Abstract
Lipid multilayer microarrays are a promising approach to miniaturize laboratory procedures by taking advantage of the microscopic compartmentalization capabilities of lipids. Here, we demonstrate a new method to pattern lipid multilayers on surfaces based on solvent evaporation along the edge where a stencil contacts a surface called evaporative edge lithography (EEL). As an example of an application of this process, we use EEL to make microarrays suitable for a cell-based migration assay. Currently existing cell migration assays require a separate compartment for each drug which is dissolved at a single concentration in solution. An advantage of the lipid multilayer microarray assay is that multiple compounds can be tested on the same surface. We demonstrate this by testing the effect of two different lipophilic drugs, Taxol and Brefeldin A, on collective cell migration into an unpopulated area. This particular assay should be scalable to test of 2000 different lipophilic compounds or dosages on a standard microtiter plate area, or if adapted for individual cell migration, it would allow for high-throughput screening of more than 50,000 compounds per plate.
Keywords: lithography, stencil, lipid, microarray, cell migration, high-throughput screening
1 Introduction
The ability for a single living cell to carry out advanced synthesis, sensing, and communication all within a micrometer scale object provides an inspiration for laboratory miniaturization. Lipids in the cell provide compartmentalization by forming a variety of organelles, vesicles, and transport carriers. Additionally, a variety of biofunctional lipids are available from both natural and synthetic sources. In order to recreate some of these capabilities in an artificial environment, we have been creating lipid multilayer micro- and nano-structures on surfaces. Previously, lipid multilayer patterns have been fabricated by dip-pen nanolithography (DPN),1–3 soft lithography (e.g. micro-contact printing and nanointaglio),4–6 photothermal patterning,7 and capillary assembly.8
Here we describe a new lipid multilayer microfabrication method that we call edge evaporation lithography (EEL) that is capable of producing linear lipid multilayer nanostructures along the edge of a stencil. This method makes use of capillary assembly8 onto a pre-patterned surface in a way similar to that carried out by Diguet et al., with a critical difference being that EEL uses an edge between a stencil and a surface as a one-dimensional template rather than controlled evaporation on a chemically patterned surface. Evaporation induced self-assembly (EISA) is a related technique based on the evaporation of a solution containing precursors to be assembled and has been used previously with lipid films.9–10 Another similar technique is capillary force lithography that combines imprint lithography with microcontact printing, or other methods of soft lithography.11–13 In contrast, EEL involves micro and nanostructure generation by solvent evaporation and allows integration of multiple materials onto the surface. These subcellular lipid multilayer structures have applications in biosensing1, 14 and high throughput screening.15–16
Small molecule microarrays are a promising approach to miniaturizing high throughput screening that could allow tens to hundreds of thousands of compounds to be tested on a single cell culture plate.17 We have previously shown that lipid multilayer microarrays with sub-cellular lateral dimensions can be used as a format for delivery of multiple lipophilic anticancer drugs to adherent cells in a microarray format, and measured cytotoxicity as a readout for efficacy. These arrays are capable of encapsulating lipophilic materials that were internalized cells without cross-contamination, as demonstrate by control cells cultured on the glass surface directly next to the lipid encapsulated drug spots did not respond significantly to the drug.15 Importantly, lipid multilayer microarrays are compatible with lipophilic compounds, while other drug screening microarrays are either limited to water soluble compounds that diffuse out of a gel into water,18 or must be covalently linked to the surface and cannot be taken up by the cells.19 At the early stages of drug discovery where high throughput screening (HTS) is used, most drug candidates at low water solubility, which is quantified by a high octanol to water partition coefficient (LogP). In standard high throughput screening, dimethyl sulfoxide (DMSO) is used as solvent to deliver compounds with high LogP values in water.20 However, this strategy cannot be used to deliver lipophilic drugs to cells from microarrays. Here, we demonstrate that EEL provides a unique solution to these miniaturization and solubility issues for cell-based small-molecule microarrays by creating a cell migration assay.
The migration of cells collectively is an important aspect of cancer metastasis,21–22 angiogenesis,23–24 wound healing,25 and organismal development.26–27 Several in vitro migration assays have been developed to assess the effects of compounds and the cellular microenvironment on the migration of cells in culture.28–29 Examples include the commonly used wound or “scratch” migration assay,25, 30–31 removable fencing assays or cell stencils,32–33 Boyden chamber assays,34 biodegradable or barriers,35–36 two phase cell-exclusion patterning,37 and microfluidic techniques.38–44 The methods listed above have the common feature that cells are cultured on a certain part of a two-dimensional substrate or three-dimensional volume and allowed to migrate into a region without cells. The number or speed of cells individually or collectively migrating into the unpopulated regions is then measured.
Cell migration assays have important applications for high throughput screening and new drug discovery. For example, thousands of compounds have been screened for their effect on cellular migration by automated microscopy of scratch assays and by Boyden chamber assays.31, 34 Several migration assays have been adapted to 96 well-plate formats with the motivation being for compatibility with high throughput screening.37, 45–46 Importantly, an innovative assay based on siRNA delivery from hydrogel microarrays followed by measurements of cell migration away from the siRNA containing spots allows siRNA screens to be carried out in a microarray format.45
In EEL, we use an elastomeric barrier or stencil to direct the precipitation of lipid and drug solutes along an edge resulting in a drug-encapsulated lipid multilayer line. We demonstrate the utility of this fabrication method to deliver lipophilic drugs to adherent cells for cell-based migration analysis (Fig 1). Unlike other migration assays, this approach makes it possible to screen different small molecule compounds and dosages on the same surface without separated compartments, and demonstrates potential scalability for high throughput screening of small lipophilic molecules. The miniaturization process described here would allow for the manufacturing of portable small molecule libraries on a chip.
