Abstract
Neutrophils and other amoeboid cells chemotax by steering their front towards chemoattractant. While Ras, Rac, Cdc42, and RhoA small GTPases all regulate chemotaxis, it has been unclear how they spatiotemporally control polarization and steering. Using fluorescence biosensors in neutrophil-like PLB-985 cells and photorelease of chemoattractant, we show that local Cdc42 signals, but not those of Rac, RhoA or Ras, precede cell turning during chemotaxis. Furthermore, preexisting local Cdc42 signals in morphologically unpolarized cells predict the future direction of movement upon uniform stimulation. Moreover, inhibition of actin polymerization uncovers recurring local Cdc42 activity pulses, suggesting that Cdc42 has the excitable characteristic of the compass activity proposed in models of chemotaxis. Globally, Cdc42 antagonizes RhoA, and maintains a steep spatial activity gradient during migration, while Ras and Rac form shallow gradients. Thus, chemotactic steering and de novo polarization are both directed by locally excitable Cdc42 signals.
Introduction
Neutrophils are professional chemotactic cells that rapidly migrate towards sites of tissue injury and infection. They initiate directed cell migration (chemotaxis) in response to sources of chemoattractants such as N-formyl-Methionine-Leucine-Phenylalanine (fMLF). Even in response to spatially uniform increases in chemoattractant, neutrophils polarize and move in a curving random walk behavior termed chemokinesis1–3. However, when such a migrating cell experiences a gradient of attractant, it gradually turns its front more often towards the higher concentration to generate a biased random walk behavior4–7. This directed gradual turning of the front of migrating cells has been termed chemotactic steering8. To computationally reproduce these two distinct directional control mechanisms, theories of chemotaxis of amoeboid cells such as neutrophils and Dictyostelium discoideum require that combined positive and negative feedback circuits generate an excitable network to produce a local compass activity9–11. Molecularly, polarization and chemotactic steering are controlled by chemoattractants such as fMLF that activate G-protein coupled receptors to regulate phosphoinositide 3-kinase (PI3K), Ras, Rac, Cdc42, RhoA and other signals, which in turn control dynamic changes in actin and myosin11–16.
Different studies have shown that PI3K, Ras, Rac, Cdc42 and RhoA can all be activated by positive feedback1,11,17–24, suggesting that each of them has the potential to be the elusive chemotactic compass in excitable network models. Although PI3K signaling initially emerged as the leading candidate among these putative compass activities11,25,26, it has since been shown that cells can chemotax in the absence of PI3K activity, albeit less effectively27,28. On the other hand, genetic studies have shown that Rac, Cdc42 or RhoA knockout leukocytes and Ras mutant Dictyostelium all have severely impaired chemotaxis18,29–33. Even though Rac has been a leading candidate to direct the steering of neutrophils34,35, the observed feedbacks for the other GTPases suggest that local Ras or Cdc42 signaling at the front or, alternatively, RhoA signaling at the cell back could be responsible for steering.
A major limitation for understanding chemotaxis has been that we do not know if and how small GTPases are spatiotemporally coordinated when neutrophils polarize, migrate, and steer towards chemoattractant. Here we show that local Cdc42 signals within the front of migrating cells direct turning towards chemoattractant to mediate the chemotactic steering behavior. We further show that basal local Cdc42 signals direct de novo polarization to mediate the chemokinesis migration behavior. Finally, we show that Cdc42 activity exhibits local excitability, a requirement for Cdc42 to be the elusive chemotactic compass in excitable network models of chemotaxis9,10.
Results
Light induced activation of chemotaxis
We investigated the spatiotemporal dynamics of small GTPase signaling in neutrophil-like PLB-985 cells by monitoring GTPase activity using stably expressed fluorescence resonance energy transfer (FRET) biosensors36. Since expression of GTPase biosensors can perturb cell migration through interactions with endogenous components, we sorted cells to achieve relatively low and consistent expression levels. Using a systematic chemotaxis assay we developed recently37, we confirmed that cells expressing each of the biosensors have approximately equal speed, chemokinesis and directionality as those of sensor-free cells (Supplementary Fig. 1a-d).
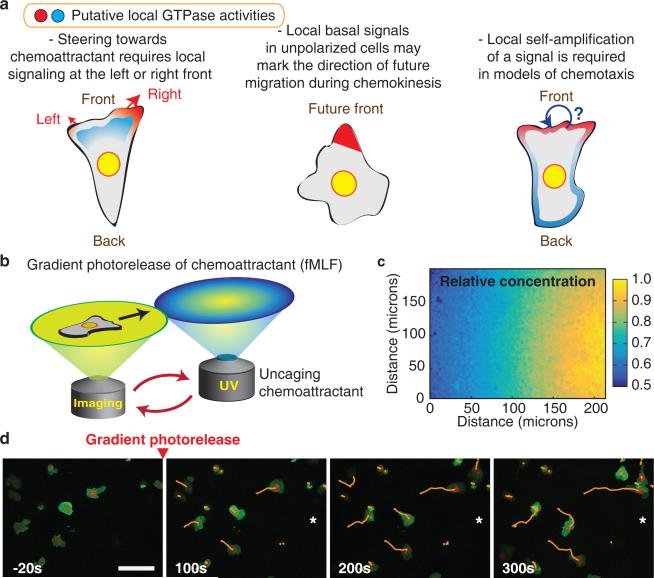
To more closely reflect a neutrophil's migration environment in vivo, we used an under agarose system which squeezes cells into a confined space where they effectively polarize and chemotax38,39. We generated gradients of chemoattractant by employing a chemically caged derivative of a fMLF (N-nitroveratryl derivative fMLF; Nv-fMLF)37,40 combined with automated ultraviolet (UV) illumination to shape chemoattractant gradients (Fig. 1b). Gradient protocols were calibrated and optimized using caged fluorescein (Fig. 1c). In response to attractant uncaging, cells activated signaling pathways (Supplementary Fig. 1e,f) and rapidly migrated in a biased random walk toward higher fMLF concentrations (Fig. 1d).
Figure 1.
Neutrophil chemotaxis controlled by automated photorelease of chemoattractant. (a) Schematic representation of the chemokinesis and chemotaxis processes. De novo polarization and chemotactic steering are key directional mechanisms. (b) Schematic figure of the microscope system used to generate gradients of the chemoattractant fMLF by light-triggered photorelease. In between imaging acquisitions, the stage is moved to deliver an uncaging pulse at a defined position relative to the imaging field of view. (c) Control experiment, visualizing the gradient of light-induced photorelease of caged fluorescein by confocal microscopy. Color bar indicates relative fluorescence intensity. (d) Movement of cells in a biased random walk in a chemoattractant gradient. A membrane marker is shown in green, a nuclear marker in red, and cell tracks are overlaid. The direction of the chemoattractant gradient is indicated with a *. Time relative to initial gradient generation is indicated. Scale bar is 50μm.
