Abstract
Crystallization of protein-protein complexes can often be problematic and therefore computational structural models are often relied upon. Such models are often generated using protein-protein docking algorithms, where one of the main challenges is selecting which of several thousand potential predictions represents the most near-native complex. We have developed a novel technique that involves the use of steered molecular dynamics (sMD) and umbrella sampling to identify near-native complexes among protein-protein docking predictions. Using this technique, we have found a strong correlation between our predictions and the interface RMSD (iRMSD) in ten diverse test systems. On two of the systems, we investigated if the prediction results could be further improved using potential of mean force calculations. We demonstrated that a near-native (<2.0 Å iRMSD) structure could be identified in the top-1 ranked position for both systems.
Keywords: protein-protein interaction, ZDOCK, steered molecular dynamics, potential of mean force, umbrella sampling
Graphical Abstract
Predicting how two proteins will bind to one another is a challenging task. This method combines protein-protein docking, steered molecular dynamics, and potential of mean force calculations to predict and evaluate protein-protein interactions.
Despite many advances in modeling, docking, and scoring, predicting protein-protein interactions is still riddled with challenges1. Selecting the final model(s) is typically considered one of the most difficult steps and is often the most critical. Here we describe a novel, physics-based, multi-step approach to identify near-native protein-protein complex structures from a set of top-ranked poses.
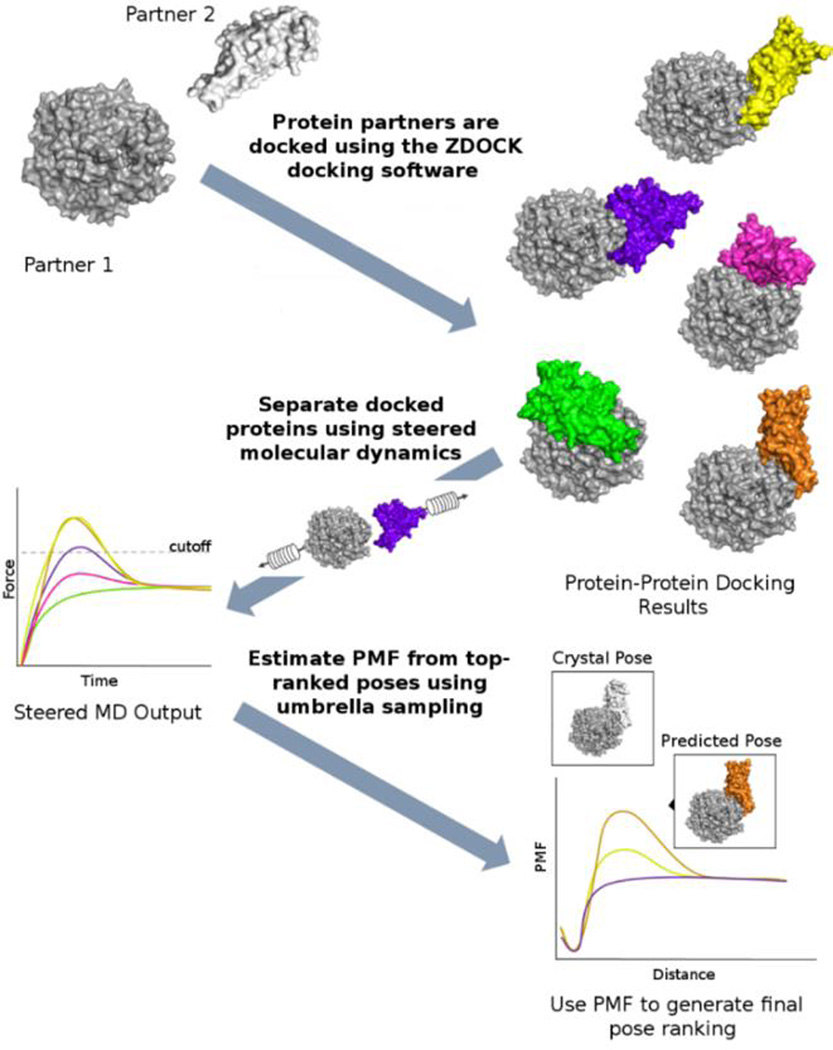
In our method, summarized in Figure 1, steered molecular dynamics (MD) simulations are used to estimate the force required to separate the partners of docked protein-protein complexes by pulling one partner away from the other. The top-10 complexes (those with the highest force required for separation) are selected for more detailed investigation using umbrella sampling. The umbrella sampling simulations combined with the weighted histogram analysis method (WHAM) provide an estimate of the potential of mean force (PMF) of protein dissociation. The difference in the PMF between the bound (starting configuration) and unbound (ending configuration) state is the calculated delta G of complex dissociation.