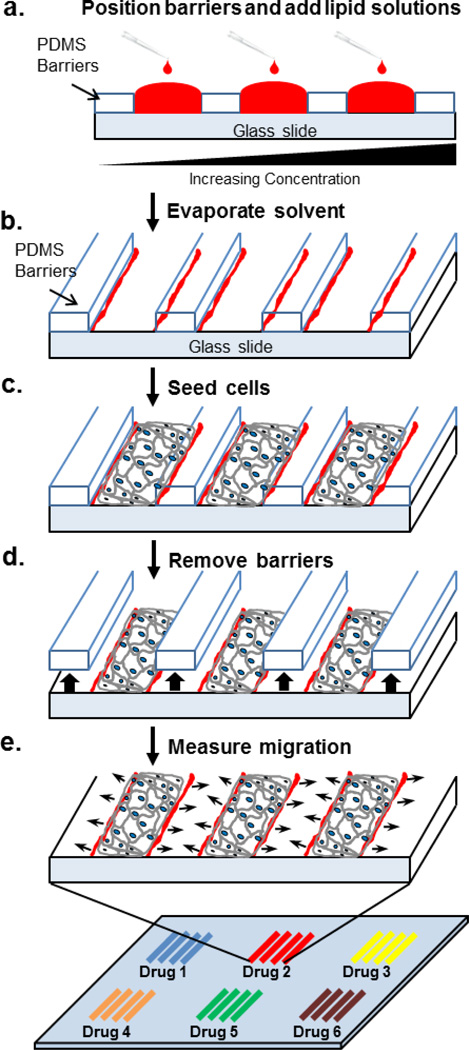
Fig 1. Schematic illustration of edge evaporation lithography and its use for microarrayed cell migration assays.
a. Polydimethylsiloxane (PDMS) barriers are placed around the lipid patterns and the spaces filled with ethanol solutions of lipid mixed with drug at various concentrations. b. The solvent evaporates leaving lipid multilayers along the edges of the PDMS barriers. c. Cells are seeded on the surface and cultured to confluence. d. barriers are removed. e. Cells are allowed to migrate and their migration rate is measured at different positions on the substrate. Many different drugs or dosages can be assayed simultaneously.
2 Materials and Methods
2.1 Preparation of lipids and hydrophobic drugs
All lipids including 1,2-dioleoyl-3-trimethylammoniumpropane (chloride salt) (DOTAP) and 1, 2-dioleoylsn- glycerol-3-phosphoethanolamine-N-(lissamine rhodamine b sulfonyl) (ammonium salt) (DOPE-rhodamine) were purchased from Avanti Polar Lipids (Alabama, USA). Docetaxel was kindly provided by Dr. Diego Zorio (Chemistry and Biochemistry Department, Florida State University). Brefeldin A (BFA) in powder form was purchased from Sigma Aldrich (Missouri, USA). To prepare lipid only solutions first, DOTAP was doped with 1 mol% DOPE-rhodamine in chloroform and dried overnight in a vacuum pump desiccators (KNF Lab, New Jersey, USA) at16 LM 15Torr 7.4 PSIG to remove solvent. Next, ethanol (100%, Sigma) was added to suspend dried lipid powder in solution. A similar procedure was used for preparing docetaxel solutions as described previously15 and modified for preparations of BFA. Docetaxel and brefeldin A powders were dissolved in ethanol to make stock solutions of 1 mg/ml and 5mg/ml, respectively. The drug solutions were diluted to the desired concentration and added with the dried lipid powder to create drug-lipid solutions.
2.2 Edge evaporation lithography (EEL)
Glass coverslips were prepared for use as substrates by first washing with detergent (Palmolive soap) and rinsing thoroughly with deionized water. Next the surfaces were subsequently rinsed in acetone, 100% ethanol, and sterile deionized ultrapure Mill-Q water (EMD Millipore, Massachusetts, USA). Coverslips were dried with a steady stream of nitrogen gas and allowed to completely dry for at least 30 minutes in a biosafety cabinet. Polydimethylsiloxane (PDMS) for creating the migration barriers was cured from the SYLGARD 184 silicone elastomer kit (Dow Corning, Michigan, USA) in a 60 °C oven overnight. PDMS strips of 15 mm long by 1 mm wide were placed between 500 – 800 µm apart on prepared glass coverslips before addition of lipid mixtures. Next, a volume of 1 µl of either lipid only or drug/lipid solutions was deposited in a PDMS channel and then the next solution is added to the adjacent channel. The array was dried overnight in a vacuum to remove residual ethanol.
2.3 Migration assay characterization
The free spaces in between PDMS stencils of the assay that contained lipid material were imaged with a G-2E/C red fluorescence filter (Nikon, Tokyo, Japan). Characterization and statistical analysis of the lipid films was performed by averaging the maximum intensity of 10 random cross sections on the edges of the barrier and in the middle of the channels. Image analysis was performed using the freeware® (NCBI, USA).
2.4 General cell culture and live/dead staining
HeLa cells (obtained from the American Type Culture Collection and maintained according to the collections guidelines) for all experiments were seeded at 2.5×105 cells/ml and grown to confluence in growth media composed of Dulbecco’s Modified Eagle growth medium (Hyclone, Massachusetts, USA) supplemented with 10% Cosmic Calf Serum (Sigma Aldrich, Missouri, USA). Cells were incubated at 37°C and 5% CO2. Trypsin with EDTA (0.25%, VWR, Pennsylvania, USA) was used for cell detachment and the medium was replaced with fresh growth medium 24 h before the experiment. Cells were stained for viability with BacLight live/dead assay (Invitrogen, California, USA) 20 minutes prior to imaging with SYTO9 and propidium iodide in Hank’s buffered saline solution (HBSS, Lonza, Basel, Switzerland). Growth media was added back and imaged on a fluorescent microscope.
2.5 Cell adhesion experiments
Lipids were dissolved in ethanol and added to each channel in increasing 10-fold amounts from 0.2 ng to 20 µg. Tests for cell attachment were performed by seeding HeLa cells into assay channel with varying amounts of lipid films. Prior to seeding for viability experiments, the cells were stained with live/dead assay as described in section 2.4. The cells were allowed to attach for two hours before washing the channels repeatedly 5 times. The number of cells per square micron area (µm2) that remained attached was counted manually. The number of dead cells was excluded because high concentrations of lipids caused dead cells to clump together and made it difficult to accurately access cell count. Experiments for cell adhesion where performed in three replicates (n = 3). Values for cell density of each treatment were determined by averaging the number of cells in 5 random 100 µm2 areas in a single image captured with a 10× objective.
