Abstract
Current phylogenetic sampling reveals that dioecy and an XY sex chromosome pair evolved once, or possibly twice, in the genus Asparagus. Although there appear to be some lineage-specific polyploidization events, the base chromosome number of 2n = 2× = 20 is relatively conserved across the Asparagus genus. Regardless, dioecious species tend to have larger genomes than hermaphroditic species. Here, we test whether this genome size expansion in dioecious species is related to a polyploidization and subsequent chromosome fusion, or to retrotransposon proliferation in dioecious species. We first estimate genome sizes, or use published values, for four hermaphrodites and four dioecious species distributed across the phylogeny, and show that dioecious species typically have larger genomes than hermaphroditic species. Utilizing a phylogenomic approach, we find no evidence for ancient polyploidization contributing to increased genome sizes of sampled dioecious species. We do find support for an ancient whole genome duplication (WGD) event predating the diversification of the Asparagus genus. Repetitive DNA content of the four hermaphroditic and four dioecious species was characterized based on randomly sampled whole genome shotgun sequencing, and common elements were annotated. Across our broad phylogenetic sampling, Ty-1 Copia retroelements, in particular, have undergone a marked proliferation in dioecious species. In the absence of a detectable WGD event, retrotransposon proliferation is the most likely explanation for the precipitous increase in genome size in dioecious Asparagus species.
Keywords: Asparagus, dioecy, sex chromosomes, transposons, genome size
Fewer than 10% of flowering plant species are dioecious—the condition where individual plants are distinctly male or female (Ainsworth 2000). Gender in some dioecious plants can be governed by a sex chromosome pair, such as in papaya (Carica papaya), white campion (Silene latifolia), persimmon, Rumex, and garden asparagus (Asparagus officinalis) (Telgmann-Rauber et al. 2007; Ming et al. 2011; Akagi et al. 2014; Hough et al. 2014). The evolution of a distinct sex chromosome pair is hypothesized to be driven by the evolution of a nonrecombining region between the X and Y (or Z and W) chromosomes, where tightly linked sex determination genes reside (Charlesworth and Charlesworth 1978). Given the repeated and independent evolution of dioecy across the angiosperm phylogeny, the transition from autosome to sex chromosome is undoubtedly governed by different sex determination genes and evolutionary processes, and consequently must be viewed in a taxon-specific context. Despite this diversity in sex chromosome evolution across the angiosperms, two particularly interesting associations can be seen in some dioecious systems coincident with variation in sexual system: the proliferation of repetitive elements, and the occurrence of one or multiple whole genome duplication (WGD) (polyploidy) events.
As a consequence of restricted recombination between regions of sex chromosomes, repetitive elements tend to persist and replicate in an unbalanced way, accumulating preferentially on hemizygous regions of Y and W chromosomes. Transposable elements can be broadly classified primarily by their means of transposition (Wicker et al. 2007); class I retrotransposons move by a “copy and paste” mechanism, and replicate through an mRNA intermediate, which ultimately results in a net increase in the element’s copy number; whereas class II transposable elements move through a DNA intermediate in a “cut and paste” fashion. Since Class I retrotransposons can range in length from 5 kb to 20 kb or longer, their proliferation can lead to drastic and rapid changes in genome size (Kidwell 2002). This accumulation, especially of active retroelements, can be clearly seen when comparing the relatively young papaya X and hermaphrodite-specific region of the Y (HSY) (VanBuren and Ming 2013). Unbalanced accumulation of transposons and other repetitive elements, paired with the inability for recombination to remove them along with other deleterious mutations, is likely a major factor that leads to the initial physical expansion and genic degeneration of a young, partially nonrecombining Y or W chromosome (Steinemann and Steinemann 1998; Bachtrog et al. 2008; Bachtrog 2013). Transposons have also been directly implicated in the evolution of sex determination genes through disruption of gene expression. In melon (Cucumis melo), a class II hAT DNA transposon insertion is responsible for promoter hypermethylation and transcriptional repression of the zinc-finger transcription factor CMWIP1, heritably inducing the transition from monoecy to gynodioecy (Boualem et al. 2008).
An association between polyploidy and transitions in sexual system across the angiosperms is most clear in the Fragaria genus, where at least four independent WGD events have occurred across all major clades, leading to an abundance of polyploid dioecious species phylogenetically placed as sister to dioecious hermaphrodites (Rousseau-Gueutin et al. 2009; Ashman et al. 2013). However, loss of dioecy has also been associated with an increase in ploidy, as seen in one clade of Mercurialis (Krähenbühl et al. 2002). The mechanisms that potentially relate WGD events to the evolution of sexually dimorphic populations are variable and poorly understood, though again owing to the extreme complexity and species-specific nature of sex chromosome and dioecy.
