Abstract
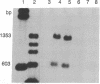
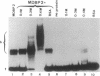
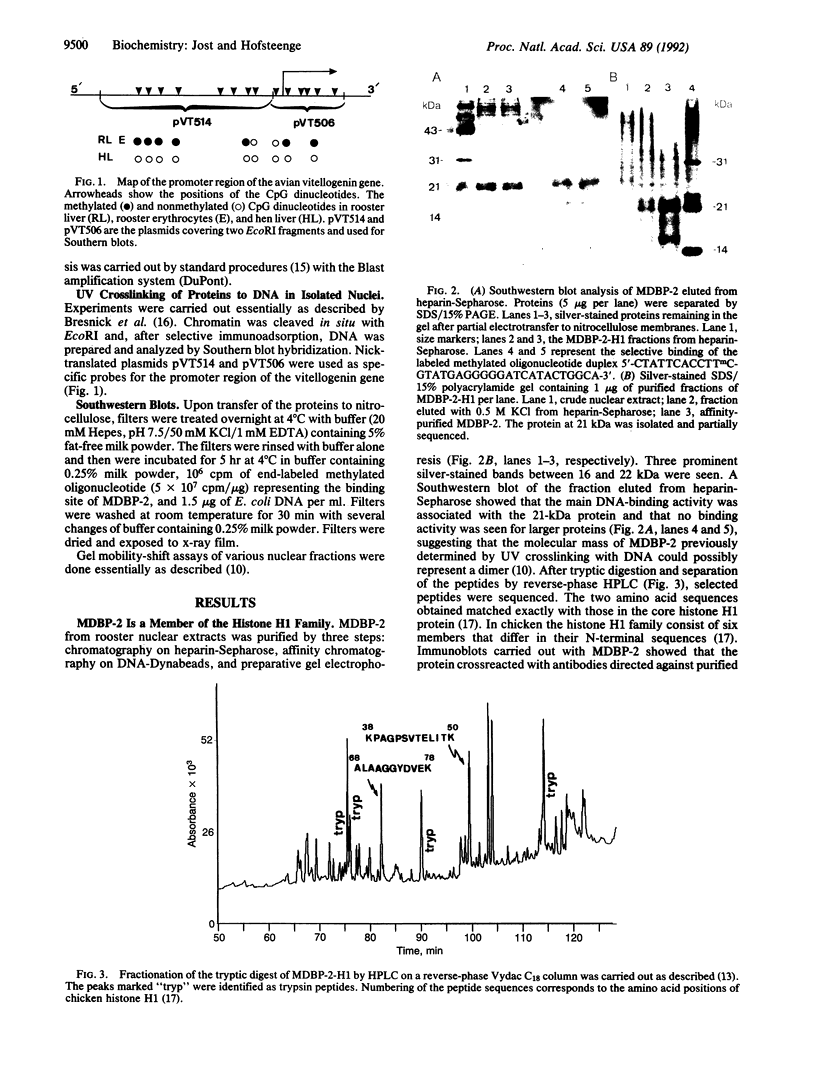
MDBP-2 is a repressor that binds preferentially to methylated DNA. Peptides derived from MDBP-2 were sequenced. The sequences of the two peptides, KPAGPS-VTELITK and ALAAGGYDVEK, are identical to those found in the chicken histone H1 core protein. In SDS/polyacrylamide gels MDBP-2 has an apparent molecular mass of 21 kDa, and antibodies directed against calf thymus total histone H1 cross-react with MDBP-2. The preferential binding of affinity-purified MDBP-2 to methylated DNA is not sequence-specific but requires a minimum length of 30 base pairs and one pair of symmetrically methylated (i.e., methylated on both strands) CpG dinucleotides. As previously shown, there is a decrease in the binding activity of MDBP-2 to methylated DNA upon estradiol treatment. Immunoblots show that upon estradiol treatment the amount of immunocrossreacting MDBP-2 protein remains unchanged. MDBP-2 enables another protein to bind DNA which by itself does not bind methylated DNA. Ultraviolet crosslinking and selective immunoadsorption assays with anti-histone H1 antibodies show that in vivo MDBP-2 preferentially binds to the methylated repressed vitellogenin gene. It is concluded that MDBP-2 may participate in the long-term silencing of genes (formation of heterochromatin) through selective binding to methylated DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauw G., Van Damme J., Puype M., Vandekerckhove J., Gesser B., Ratz G. P., Lauridsen J. B., Celis J. E. Protein-electroblotting and -microsequencing strategies in generating protein data bases from two-dimensional gels. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7701–7705. doi: 10.1073/pnas.86.20.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandt W. F., Strickland W. N., Strickland M., Carlisle L., Woods D., von Holt C. A histone programme during the life cycle of the sea urchin. Eur J Biochem. 1979 Feb 15;94(1):1–10. doi: 10.1111/j.1432-1033.1979.tb12864.x. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992 Jan 25;20(2):273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Cole R. D. Species and organ specificity in very lysine-rich histones. J Biol Chem. 1968 Sep 10;243(17):4500–4505. [PubMed] [Google Scholar]
- Böhm E. L., Strickland W. N., Strickland M., Thwaits B. H., van der Westhuizen D. R., von Holt C. Purification of the five main calf thymus histone fractions by gel exclusion chromatography. FEBS Lett. 1973 Aug 15;34(2):217–221. doi: 10.1016/0014-5793(73)80797-5. [DOI] [PubMed] [Google Scholar]
- Cole R. D. A minireview of microheterogeneity in H1 histone and its possible significance. Anal Biochem. 1984 Jan;136(1):24–30. doi: 10.1016/0003-2697(84)90303-8. [DOI] [PubMed] [Google Scholar]
- Coles L. S., Robins A. J., Madley L. K., Wells J. R. Characterization of the chicken histone H1 gene complement. Generation of a complete set of vertebrate H1 protein sequences. J Biol Chem. 1987 Jul 15;262(20):9656–9663. [PubMed] [Google Scholar]
- Croston G. E., Kerrigan L. A., Lira L. M., Marshak D. R., Kadonaga J. T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991 Feb 8;251(4994):643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- Geiser M., Mattaj I. W., Wilks A. F., Seldran M., Jost J. P. Structure and sequence of the promoter area and of a 5' upstream demethylation site of the estrogen-regulated chicken vitellogenin ii gene. J Biol Chem. 1983 Jul 25;258(14):9024–9030. [PubMed] [Google Scholar]