2.6 Cell migration assay of HeLa cells
The assays were carried out on glass coverslips placed in wells of 6-well plates for cell culture. Prior to seeding for viability experiments, the cells were stained with the live/dead assay as described in section 2.4. For experiments, cells were seeded onto the prepared glass coverslips with arrays by gently pipetting 1 ml of cells in suspension at a concentration of 2.5×105 cells/ml directly over each PDMS channel to allow cells to settle in them at consistent density. The same method was used to seed other parts of the slide for use as control areas. Cells were allowed to settle for 1–2 hours before the PDMS barriers were removed to promote cell migration. After barrier removal, the coverslips were washed 3 times with HBSS and replaced with fresh growth media to remove free lipids or drugs. The cells were incubated over the patterned areas for up to 24 hours. The width of each cell strip was measured manually by taking at least random 20 measurements using Nikon Elements 4.0 analysis software and the average migration rate for each strip was determined with the following equation: Migration Rate (µm/hr) = (WidthT=Final – WidthT=Initial)/Total Time. Width is the average width in micrometers (µm) of the cell monolayer measured either at the beginning of the experiment (T = 0 hrs) or at the end (T = 24 hrs).
2.7 Microscopy and time-lapse live cell imaging
The images for cell migration were captured on a Nikon Ti Eclipse (Tokyo, Japan) inverted microscope with 4× (NA = 0.13), 10× (NA = 0.3), and 20× (NA = 0.45) objectives. Images for cell migration were taken after barrier removal and once again 24 hours after the incubation period in phase contrast. Fluorescent filters used were B-2E/C and G-2E/C (Nikon, Tokyo, Japan) for the dyes used in this work. Time-lapse live cell imaging was performed in a Viva View FL Incubator Fluorescent Microscope (Olympus, Tokyo, Japan) using standard cell incubation conditions.
2.8 Statistical Analysis
A student t-test was performed to determine statistical significance between means of each sample (p-value < 0.05 or 0.01, as stated). Error bars in figures denote standard error of the mean unless otherwise stated. All experiments involving cell culture were performed at least in triplicate.
3 Results and Discussion
3.1 Characterization of lipid films for cell migration assay
EEL involves the production of a thin lipid multilayer nanostructured film along the edge of a stencil (Fig. 1a–b). We used PDMS as an elastomeric stencil to direct the precipitation of lipid and drug solutes along an edge resulting in a drug-encapsulated lipid multilayer film that can deliver lipophilic drugs to adherent cells for migration assays (Fig. 1c–e). A video of the lines that form at the edge as the solvent evaporates is shown in supplementary video 1. This technique is compatible with tissue culture plastic for use in cell culture microplates as well as glass surfaces. Glass was chosen as a substrate for its superior imaging properties. We were able to achieve a throughput of 14 tests on a standard glass coverslip (4.84 cm2) which is about 3 tests per cm2 by manually positioning the PDMS barriers as a proof-of-concept. The barriers are 1 cm long and can fit inside a well for 24-well microplates (~1.56 cm diameter) but are too large in size for wells of a 96-well microplate (~0.64 cm diameter). However, up to 5 tests can be performed in each well of a 24-well plate for a total number of 120 tests (25% more tests than a 96-well microplate). Although this is a simple procedure that doesn’t require photolithography, a drawback of this assay is that the PDMS stencils are aligned by hand, limiting the throughput. This limitation can be avoided by creating a large stencil with equal spacing using photolithography, and possibly decreasing the dimensions of the features in the stencil. This strategy has been adopted from Poujade et al., which demonstrated that minimal damage is done to cells from peeling on the PDMS stencil when compared to more commonly used techniques such as the scratch migration assay.33 Achieving barrier dimensions of 5 mm long by 0.5 mm wide and channels widths of 0.5 mm would make this assay compatible with 96-well microplates can achieve 2000 tests on the area of a standard microplate, or approximately 20-fold increase over the current 96-well format. Furthermore, had the microarray migration assay described by Onuki-Nagasaki et al.45 used with lipid multilayers, then the assay could be scaled to more than 50,000 tests per microplate making it possible to carry out a high throughput screen on a single plate.
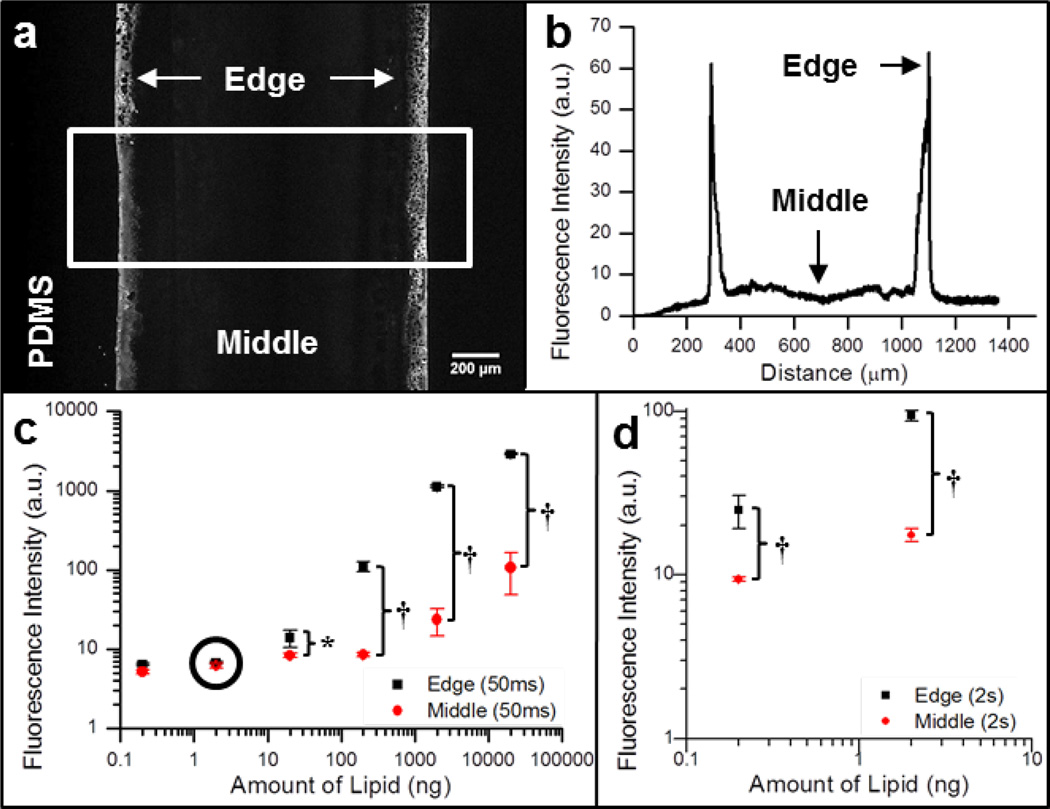
We determined the optimal thickness of lipid films using fluorescent analysis shown in Fig 2. Fluorescence can capture the films in contact with PDMS barriers, an advantage over AFM analysis because the barriers would have to be removed and can potentially deform film shape seen in Fig S1d. Previously, we have shown that fluorescent intensity of DOPE-rhodamine doped lipids is directly correlated to lipid multilayer height or thickness.47 A proportional relationship was also observed from these lipid films between sensitivity (fluorescence intensity versus camera exposure time) and the amount of lipids added between the PDMS stencils (Supplementary Fig S1). We found that the thickness of the lipid multilayers can be controlled by the amount of lipids added between the stencils (Fig 2c). The initial concentration of lipid determined how the lipids dried within the PDMS channels. Adding 200 ng of lipid or higher caused excess lipids to be dried within the middle of the PDMS stencils in addition to thicker multilayers on the edges of the stencil. However at lower lipids amounts (less than 200 ng), we could only detect lipids dried to the edges of the barriers (Fig 2b). This control of multilayer thickness was important when cells were added to the assay.