Garden asparagus (A. officinalis L.) is a particularly useful dioecious plant for studying sex chromosome evolution given that it has cytologically homomorphic X and Y sex chromosomes, suggesting that the transition from hermaphroditism to dioecy was recent (Telgmann-Rauber et al. 2007; Kubota et al. 2012). Coincident with the evolution of dioecy was a range shift from South Africa into North Africa, Europe, and Asia (Štajner et al. 2002; Kubota et al. 2012; Norup et al. 2015). It was previously reported that dioecious Asparagus species tend to have larger genomes than hermaphrodites, but there was no evidence supporting a WGD event that separates the dioecious species from the hermaphrodites (Kuhl et al. 2005). The base chromosome number of 2n = 2× = 20 is generally consistent across the genus, except for instances of very recent polyploidization in some species (Kanno and Yokoyama 2011). These findings suggest one of two hypotheses may be responsible for an increase in genome size: one possibility is that a WGD occurred in the last common ancestor of all dioecious species, followed by a chromosome fusion or reduction; and another possibility is that repetitive DNA has proliferated to drive the increase in the genome sizes of dioecious species. Here, we test both hypotheses by first leveraging transcriptome assemblies for one hermaphroditic and one dioecious species to identify the relative timing of WGD events in the genus Asparagus. We then use shallow Roche 454 whole genome shotgun sequencing from four hermaphrodites and four dioecious species that are sampled from across the phylogeny to assess the repetitive element content of each species in relation to its genome size.
Materials and Methods
Flow cytometry genome size estimation
The genome sizes of A. officinalis, A. virgatus, and A. asparagoides were estimated by flow cytometry at the Benaroya Research Institute at Virginia Mason in Seattle, WA. Nuclei isolations from a single mature leaf were analyzed in three technical replicates for each species. The genome sizes of A. aphyllus, A. stipularis, and A. falcatus were estimated by flow cytometry using the known genome size of A. officinalis (1C-value = 1.37 pg) as a reference standard. Ten plants for each species, grown in a greenhouse, were sampled, and three randomly selected plants were analyzed. The analysis was carried out with the Partec PAS flow cytometer (Partec, http://www.partec.de/), equipped with a mercury lamp. Fully expanded leaves (0.1 g) were chopped in 300 μl nuclei extraction buffer (CyStain ultraviolet Precise P Nuclei Extraction Buffer; Partec, Münster, Germany) for 30–40 sec. The solution was filtered through a 30-mm Cell-Trics disposable filter (Partec), and 1.2 ml of staining solution containing 4,6-diamidino-2- phenylindole was added. The relative fluorescence intensity of stained nuclei was measured on a linear scale, and 4000–5000 nuclei for each sample were analyzed (Galbraith et al. 1998). DNA content histograms were generated using the Partec software package (FloMax). Given that the X and Y chromosomes in garden asparagus (A. officinalis) are cytologically homomorphic (Deng et al. 2012), representing a lack of degeneration and the relatively young age of Y, we did not discern between potential sex differences in the dioecious species.
Transcriptome-based Ks analysis
Transcriptomes from dioecious A. officinalis and hermaphroditic A. asparagoides were used to infer a putative WGD event in the genus Asparagus. The transcriptome assembly and translation for A. officinalis was taken from Harkess et al. (2015) (http://datadryad.org/resource/doi:10.5061/dryad.92c60). We generated leaf RNA-Seq for A. asparagoides by first isolating total RNA from mature leaf tissue using a Qiagen RNeasy Plant Mini kit. Total RNA quantity and quality was assessed using an RNA Nano chip on the Bioanalyzer 2100. A sequencing library was generated using the TruSeq RNA Library Prep Kit v2 (Illumina) according to the manufacturer’s instructions, using 1 μg of total RNA input. The library was sequenced with paired-end 100-nt reads on an Illumina HiSeq2000, generating 55,686,513 read pairs (nearly 11 Gb of data). Reads were quality trimmed using Trimmomatic (v0.32), removing sequencing adapters, and clipping 3′ and 5′ read ends with a quality score lower than Phred 5. Cleaned reads were assembled using Trinity (r20140717) with default parameters. We filtered transcript isoforms with low support by removing isoforms with < 0.01% of the Trinity gene subcomponent read support. Coding sequence and peptide translations were inferred using TransDecoder (r20140704) with default settings. Raw sequence reads for A. asparagoides have been deposited under BioProject PRJNA326431.
Using a pipeline from McKain et al. (2012; https://github.com/mrmckain/FASTKs), we first identified putative paralogs in each filtered transcriptome assembly using all vs. all blastn (1e–40 cutoff). Peptide sequences for hit pairs longer than 100 amino acids were aligned using MUSCLE (v3.8.31), then codon alignments were inferred using PAL2NAL (v13) (Suyama et al. 2006). For each paralog pair, Ks was calculated using CodeML in PAML (Yang 2007) (v4.8).
454 pyrosequencing and transposon quantification
Whole genomic DNA was extracted from four hermaphroditic and four dioecious species using a CTAB method (Doyle and Doyle 1987). Sequencing libraries were prepared using the Roche 454 GS FLX Titanium library preparation kit according to the manufacturer’s instructions. Raw reads were first deduplicated to remove probable emulsion PCR sequencing artifacts, then filtered to remove reads < 100 nt long. Read names from all species were first prepended with a unique species identifier, and concatenated. The RepeatExplorer (v0.9.7.4) pipeline (http://www.repeatexplorer.org) was then used to cluster, assemble, and annotate all filtered shotgun reads against a custom garden Asparagus RepeatMasker database (see below) using otherwise default settings. Clustering and heatmap production of the 100 largest transposon clusters was performed using heatmap.2 in the gplot package in R (v3.2.1) using default settings; a distance matrix was generated using Euclidean distances, and hierarchical clustering was performed using “complete” clustering.