Figure 1.
First, the target proteins (Partner 1 and Partner 2) are docked using ZDOCK, resulting in several thousand poses. The top-100 poses are selected based on the internal ZDOCK scoring function. These poses are then separated using sMD, and the force required for separation is computed. The top-10 poses from the force calculations are then re-ranked using by PMF.
A set of ten diverse protein-protein complexes was used to evaluate our method (Table 1). From ~54,000 poses produced using ZDOCK2, a set of ~100 representative poses were selected. The selected poses were then evaluated using steered MD and five standard scoring functions, zrank13, zrank23, zdock2, irad4, and a custom potential based on van der Waals, electrostatics and knowledge-based terms5, herein referred to as “stats”. The scoring functions were independently evaluated using the interface RMSD (iRMSD), a commonly used metric to evaluate protein-protein docking poses6.
Table 1.
Summary of systems used to evaluate scoring functions. Those shown in bold were used in the PMF calculations.
| System (PDB ID) | Residues and Chainsa | IRMSD Range of Docking Resultsb |
Number of Poses Tested |
Reference |
|---|---|---|---|---|
| Ubiquitin ligase and ubiquitin (2OOB) |
Total: 113 Pull: 42 Stationary: 71 |
1.62Å–9.97Å | 96 | 7 |
| Trypsin and CMTI-I peptide inhbitor (1PPE) |
Total: 274 Pull: 29 Stationary: 245 |
0.65Å–9.67Å | 100 | 8 |
|
Antibody and antigen (1VFB) |
Total: 352 Pull: 129 Stationary: 107 & 116 |
1.32Å–9.98Å | 100 | 9 |
|
aminoacyl-tRNA synthetase and tRNA aminoacylation cofactor Arc1p (2HRK) |
Total: 282 Pull: 102 Stationary: 180 |
1.96Å–9.94Å | 98 | 10 |
| Ribonuclease A and a peptide inhibitor (1DFJ) |
Total : 579 Pull: 124 Stationary: 455 |
1.19Å–9.81Å | 99 | 11 |
| Ferredoxin-NADP Reductase and ferredoxin (1EWY) |
Total: 395 Pull: 98 Stationary:297 |
1.43 Å–9.98 Å | 100 | 12 |
| Matriptase and aprotinin (1EAW) |
Total: 299 Pull:58 Stationary: 241 |
0.74 Å –9.89 Å | 100 | 13 |
| SARS-receptor binding domain and receptor (2AJF) |
Total: 777 Pull:180 Stationary:597 |
1.63Å–10.0 Å | 98 | 14 |
| Ecotin and trypsin (1EZU) |
Total:365 Pull:142 Stationary:223 |
1.37 Å – 9.99 Å | 97 | 15 |
| Uracil-DNA Glycosylase and its protein inhibitor (1UDI) |
Total:207 Pull:83 Stationary: 124 |
1.15 Å – 9.89 Å | 100 | 16 |
“Pull” refers to the length of the chain that was pulled during the steered MD simulation, “Stationary” refers to the chain that was restrained during the steered MD simulation.
Results were pre-filtered to remove any poses above 10 Å.