Steep polarization of Cdc42 and RhoA activities – and broader patterns of Ras and Rac activities – at the leading edge of chemotaxing neutrophil-like cells
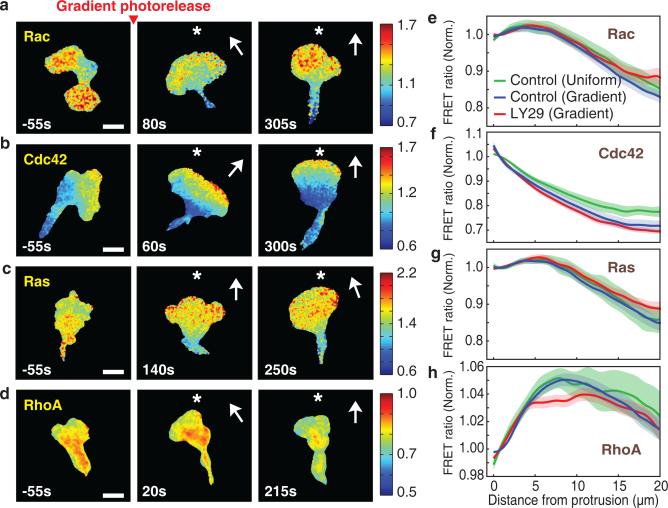
We measured spatial activity gradients of Ras, Rac, Cdc42 and RhoA in chemotaxing cells as a function of distance backwards from the protruding leading edge (Supplementary Fig. 2a). We averaged spatial profiles over time and then normalized to the levels at the front edge (Supplementary Fig. 2b,c). Consistent with earlier polarity studies1, we confirmed that Rac and Cdc42 activities were higher at the front of cells, while RhoA activity was lower at the front (Fig. 2a,b,d). However, the activity gradient of Rac was much less steep than that of Cdc42 (Fig. 2a,b and Supplementary Video 1,2) and the activity of Cdc42 peaked at the leading edge (Fig. 2f), while Rac activity peaked 5 micrometers behind (Fig. 2e). The activity of Ras, an important regulator of Dictyostelium chemotaxis and a regulator of PI3K and other processes in neutrophils41, was polarized with a similarly shallow gradient as Rac (Fig. 2c,g and Supplementary Video 3). Finally, we observed a steep depletion of RhoA activity at the leading edge with an activity gradient that was inverse at the front to that of Cdc42 (Fig. 2d,h and Supplementary Video 4). Interestingly, these intracellular GTPase signaling gradients are largely independent of whether or not an external chemoattractant gradient is present (Fig. 2e-h).
Figure 2.
Cdc42 and opposing RhoA activities are steeply polarized at the front. (a-d) Spatial activity profiles of Rac (a), Cdc42 (b), Ras (c), and RhoA (d) in a light-induced gradient of fMLF gradient (approximately 8% across a cell). Time relative to gradient stimulation is marked and the direction of the chemoattractant gradient is indicated with a *. Color bars indicate the range of biosensor FRET ratios. Scale bar is 10μm. (e-h) Relative activities of Rac (e), Cdc42 (f), Ras (g), and RhoA (h) in migrating cells as a function of distance backwards from the leading edge. Values are normalized to the levels at the front edge. Error bars indicate ± s.e.m. of n=10 (Rac control uniform), n=18 (Rac control gradient), n=18 (Rac LY29 (50μM) gradient), n=9 (Cdc42 control uniform), n=26 (Cdc42 control gradient), n=16 (Cdc42 LY29 (50μM) gradient), n=13 (Ras control uniform), n=19 (Ras control gradient), n=23 (Ras LY29 (50μM) gradient), n=10 (RhoA control uniform), n=44 (RhoA control gradient), and n=14 (RhoA LY29 (50μM) gradient) cells. Time averaged gradients are shown including all cells measured for each GTPase.
We further tested whether the spatial gradients of the GTPase activities are dependent on PI3K activity by repeating the above experiment in the presence of the PI3K inhibitor LY294002 (LY29), which blocked the membrane translocation of a PIP3-responsive PH domain of Akt (PHAkt) (Supplementary Fig. 3a,b). Consistent with previous findings that PI3K is not required for chemotaxis27,28, the relative activity profiles of all four GTPases were unaffected (Fig. 2e-h and Supplementary Fig. 3c). Thus, the protrusive leading edge is marked by steep opposing local gradients of Cdc42 and RhoA activities, with more broadly extending gradients of Rac and Ras activities, and our results suggest that these intracellular gradients are generated by cell-intrinsic and PI3K-independent feedback mechanisms.
Local Cdc42 signals within the cell front precede turning towards chemoattractant
As neutrophils polarize and migrate, their sensitivity to chemoattractant is high in the front and suppressed in the back42. To turn towards a chemoattractant source, neutrophils must therefore compute chemoattractant concentration as a left versus right difference across the cell front4,6,9. While exogenous local activation of Rac at the front has been shown to induce cell turning48, it is not yet known whether the endogenous mediator of steering is indeed Rac. Per definition, the endogenous mediator of steering must transiently increase in the left or right side of the front before cells turn during chemotactic steering.
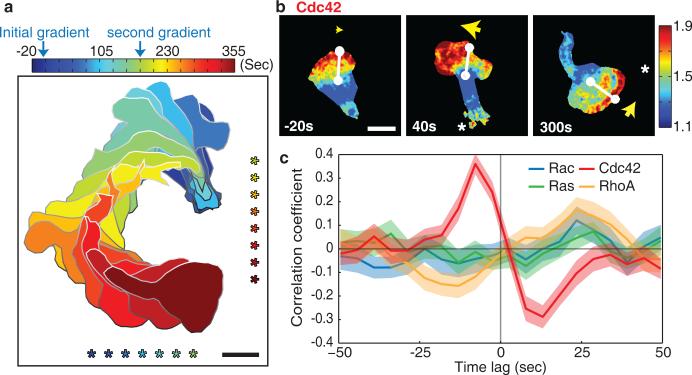
In order to observe a large number of turning events under standardized conditions, we induced efficient turning by switching the direction of an fMLF gradient from the bottom to the right of the imaging area (Fig. 3a). We then performed a temporal cross-correlation analysis between (i) the GTPase activity difference between the left and right sides of the cell front and (ii) the angular change in the direction of migration (see Methods for details). Strikingly, only local elevation of Cdc42 signals preceded turning of migrating cells, with a temporal offset of about 7.5 seconds (Fig. 3b,c and Supplementary Video 5). No such correlation was observed for local Ras or Rac signaling (Fig. 3c and Supplementary Fig. 4a,b). RhoA showed an opposing albeit weaker correlation, with depletion of its activity preceding turning by a similar time (Fig. 3c and Supplementary Fig. 4c). We confirmed our results with an alternate analysis modeled after a strategy used in fibroblasts45 by defining a “signaling vector” as a weighted average of vectors pointing towards cell edge regions of high GTPase activity. The direction of this signaling vector correlated with future cell turning only for Cdc42 activity (Supplementary Fig. 4d,e). These results suggest that of the four small GTPases, only higher Cdc42 and possibly lower RhoA activities fit the required activity patterns for chemotactic steering of cells.
Figure 3.
Endogenous local Cdc42 signal differences between the left and right cell front direct turning towards chemoattractant. (a) Masks of the shape and position of a cell are overlaid for a series of timepoints as the cell was exposed to changing chemoattractant gradient conditions to induce turning. The direction of the fMLF gradient is marked *. The time sequence is color coded. Scale bar is 10μm. (b) Cdc42 activity in a cell during chemotactic turning induced by switching the location of the photorelease of Nv-fMLF. A white line connects the centroid of the cell to the center of the cell front. A yellow arrow marks the cell front. Cdc42 activity is represented as a color scale. Time relative to stimulation is indicated and the direction of the chemoattractant gradient is indicated with a *. Scale bar is 10μm. (c) Comparison of the temporal cross-correlation analysis between different GTPase activities between the left and right sides of the cell front and the angular change in direction of migration. Error bars indicate ± s.e.m. of n=41 (Rac), n=47 (Cdc42), n=42 (Ras), and n=47 (RhoA) cells.