- Harrison J. J., Schwoch G., Schweppe J. S., Jungmann R. A. Phosphorylative modification of histone H1 subspecies following isoproterenol and N6,O2'-dibutyryl cyclic AMP stimulation of rat C6 glioma cells. J Biol Chem. 1982 Nov 25;257(22):13602–13609. [PubMed] [Google Scholar]
- Hofsteenge J., Kieffer B., Matthies R., Hemmings B. A., Stone S. R. Amino acid sequence of the ribonuclease inhibitor from porcine liver reveals the presence of leucine-rich repeats. Biochemistry. 1988 Nov 15;27(23):8537–8544. doi: 10.1021/bi00423a006. [DOI] [PubMed] [Google Scholar]
- Jost J. P., Munch O., Andersson T. Study of protein-DNA interactions by surface plasmon resonance (real time kinetics). Nucleic Acids Res. 1991 May 25;19(10):2788–2788. doi: 10.1093/nar/19.10.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost J. P., Saluz H. P., Pawlak A. Estradiol down regulates the binding activity of an avian vitellogenin gene repressor (MDBP-2) and triggers a gradual demethylation of the mCpG pair of its DNA binding site. Nucleic Acids Res. 1991 Oct 25;19(20):5771–5775. doi: 10.1093/nar/19.20.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kinkade J. M., Jr Qualitative species differences and quantitative tissue differences in the distribution of lysine-rich histones. J Biol Chem. 1969 Jun 25;244(12):3375–3386. [PubMed] [Google Scholar]
- Mayer R. E., Hendrix P., Cron P., Matthies R., Stone S. R., Goris J., Merlevede W., Hofsteenge J., Hemmings B. A. Structure of the 55-kDa regulatory subunit of protein phosphatase 2A: evidence for a neuronal-specific isoform. Biochemistry. 1991 Apr 16;30(15):3589–3597. doi: 10.1021/bi00229a001. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Pawlak A., Bryans M., Jost J. P. An avian 40 KDa nucleoprotein binds preferentially to a promoter sequence containing one single pair of methylated CpG. Nucleic Acids Res. 1991 Mar 11;19(5):1029–1034. doi: 10.1093/nar/19.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R., Cole R. D. Histone H1 subfractions and H10 turnover at different rates in nondividing cells. Biochemistry. 1982 Feb 2;21(3):456–460. doi: 10.1021/bi00532a006. [DOI] [PubMed] [Google Scholar]
- Saluz H. P., Feavers I. M., Jiricny J., Jost J. P. Genomic sequencing and in vivo footprinting of an expression-specific DNase I-hypersensitive site of avian vitellogenin II promoter reveal a demethylation of a mCpG and a change in specific interactions of proteins with DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6697–6700. doi: 10.1073/pnas.85.18.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluz H. P., Jiricny J., Jost J. P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura A., Sapp M., Rodriguez-Campos A., Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol. 1989 Dec;9(12):5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen R. H., Cole R. D. Histone biosynthesis in the mammary gland during development and lactation. J Biol Chem. 1969 Sep 25;244(18):4878–4887. [PubMed] [Google Scholar]
- Wang R. Y., Zhang X. Y., Khan R., Zhou Y. W., Huang L. H., Ehrlich M. Methylated DNA-binding protein from human placenta recognizes specific methylated sites on several prokaryotic DNAs. Nucleic Acids Res. 1986 Dec 22;14(24):9843–9860. doi: 10.1093/nar/14.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Zhang D. L., Ehrlich K. C., Supakar P. C., Ehrlich M. A plant DNA-binding protein that recognizes 5-methylcytosine residues. Mol Cell Biol. 1989 Mar;9(3):1351–1356. doi: 10.1128/mcb.9.3.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg J. M., Alva G. P., Jost J. P. Characterisation of a genomic clone covering the structural mouse MyoD1 gene and its promoter region. Nucleic Acids Res. 1991 Dec 11;19(23):6433–6439. doi: 10.1093/nar/19.23.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]