Fig 2. Control of multilayer thickness in EEL by solute concentration.
a. Fluorescence images of multilayer film arrays with 2 ng of lipids (selection in white used for intensity plot profile). b. Plot profile graph of the fluorescence intensity averaged across the horizontal cross section outlined in panel a taken at a high 2 second exposure time. c. Graph of fluorescence intensity as a function of concentration at the edge and middle of channel. The circled point indicates the amount shown in panels a and b. d. Analysis of fluorescence intensity as a function of amount of lipid from data captured at 2 second exposure for 0.2 and 2 ng of lipid to highlight the difference between edge and middle intensities. It is important to note that the fluorescence data used for analysis in panels c and d were taken at different exposure times. When the exposure time is optimized then there is a clear distinction between the intensity of edge and middle points shown in panel b. Data is expressed as standard error of the mean (n = 10). The * and † represent significance for p < 0.05 and p < 0.01, respectively.
3.2 Cell adhesion on different lipid multilayer thicknesses
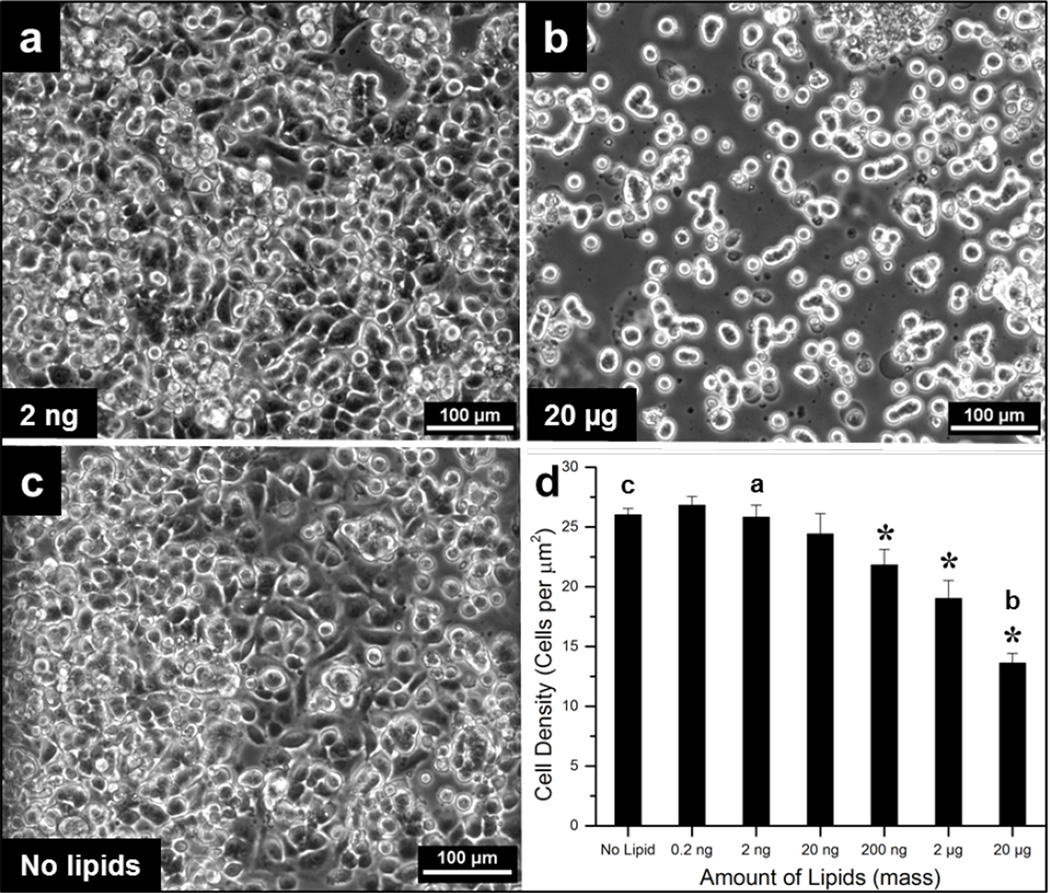
The effect of lipid multilayer thickness on cell adhesion was determined and is shown in Fig 3. HeLa cells were adherent to lipid films created from 20 ng or less but began to adhere significantly less (p < 0.05) at higher concentrations compared to untreated glass (Fig 3c). Cells grown on lipid multilayers created from 200 ng of lipids or more exhibited abnormal morphology and appeared dead compared to thinner multilayers (Fig 3a and 3b). This toxic effect could be attributed from the cationic lipid DOTAP that was used, which has been shown to be cytotoxic at high concentrations.48 It should be noted that at low concentrations, DOTAP is nontoxic and has been shown to be a suitable carrier molecule for cellular uptake.15 These findings corroborate other studies that cells have poor adhesion to certain material surfaces such as lipid bilayers.49 High concentrations of lipid on the surface could also influence the ability of cells to attach to the surface normally by disrupting the ability of adhesion proteins to interact with the surface. Additionally, higher numbers of dead cells were observed over regions with larger amounts of lipid (data not shown). Therefore, depositing 20 ng of lipid or less is suitable for cell studies than larger amounts based on the cell adhesion data.