Only local basal Cdc42 activity predicts the future direction of polarization
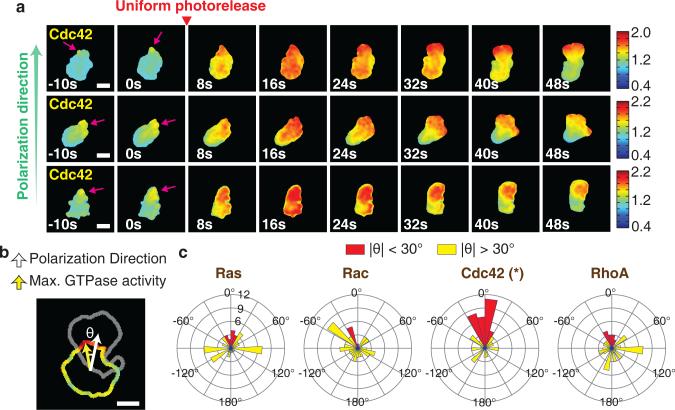
Before neutrophils start to migrate during chemokinesis or chemotaxis, the unpolarized cell must break symmetry. We investigated the spatial dynamics of GTPase activities during de novo polarization of PLB-985 cells using uniform photorelease of Nv-fMLF to trigger a chemokinesis response (Supplementary Fig. 5a). We first investigated GTPase activities in morphologically unpolarized cells and observed locally fluctuating activities for all GTPases (Supplementary Fig. 5b). Using time-lapse analysis, we noticed that small regions of elevated Cdc42 activity prior to stimulation appeared to mark the cell's eventual direction of polarization (Fig. 4a and Supplementary Video 6). To quantify this observation, we measured the angle between the site of maximal GTPase activity at the cell periphery before stimulation and the eventual direction of polarization after stimulation (Fig. 4b). Pre-existing asymmetry in Cdc42 activity was highly predictive of the future direction of cell polarization. In contrast, asymmetry in Ras, Rac, and RhoA activity showed no predictive power (Fig. 4c). The same analysis showed a clear correlation between the lowest peripheral Cdc42 activity and the future back orientation, but there was no correlation between the lowest RhoA activity and the future front (Supplementary Fig. 6a). This suggests that fluctuating Cdc42 activities direct symmetry breaking and de novo polarization upon induction of chemokinesis. Thus, rather than relying on two separate mechanisms, leukocytes employ Cdc42 signals for both the chemotactic steering of already polarized migrating cells and the de novo polarization of unpolarized cells.
Figure 4.
Basal Cdc42 activity fluctuations in morphologically unpolarized cells predict the future direction of cell polarization. (a) Examples of morphologically unpolarized cells with asymmetric Cdc42 activity prior to stimulation (left panels). Pink arrows indicate the site of pre-existing Cdc42 activity at cell periphery. Cdc42 activity is indicated by the color scale. Scale bar is 10μm. (b) Schematic representation for analyzing the angular difference between direction of movement and direction of the maximal peripheral GTPase activity prior to stimulation. Shown are the contour of the edge of a cell before stimulation, colored according to the FRET ratio, overlaid with the contour (in gray) of the same cell after polarization and initial movement. Scale bar is 10μm. (c) Rose plots showing distributions of the angle between the maximal local GTPase activity before stimulation and the subsequent migration direction. Red color indicates angles between -30 and 30 degrees. P-values were calculated using the sign test applied to the cosine of the angles. * indicates p-value is less than 0.0001. n=68 (Rac), n=69 (Cdc42), n=63 (Ras), and n= 65 (RhoA) cells.
Pulsatile and spatially focused Cdc42 activity reveals characteristics of a locally excitable network
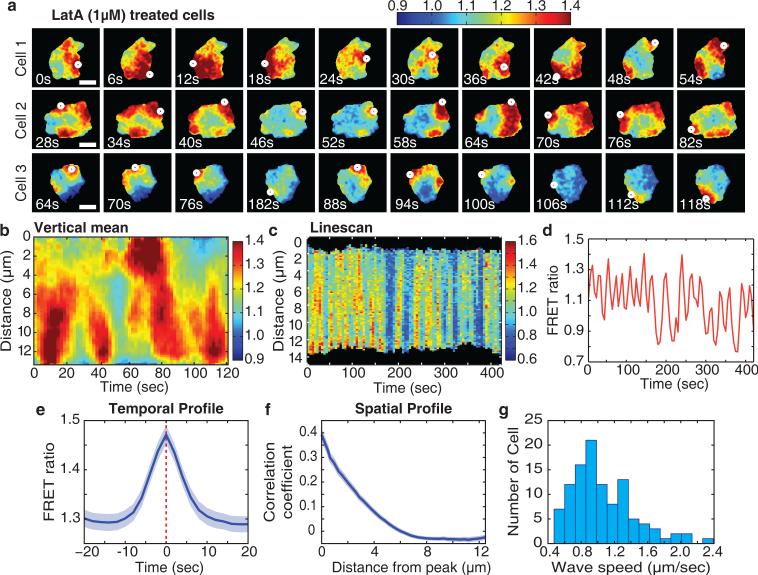
Most models for chemotaxis require that the compass activity directing the turning of migrating cells is locally amplified within an excitable network9,10,46–49. Since the actin cytoskeleton and small GTPase signals are connected by feedback16,26, we determined whether actin polymerization is necessary for local Cdc42 increases. Surprisingly, upon inhibition of actin polymerization using Latrunculin A (LatA), we observed high amplitude local pulses of Cdc42 activity that changed their intensity and location over time even in the absence of chemoattractant (Fig. 5a,b and Supplementary Video 7). This behavior of Cdc42 was independent of PI3K (Supplementary Fig. 7) and reminiscent of pulsatile activations of Ca2+ and Erk signaling that involve autocatalytic positive feedback paired with a slower negative feedback50,51. The pulsatile behavior of Cdc42 activation persisted for long time periods (Fig. 5c,d) with a typical pulse duration of about 10 seconds (Fig. 5e), a median time between peaks of 25 seconds (Fig. 5d) and a local spatial diameter of about 5 microns (Fig. 5f). Further indicative of a locally excitable system, we observed in many cells Cdc42 activation waves that propagated across the length of a cell at a speed of about 1.1 micrometer/second (Fig. 5a,b,g).
Figure 5.
Locally pulsatile Cdc42 activity in the absence of actin polymerization argue for the existence of an excitable network. (a) Examples of locally pulsatile Cdc42 activity revealed by treating cells with LatA (1μM). Cdc42 activity is indicated by the color scale. White dots mark the location of maximum Cdc42 activity. The images were taken every 2 seconds for 120 seconds. Scale bar is 10μm. (b) Kymographs of Cdc42 activity for a cell 1 in Figure 5a treated with LatA (1μM), showing different time scales. The Cdc42 activity was averaged over the vertical (y-axis) direction to get a one dimensional profile for each timepoint. For this analysis, images were taken at 2 second intervals. Cdc42 activity is indicated by the color scale. (c) Kymograph of a line scan Cdc42 activity in a LatA (1μM)-treated cell. For this analysis, images were taken every 5 seconds for 420 seconds. Cdc42 activity is indicated by the color scale. (d) Quantitative measurements of local Cdc42 activity in LatA (1μM)-treated cells. Shown is a temporal trace for a selected 5 μm square region within an individual cells. For this analysis, images were taken every 5 seconds for 420 seconds. (e) Temporal profile of peaks of Cdc42 activity in LatA (1μM)-treated cells. Peaks of Cdc42 activity were automatically detected from timecourses of activity in 6 μm square regions within cells. The peaks were aligned at their maxima and then averaged. For this analysis, images were taken at 2 second intervals. Error bars indicate ± s.e.m. of n=66 cells. (f) Spatial autocorrelation of Cdc42 activity in LatA (1μM)-treated cells. For this analysis, images were taken at 2 second intervals. Error bars indicate ± s.e.m. of n=85 cells. (g) Histogram of instantaneous speeds of Cdc42 waves. n=107 cells. For this analysis, images were taken at 2 second intervals.
The observation of actin-independent Cdc42 pulsatile activation was surprising since previous models of chemotaxis included actin polymerization as part of positive feedback-driven local excitatory loops16,26. As local Cdc42 signals do not spatially propagate in the absence of LatA, actin polymerization may instead have a role in locally stabilizing pulses of Cdc42 activity. Thus, rather than mediating a gradual increase in activity, signaling from the receptor may trigger local pulses of Cdc42 activity, a characteristic feature of the chemotactic compass in excitable network models.