To improve the annotations of repetitive element clusters generated through the RepeatExplorer pipeline, instead of utilizing default RepeatMasker Viridplantae libraries, we generated a much higher coverage of 454 reads for A. officinalis to build a comprehensive database of annotated exemplar repeats for the Asparagus genus. A custom garden Asparagus RepeatMasker database was generated using similar methodology. A total of 893,623 454 FLX Titanium reads were generated from leaf tissue of a doubled haploid (YY) garden asparagus individual. Reads were more stringently filtered to a 150 nt minimum length. The same version of RepeatExplorer was then run, and the resulting cap3 consensus assemblies for each cluster were annotated using RepeatClassifier, part of the RepeatModeler (v1.0.8) suite, with default settings. A total of 22,361 sequences greater than 150nt in length and with annotations was retained for annotating all repetitive element clusters, and are available in Dryad (http://dx.doi.org/10.5061/dryad.1k450). Raw 454 shotgun sequence data for all individuals have also been deposited in Dryad.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Genome size increases in dioecious Asparagus
Genome sizes and ploidy vary greatly across the order Asparagales, with 1C values ranging from 0.3 pg to 88.2 pg (Leitch et al. 2010). Diploid dioecious Asparagus species have been reported as having genome sizes nearly double the size of diploid hermaphroditic congeners (Štajner et al. 2002; Fukuda et al. 2005; Kubota et al., 2012). We first confirmed this by generating or supplementing published genome size estimations for eight Asparagus species, four hermaphrodites, and four dioecious species, sampled across all major clades of the Asparagus phylogeny (Kubota et al. 2012) (Table 1). All individuals have been documented as diploids (2n = 2× = 20) except for A. maritimus, a hexaploid (Štajner et al. 2002; Kanno and Yokoyama 2011). Flow cytometry-derived genome sizes (pg/1C) for hermaphrodites range from 0.72 to 1.06, while dioecious species range from 1.09 to 1.37. Dioecious species tend to have larger genome sizes than hermaphroditic species (unpaired t-test, P = 0.0173). An outlier is the hermaphrodite A. asparagoides, with a relatively large genome size (1C = 2.40; Dixon’s Q test, P = 0.074).
Table 1. Genome sizes, 454 pyrosequencing, and repetitive element clustering.
| Species | Sexual System | Picograms/Nucleus (Mean ± SD) | 1C Value | Raw Reads | Filtered Reads | Clustered Reads (%) |
|---|---|---|---|---|---|---|
| A. officinalis | Dioecious | 2.74 ± 0.044 | 1.37 | 29,677 | 26,525 | 54.4% |
| A. maritimus | Dioecious | 7.87 ± 0.204a | 1.31 | 49,616 | 45,036 | 53.7% |
| A. aphyllus | Dioecious | 2.49 ± 0.007 | 1.25 | 47,322 | 42,808 | 58.9% |
| A. stipularis | Dioecious | 2.17 ± 0.005 | 1.09 | 30,405 | 27,911 | 56.4% |
| A. falcatus | Hermaphrodite | 2.11 ± 0.007 | 1.06 | 26,836 | 24,304 | 60.4% |
| A. virgatus | Hermaphrodite | 1.66 ± 0.055 | 0.83 | 45,043 | 41,053 | 45.0% |
| A. pyrimidalis | Hermaphrodite | 1.44 ± 0.037a | 0.72 | 56,197 | 51,293 | 53.8% |
| A. asparagoides | Hermaphrodite | 4.80 ± 0.062 | 2.40 | 41,952 | 37,435 | 59.2% |
| Sum | 247,755 | 224,804 | ||||
| Average | 41,293 | 37,467 |
Data from Štajner et al. (2002).
No evidence for a dioecy-specific polyploidy event
We employed a phylogenomics approach to test whether a WGD event separates the dioecious and hermaphroditic species in Asparagus. Transcriptome assemblies were generated for two species sampled broadly across the phylogeny: a basal diploid hermaphrodite (A. asparagoides; 2n = 2× = 20), and diploid dioecious garden asparagus (A. officinalis; 2n = 2× = 20). Intraspecific paralog pairs and interspecific orthologous gene pairs were inferred to generate Ks (synonymous substitution rate) distributions, and assess the relative timing of WGD event relative to speciation events (Blanc and Wolfe 2004; Cui et al. 2006; McKain et al. 2012; Doyle and Egan 2010). Despite being an outlier in terms of genome size, A. asparagoides was utilized for the comparison given that it is a basal member of the genus, shares the same diploid chromosome count as A. officinalis, and that transcriptome-based Ks analyses are independent of genome size.