A prediction was considered “good” if the iRMSD ≤ 2.0 Å, a pose was considered “acceptable” if the iRMSD ≤ 4.0 Å, and a prediction was considered “poor” if the iRMSD > 4.0 Å.
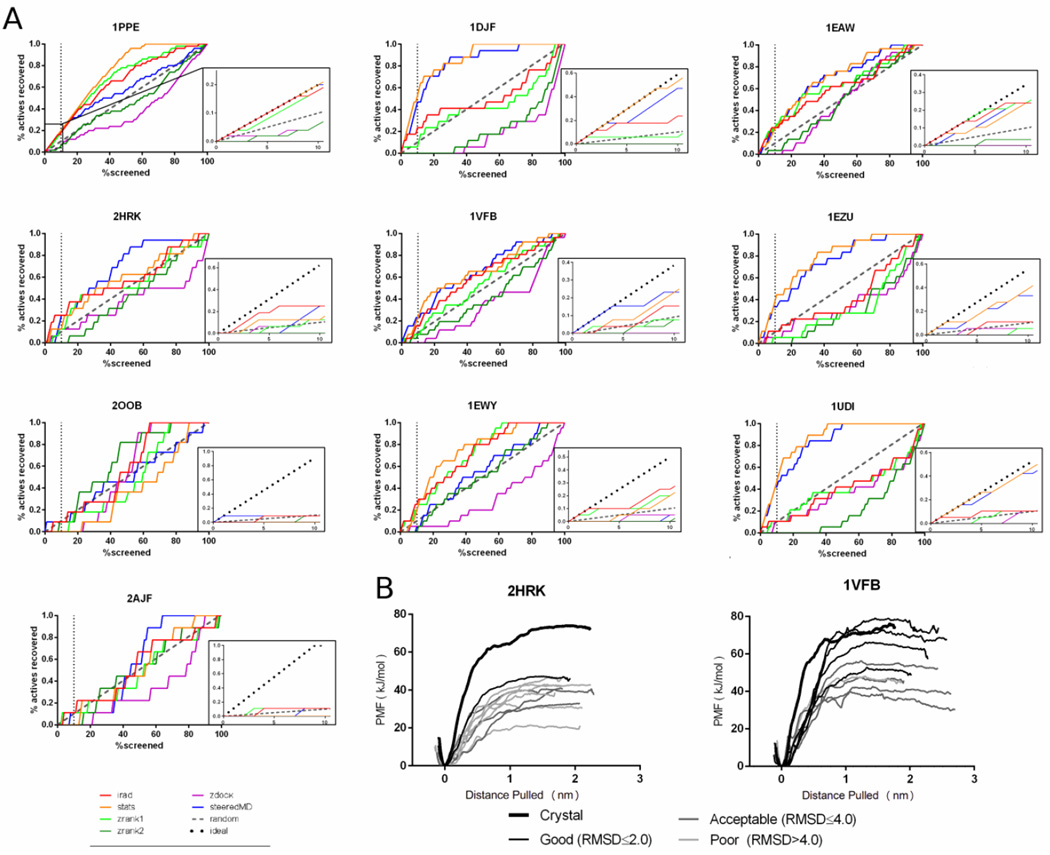
Plots showing the number of actives recovered versus the percentage of complexes screened are shown in Figure 2A (tabulated values are shown in Supplemental Table 1). Steered MD produced the best results of any scoring scheme tested producing at least one good pose for 7/10 systems tested and an acceptable pose for 10/10 systems within the top-10 predictions. The irad and stats scoring functions performed similarly to sMD, both produced good predictions in 6/10 cases and acceptable poses were predicted from 10/10 and 8/10 poses respectively. In terms of enrichment, steered MD and stats perform the best. This is especially apparent in 1DJF, 1EZU, and 1UDI, where steered MD and stats significantly outperform the other scoring functions. Furthermore, both perform perfectly or nearly perfectly in 4/10 systems (1PPE, 1DJF, 1EAW, and 1UDI) as shown in Figure 2A (dotted line in inset indicates perfect prediction).
