Abstract
Candida glabrata is an important human fungal pathogen whose incidence continues to rise. Because many clinical isolates are resistant to azole drugs, the drugs of choice to treat such infections are members of the echinocandin family, although there are increasing reports of resistance to these drugs as well. In efforts to better understand the genetic changes that lead to altered responses to echinocandins, we screened a transposon-insertion library of mutants for strains to identify genes that are important for cellular responses to caspofungin, a member of this drug family. We identified 16 genes that, when disrupted, caused increased tolerance, and 48 genes that, when disrupted, caused increased sensitivity compared to the wild-type parental strain. Four of the genes identified as causing sensitivity are orthologs of Saccharomyces cerevisiae genes encoding proteins important for the cell wall integrity (CWI) pathway. In addition, several other genes are orthologs of the high affinity Ca2+ uptake system (HACS) complex genes. We analyzed disruption mutants representing all 64 genes under 33 different conditions, including the presence of cell wall disrupting agents and other drugs, a variety of salts, increased temperature, and altered pH. Further, we generated knockout mutants in different genes within the CWI pathway and the HACS complex, and found that they too exhibited phenotypes consistent with defects in cell wall construction. Our results indicate that small molecules that inhibit the CWI pathway, or that the HACS complex, may be an important means of increasing the efficacy of caspofungin.
Keywords: caspofungin, echinocandins, cell wall integrity pathway, high affinity calcium uptake system
Candida glabrata is a serious human pathogen; it is estimated that ∼20% of systemic candidiasis infections in the US are now caused by C. glabrata, making it the second most frequent cause after C. albicans (Richardson and Lass-Florl 2008). Immunocompromised patients, those receiving chemotherapy, immunosuppressive drugs, or infected with HIV/AIDS are particularly susceptible (Kale and Johnson 2005; Richardson and Lass-Florl 2008).
Treatment of C. glabrata infections is hampered by a dearth of effective drugs. Until recently, treatment of fungal infections was limited to amphotericin B and azoles, both of which target ergosterol—a key component of the fungal cellular membrane. However, known off-target effects, including organ toxicity, limit their effectiveness. Perhaps more problematic is the growing number of C. glabrata clinical isolates that are resistant to azoles, which has made this family of drugs ineffective against this pathogen (Krcmery and Barnes 2002). As a result, a significant fraction of patients, upwards of 35–40%, suffering from systemic C. glabrata infections die annually (Krcmery and Barnes 2002; Pfaller and Diekema 2004).
The development of the echinocandin class of drugs has helped to fulfill the need for more efficacious and safer antifungal drugs. This class exerts its fungicidal effects by disrupting cell wall synthesis, an ideal target because no comparable structure is present in human cells. Specifically, echinocandins (e.g., caspofungin, micafungin, etc.) impact fungal cell wall synthesis by inhibiting the β-glucan synthases encoded by FKS1, FKS2, and, to a lesser extent, FKS3 (Sucher et al. 2009), and represent the newest and most promising treatment modality available. Nevertheless, there are increasing reports of clinical resistance, chiefly as a result of “hot-spot” mutations in FKS1, and especially FKS2 (Arendrup et al. 2012, 2013; Duran-Valle et al. 2012; Katiyar et al. 2012; Singh-Babak et al. 2012; Borghi et al. 2014; Pham et al. 2014). Echinocandin resistance in some Candida species, especially C. albicans is correlated with chitin overexpression (Walker et al. 2008, 2013). Further, other work has shown that stress from anti-fungals causes genetic instability in Candida species and other fungal pathogens, leading to multi-drug resistance (Shor and Perlin 2015).
C. glabrata, despite the genus name, is more closely related to Saccharomyces cerevisiae than to C. albicans (Kaur et al. 2005; Roetzer et al. 2011). Much of what is known about the genetics underpinning synthesis and maintenance of the cell wall comes from numerous studies in S. cerevisiae (Lesage and Bussey 2006; Orlean 2012), but there is an increasing focus on this pathway in pathogenic fungi as well (Dichtl et al. 2016). The fungal cell wall forms the outermost boundary for maintaining cell shape and permeability to macromolecules. Mechanical strength is provided by large complex macromolecules including glucans, chitin, and mannoproteins. The cell wall is dynamic, changing at different stages in the yeast life cycle (growth and budding), and in response to external cues (induction of sporulation among others) (Lesage and Bussey 2006; Orlean 2012). In C. glabrata, the cell wall appears to be similar, although generally thicker [up to 200 nm in C. glabrata (de Groot et al. 2008) compared to 120 nm in S. cerevisiae (Klis et al. 2014)] with a higher mannose/glucose ratio (0.81 compared to 0.57; de Groot et al. 2008), and 50% more protein than in S. cerevisiae cell walls (de Groot et al. 2008). In addition, to support the pathogenic lifestyle of C. glabrata, one of the key evolutionary adaptations is the expansion of the number of genes encoding cell surface adhesin proteins, which mediate interactions between the yeast cells and the host (Roetzer et al. 2011).