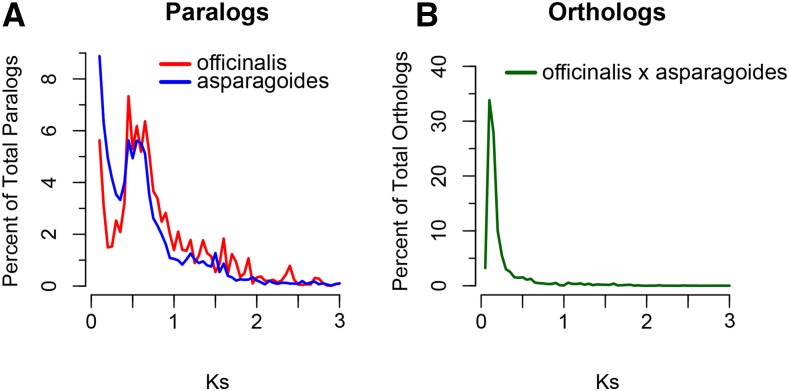
Transcriptome assembly and translation results for the two species are presented in Supplemental Material, Table S1. One distinct, shared, polyploidization event (Ks ∼0.5) was inferred from the Ks frequency distribution of paralogous pairs in both Asparagus species (Figure 1 and Table S2). Additionally, orthologous pairs exhibit a Ks peak close to 0, representing low divergence, and suggestive of recent diversification of species and/or similar mutation rates. Comparison of orthologs and paralogs demonstrates that at least one detectable genome duplication event occurred before the diversification of the Asparagus genus (Figure 1).
Figure 1.
Transcriptome-based Ks frequency distributions for (A) paralogous, and (B) orthologous pairs of dioecious A. officinalis and hermaphroditic A. asparagoides. Paralogous and orthologous Ks distributions suggest a shared whole genome duplication event at Ks ∼0.5 that occurred before the diversification of the Asparagus genus.
A major limitation with Ks analyses is that more recent duplication events are difficult to detect (Blanc and Wolfe 2004; Cui et al. 2006). This issue is exacerbated when using de novo transcriptome assemblies, where recently duplicated paralogs can be computationally mistaken as alleles, and incorrectly collapsed into a single transcript sequence during the assembly process. Gene duplication, in addition to sequencing and assembly errors, can contribute to a high frequency of low Ks gene pairs. Given that the A. officinalis and A. asparagoides ortholog peak overlaps with the left-hand shoulder in the paralog Ks plot, the plots alone do not allow us to unambiguously reject recent WGDs within the Asparagus genus. Given that there are no current age estimates for the divergence of the Asparagus lineage, we cannot exclude the possibility that a more recent duplication event, such as one that may have co-occurred with the evolution of dioecy, could be undetectable by transcriptome data. However, such a whole genome duplication event would need to be followed by multiple chromosome fusion or loss events to reduce the chromosome number back to 2n = 2× = 20 found in most karyotyped dioecious Asparagus species (Kanno and Yokoyama 2011).
Lineage-specific expansion of transposable elements
Given the lack of evidence that ancient polyploidy was responsible for the larger genome sizes of dioecious Asparagus species relative to hermaphroditic species, we assessed the alternative hypothesis that the genome size increases in dioecious species were at least partly due to transposon amplification. We utilized whole genome shotgun sequence reads to assess the repetitive content of hermaphrodite and dioecious Asparagus species using the RepeatExplorer Galaxy server (http://www.repeatexplorer.org). Briefly, this method utilizes all-by-all read comparisons followed by Louvain clustering (Blondel et al. 2008) to place reads into unbiased clusters of putative high copy elements, followed by a RepeatMasker annotation and cap3 assembly (Huang and Madan 1999).
A total of 327,048 raw reads were sequenced for the eight genomes using Roche 454 FLX chemistry, with genome coverages ranging from 0.0051× to 0.0234× (Table S3). After removing duplicate reads that were likely clonal, 321,865 total reads remained for analysis. To improve clustering, we then removed reads < 100 nt long, yielding a filtered set of 296,365 reads (mean = 37,047 reads per species) with a mean length of 321 nt. The filtered set of reads was concatenated and clustered using the RepeatExplorer pipeline, placing 162,435 reads into 29,643 repetitive element clusters (Table 1). Repetitive element clusters were filtered by read count, requiring at least 0.01% of the total filtered reads (30 reads), amounting to 336 clusters for downstream analysis. These clusters were annotated against a custom RepeatMasker database generated with additional data for dioecious A. officinalis. For a given cluster of repetitive elements, the repetitive fraction of each species’ genome was calculated as the number of a given species’ reads in a cluster, divided by the total number of reads sequenced for that species, represented as a percentage.
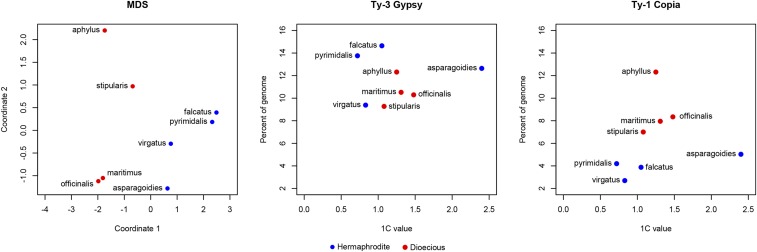
Multidimensional scaling (MDS) analysis of the genomic proportions for all clusters shows that dioecious and hermaphroditic species form two distinct clusters (Figure 2). In general, Gypsy and Copia retrotransposons dominate the genomic landscape for all sampled Asparagus species (Figure 3). In all four dioecious species, Gypsy retrotransposons occupy a larger percentage of each genome than in the hermaphrodites, although Copia elements have distinctly expanded in the dioecious species (Figure 2). This suggests that both Gypsy and Copia elements increased in copy number in the dioecious species, and the proliferation of Copia elements was a more substantial contributor to the expansion of dioecious genome sizes. Taken together, the lack of evidence for recent WGDs described in Kuhl et al. (2005), paired with the increased abundance of Copia elements in dioecious species, supports our hypothesis that there are no recent WGDs in the Asparagus genus.