Fig 3. Cell adhesion to lipid films at low amounts below 20 ng.
a. Micrograph in phase contrast of adherent HeLa cells in channels created from lipid amounts of 2 ng. Carrying out EEL with this amount of lipid allows for HeLa cells to spread out and attach. b. Micrograph in phase contrast of HeLa cells poorly spread out and attached on substrate coated with lipid amounts greater than 20 µg. A higher multilayer thickness results in cells looking balled up and not spread out leading to cell death. c. Phase contrast image of cells without any lipid treatment for comparison. d. Analysis of adherent cell density versus lipid concentration solution used to form films in assay channels (lipid amounts used in panels a – c are indicated). The * represents significant difference from control (p < 0.05). Images and data were collected after 2 hours of incubation. Experiments were performed in triplicate (n = 3) and data is expressed as standard error of the mean.
3.3 Cell migration on different lipid multilayer thicknesses
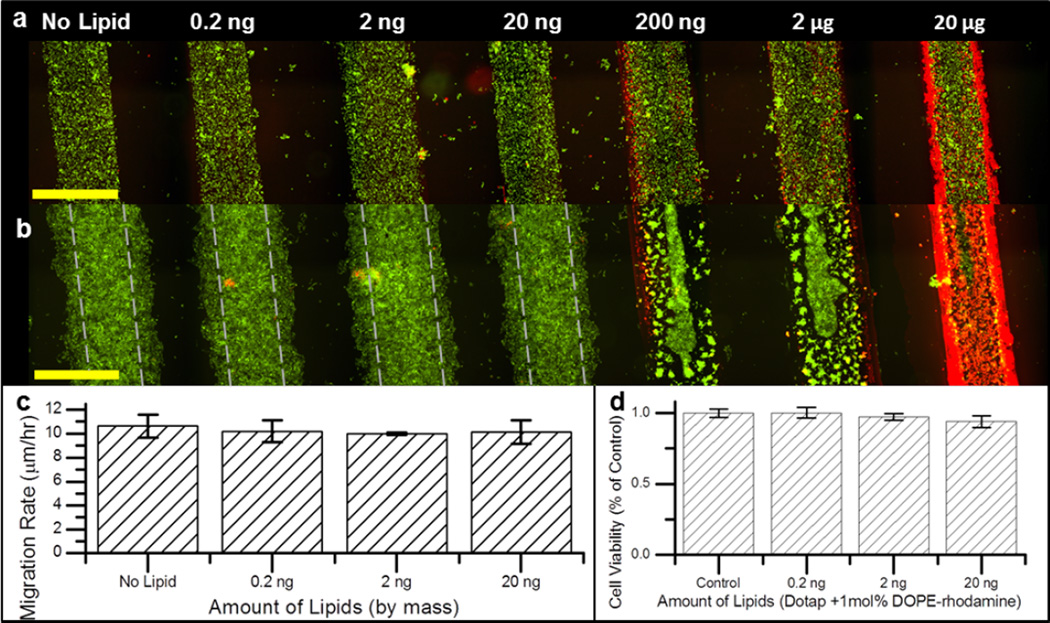
The effect of lipid multilayer thickness on HeLa cell migration was determined in Fig 4. Phase contrast and fluorescent micrographs were taken immediately after PDMS barrier removal and 24 hours after to measure the migration rate in response to multilayer thickness (Fig 4a and 4b). Cell strips exposed to no lipids were able to close the free space in between strip after about 4 days in culture (Supplementary Fig S4). It was observed that, the cells in the strips may not be confluent at early time points (Fig 4a), however these cells spread and become in contact after several hours (Fig 3a and 3c). To visualize the amount of lipid on the surface, DOPE-rhodamine lipid was doped at 1 mol% with the DOTAP lipid. The migration rate of HeLa cells was not significantly (p < 0.05) affected by lipid thickness in channels created from lipids of 20 ng or less but were significantly hindered at higher concentrations (Fig 4c). Lipid film uniformity could also affect cell migration, however we observed that standard error calculated from migration rate were lower than 10% of the mean across the whole strip. Excess lipid on the surface either reduced cell attachment or triggered cell death which significantly reduced the ability of the cell monolayer to migrate. Therefore, we used 20 ng of lipid for all migration assays because this amount was found to have minimal cytotoxicity and no significant effect on HeLa migration rate. The effect of starting cell monolayer strip width on cell migration rate was investigated with NIH3T3 fibroblast cells and the results indicated no significant correlation between start strip width (between 250 – 1300 µm) and the strips migration rate up to 24 hours (Fig S4).
Fig 4. Cell migration can be measured from films made with lipid amounts of 20 ng and below.
a. Initial HeLa cell epithelial sheets before migration; and b. 24 hours after migration (lipid spots shown in red are doped with 1 mol percent DOPE-rhodamine and cells shown in green stained with SYTO9). Track marks in panel b denote initial cell monolayer width in a. c. Migration rate of HeLa cells over 24 hours at 0, 0.2 ng, 2 ng, and 20 ng. d. HeLa cell viability expressed as per cent of control of cells grown over lipid films for 24 hours. Images in a and b are 6×3 stitched micrograph images captured with a motorized stage. Experiments for migration and viability performed in triplicate and data is expressed as standard error of the mean. Scale bars are 1000 µm.
