7 protein expression, in keeping with previous reports for nonmatched cohorts [26], [27], [28], and points to an association between AR-V7 expression and poor prognosis.
5. Conclusions
This is the first longitudinal report demonstrating increasing AR-V7 protein expression in matched same-patient biopsies before and after endocrine treatment from patients with emerging resistance. The data indicate that AR-V7 protein expression increases substantially following castration in many but not all CRPCs, and that AR-V7 levels are associated with poorer outcome, regardless of AR-FL protein expression. However, because the rabbit monoclonal antibody EP343 is not completely specific to AR-V7, its future use to guide therapeutic choice is best pursued alongside AR-FL protein expression. These data support the claim that AR-V7 is important in driving treatment resistance in this disease. Moreover, our finding that AR-V7 expression decreases in some tumours during the emergence of resistance indicates that AR-V7 expression is not the sole mechanism underlying treatment resistance in CRPC.
Author contributions: Johann S. de Bono had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Welti, Rodrigues, Sharp, de Bono, Plymate.
Acquisition of data: Welti, Rodrigues, Sharp, Sun, Riisnaes, Figueiredo, Zafeiriou, Rescigno.
Analysis and interpretation of data: Welti, Rodrigues, Sharp, de Bono, Plymate.
Drafting of the manuscript: Welti, Rodrigues, Sharp, de Bono, Plymate.
Critical revision of the manuscript for important intellectual content: Welti, Rodrigues, Sharp, Sun, Riisnaes, Figueiredo, Zafeiriou, Rescigno, de Bono, Plymate.
Statistical analysis: Sharp, Lorente.
Obtaining funding: de Bono, Plymate.
Administrative, technical, or material support: Welti, Rodrigues, Sharp, Sun, Riisnaes, Figueiredo, Zafeiriou, Rescigno, de Bono, Plymate.
Supervision: de Bono, Plymate.
Other: None.
Financial disclosures: Johann S. de Bono certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: Study funding was received from the Department of Defence (DoD grant no. UWSC7395), Movember/Prostate Cancer UK (grant no. CEO013-2-002), the Prostate Cancer Foundation, and an Experimental Cancer Medical Centre (ECMC) grant from Cancer Research UK and the Department of Health (ref. C51/A7401). The sponsors played a role in the collection, management, analysis, and interpretation of the data.
Acknowledgments: The authors acknowledge NHS funding to the NIHR Biomedical Research Centre at the Royal Marsden and The Institute of Cancer Research. Adam Sharp was funded by a Medical Research Council Fellowship. Stephen Plymate was supported by the NCI Pacific Northwest Prostate Cancer Specialized Program of Research Excellence (2 P50 CA 097186-07), NIH P01 CA163227, US Army Medical Research and Material Command Prostate Cancer Research Program W81XWH-13-2-0093, and a Department of Veterans Affairs Merit Award. We wish to thank Colm Morrissey and Eva Corey, UW Department of Urology, for LuCaP xenograft tissues.
Associate Editor: James Catto
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.eururo.2016.03.049.
Contributor Information
Johann S. de Bono, Email: johann.de-bono@icr.ac.uk.
Stephen R. Plymate, Email: splymate@uw.edu.
Appendix A. Supplementary data
References
- 1.Dehm S.M., Schmidt L.J., Heemers H.V., Vessella R.L., Tindall D.J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nadiminty N., Tummala R., Liu C. NF-κB2/p52 induces resistance to enzalutamide in prostate cancer: role of androgen receptor and its variants. Mol Cancer Ther. 2013;12:1629–1637. doi: 10.1158/1535-7163.MCT-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis E.S., Lu C., Wang H. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu R. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferraldeschi R., Welti J., Luo J., Attard G., de Bono J.S. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene. 2015;34:1745–1757. doi: 10.1038/onc.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attard G., Reid A.H., Yap T.A. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26:4563–4571. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 7.Scher H.I., Fizazi K., Saad F. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 8.de Bono J.S., Logothetis C.J., Molina A. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan C.J., Smith M.R., de Bono J.S. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu R., Lu C., Mostaghel E.A. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ware K.E., Garcia-Blanco M.A., Armstrong A.J., Dehm S.M. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr Relat Cancer. 2014;21:T87–T103. doi: 10.1530/ERC-13-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L.L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenster G., van der Korput H.A., van Vroonhoven C., van der Kwast T.H., Trapman J., Brinkmann A.O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 14.Cao B., Qi Y., Zhang G. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Chan S.C., Brand L.J., Hwang T.H., Silverstein K.A., Dehm S.M. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostaghel E.A., Marck B.Y., Plymate S.R. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res. 2011;17:5913–5925. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson P.A., Chen Y.F., Balbas M.D. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu D., Zhan Y., Qi Y. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res. 2015;75:3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan S.C., Selth L.A., Li Y. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43:5880–5897. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budd W.T., Seashols-Williams S.J., Clark G.C. Dual action of miR-125b as a tumor suppressor and oncomiR-22 promotes prostate cancer tumorigenesis. PLoS One. 2015;10:e0142373. doi: 10.1371/journal.pone.0142373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G., Sprenger C., Wu P.J. MED1 mediates androgen receptor splice variant induced gene expression in the absence of ligand. Oncotarget. 2015;6:288–304. doi: 10.18632/oncotarget.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchini D., Omlin A., Pezaro C. First-in-human phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br J Cancer. 2013;109:2579–2586. doi: 10.1038/bjc.2013.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Detre S., Saclani Jotti G., Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferraldeschi R., Welti J., Powers M.V. Second-generation HSP90 inhibitor Onalespib blocks mRNA splicing of androgen receptor variant 7 in prostate cancer cells. Cancer Res. 2016;76:2731–2742. doi: 10.1158/0008-5472.CAN-15-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakazawa M., Antonarakis E.S., Luo J. Androgen receptor splice variants in the era of enzalutamide and abiraterone. Horm Cancer. 2014;5:265–273. doi: 10.1007/s12672-014-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornberg E., Ylitalo E.B., Crnalic S. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu Y., Dai B., Ye D. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efstathiou E., Titus M., Wen S. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol. 2015;67:53–60. doi: 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.