Figure 2.
Multidimensional scaling (MDS) and relationship of genome size to Gypsy and Copia retroelement content for both dioecious and hermaphroditic genomes. Blue dots represent hermaphroditic species, while red dots represent dioecious species.
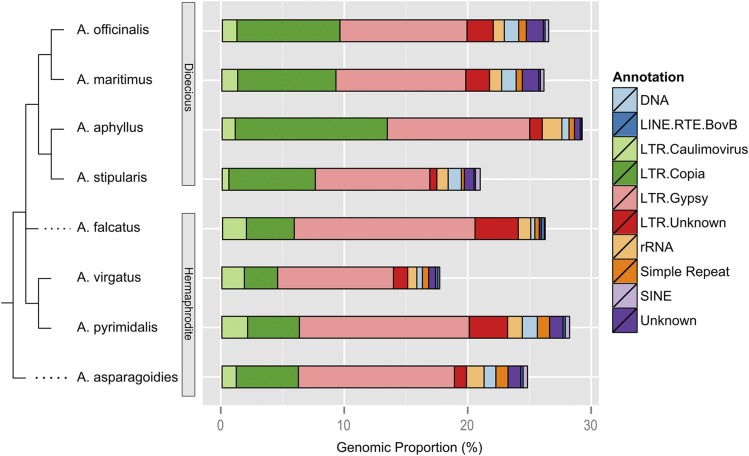
Figure 3.
Cladogram of Asparagus species relationships with high copy repetitive elements. High copy elements refer to clusters with greater than 0.01% of the total read count in the multispecies analysis, able to be most confidently annotated against the custom A. officinalis repetitive element database. DNA transposons from several families were collapsed into a single annotation class.
We identified 46 repetitive element clusters that were private to the dioecious species, and 37 clusters that were private to all hermaphroditic species. In the dioecious species, 26 clusters were Gypsy, and seven clusters were Copia, whereas in the hermaphroditic species, 12 clusters were Gypsy, and 11 clusters were Copia. This suggests that there is active turnover of transposable elements in the Asparagus genus, perhaps coincident with the evolution of dioecy and a sex chromosome. Additionally, it is possible that a small number of Copia elements may be largely responsible for the genome size expansion in dioecious species, but this would require whole genome assemblies and annotations as RepeatExplorer is limited in ability to finely delimit elements.
One caveat for performing a single repeat clustering analysis including all species (as opposed to individually analyzing each species) is that low frequency or moderately diverged sequences from phylogenetically distant species may not cluster. Additionally, there could be less power for detecting species-specific transposon family proliferations. Consequently, these estimates of repetitive element content are certainly underestimates of the total proportion of repetitive element content in each species’ genome. To understand the level of difference in these two analysis types, we generated 893,623 additional 454 shotgun reads (mean length 526 nt) for a mature double haploid YY A. officinalis individual, and ran the RepeatExplorer pipeline with this single species. The repeat content was estimated at 71.1%, much greater than the 54.4% that was estimated by concatenating eight species in a single analysis. This result suggests that the genomic proportions of transposons estimated through multispecies read clustering in this study should be interpreted as being underestimates, biased toward high copy elements with lower divergence between species, and used mostly for comparisons of high copy element percentages between species. The advantage of this analysis is that direct comparisons for a given transposon cluster can be assessed across all species, without the need to perform additional clustering between species. Without genome sequences and assemblies for several hermaphroditic and dioecious Asparagus species, including high quality repetitive element annotations and length distributions, we cannot quantify with certainty the nucleotide contribution that each transposable element class contributes to genome size increase.
The method of repeat quantification and sequence read type also largely affects the estimated proportion of repetitive elements. Repetitive element content has previously been estimated for A. officinalis in at least three separate studies. Vitte et al. (2013) directly annotated garden Asparagus Bacterial Artificial Chromosome (BAC) assemblies for transposon content. By comparing the sequence alignment identity of intact long terminal repeats (LTRs) from retroelements, and applying a clock estimation from rice retroelement divergence (Ma and Bennetzen 2004), Vitte et al. (2013) estimated that the majority of the Asparagus genome is comprised of young, recently inserted (< 6 million yr ago) and nested retroelements. Li et al. (2014) took a high-throughput sequencing approach, and inferred that the garden Asparagus genome is 53% repetitive by de novo assembling genomic paired-end 100 nt Illumina reads into a ∼400 Mbp assembly with a scaffold N50 of 1504 nt. Hertweck (2013) took a similar approach with 80-bp Illumina read data, and independently estimated 47% of the garden Asparagus genome as comprising repetitive elements. We hypothesize that our much higher estimation of 71.1% repetitive content is due largely to the increased detection power coming with longer 454 reads relative to 80–100 bp Illumina reads, and our use of RepeatExplorer’s unique assembly-free, graph-based clustering, and annotation of individual long reads.