Figure 2.
A) Plot describing the recovery of acceptable poses (iRMSD ≤4.0) versus number of poses screened for each scoring function. Insets represent show an expanded plot of the initial 10% of complexes screened. B) PMF curves from umbrella sampling in 2HRK (left) and 1VFB (right). The crystal structures are shown in the bolded black line. The poses have been color coded from dark to light, where the darkest lines represent low iRMSD structures (≤2.0 Å), the slightly lighter lines represent acceptable structures (IRMSD ≤4.0Å), and the lightest lines represent unacceptable structures (iRMSD> 4.0Å).
In general, most scoring functions that were tested produced a good or acceptable pose within the top-10 predictions, but oftentimes the top-10 predictions also included several poor poses. The inclusion of poor poses is less detrimental if they are ranked below good or acceptable poses, but this was not always the case. For instance, in 2HRK, only a single good pose (iRMSD: 1.98 Å) was identified in the top-10 predictions by steered MD and this pose was ranked 10th overall. Furthermore the three acceptable poses (iRMSDs: 2.49 Å, 2.13 Å, and 2.86 Å) were also ranked poorly (7th, 8th and 9th, respectively). In a blind prediction scenario, this type of result could easily lead to an unproductive final model. Thus we attempted to further refine the top-10 predictions using umbrella sampling.
Ideally, in cases such as 2HRK, re-scoring using PMF will result in the low iRMSD structures being re-ranked closer to the top. Alternatively, the 1VFB dataset contains several successful poses that are ranked near the top and 7 out of the top 10 poses are acceptable (iRMSD ≤ 4.0 Å). To ensure that re-ranking by PMF does not alter a successful screen, the top-10 predicted complexes from the 1VFB systems were also re-scored using umbrella sampling.
Umbrella sampling is a technique where overlapping MD trajectories are utilized to produce an estimate of the potential of mean force (PMF) along a pre-defined reaction coordinate, in this case the distance describing the dissociation of the two protein units along the vector created by the centers of mass of each unit. These calculations, although computationally expensive, may provide a more accurate quantification of protein-protein interactions compared to steered MD alone. As a proof of concept, we selected the top-10 structures from 2HRK and 1VFB and used umbrella sampling to re-rank these structures.
In both cases, re-ranking the top-10 poses using the PMF calculated by umbrella sampling improved the results. In 2HRK, the lowest iRMSD complex (1.98 Å) rose from a 10th place when ranked by steered MD alone to 1st when using PMF (Figure 2B-left panel). Likewise in the 1VFB dataset, the 9.84Å structure fell from 1st ranked in the steered MD ranking down to one of the lowest ranked structures when ranked by PMF (Figure 2B–right panel). In addition, in the 1VFB dataset all good poses (iRMSD ≤ 2.0Å) were ranked in the top-4 highest positions using PMF (Figure 2B–right panel).
As a comparison, we also calculated the PMF of the crystal structures (shown in bolded black lines in Figure 2B). In the case of 1VFB, the calculated PMF of crystal structure was in agreement with the low iRMSD (≤ 2.0Å) structures. This finding suggests that the crystal structure and accurately predicted poses demonstrate similar behavior in the calculations. However, in the case of 2HRK, the PMF of the crystal structure was ~30 kJ/mol larger than the best ranked structure (1.98 Å). One possible explanation for this finding is that not all crystal contacts are adequately reproduced in the docked results. One possible explanation for this observation is interfacial waters.