Cdc42 and RhoA polarize earlier than Ras and Rac, coincident with the first polarized membrane protrusion
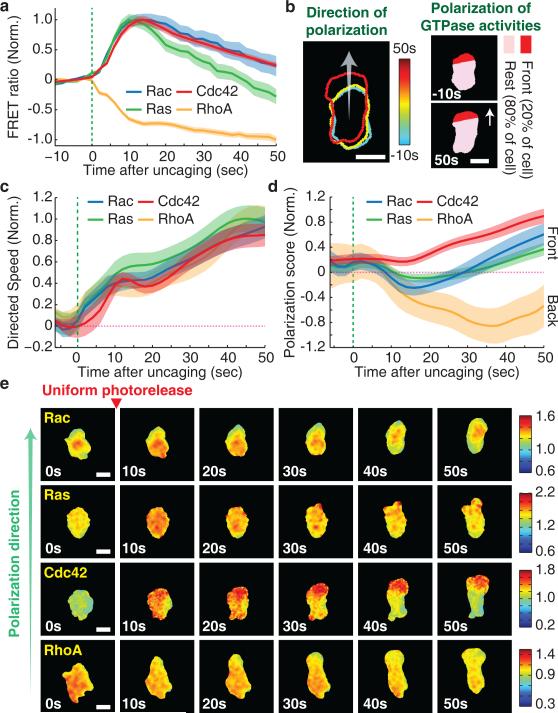
To gain insight into how Cdc42 might direct cell steering downstream of receptor activation, we monitored the spatiotemporal signaling dynamics of the four GTPases. We first found that upon uniform stimulation of unpolarized cells, the activities of Ras, Rac, and Cdc42 increased over a ten second interval with almost identical kinetics and that the activity of RhoA decreased during the same interval (Fig. 6a). However, it takes cells approximately 50 seconds to reach a steady state migration speed (Supplementary Fig. 8a). We therefore examined the spatial activity profiles of the GTPase signals over this time frame, and we found markedly different dynamics. We computationally defined the direction of polarization and quantified polarization of GTPase activities by measuring the difference in activity between the site of the eventual cell front and the cell body as a function of time (Fig. 6b). As an important control, we showed that the time course of the initial protrusion event was independent of the expression of the sensors (Fig. 6c). We found that Cdc42 and RhoA activities polarized simultaneously with the initial onset of polarized membrane protrusion, approximately 20 seconds before a polarization of Ras and Rac could be observed (Fig. 6d,e). Rac activity showed initially a slight relative depletion of its activity in the nascent protrusion, before polarizing maximally towards the front with a delay. In contrast, the maximal induction of Cdc42 and RhoA gradients correlated with cells reaching their steady state speed, providing further support for a central role of Cdc42 and potentially RhoA in de novo polarization.
Figure 6.
Cdc42 and RhoA activities polarize before Rac and Ras activities during de novo polarization and induced chemokinetic movement. (a) Kinetics of cell-averaged GTPase activities after uniform photorelease. Values were normalized by the initial and maximum activity. Green line marks the start of gradient generation by photorelease. Error bars indicate ± s.e.m. of n=30 (Rac), n=35 (Ras), n=31 (Cdc42), and n=67 (RhoA). (b) Left, the contours of the cell edge are overlaid for several color coded timepoints during polarization and movement. Arrow indicates the computationally determined polarization direction. Right, schematics explaining the analysis of the polarization of GTPase activities. The front 20% of cell pixels were determined automatically based on the polarization direction. Scale bar is 10μm. (c) Overlay of timecourses of directed speed for cells with FRET biosensors during de novo polarization in Figure 6d. Directed speed was measured as the rate of movement of the cell centroid in the direction of eventual cell polarization. Each curve is labeled according to the FRET biosensor expressed in the corresponding cells. Green dotted line marks the time of chemoattractant release. Error bars indicate ± s.e.m. of n=26 (Rac), n=28 (Ras), n=33 (Cdc42), and n=26 (RhoA) cells. (d) Timecourse of polarization of GTPase activity (scored by FRET ratio in the front 20% of pixels minus the ratio in the back 80% of pixels) after uniform photorelease of Nv-fMLF. Values were normalized by the maximum (Rac, Ras, and Cdc42) or minimum (RhoA) activity. Green dotted line marks the time of chemoattractant release. Error bars indicate ± s.e.m. of n=26 (Rac), n=28 (Ras), n=33 (Cdc42), and n=26 (RhoA) cells. (e)Time-course of GTPase activities as the cell polarize after photorelease. Color bars indicate the range of biosensor FRET ratios. Scale bar is 10μm.
Local activation of Cdc42 and inactivation of RhoA are correlated in space and time with local membrane protrusion
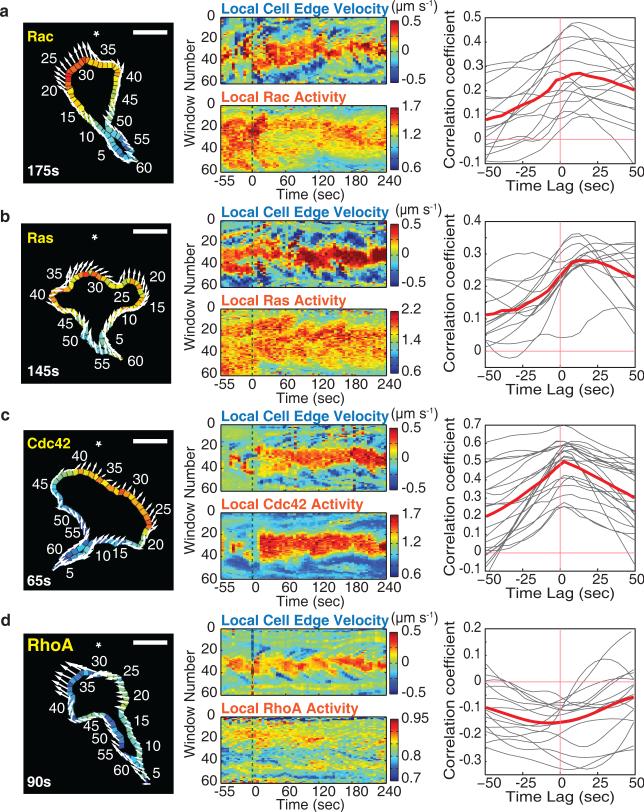
We further examined the relationship between local GTPase activities and membrane protrusion in steadily moving cells by performing a spatial correlation analysis. We parameterized the edge of the cell into 60 evenly spaced regions (Supplementary Video 2) and measured FRET signals for each local region at each time point, as well as the local protrusion or retraction rate of the corresponding cell edge segment between frames. In a spatial kymograph analysis, we found that elevated Cdc42 activity and depleted RhoA activity correlated tightly in space with membrane protrusion, whereas Ras and Rac activities were increased over a much broader region than the protruding membrane segment (Fig. 7a-d; middle). We next performed a temporal cross-correlation analysis that has previously been performed for RhoA, Rac and Cdc42 in slow migrating and stationary fibroblasts52. Rac activity followed protrusion (Fig. 7a), similar to the results for Rac in fibroblasts. Local Ras activity, which was not analyzed in fibroblasts, also followed with a lag after protrusion (Fig. 7b). However, our results in the fast moving neutrophil-like cells differed from the results in the much slower fibroblasts52,53 in that the activity of Cdc42 and reduced RhoA activity were both correlated with local protrusion with little or no time offset (Fig. 7c,d). The slower moving fibroblasts may have increased rather than decreased RhoA activity at the leading edge since they are known to exhibit much more prominent oscillatory membrane retraction events. Together with the close spatial antagonism between Cdc42 and RhoA in Figure 2 and 6, this suggests that locally increased Cdc42, supported by the absence of local RhoA activity, directs the sites of local protrusions also for the forward movement of neutrophils.
Figure 7.