Transposon clustering yields phylogenetic signal
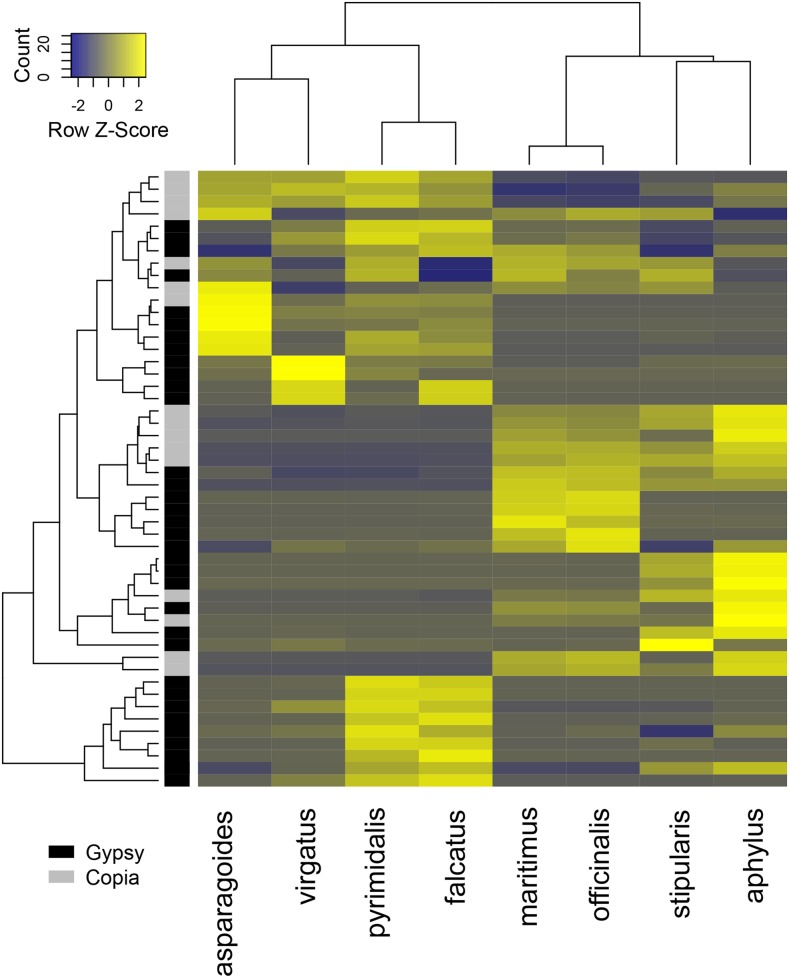
Clustering of the genomic proportions for the 100 largest Gypsy and Copia retrotransposon clusters also reveals phylogenetic signal in the data (Figure 4). The deepest branch divides the hermaphroditic and dioecious species from each other, and all species are paired with their closest phylogenetic neighbor given the current phylogeny and sampling from Kubota et al. (2012), with the exception of the earliest diverging species in the genus, A. asparagoides. The genomic proportions of repetitive elements have been used to identify phylogenetic signal in several plant species with species relationships that have been difficult to resolve with traditional low copy gene sequencing (Dodsworth et al. 2014). While our clustering approach may be less able to detect low- and medium-frequency repeats compared to the approach of Dodsworth et al. (2014), here we show a complementary analysis that yields similar results using high copy transposon clusters.
Figure 4.
Heatmap clustering of 100 largest Gypsy or Copia element clusters. Rows represent individual clusters, annotated as Gypsy (black), and Copia (gray).
Recently, Norup et al. (2015) proposed two origins of dioecy within Asparagus, providing an alternative to the previously hypothesized single origin (Kubota et al. 2012). Our sampling includes species derived from both of the hypothesized origins of dioecy from Norup et al. (2015), which indicates that dioecy evolved in one clade that includes A. officinalis and A. maritimus, as well as another clade that includes A. stipularis and A. aphyllus. In the case of multiple origins of dioecy, without hermaphroditic outgroup species for each origin, our limited sampling does not allow us to describe the potentially different repetitive element radiations in the two dioecious clades. Further, it is possible that transposon proliferation and genome size increase occurred in the common ancestor of both dioecious lineages, predating the origin of dioecy. Rigorous testing of a general relationship between transposon activity and the origin of sex chromosomes will come with future meta-analyses including data from this and other comparative studies of transposon activity in hermaphrodite and dioecious lineages.