3.4 Effect of docetaxel and BFA from lipid multilayer films on cell migration
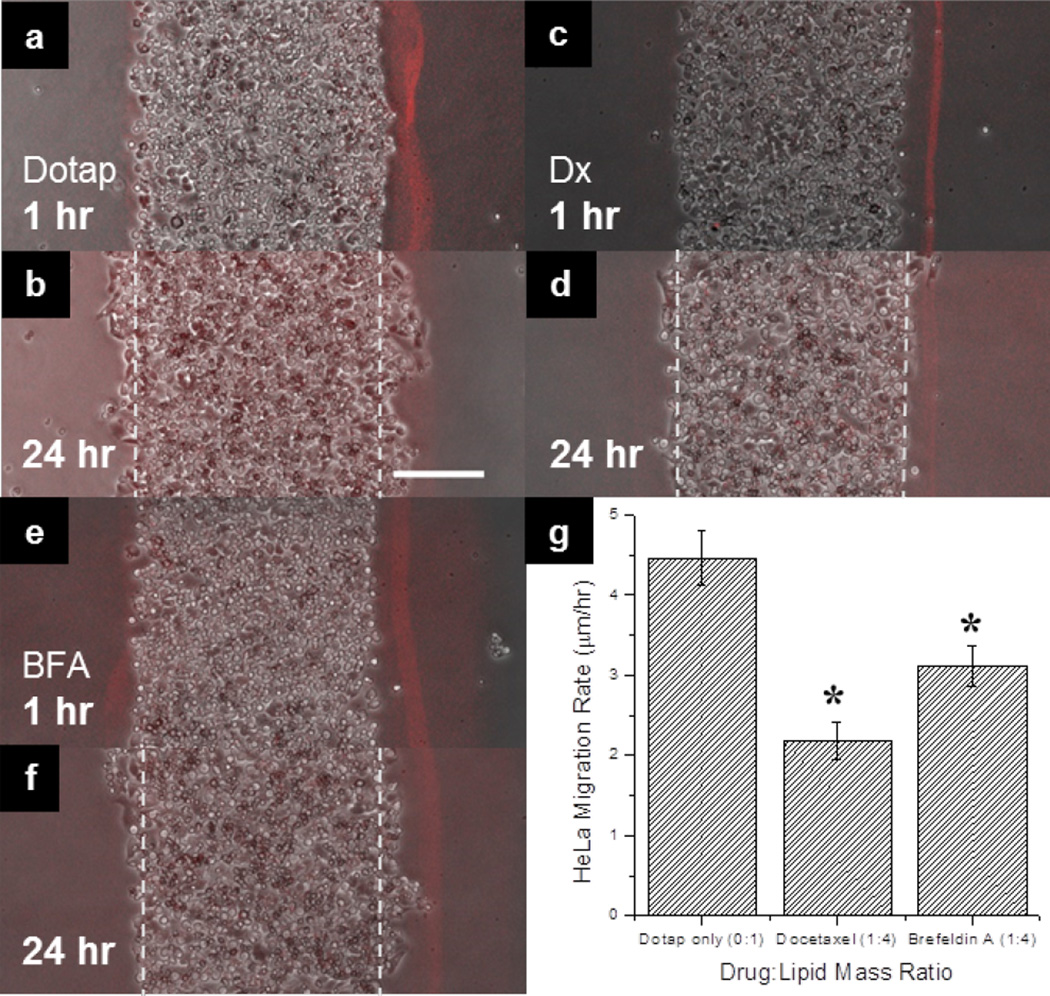
We used EEL to demonstrate a new assay to investigate the effect of two lipophilic drugs on cell migration of HeLa cells in a microarray format. The drugs were: (1) the antimicrotubule drug docetaxel and (2) Brefeldin A (BFA) that inhibits intracellular protein transport. Docetaxel has a logP of 4.1, and BFA that has a logP of 1.61.50–51 Docetaxel and BFA were delivered into HeLa cells by uptake from encapsulated lipid films (Fig 5 and Fig S3). We have shown that HeLa cell migration is also inhibited by BFA and docetaxel which suggests that docetaxel and other taxol derivatives could be used to target different cell processes for cancer therapies. Additionally, three different docetaxel to lipid ratios (by mass) of 1:10, 1:4 and 1:2 were assayed at once on the same array and were found to significantly (p < 0.05) reduce collective cell migration compared to the control group (Fig S2). Furthermore, these results indicated that docetaxel delivered by encapsulated lipid films reduced the migration rate of the cells dose-dependently over 24 hours due to disruption of microtubule dynamics by docetaxel (Supplementary Fig S2). Negative controls for drug treatments include migration niches without lipids, as well as lipids with no drug and lipids at low concentrations of drugs (Fig 4, Supplementary Fig S2). The results indicate that the lipids do not significantly impede migration. Also, we cannot confidently conclude that the lipids may affect drug efficacy by some unknown interaction and such conclusions would need further investigation. Supplementary Fig S3 shows high magnification images of cells migrating from the edge, and Supplementary videos 2 and 3 show live cell imaging of separate monolayers “healing” the open space in this assay. Although cell migration is not expected to be linear and live cell imaging would be ideal for high content screening, the relative migration at a certain time point can be used to assay for drug efficacy for high throughput screening purposes by comparison to a control.
Fig 5. Effect of Docetaxel and Brefeldin A on HeLa cell migration.
a. A merged micrograph of HeLa cell strip (in phase contrast) in contact with a Dotap only fluorescent lipid film (doped with 1 mol% DOPE-rhodamine) after 1 hour after PDMS barriers were removed and b. after 24 hours. c. A merged image of HeLa strip incubated with a docetaxel (Dx) encapsulated fluorescent lipid film after 1 hour and d. 24 hours. e. A merged image of HeLa strip incubated with a brefeldin A (BFA) encapsulated lipid film after 1 hour and f. 24 hours. g. Analysis of HeLa migration rate in microns per hour as a function of drug treatment from lipid multilayer films. Track marks in panels b, d, and f denote initial cell monolayer width in a, c, and e, respectively. The * represents significant difference from Dotap only control (p < 0.05). Data for each treatment was performed in triplicate twice (n = 6) and is expressed as standard error of the mean. Scale bar in panel b. is 200 µm for panels a–f.
One unique capability of this lipid-based surface delivery system over other existing assays is that migrating cells on the edge of the barrier region are exposed locally to lipid encapsulated drug. Although cell displacement and coordination within the cell strip is complex, epithelial cells have been found to proliferate largely within the interior region, while the outer layer of cells are more actively involved in migration and monolayer expansion.33 Locally delivering drugs to the outer region could provide more insights into monolayer growth mechanisms. Additionally, this assay allows for the study of poorly water soluble compounds such as docetaxel and BFA on cell movement following drug delivery into the cells. Different amounts or types of compounds can also be tested at the same time in parallel in a microarray format. Furthermore, this assay requires smaller amounts of drug per assay as compared to a standard scratch migration assay which requires dissolving the drug at certain concentrations in each micro-well.
4 Conclusions
We present edge evaporation lithography (EEL) as a new method to fabricate lipid-based drug delivery microarrays suitable for the investigation of the effect of the hydrophobic compounds docetaxel and BFA on HeLa cell migration. Our results demonstrate in vitro that docetaxel and BFA delivered into cells locally from surface supported lipid films significantly inhibited cellular migration. This novel approach not only will allow delivery and subsequent study of the effects of poorly water soluble drugs on cell migration, structures and functions but will also allow in vitro screening of a variety of different drugs for their effects on cells. This migration assay is unique in that multiple different compounds and dosages can be screened on the same surface, suitable for high throughput screening microarrays.
Supplementary Material
Acknowledgments
SL and NV thank Tom Keller for the helpful discussion. This work was supported by the National Institutes of Health grant R01 GM107172-01.