The hydration site analysis program WATsite17, was used to compare the number of hydration sites in or immediately adjacent to the interface of the protein-protein complex based on the x-ray conformation and pose with lowest iRMSD for 2HRK and 1VFB. A comparison between the 2HRK crystal structure and the equilibrated lowest iRMSD pose (1.98Å) revealed that not all contact-mediating hydration sites in the x-ray structure of the protein-protein interface were conserved in the low iRMSD pose (Supplemental Figure 1). Whereas 16 contact-mediating hydration sites were identified in the protein interface of the x-ray structure only 12 were found in the low iRMSD pose (Supplementary Table 2). Repeating this analysis for 1VFB revealed that the same number of contact-mediating waters were identified in low iRMSD and x-ray structure supporting the observation that the PMF of the crystal structure and good poses were approximately equal. Thus, important water-mediated interactions are lost for the low iRMSD pose resulting in reduced complex stability compared to the x-ray structure of the complex.
To the best of our knowledge, this is the first time that steered MD and PMF calculations have been used to evaluate protein-protein docking poses. Furthermore the use of explicit solvent MD simulations allows for the incorporation of waters into the interface which are accounted for in our procedure, a feature that is very rarely included in traditional docking and scoring methods.
Despite the limited number of test cases, we believe the proposed stepwise method to be a promising approach, although there are some important considerations about the limitations of this method. Importantly, the time required for calculation of PMF profiles could present a significant limitation. In practice, we suggest that a more rapid scoring function might be used as a pre-filter, prior to implementing the more computationally demanding umbrella sampling (both the stats and the irad scoring functions performed exceptionally well in our hands). In addition to the time required to calculate the PMF profiles, the calculations are sensitive to the reproducibility of the interactions in the interface. In an ideal case, protein partners would not change conformation upon binding and interface interactions would be strictly between partners (i.e. not mediated by water or other co-factors). Caution should be exercised in cases where drastic conformational changes are thought to occur or in cases where protein interactions are extensively mediated by other molecules. Methods such as principal component analysis (PCA), may be employed to determine the best vector for sMD simulations in cases of intricate interfaces or where significant conformational change is anticipated
In summary, the use of steered MD and umbrella sampling in ranking protein-protein docking conformations represents a novel approach in this field and has been found to be successful in the test cases presented here and elsewhere18. While there are some limitations to this approach, notably the computational cost, we believe that this approach may prove useful in a range of systems and be a complimentary approach to the currently used scoring functions for protein-protein docking.
Methods
Only a general outline of the procedure and tools used has been included here, a detailed methods section has been included in the supplemental information.
All protein systems were docked using the ZDOCK algorithm producing ~54,000 conformations. From these 100 representative conformations were selected for steered MD. Gromacs 4.6.1 was used to prepare and equilibrate each system prior to sMD. From the sMD simulations, the total force was computed as the difference between the lowest and highest recorded force for each simulation. Umbrella sampling was performed on the top-10 structures from 1VFB and 2HRK and the g_wham program from Gromacs was used to estimate the PMF using the sampled windows. WATsite was used in the interfacial water analysis.
Supplementary Material
Acknowledgments
MAL and LJK acknowledge support from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM092855). LJK acknowledges support from the Graduate Assistantship in Areas of National Need (GAANN) Fellowship from the US Department of Education. DK and JER acknowledge support from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM097528) and the National Science Foundation (IIS1319551, DBI1262189, IOS1127027).
Footnotes
((Additional Supporting Information may be found in the online version of this article.))