Cdc42 and opposing RhoA activities correlate spatiotemporally with local protrusion during cell migration. (a-d) Left, example image of local GTPase activities measured in 60 segmented regions along the cell edge. Arrows indicate the movement of the corresponding edge segments between sequential images. The direction of the chemoattractant gradient is indicated with a *. Time after stimulation is marked. Scale bar is 10μm. Middle, spatial and temporal activity maps of edge segment protrusion dynamics against local GTPase activities. Color bars indicate protrusion-retraction dynamics and activation level of FRET biosensors. Green dotted lines mark the start of gradient generation by photorelease. Right, temporal cross-correlation analysis between edge protrusion and the respective GTPase activities averaged over all sampling segments as a function of time offset. Negative time lag values indicate GTPase activity preceding protrusion activity. The gray curves are data for individual cells, and the red curves represent the mean for all cells measured. n=15 (Rac), n=14 (Ras), n=22 (Cdc42), and n=15 (RhoA) cells.
Cdc42 antagonizes RhoA activity
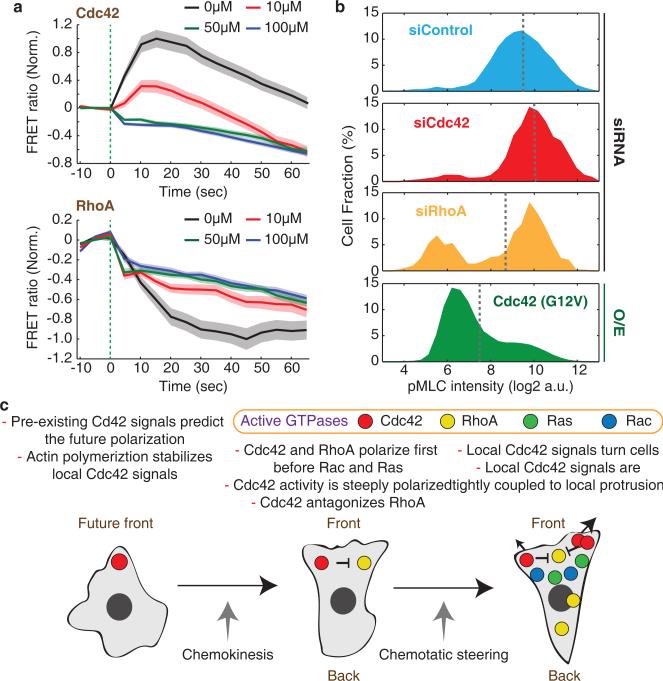
Since Cdc42 and RhoA activities were spatiotemporally anticorrelated in the front region, we investigated whether Cdc42 may suppress RhoA activity. Such a mechanism was plausible based on a study in endothelial cells that showed that Cdc42 activation inhibited RhoA but not vice versa54. We stimulated cells by photorelease of Nv-fMLF in the presence or absence of increasing concentrations of ZCL278, an inhibitor that is specific for Cdc42 over Ras and Rac55 (Supplementary Fig. 8b), and measured the kinetics of Cdc42 and RhoA responses. Concentrations of ZCL278 that partially or completely blocked activation of Cdc42 resulted in reduced RhoA activity changes in response to fMLF (Fig. 8a), suggesting that Cdc42 is inhibiting RhoA.
Figure 8.
Cdc42 antagonizes RhoA activity. (a) Effect of the Cdc42 inhibitor ZCL278 on the kinetics of cell-averaged Cdc42 (Top) and RhoA (Bottom) activities in response to chemoattractant photoreleasae. Values were normalized by the initial and maximum activity. Green dotted lines mark the time of chemoattractant release. Error bars indicate ± s.e.m. of n=73 (Cdc42 0μM), n=75 (Cdc42 10μM), n=89 (Cdc42 50μM), n=114 (Cdc42 100μM), n=88 (RhoA 0μM), n=83 (RhoA 10μM), n=104 (RhoA 50μM), and n=130 (RhoA 100μM) cells. Of note, we observed a small but immediate apparent decrease in the observed FRET ratios for both sensors that we believe is an artifact from interaction between UV light and the ZCL278 compound. (b) Histogram of phosphomyosin light chain (pMLC) intensities measured in individual cells by immunofluorescence for control cells and cells perturbed by Cdc42 or RhoA knockdown or expression of a constitutive active Cdc42. Gray dotted lines mark mean pMLC intensities. n=8635 (siControl), n=2419 (siCdc42), n=634 (siRhoA), and n=22201 (Cdc42 (G12V)) cells. (c) Proposed model of GTPase activation for neutrophil polarization. Cdc42 activity has an autocatalytic character and locally inhibits RhoA activity to define and steer the front of the cell. Ras and Rac polarize later and support polarization.
We further investigated a link from Cdc42 to RhoA by using immunofluorescence to measure levels of phospho-Myosin Light Chain (pMLC), a main output of RhoA signaling56, under conditions where we perturbed either Cdc42 or RhoA activity (Supplementary Fig. 8c). A histogram of the single cell pMLC levels showed a bimodal distribution, consistent with a bistable regulation of the upstream small GTPase signaling system (Fig. 8b). Cdc42 knockdown and incubation with ZCL278 each led to an increases in pMLC, whereas RhoA knockdown caused a reduction of pMLC (Fig. 8b and Supplementary Fig. 8d). Expression of a constitutively active Cdc42 (G12V) resulted in a strong reduction in pMLC levels (Fig. 8b). These observation support the hypothesis that Cdc42 antagonizes myosin-mediated contraction in neutrophils by locally inactivating RhoA1, arguing that Cdc42 is the primary GTPase that directs chemotactic steering.
Discussion
Our results suggest that Cdc42, rather than the other likely candidates, PIP3, Rac, Ras, or RhoA, acts as the compass activity that directs chemotactic steering. Furthermore, our finding that Cdc42 signals exhibit local excitability fulfills a key requirement of many models of symmetry breaking, polarization, and chemotactic steering17,46,47,57. Our results were surprising given that direct homologs of Cdc42 are absent in Dictyostelium58, an organism with chemotactic behavior very similar to that of neutrophils. However, locally excitable Cdc42 signals have been shown to orient mating protrusions in the Saccharomyces cerevisiae48,49,59. A necessary role for Cdc42 in chemotaxis is also supported by previous studies in leukocytes showing that knockout of Cdc42 or expression of dominant negative Cdc42 results in multiple fronts, reduced persistence and suppressed chemotaxis16,29,60.
One of the most intriguing aspects of chemotaxis is that cells typically respond to changing attractant gradients with gradual turning while maintaining cell polarity, rather than with abrupt changes of direction. This observation can be explained if cell polarity is bistable, generating core intracellular signaling gradients independent of external chemoattractant gradients, and if polarized cells steer by sensing differences in chemoattractant concentration across the cell front. Consistent with this framework, the sensitivity of receptor signaling in leukocytes is restricted to the front42 and abstract models introducing a receptor-controlled left versus right steering signal can recreate the biased random walks of chemotaxing cells4,6. Our study provides two major insights into this process. First, we found that the shape of the intracellular gradients of Cdc42, Rac, Ras, and RhoA activities are nearly identical whether or not cells are in a chemoattractant gradient (Fig. 2e-h). Thus, control of polarization and migration is indeed likely to be bistable, with intracellular signaling gradients determined primarily by cell-intrinsic feedback connections. Second, we found that local Cdc42 activity, and the linked suppression of RhoA activity, are the key molecular signals downstream of receptor inputs that mediate left versus right steering towards the chemoattractant source. Nevertheless, rather than driving a steady graded bias, receptor input is integrated into a locally excitable Cdc42 network to generate pulses of activity that can direct turning in the context of stable cell polarity.