Footnotes
Supplemental Material: The online version of this article (DOI: 10.1515/nanofab-2015-0004) offers supplementary material.
Contributor Information
Nicholas Vafai, Department of Biological Sciences, Florida State University, Tallahassee, FL, 32306-4370, USA.
Troy W. Lowry, Department of Physics, Florida State University, Tallahassee, FL, 32306-4350, USA
Korey A. Wilson, Department of Biological Sciences, Florida State University, Tallahassee, FL, 32306-4370, USA
Michael W. Davidson, National High Magnetic Field Laboratory, 1800 East Paul Dirac Drive, Florida State University, Tallahassee, FL 32310-3706, USA
Steven Lenhert, Department of Biological Sciences, Florida State University, Tallahassee, FL, 32306-4370, USA.
References
- 1.Lenhert S, Brinkmann F, Laue T, Walheim S, Vannahme C, Klinkhammer S, et al. Lipid multilayer gratings. Nat. Nanotechnol. 2010;5:275–279. doi: 10.1038/nnano.2010.17. [DOI] [PubMed] [Google Scholar]
- 2.Lenhert S, Sun P, Wang YH, Fuchs H, Mirkin CA. Massively parallel dip-pen nanolithography of heterogeneous supported phospholipid multilayer patterns. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 3.Sekula S, Fuchs J, Weg-Remers S, Nagel P, Schuppler S, Fragala J, et al. Multiplexed Lipid Dip-Pen Nanolithography on Subcellular Scales for the Templating of Functional Proteins and Cell Culture. Small. 2008;4:1785–1793. doi: 10.1002/smll.200800949. [DOI] [PubMed] [Google Scholar]
- 4.Lowry TW, Kusi-Appiah A, Guan J, Van Winkle DH, Davidson MW, Lenhert S. Materials Integration by Nanointaglio. Adv. Mater. Int. 2014;1:1–5. doi: 10.1002/admi.201300127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majd S, Mayer M. Generating Arrays with High Content and Minimal Consumption of Functional Membrane Proteins. J. Am. Chem. Soc. 2008;130:16060–16064. doi: 10.1021/ja8055485. [DOI] [PubMed] [Google Scholar]
- 6.Nafday OA, Lowry TW, Lenhert S. Multifunctional Lipid Multilayer Stamping. Small. 2012;8:1021–1028. doi: 10.1002/smll.201102096. [DOI] [PubMed] [Google Scholar]
- 7.Mathieu M, Schunk D, Franzka S, Mayer C, Hartmann N. Temporal stability of photothermally fabricated micropatterns in supported phospholipid multilayers. J. Vac. Sci. Technol. A. 2010;28:953–957. [Google Scholar]
- 8.Diguet A, Le Berre M, Chen Y, Baigl D. Preparation of Phospholipid Multilayer Patterns of Controlled Size and Thickness by Capillary Assembly on a Microstructured Substrate. Small. 2009;5:1661–1666. doi: 10.1002/smll.200900368. [DOI] [PubMed] [Google Scholar]
- 9.Brinker CJ, Lu YF, Sellinger A, Fan HY. Evaporationinduced self-assembly: Nanostructures made easy. Adv. Mater. 1999;11:579–585. [Google Scholar]
- 10.Yuan B, Xing LL, Zhang YD, Lu Y, Mai ZH, Li M. Self-assembly of highly oriented lamellar nanoparticlephospholipid nanocomposites on solid surfaces. J. Am. Chem. Soc. 2007;129:11332–11333. doi: 10.1021/ja074235n. [DOI] [PubMed] [Google Scholar]
- 11.Cai YJ, Zhao Z, Chen JX, Yang TL, Cremer PS. Deflected Capillary Force Lithography. ACS Nano. 2012;6:1548–1556. doi: 10.1021/nn2045278. [DOI] [PubMed] [Google Scholar]
- 12.Jeong HE, Kwak R, Khademhosseini A, Suh KY. UV-assisted capillary force lithography for engineering biomimetic multiscale hierarchical structures: From lotus leaf to gecko foot hairs. Nanoscale. 2009;1:331–338. doi: 10.1039/b9nr00106a. [DOI] [PubMed] [Google Scholar]
- 13.Suh KY, Kim YS, Lee HH. Capillary force lithography. Adv. Mater. 2001;13:1386–1389. [Google Scholar]
- 14.Anrather D, Smetazko M, Saba M, Alguel Y, Schalkhammer T. Supported membrane nanodevices. J. Nanosci. Nanotechnol. 2004;4:1–2. doi: 10.1166/jnn.2004.226. [DOI] [PubMed] [Google Scholar]
- 15.Kusi-Appiah AE, Vafai N, Cranfill PJ, Davidson MW, Lenhert S. Lipid multilayer microarrays for in vitro liposomal drug delivery and screening. Biomaterials. 2012;33:4187–4194. doi: 10.1016/j.biomaterials.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Majd S, Mayer M. Hydrogel stamping of arrays of supported lipid bilayers with various lipid compositions for the screening of drug–membrane and protein–membrane interactions. Angew. Chem. 2005;117:6855–6858. doi: 10.1002/anie.200502189. [DOI] [PubMed] [Google Scholar]
- 17.Diaz-Mochon JJ, Tourniaire G, Bradley M. Microarray platforms for enzymatic and cell-based assays. Chem. Soc. Rev. 2007;36:449–457. doi: 10.1039/b511848b. [DOI] [PubMed] [Google Scholar]
- 18.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tourniaire G, Collins J, Campbell S, Mizomoto H, Ogawa S, Thaburet JF, et al. Polymer microarrays for cellular adhesion. Chem. Comm. 2006:2118–2120. doi: 10.1039/b602009g. [DOI] [PubMed] [Google Scholar]
- 20.Balakin KV, Savchuk NP, Tetko IV. In silico approaches to prediction of aqueous and DMSO solubility of drug-like compounds: Trends, problems and solutions. Curr. Med. Chem. 2006;13:223–241. doi: 10.2174/092986706775197917. [DOI] [PubMed] [Google Scholar]
- 21.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion - Migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat. Rev. Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 22.Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin. Cancer Biol. 2012;22:187–193. doi: 10.1016/j.semcancer.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Eilken HM, Adams RH. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr. Opin. Cell Biol. 2010;22:617–625. doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Griffioen AW, Molema G. Angiogenesis: Potentials for pharmacologic intervention in the treatment of cancer, cardiovascular diseases, and chronic inflammation. Pharmacol. Rev. 2000;52:237–268. [PubMed] [Google Scholar]