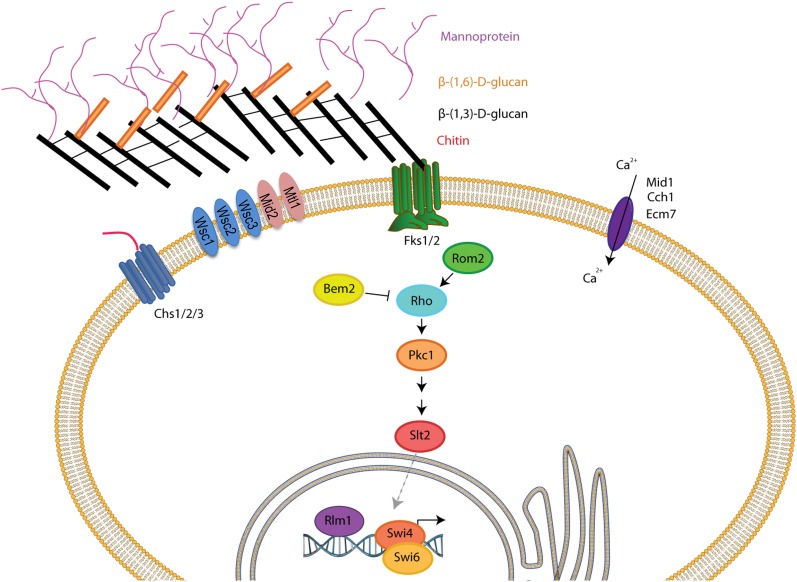
In S. cerevisiae, defects in or damage to the cell wall results in osmotic instability in turn activating the cell wall integrity (CWI) pathway, which ultimately controls transcription of cell wall-related genes such as FKS2/GSC2 (Levin 2011) (see Figure 1). Sensor-transducer proteins such as Slg1/Wsc1, Wsc2, Wsc3, Mid2, and Mtl1 detect damage (Kock et al. 2015). These signal to Rom1/2 (Ozaki et al. 1996; Manning et al. 1997), and guanine nucleotide exchange factors (GEFs) for Rho1 (Madden and Snyder 1998). Rho1 then activates the protein kinase Pkc1 (Antonsson et al. 1994; Watanabe et al. 1994; de la Fuente and Portillo 2000; Valdivia and Schekman 2003), which initiates a kinase cascade, in turn activating the kinase Bck1 (a MAPKKK), which then activates Mkk1/2 (MAPKKs) finally activating the MAP kinase Slt2/Mpk1 (Reinoso-Martin et al. 2003; Levin 2011). Slt2 phosphorylates several transcription factors, including Rlm1 (Jung et al. 2002) and Swi4/6 [(Nasmyth and Dirick 1991; Siegmund and Nasmyth 1996), also called SBF], turning on the expression of genes encoding proteins responsible for synthesis of the cell wall (Figure 1) (Levin 2011). S. cerevisiae strains lacking components of the CWI pathway, in particular strains lacking PKC1, BCK1, and SLT2, display pronounced sensitivity to caspofungin, demonstrating that the CWI pathway is required for caspofungin tolerance (Reinoso-Martin et al. 2003; Markovich et al. 2004). The high affinity calcium uptake system (HACS; Muller et al. 2001), a complex of at least three proteins, Mid1, Cch1, and Ecm7 (Martin et al. 2011), also signals to some of the genes responsible for cell wall synthesis via the calcinuerin-regulated transcription factor, Crz1 (Levin 2011; Martin et al. 2011). Thus regulation of cell wall biosynthesis is multi-layered, and receives information from a number of different pathways. Similar pathways exist in many pathogenic fungi of interest, including C. glabrata, other Candida species, Aspergillus, Cryptococcus, and Pneumocystis (Dichtl et al. 2016).
Figure 1.
The cell wall, the cell integrity pathway, and the HACS complex (adapted from Levin 2011). Fks1 and Fks2 (green transmembrane proteins) are responsible for β-glucan synthesis {illustrated by black [β-(1,3)-d-glucan] and gold [β-(1,6)-d-glucan] bars} and are the targets for caspofungin action. The glucans form the inner cell wall. Mannoproteins are attached to the glucan layer and form the outer cell wall. Note that, in reality, the cell wall is closely apposed to the plasma membrane, containing the stress sensors, the three Wsc proteins, Mid2, and Mtl1. The chitin synthases, Chs1, Chs2, and Chs3 (dark blue), synthesize the chitin polymer that is found primarily at the bud neck between mother and daughter cells, and at bud scars on mother cells. Proteins of the cell wall integrity pathway, initiated by activation of the monomeric G protein Rho1, are illustrated in the center of the cartoon, resulting in activation of the transcription factors, Rlm1, and Swi4/Swi6. The plasma membrane complex for high affinity Ca2+ uptake, Cch1, Mid1, and Ecm7 is also illustrated here (dark purple).