As an additional clue for understanding the connections between cell steering and polarity, we observed that the fast-acting locally excitable Cdc42 circuit was often not sufficient to define a single front. Upon cell stimulation when there was no clear preexisting asymmetry in Cdc42 activity, we frequently observed two initial protrusions at opposing ends of the cell, each marked by elevated Cdc42 activity. In all cases, one of the two protrusions gradually disappeared within approximately 60 seconds, leaving only one Cdc42-marked cell front remaining (Supplementary Fig. 6b). This suggests that locally excitable Cdc42 signals drive the generation of one or more initial protrusive fronts, but additional mechanisms are needed to restrict Cdc42 activity to a single front. Most models of chemotaxis include such global mechanisms to restrict a local compass activity9,61–63.
Previous studies showed that Rac is necessary for the migration and chemotaxis of neutrophils18,30,31, and that Rac, rather than Cdc42, induces most of the Arp2/3-mediated branched actin polymerization required for motility64,65. Our finding that Rac activity is elevated over a broad spatial region supports a previous observation that waves of actin polymerization, mediated by the Rac-regulated WAVE complex, frequently start in the middle of the cell and move towards the front66. The distinct spatial dynamics of Rac and Cdc42 suggest that these waves of actin polymerization must coordinate at the front with sites of locally high Cdc42 and low RhoA activity to cooperatively promote local membrane protrusion.
Polarized Rac activity has been proposed to result from positive feedback between Rac and actin polymerization26,67. We observed correlation between protrusion and Rac activity (Fig. 7a) that supports the existence of this positive feedback. However, the increase in Rac activity is delayed by about 10 seconds after protrusion and it persists for tens of seconds afterwards. Thus, our results argue that this positive feedback acts on a slower timescale and over a larger region in space than the locally excitable Cdc42 circuit. We also found that Rac and Ras have nearly identical spatial and temporal activity profiles, suggesting that they may be part of the same global positive feedback circuit.
Our study argues that locally excitable Cdc42 activity acts in opposition to RhoA to direct both chemotactic steering and de novo polarization. In contrast, Rac may act across a broader region to drive actin polymerization and promote stable polarization. At a conceptual level, a network combining a fast local Cdc42-based positive feedback with slower, broader Rac- and Ras-based positive feedback is an appealing circuit design to create a stable but steerable polarity system.
Methods
Cell culture
PLB-985 cells were obtained from the laboratory of Dr. Orion Weiner and cultured in RPMI 1640 medium with L-glutamine and 25mM HEPES (Invitrogen catalog #22400) supplemented with 10% heat-inactivated fetal bovine serum, L-glutamine, penicillin, and streptomycin at 37 in a humidified atmosphere of 5% CO2. Cells were differentiated for 6 days in medium supplemented with 1.3% DMSO prior to each experiment. The PLB-985 cell line has been reported as being a misidentified subline of the HL-60 cell line in the ICLAC database. We have not determined whether this is the case for our cultures. However, both PLB-985 and HL-60 cells are established cell line models for human neutrophils, and the protocols used for the two cell lines are the same. We chose to use PLB-985 rather than HL-60 cells because they exhibited slightly better motility and chemotaxis in our experimental conditions.
Constructs
The Ras, Rac, Cdc42, and RhoA sensors were described previously36. The PI3K biosensor was the PH domain of Akt (2-147) fused to YFP, and it was cloned into pPBbsr, which harbors blasticidin S-resistant gene68. To generate stable cell lines, constructs containing the sensors within the piggyBac transposon were introduced into cells by electroporation and integrated using the piggyBac transposase68. Histone 2B fused to mCherry was cloned into the CSII-EF lentiviral vector, and introduced into cells by viral transduction.
Compounds and inhibitors
The chemical inhibitors used in this study were: LY294002 (Cayman) and ZCL278 (XcessBio). The caged derivative of fMLF was synthesized as described40. Fluo-3 AM and CMNB-Caged fluorescein (Fluorescein bis-(5-Carboxymethoxy-2-Nitrobenzyl) Ether, Dipotassium Salt) were obtained from Invitrogen.
Live cell imaging and Image analysis
Cells were plated in glass bottom 96-well plates (Greiner Bio One catalog #655892) and covered with a liquid imaging medium containing 1.5% low melting temperature agarose containing 100nM of Nv-fMLF. The agarose was allowed to solidify before the cells were moved to 37°C for imaging. Cells were imaged using a custom-assembled spinning disc confocal/epifluorescence microscope system built on a Zeiss Axiovert 200M microscope with a 40X (1.3 N.A.) objective at 37°C. In order to avoid artifacts from cell movement between sequential CFP and FRET image acquisitions, we used an Optical Insights Dual-View beam splitter (Photometrics) for simultaneous acquisition. For our gradient photorelease, images were taken at 5 sec intervals with 300 msec photorelease at a distance of 300 µm every 3 frames. To measure polarization kinetics, images were taken at 2 sec intervals.
Image alignment
All images for ratiometric FRET measurements were taken using an Optical Insights Dual-View beam splitter (Photometrics) for simultaneous acquisition of CFP and YFP emission light from a common excitation pulse. The two images were collected on two halves of a camera and were computationally registered to allow ratiometric calculations. We found that perfect registration of the two images could not be achieved by simple alignment using translations and rotations, but instead small stretch and shear terms were also required. Therefore, we designed custom software using MATLAB to achieve perfect registration of the two images throughout the entire field of view. This software computes a coordinate-mapping function between the two image channels using images taken of a grid-patterned reference slide. Local alignments are computed piecewise across the field of view, and the alignments are used to fit one global coordinate mapping function of the form:
where Cij are the fitted parameters. Pixel intensities for the CFP image were then mapped to the YFP coordinate system by linear interpolation.
Background subtraction, cell segmentation, and cell tracking
Images were first processed with local background subtraction in the following manner: An initial conservative segmentation was performed to define a set of background pixels excluding pixels with significant signal from cells or other fluorescent objects. The background intensity was then taken to be the median intensity of neighboring background pixels. The sum of the CFP and FRET images (after background subtraction) was then used to compute cell masks. The sum of the two channels was used because it has better signal-to-noise than either individual channel, and its value is less sensitive to differences in FRET efficiency. Cells were first crudely identified as large nonbackground objects, and then masks were computed locally for each cell. The mask was computed by first smoothing the image with Gaussian filter of width 2 pixels (0.32 microns) to reduce pixel noise, followed by application of an unsharp mask to enhance edges, and finally automatic thresholding to define cell masks. Cells were tracked from frame to frame using a nearest neighbor method based on cell centroid positions.
Analysis of de novo polarization
We selected for analysis cells which showed no significant movement prior to stimulation with chemoattractant photorelease. A single direction of polarization was defined for each cell as the angle of cell centroid movement for the first time that the centroid moved persistently in the same direction over five consecutive frames. Directed displacement (a measure of cell movement in the polarization direction) was computed as the dot product between the displacement vector (the difference between cell centroids from adjacent frames) and a unit vector in the direction of polarization. GTPase polarization was measured as the sum of the FRET intensities in the frontmost 20% of pixels in the cell polarization direction divided by sum of the corresponding CFP intensities, minus the analogous ratio for the backmost 80% of the cell's pixels (see Fig. 6b). Other measures of GTPase polarization gave similar results.
Analysis of predictive power of GTPase activity for future direction of cell polarization
Only cells that were not moving prior to stimulation and moved significantly after stimulation were included for analysis. Cells that had moved a total distance (centroid to centroid displacement) of less than 2 pixels (here 0.64 microns, as these images were acquired with a bin size of 2) over the ten seconds of imaging prior to stimulation were considered nonmoving. For each cell, the angle between the direction of maximal biosensor FRET signal at the cell periphery and the eventual direction of cell polarization was measured. The FRET signal at the cell periphery was computed around the entire cell periphery at one degree intervals (angle between a vector extending from the cell centroid and a reference direction). Each FRET ratio value was computed as the sum of the FRET intensities divided by the sum of CFP intensities over a window including all pixels within 1.6 microns of the cell edge and within 10 degrees of the direction of interest. The direction of maximal FRET ratio was then identified. The direction of cell polarization was defined by the difference between the cell centroid immediately after stimulation, and the cell centroid 2 minutes after stimulation.