Compared to Asparagus, similar cases of lineage-specific transposon expansion have been found in the Asteraceae, where a small number of Gypsy families have been expanding since the branch leading to the Asteraceae (Staton and Burke 2015). We hypothesize that the proliferation of both Gypsy and Copia retroelements in dioecious lineages is associated with two coincident events in Asparagus evolution: range expansion and the origin of dioecy. As others have documented, range expansion out of South Africa is associated with a transition of ancestrally hermaphroditic Asparagus species to dioecy within a clade distributed across Europe and Asia. (Štajner et al. 2002; Kuhl et al. 2005; Kanno and Yokoyama 2011; Kubota et al. 2012; Norup et al. 2015). Founder populations formed during this range expansion with small effective population sizes may have been especially susceptible to weakly deleterious transposon proliferation due to the reduced strength of purifying selection relative to populations with large effective sizes (Lynch et al. 2011). In addition, the origin of sex chromosomes alone may have promoted proliferation of retrotransposons. Suppressed recombination within the region of the sex chromosomes where gender determination genes reside in the first dioecious Asparagus species may have harbored active retrotransposons. An excess of repetitive elements can be found in the nonrecombining regions of several plant Y chromosomes. For instance, the Y chromosomes in both Silene and papaya can be replete with, or entirely lacking, tandem arrays and LTR retroelements that distinguish them from both the X and other autosomes (Pritham et al. 2003; Filatov et al. 2009; VanBuren and Ming 2013). Recombination is selected against in these sex determination regions of a sex chromosome, given that recombination could break apart genes influencing male and female function, leading to the formation of neuters. As a consequence of nonrecombination, portions of Y (or W) chromosomes are particularly susceptible to the effects of Müller’s ratchet—an evolutionary process leading to the accumulation of slightly deleterious elements that can be accelerated in small effective population sizes (Charlesworth and Langley 1989; Moran 1996). Further, ectopic exchange (recombination at nonhomologous sites) is also predicted to control the proliferation of transposable elements in genomes by creating deleterious chromosomal rearrangements. While selected against in recombining portions of the genome, the rate of fixed ectopic exchange in nonrecombining regions of the Y is expected to be lower or zero, potentially leading to the proliferation of TE copy number through relaxed selection (Charlesworth and Langley 1989). This selection on young sex chromosomes may drive the maintenance and proliferation of repetitive elements, and, in concert with faster mutation rates and background selection, may lead to the initial expansion and subsequent degeneration of sex chromosomes (Engelstädter 2008). While the accumulation of repetitive elements on sex chromosomes has been well studied, understanding the spread of these elements to autosomes, and the subsequent contributions to genome size increase, will require additional comparative genomic analyses.
Supplementary Material
Acknowledgments
The authors would like to thank the editor and two anonymous reviewers for comments and insight that substantially improved the manuscript. This work was funded by the National Science Foundation (DEB 0841988).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.030239/-/DC1
Communicating editor: S. I. Wright
Literature Cited
- Ainsworth C., 2000. Boys and girls come out to play: the molecular biology of dioecious plants. Ann. Bot. (Lond.) 86: 211–221. [Google Scholar]
- Akagi T., Henry I. M., Tao R., Comai L., 2014. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346: 646–650. [DOI] [PubMed] [Google Scholar]
- Ashman T.L., A. Kwok, and B. C. Husband, 2013. Revisiting the dioecy-polyploidy association: alternate pathways and research opportunities. Cytogenet. Genome Res. 140: 241–255. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Hom E., Wong K. M., Maside X., de Jong P., 2008. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 9: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K. H., 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel V. D., Guillaume J., Lambiotte R., Lefebvre E., 2008 Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008: P10008. [Google Scholar]
- Boualem A., Fergany M., Fernandez R., Troadec C., Martin A., et al. , 2008. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321: 836–838. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997. [Google Scholar]
- Charlesworth, B., and C. H. Langley, 1989 Population genetics of transposable elements in Drosophila, pp. 150–176 in Evolution at the molecular level, edited by R. Selander, A. Clark, and T. Whittam. Sinauer, Sunderland, MA. [Google Scholar]
- Cui L., Wall P. K., Leebens-Mack J. H., Lindsay B. G., Soltis D. E., et al. , 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C.-L. L., Qin R.-Y. Y., Wang N.-N. N., Cao Y., Gao J., et al. , 2012. Karyotype of Asparagus by physical mapping of 45S and 5S rDNA by FISH. J. Genet. 91: 209–212. [DOI] [PubMed] [Google Scholar]
- Dodsworth S., Chase M. W., Kelly L. J., Leitch I. J., Macas J., et al. , 2014. Genomic repeat abundances contain phylogenetic signal. Syst. Biol. 64: 112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Doyle J. J., Egan A. N., 2010. Dating the origins of polyploidy events. New Phytol. 186: 73–85. [DOI] [PubMed] [Google Scholar]