References and Notes
- 1.Moal I, Torchala M, Bates P, Fernandez-Recio J. The scoring of poses in protein-protein docking: current capabilities and future directions. BMC Bioinform. 2013;14(1):286. doi: 10.1186/1471-2105-14-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R, Li L, Weng Z. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52(1):80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 3.Pierce B, Weng Z. ZRANK: Reranking protein docking predictions with an optimized energy function. Proteins. 2007;67(4):1078–1086. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- 4.Vreven T, Hwang H, Weng Z. Integrating atom-based and residue-based scoring functions for protein–protein docking. Protein Sci. 2011;20(9):1576–1586. doi: 10.1002/pro.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquivel-Rodríguez J, Yang YD, Kihara D. Multi-LZerD: Multiple protein docking for asymmetric complexes. Proteins. 2012;80(7):1818–1833. doi: 10.1002/prot.24079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Lensink MF, Méndez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69(4):704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]; (b) Pons C, Grosdidier S, Solernou A, Pérez-Cano L, Fernández-Recio J. Present and future challenges and limitations in protein–protein docking. Proteins. 2010;78(1):95–108. doi: 10.1002/prot.22564. [DOI] [PubMed] [Google Scholar]
- 7.Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, Park M, Gehring K. Structural Basis for Ubiquitin-Mediated Dimerization and Activation of the Ubiquitin Protein Ligase Cbl-b. MolCell. 2007;27(3):474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Bode W, Greyling HJ, Huber R, Otlewski J, Wilusz T. The refined 2.0 Å X-ray crystal structure of the complex formed between bovine β-trypsin and CMTI-I, a trypsin inhibitor from squash seeds (Cucurbita maxima) Topological similarity of the squash seed inhibitors with the carboxypeptidase A inhibitor from potatoes. FEBS Lett. 1989;242(2):285–292. doi: 10.1016/0014-5793(89)80486-7. [DOI] [PubMed] [Google Scholar]
- 9.Bhat TN, Bentley GA, Boulot G, Greene MI, Tello D, Dall'Acqua W, Souchon H, Schwarz FP, Mariuzza RA, Poljak RJ. Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc Natl Acad Sci. 1994;91(3):1089–1093. doi: 10.1073/pnas.91.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simader H, Hothorn M, Köhler C, Basquin J, Simos G, Suck D. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes. Nucleic Acids Res. 2006;34(14):3968–3979. doi: 10.1093/nar/gkl560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374(6518):183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 12.Morales R, Kachalova G, Vellieux F, Charon M-H, Frey M. Crystallographic studies of the interaction between the ferredoxin-NADP+ reductase and ferredoxin from the cyanobacterium Anabaena: looking for the elusive ferredoxin molecule. Acta Crystallogr. Sect. D. 2000;56(11):1408–1412. doi: 10.1107/s0907444900010052. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, Madison EL. Catalytic Domain Structures of MT-SP1/Matriptase, a Matrix-degrading Transmembrane Serine Proteinase. J. Biol. Chem. 2002;277(3):2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Li W, Farzan M, Harrison SC. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 15.Gillmor SA, Takeuchi T, Yang SQ, Craik CS, Fletterick RJ. Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases1. J Mol Biol. 2000;299(4):993–1003. doi: 10.1006/jmbi.2000.3812. [DOI] [PubMed] [Google Scholar]
- 16.Savva R, Pearl LH. Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase–uracil glycosylase inhibitor protein complex. Nature Struct Biol. 1994;(2):752–757. doi: 10.1038/nsb0995-752. [DOI] [PubMed] [Google Scholar]
- 17.Hu B, Lill MA. WATsite: Hydration Site Prediction Program with PyMOL Interface. J Comp Chem. 2014;35(16):1255–1260. doi: 10.1002/jcc.23616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Meyer AG, Sawyer SL, Ellington AD, Wilke CO. Analyzing Machupo virus-receptor binding by molecular dynamics simulations. PeerJ PrePrints. 2014;2:e138v3. doi: 10.7717/peerj.266. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cheung LS-L, Shea DJ, Nicholes N, Date A, Ostermeier M, Konstantopoulos K. Characterization of Monobody Scaffold Interactions with Ligand via Force Spectroscopy and Steered Molecular Dynamics. Scientific Reports. 2015;5:8247. doi: 10.1038/srep08247. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Allen W, Wiley M, Myles K, Adelman Z, Bevan D. Steered molecular dynamics identifies critical residues of the Nodamura virus B2 suppressor of RNAi. J Mol Model. 2014;20(3):1–10. doi: 10.1007/s00894-014-2092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Rodriguez RA, Yu L, Chen LY. Computing Protein–Protein Association Affinity with Hybrid Steered Molecular Dynamics. Journal of Chemical Theory and Computation. 2015;11(9):4427–4438. doi: 10.1021/acs.jctc.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.