Analysis of intracellular spatial patterns of biosensor signals
To determine the spatial pattern of biosensor signals in polarized, moving cells, we segmented and tracked cells as described above. We computed ratiometric FRET values by first smoothing background-subtracted CFP and FRET images with a disk filter of radius 5 pixels (0.8 microns) to reduce pixel noise (and only using pixels within the cell mask for smoothing). We then divided the FRET intensities by the corresponding CFP intensities to get ratio values. We included for analysis frames for which the cell was moving consistently from the frame before analysis to the frame after analysis. We defined protrusion pixels from frame to frame by subtracting the cell masks. We then defined the protruding front of the cell as the largest connected region of boundary pixels that were in the protrusion pixels for the current frame relative to the previous frame, and within one pixel of the protrusion pixels in the following frame. For every pixel in the cell mask, we then defined its distance from the protruding edge of the cell as the path length (in microns) of the shortest path within the cell mask from that pixel to protruding front edge pixels (using the Matlab function bwdistgeodesic). We then computed the mean FRET ratio as a function of distance from the protruding edge of the cell for each biosensor.
Analysis of correlations between cell edge dynamics and biosensor signals
From cell masks, the cell edge was parametrized into 300 roughly equally spaced points on the cell boundary. From these points, 60 windows were defined for analysis, each with a depth of 10 pixels (1.6 microns) into the cell, and centered on every fifth cell boundary point. Pixels within the cell within 1.6 microns of the cell edge were unambiguously associated with windows by assigning each pixel to the closest window defining cell boundary point. For each window, the FRET ratio was computed as the sum of the FRET intensities divided by the sum of the CFP intensities over all pixels in the window. The local protrusion/retraction velocity of the cell edge was determined by tracking the boundary points from frame to frame by minimization of the squared sum of distances between matched points while preserving the ordering of points along the cell periphery. The protrusion/retraction velocity for each boundary point was computed as the dot product of the point's movement vector with a unit vector normal to cell boundary, divided by the time step in between frames. The velocity for each window was the mean of the velocities for the five boundary points in the window. The above calculations gave a FRET measurement for each window at each time point, and an edge velocity measurement for each window for each step in between sequential frames. Cross-correlation values were then computed as a function of offset in time using Pearson's correlation.
Analysis of correlations between asymmetry in biosensor signals across the cell front and cell turning
The same images, cell segmentation, local windows, and protrusion/retraction analysis were used as in the above analysis. Additionally, the center of the cell front was defined for each frame using the map of protrusion values (such as those shown in Fig. 7a-d; middle) smoothed by a smoothing filter with triangular shape in both the time and space dimension spanning a total of 3 time points and 7 windows. After the smoothing, the center of the cell front was defined as the window farthest from a negative protrusion value (i.e. the center of the largest protrusion). For an individual frame, the center of the cell front was defined based on protrusion between that frame and the following frame. Asymmetry in the FRET signal was computed as the mean FRET signal over the 10 windows to the left of this center window minus the mean FRET signal over the 10 windows to the right of the center window. To reduce noise, the cell centroid positions and center of the cell front positions were smoothed using Lowess regression. Instantaneous cell direction was defined by the vector from the cell centroid to the center of the cell front. The turning angle in between frames was then defined as the angular turn in the cell direction vector between consecutive frames. Temporal cross-correlations were then computed as a function of offset in time between asymmetry in biosensor signal and cell turning.
Analysis of correlations between a signaling vector and cell turning
The same data and cell centroid positions were used as in the above turning analysis. The same cell edge windows were used as in the edge dynamics analysis. A schematic of this analysis is depicted in Supplementary Fig. 4d. For each frame, a cell signaling vector was computed using all edge windows with a FRET ratio greater than the mean FRET ratio over all windows. The signaling vector was defined as the vector sum of the vectors pointing from the cell centroid to the center of each included window, weighted by the FRET ratio for that window minus the mean FRET ratio over all windows. Cell directionality was defined frame-to-frame intervals using cell movement vectors pointing from the earlier cell centroid position to the later cell centroid position. A cell turning angle was defined for each frame as the angle between the preceding cell direction vector and the following cell direction vector. A signaling angle was defined for each frame as the angle between the preceding cell direction vector and the signaling vector in that frame. In each case, an angle of zero constitutes straight ahead and positive and negative angles indicate turns to the left or right. Cross-correlations for different temporal offsets were computed between the cell turning angles and the signaling angles for each GTPase biosensor.
siRNA electroporation and Western blotting
At 3 days after differentiation, PLB-985 cells were spun down and resuspended in extracellular buffer (5 mM KCl, 125 mM NaCl, 1.5 mM CaCl2, 1.5 mM MgCl2, 10 mM Glucose, 20 mM HEPES, pH 7.4). 0.9 uM Control (Dharmacon, D-001206-14-05), Cdc42 (Dharmacon, M-005057-01-0005), and RhoA (Dharmacon, E-003860-00-0005) siRNA pools were mixed with the cell suspension and were introduced into cell by parallel electroporation using a custom-built 96-well electroporation device69. Cells were then incubated 3 days more with differentiation media in 96 well plate. For western blot, whole-cell extracts were prepared and resolved by 4%-10% gradient SDS-PAGE. The proteins were then transferred to a nitrocellulose membrane. Primary antibodies, anti-Cdc42 (Cell Signaling Technology, 2462, 1:1,000), anti-RhoA (Cell Signaling Technology, 2117, 1:1,000), and anti-GAPDH (Cell Signaling Technology, 5174, 1:1,000), were incubated overnight at 4°C. Bound antibodies were visualized with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) and the ECL system (Millipore).
Immunofluorescence
After fixation in PBS containing 4% formaldehyde, cells were washed three times in PBS, permeabilized and blocked with 0.1% Triton X-100, 1% BSA, 10% FBS, and 0.01% NaN3 for 1 hr at 25 °C, and stained overnight at 4 °C with anti-phospho Myosin Light Chain 2 (Ser19) (Cell Signaling Technology, 3671, 1:200). Primary antibody was visualized using Alexa Fluor 488 (Life Technology, A11034, 1:1,000). Images were taken on an ImageXpress Micro (Molecular Devices) with 20X (0.75 N.A.) objective.
Statistics & reproducibility
For Fig. 4c and Supplemental Fig. 6a, we used the sign test to test whether 90 degrees minus the median angle between the direction of cell movement and the direction of maximal (or minimal) FRET signal was greater or less than zero. The sign test is a nonparametric test that makes minimal assumptions and tests whether the median of a set of numbers is significantly different from zero. For Supplemental Fig. 8b, we used the Mann-Whitney U test (ranksum in MATLAB) to test whether the normalized FRET ratios measured at later time points differed from the values at time zero. The U test is nonparametric and is appropriate when values are not known to be normally distributed. All of the results in this paper were independently replicated at least four time except Fig. 8b, Supplementary Fig. 1b, Supplementary Fig. 1c, Supplementary Fig. 1f, Supplementary Fig. 3b, Supplementary Fig. 8b, and Supplementary Fig. 8d for which two independent experiments were performed.
Supplementary Material
Acknowledgements
We thank to K. Aoki and M. Matsuda for GTPase sensors; Arnold Hayer, Damien Garbett, and Amy Winans for critical reading of the manuscript and helpful discussions; and the Stanford Shared FACS Facility for cell sorting and the NIGMS for funding.
Footnotes
Author Contributions
H. W. Y., S. C. and T. M. designed the experiments. H. W. Y. and S. C. performed the experiments and analyzed the data. H. W. Y., S. C. and T. M. interpreted the data. H. W. Y., S. C. and T. M. wrote the paper.
Code availability
The Matlab scripts for analysis described in this paper are available on Github at https://github.com/MeyerLab/Chemotaxis_Analysis.