- Engelstädter J., 2008. Muller’s ratchet and the degeneration of Y chromosomes: a simulation study. Genetics 180: 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov D. A., E. C. Howell, C. Groutides, and S. J. Armstrong, 2009. Recent spread of a retrotransposon in the Silene latifolia genome, apart from the Y chromosome. Genetics 181: 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Ashizawa H., Suzuki R., Ochiai T., Nakamura T., et al. , 2005. Molecular phylogeny of the genus Asparagus (Asparagaceae) inferred from plastid petB intron and petD–rpoA intergenic spacer sequences. Plant Spec. Biol. 20: 121–132. [Google Scholar]
- Galbraith D. W., Lambert G. M., Macas J., Doležel J., 1998. Analysis of nuclear DNA content and ploidy in higher plants, pp. 7.6.1–7.6.21 in Current protocols in cytometry, edited by J. P. Robinson, Z. Darzynkiewicz, P. N. Dean, L. G. Dressler, A. Orfao et al John Wiley & Sons, New York. [DOI] [PubMed] [Google Scholar]
- Harkess A., Mercati F., Shan H. Y., Sunseri F., Falavigna A., et al. , 2015. Sex-biased gene expression in dioecious garden asparagus (Asparagus officinalis). New Phytol. 207: 883–892. [DOI] [PubMed] [Google Scholar]
- Hertweck K. L., 2013. Assembly and comparative analysis of transposable elements from low coverage genomic sequence data in Asparagales. Genome 56: 487–494. [DOI] [PubMed] [Google Scholar]
- Hough J., Hollister J. D., Wang W., Barrett S. C. H., Wright S. I., 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc. Natl. Acad. Sci. USA 2014: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A., 1999. CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno, A., and J. Yokoyama, 2011 Asparagus, pp. 23–42 in Wild Crop Relatives: Genomic and Breeding Resources, edited by C. Kole. Springer, Berlin. [Google Scholar]
- Kidwell M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63. [DOI] [PubMed] [Google Scholar]
- Krähenbühl M., Yuan Y. M., Küpfer P., 2002. Chromosome and breeding system evolution of the genus Mercurialis (Euphorbiaceae): implications of ITS molecular phylogeny. Plant Syst. Evol. 234: 155–169. [Google Scholar]
- Kubota S., Konno I., Kanno A., 2012. Molecular phylogeny of the genus Asparagus (Asparagaceae) explains interspecific crossability between the garden asparagus (A. officinalis) and other Asparagus species. Theor. Appl. Genet. 124: 345–354. [DOI] [PubMed] [Google Scholar]
- Kuhl J. C., Havey M. J., Martin W. J., Cheung F., Yuan Q., et al. , 2005. Comparative genomic analyses in Asparagus. Genome 1060: 1052–1060. [DOI] [PubMed] [Google Scholar]
- Leitch I. J., Beaulieu J. M., Chase M. W., Leitch A. R., Fay M. F., 2010. Genome size dynamics and evolution in monocots. J. Bot. 2010: 1–18. [Google Scholar]
- Li S. F., Gao W. J., Zhao X. P., Dong T. Y., Deng C. L., et al. , 2014. Analysis of transposable elements in the genome of Asparagus officinalis from high coverage sequence data. PLoS One 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Bobay L.-M., Catania F., Gout J.-F., Rho M., 2011. The repatterning of eukaryotic genomes by random genetic drift. Annu. Rev. Genomics Hum. Genet. 12: 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J. L., 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain M. R., Wickett N., Zhang Y., Ayyampalayam S., McCombie W. R., et al. , 2012. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Am. J. Bot. 99: 397–406. [DOI] [PubMed] [Google Scholar]
- Ming R., Bendahmane A., Renner S. S., 2011. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62: 485–514. [DOI] [PubMed] [Google Scholar]
- Moran N. A., 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norup M. F., Petersen G., Burrows S., Bouchenak-Khelladi Y., Leebens-Mack J., et al. , 2015. Evolution of Asparagus L. (Asparagaceae): Out-of-South-Africa and multiple origins of sexual dimorphism. Mol. Phylogenet. Evol. 92: 25–44. [DOI] [PubMed] [Google Scholar]
- Pritham E. J. E., Zhang Y. H., Feschotte C., Kesseli R. R. V., 2003. An Ac-like transposable element family with transcriptionally active Y-linked copies in the white campion, Silene latifolia. Genetics 165: 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M., Gaston A., A. Aïnouche, M. L. Aïnouche,, K. Olbricht et al, 2009. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): New insights from phylogenetic analyses of low-copy nuclear genes. Mol. Phylogenet. Evol. 51: 515–530. [DOI] [PubMed] [Google Scholar]
- Štajner N., Bohanec B., Javornik B., 2002. Genetic variability of economically important Asparagus species as revealed by genome size analysis and rDNA ITS polymorphisms. Plant Sci. 162: 931–937. [Google Scholar]
- Staton S. E., Burke J. M., 2015. Evolutionary transitions in the Asteraceae coincide with marked shifts in transposable element abundance. BMC Genomics 16: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann M., Steinemann S., 1998. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica 102–103: 409–420. [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P., 2006. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue): W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgmann-Rauber A., Jamsari A., Kinney M. S., Pires J. C., Jung C., 2007. Genetic and physical maps around the sex-determining M-locus of the dioecious plant Asparagus. Mol. Genet. Genomics 278: 221–234. [DOI] [PubMed] [Google Scholar]
- VanBuren R., Ming R., 2013. Dynamic transposable element accumulation in the nascent sex chromosomes of papaya. Mob. Genet. Elements 3: e23462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C., Estep M. C., Leebens-Mack J., Bennetzen J. L., 2013. Young, intact and nested retrotransposons are abundant in the onion and Asparagus genomes. Ann. Bot. (Lond.) 112: 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Sabot F., Hua-Van A., Bennetzen J. L., Capy P., et al. , 2007. A unified classification system for eukaryotic transposable elements. Natl. Rev. 8: 973–982. [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.