- 25.Witte MB, Barbul A. General principles of wound healing. Surg. Clin. North Am. 1997;77:509–528. doi: 10.1016/s0039-6109(05)70566-1. [DOI] [PubMed] [Google Scholar]
- 26.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev. Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Weijer CJ. Collective cell migration in development. J. Cell Sci. 2009;122:3215–3223. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- 28.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 29.Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 30.van Horssen R, ten Hagen TLM. Crossing Barriers: The New Dimension of 2D Cell Migration Assays. J. Cell. Physiol. 2011;226:288–290. doi: 10.1002/jcp.22330. [DOI] [PubMed] [Google Scholar]
- 31.Yarrow JC, Totsukawa G, Charras GT, Mitchison TJ. Screening for cell migration inhibitors via automated microscopy reveals a rho-kinase inhibitor. Chem. Biol. 2005;12:385–395. doi: 10.1016/j.chembiol.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Lenhert S, Meier MB, Meyer U, Chi LF, Wiesmann HP. Osteoblast alignment, elongation and migration on grooved polystyrene surfaces patterned by Langmuir-Blodgett lithography. Biomaterials. 2005;26:563–570. doi: 10.1016/j.biomaterials.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 33.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, et al. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin KD, Lee MY, Shin DS, Lee S, Son KH, Koh S, et al. Blocking tumor cell migration and invasion with biphenyl isoxazole derivative KRIBB3, a synthetic molecule that inhibits Hsp27 phosphorylation. J. Biol. Chem. 2005;280:41439–41448. doi: 10.1074/jbc.M507209200. [DOI] [PubMed] [Google Scholar]
- 35.Attoub S, Hassan AH, Vanhoecke B, Iratni R, Takahashi T, Gaben A-M, et al. Inhibition of cell survival, invasion, tumor growth and histone deacetylase activity by the dietary flavonoid luteolin in human epithelioid cancer cells. Eur. J. Pharmacol. 2011;651:18–25. doi: 10.1016/j.ejphar.2010.10.063. [DOI] [PubMed] [Google Scholar]
- 36.Gough W, Hulkower KI, Lynch R, McGlynn P, Uhlik M, Yan L, Lee JA. A Quantitative, Facile, and High-Throughput Image-Based Cell Migration Method Is a Robust Alternative to the Scratch Assay. J. Biomol. Screen. 2011;16:155–163. doi: 10.1177/1087057110393340. [DOI] [PubMed] [Google Scholar]
- 37.Tavana H, Kaylan K, Bersano-Begey T, Luker KE, Luker GD, Takayama S. Rehydration of Polymeric, Aqueous, Biphasic System Facilitates High Throughput Cell Exclusion Patterning for Cell Migration Studies. Adv. Funct. Mater. 2011;21:2920–2926. doi: 10.1002/adfm.201002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 39.Conant CG, Nevill JT, Schwartz M, Ionescu-Zanetti C. Wound Healing Assays in Well Plate-Coupled Microfluidic Devices with Controlled Parallel Flow. J. Lab. Autom. 2010;15:52–57. [Google Scholar]
- 40.Huang XW, Li L, Tu Q, Wang JC, Liu WM, Wang XQ, et al. On-chip cell migration assay for quantifying the effect of ethanol on MCF-7 human breast cancer cells. Microfluid. Nanofluid. 2011;10:1333–1341. [Google Scholar]
- 41.Kim BJ, Wu MM. Microfluidics for Mammalian Cell Chemotaxis. Ann. Biomed. Eng. 2012;40:1316–1327. doi: 10.1007/s10439-011-0489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu TJ, Lin BC, Qin JH. Carcinoma-associated fibroblasts promoted tumor spheroid invasion on a microfluidic 3D co-culture device. Lab Chip. 2010;10:1671–1677. doi: 10.1039/c000022a. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Zhu J, Deng C, Xing WL, Cheng J. An automatic and quantitative on-chip cell migration assay using self-assembled monolayers combined with real-time cellular impedance sensing. Lab Chip. 2008;8:872–878. doi: 10.1039/b804130j. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Kim M-C, Marquez M, Thorsen T. High-density microfluidic arrays for cell cytotoxicity analysis. Lab Chip. 2007;7:740–745. doi: 10.1039/b618734j. [DOI] [PubMed] [Google Scholar]
- 45.Onuki-Nagasaki R, Nagasaki A, Hakamada K, Uyeda TQP, Fujita S, Miyake M, Miyake J. On-chip screening method for cell migration genes based on a transfection microarray. Lab Chip. 2008;8:1502–1506. doi: 10.1039/b803879a. [DOI] [PubMed] [Google Scholar]
- 46.Timm DM, Chen J, Sing D, Gage JA, Haisler WL, Neeley SK, et al. A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis. Sci. Rep. 2013;3:1–8. doi: 10.1038/srep03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nafday OA, Lenhert S. High-throughput optical quality control of lipid multilayers fabricated by dip-pen nanolithography. Nanotechnology. 2011;22:1–7. doi: 10.1088/0957-4484/22/22/225301. [DOI] [PubMed] [Google Scholar]
- 48.Groves JT, Mahal LK, Bertozzi CR. Control of cell adhesion and growth with micropatterned supported lipid membranes. Langmuir. 2001;17:5129–5133. [Google Scholar]
- 49.Tang F, Hughes JA. Synthesis of a single-tailed cationic lipid and investigation of its transfection. J. Control. Release. 1999;62:345–358. doi: 10.1016/s0168-3659(99)00158-3. [DOI] [PubMed] [Google Scholar]
- 50.Fayad W, Rickardson L, Haglund C, Olofsson MH, D’Arcy P, Larsson R, et al. Identification of Agents that Induce Apoptosis of Multicellular Tumour Spheroids: Enrichment for Mitotic Inhibitors with Hydrophobic Properties. Chem. Biol. Drug Des. 2011;78:547–557. doi: 10.1111/j.1747-0285.2011.01170.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhu J-W, Nagasawa H, Nagura F, Mohamad SB, Uto Y, Ohkura K, Hori H. Elucidation of strict structural requirements of Brefeldin A as an inducer of differentiation and apoptosis. Bioorg. Med. Chem. 2000;8:455–463. doi: 10.1016/s0968-0896(99)00297-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.