References
- 1.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Niggli V. Signaling to migration in neutrophils: importance of localized pathways. Int. J. Biochem. Cell Biol. 2003;35:1619–1638. doi: 10.1016/s1357-2725(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 3.Becker EL. Stimulated neutrophil locomotion: chemokinesis and chemotaxis. Arch. Pathol. Lab. Med. 1977;101:509–13. [PubMed] [Google Scholar]
- 4.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev. Cell. 2005;8:215–27. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu. Rev. Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 6.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 7.Alt W. Biased random walk models for chemotaxis and related diffusion approximations. J. Math. Biol. 1980;9:147–77. doi: 10.1007/BF00275919. [DOI] [PubMed] [Google Scholar]
- 8.Wells WA. Chemotaxis by local steering. J. Cell Biol. 2005;168:674–675. [Google Scholar]
- 9.Iglesias P. a., Devreotes PN. Biased excitable networks: How cells direct motion in response to gradients. Curr. Opin. Cell Biol. 2012;24:245–253. doi: 10.1016/j.ceb.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M, et al. Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis. Nat. Commun. 2014;5:5175. doi: 10.1038/ncomms6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickert P, Weiner OD, Wang F, Bourne HR, Servant G. Leukocytes navigate by compass: roles of PI3Kgamma and its lipid products. Trends Cell Biol. 2000;10:466–73. doi: 10.1016/s0962-8924(00)01841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokoch GM. Chemoattractant signaling and leukocyte activation. Blood. 1995;86:1649–60. [PubMed] [Google Scholar]
- 13.Charest PG, Firtel R. a. Big roles for small GTPases in the control of directed cell movement. Biochem. J. 2007;401:377–90. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 15.Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan S, et al. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J. Cell Biol. 2003;160:375–85. doi: 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner OD, et al. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 2002;4:509–13. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun CX, et al. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood. 2004;104:3758–3765. doi: 10.1182/blood-2004-03-0781. [DOI] [PubMed] [Google Scholar]
- 19.Huang C-H, Tang M, Shi C, Iglesias P. a, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat. Cell Biol. 2013;15:1307–16. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell. 2003;114:215–27. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- 21.Wong K, Pertz O, Hahn K, Bourne H. Neutrophil polarization: spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3639–44. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, et al. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–14. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 23.Kitzing TM, et al. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina F, et al. Activated RhoA Is a Positive Feedback Regulator of the Lbc Family of Rho Guanine Nucleotide Exchange Factor Proteins. J. Biol. Chem. 2013;288:11325–11333. doi: 10.1074/jbc.M113.450056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourne HR, Weiner O. A chemical compass. Nature. 2002;419:21. doi: 10.1038/419021a. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, et al. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 2002;4:513–8. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 27.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 2007;17:813–7. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Afonso PV, Parent CA. PI3K and chemotaxis: a priming issue? Sci. Signal. 2011;4:pe22. doi: 10.1126/scisignal.2002019. [DOI] [PubMed] [Google Scholar]
- 29.Lämmermann T, et al. Cdc42-dependent leading edge coordination is essential for interstitial dendritic cell migration. Blood. 2009;113:5703–5710. doi: 10.1182/blood-2008-11-191882. [DOI] [PubMed] [Google Scholar]
- 30.Roberts AW, et al. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 1999;10:183–96. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- 31.Glogauer M, et al. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J. Immunol. 2003;170:5652–7. doi: 10.4049/jimmunol.170.11.5652. [DOI] [PubMed] [Google Scholar]
- 32.Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell. 2006;17:4543–50. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 2004;167:505–18. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, et al. Identifying network motifs that buffer front-to-back signaling in polarized neutrophils. Cell Rep. 2013;3:1607–1616. doi: 10.1016/j.celrep.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo SK, et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell. 2010;18:226–36. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–56. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins SR, et al. Using light to shape chemical gradients for parallel and automated analysis of chemotaxis. Mol. Syst. Biol. 2015;11:804. doi: 10.15252/msb.20156027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat. Immunol. 2008;9:960–9. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- 39.Renkawitz J, Sixt M. Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep. 2010;11:744–50. doi: 10.1038/embor.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirrung MC, Drabik SJ, Ahamed J, Ali H. Caged chemotactic peptides. Bioconjug. Chem. 2000;11:679–681. doi: 10.1021/bc000001g. [DOI] [PubMed] [Google Scholar]
- 41.Zheng L, Eckerdal J, Dimitrijevic I, Andersson T. Chemotactic peptide- induced activation of Ras in human neutrophils is associated with inhibition of p120-GAP activity. J. Biol. Chem. 1997;272:23448–54. doi: 10.1074/jbc.272.37.23448. [DOI] [PubMed] [Google Scholar]
- 42.Zigmond SH. Cell polarity: an examination of its behavioral expression and its consequences for polymorphonuclear leukocyte chemotaxis. J. Cell Biol. 1981;89:585–592. doi: 10.1083/jcb.89.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh M, et al. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–6. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 44.Gilbert SH, Perry K, Fay FS. Mediation of chemoattractant-induced changes in [Ca2+]i and cell shape, polarity, and locomotion by InsP3, DAG, and protein kinase C in newt eosinophils. J. Cell Biol. 1994;127:489–503. doi: 10.1083/jcb.127.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiger MC, Ahmed S, Welf ES, Haugh JM. Directional persistence of cell migration coincides with stability of asymmetric intracellular signaling. Biophys. J. 2010;98:67–75. doi: 10.1016/j.bpj.2009.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chau AH, Walter JM, Gerardin J, Tang C, Lim W. a. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell. 2012;151:320–32. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschuler SJ, Angenent SB, Wang Y, Wu LF. On the spontaneous emergence of cell polarity. Nature. 2008;454:886–9. doi: 10.1038/nature07119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butty A-C, et al. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–76. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–5. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 50.Meyer T, Stryer L. Molecular model for receptor-stimulated calcium spiking. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5051–5. doi: 10.1073/pnas.85.14.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shankaran H, Wiley HS. Oscillatory dynamics of the extracellular signal- regulated kinase pathway. Curr. Opin. Genet. Dev. 2010;20:650–655. doi: 10.1016/j.gde.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–72. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 54.Moreau V, Tatin F, Varon C, Génot E. Actin can reorganize into podosomes in aortic endothelial cells, a process controlled by Cdc42 and RhoA. Mol. Cell. Biol. 2003;23:6809–6822. doi: 10.1128/MCB.23.19.6809-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friesland A, et al. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1261–6. doi: 10.1073/pnas.1116051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amano M, et al. Phosphorylation and Activation of Myosin by Rho-associated Kinase (Rho-kinase). J. Biol. Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 57.Howell AS, et al. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 2011;21:740–6. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J. Cell Biol. 1998;141:1147–57. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Houk AR, et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–88. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Postma M, Van Haastert PJ. A diffusion-translocation model for gradient sensing by chemotactic cells. Biophys. J. 2001;81:1314–1323. doi: 10.1016/S0006-3495(01)75788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jilkine A, Edelstein-Keshet L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput. Biol. 2011;7:e1001121. doi: 10.1371/journal.pcbi.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glogauer M, Hartwig J, Stossel T. Two pathways through Cdc42 couple the N-formyl receptor to actin nucleation in permeabilized human neutrophils. J. Cell Biol. 2000;150:785–96. doi: 10.1083/jcb.150.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiner OD, Marganski W. a., Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:2053–2063. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peyrollier K, et al. A role for the actin cytoskeleton in the hormonal and growth- factor-mediated activation of protein kinase B. Biochem. J. 2000;352(Pt 3):617–22. [PMC free article] [PubMed] [Google Scholar]
- 68.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guignet EG, Meyer T. Suspended-drop electroporation for high-throughput delivery of biomolecules into cells. Nat. Methods. 2008;5:393–5. doi: 10.1038/nmeth.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.