In this study, we screened a collection of ∼ 27,000 C. glabrata strains created by insertion of a transposable element derived from Tn7, covering about 75% of the nonessential genes in the genome, for altered sensitivity to caspofungin (Castano et al. 2003). We identified 16 genes that, when disrupted, cause decreased susceptibility compared to wild type, and 48 genes that cause increased sensitivity. Isolated from this latter group were several C. glabrata genes that are putative orthologs of genes encoding proteins in the CWI pathway, and of the HACS complex of S. cerevisiae. When we made complete gene knockouts of the CWI and HACS genes isolated from the screen, as well as a number of other putative CWI orthologs, the strains exhibited cell wall defects, confirming that these genes encode proteins important for cell wall biosynthesis in C. glabrata.
Materials and Methods
Materials
Media reagents and chemicals were purchased from Thermo Fisher Scientific (Pittsburgh, PA) unless otherwise noted.
Yeast strains and media
The collection of 27,000 C. glabrata Tn7 URA3 hph KmR R6Kγ (Tn7 UKR-H) mutagenized strains used in the screen were obtained from Brendan Cormack (Castano et al. 2003). The parental strain for this collection is BG14 (ura3Δ(-85 + 932)::Tn903NeoR) derived from the wild-type strain BG2 (Cormack and Falkow 1999). Strain BG2 was used as the reference strain for optimizing growth, drug treatment, and zymolyase assays. Yeast were grown at 30° in YPD medium (1% yeast extract, 2% Bacto-peptone, and 2% glucose) unless otherwise stated (Adams et al. 1997). Synthetic dextrose (SD) medium lacking uracil was used for selection of yeast transformants (Adams et al. 1997). To make solid media, 1.5% or 2% (w/v) agar was added. Strains were stored in 15% glycerol at –80°.
Identification of the Tn7 insertion site in C. glabrata genomic DNA
The location of the transposon in each strain of interest was determined by sequencing DNA directly flanking either side of the transposable element. Specifically, genomic DNA was extracted (Adolph 1996), resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), digested with XbaI (Promega, Madison, WI), and ligated by combining 1 µg of cut genomic DNA with T4 DNA ligase (New England Biolabs).
Escherichia coli strain BW23473 (Δlac-169 robA1 creC510 hsdR514 ΔuidA::pir endA recA1; Yale E.coli Stock Center, New Haven, CT) was used to maintain the Tn7-containing plasmids, which have an R6Kγ origin of replication requiring expression of the pir gene (Metcalf et al. 1996). Electrocompetent BW23473 were electroporated using a Gene Pulser (Bio-Rad, Hercules, CA) with the ligation mixture, then incubated in SOC medium (Hanahan 1983) for 1 hr at 37°, then plated on LB agar (Bertani 1951) containing 50 µg/ml kanamycin. Plasmids were isolated using the QIAprep Spin Miniprep kit (Qiagen, Valencia, CA).
To identify the genomic DNA flanking the transposable element in mutant strains, plasmids were sequenced outward from the transposon using the following primers: Tn7 Left (5′-ATAATCCTTAAAAACTCCATTTCCACCCCTCCCAG-3′), and Tn7 Right (5′-GACTTTATTGTCATAGTTTAGATCTATTTTGTTCAG-3′) (Green et al. 2012). Sequencing was performed by Genewiz (South Plainfield, NJ). The sequences obtained were compared to data in the Candida Genome Database (http://candidagenome.org; Inglis et al. 2012).
As a control, the presence of the Tn7 insertion was verified by PCR of genomic DNA. Oligonucleotides complementary to genomic DNA immediately up and downstream of the identified insertion site were used for PCR; amplification across the Tn7 insertion generates a larger band than the wild-type gene. PCR was also performed using one oligonucleotide specific to genomic DNA upstream of the insertion, and one specific to URA3 within the Tn7 element; only PCR of DNA containing the transposon results in a band. Strains with evidence of a wild-type gene were eliminated from further consideration.
Growth assays
Cells were grown in YPD medium at 30° to OD600 0.8, then diluted 25-fold into a final volume of 0.25 ml in YPD containing varying concentrations of caspofungin (Merck, Whitehouse Station, NJ) in a 96-well plate, and placed at 30° under medium shaking. The plates were read at OD600 every 15 min for up to 10 hr using a GloMax microtiter plate spectrophotometer (Promega). The maximal specific growth rate (κmax) of each culture was calculated by fitting an exponential regression over the experimental points. These points were selected to yield a correlation coefficient (R2) higher than 0.9 when possible (in cultures where cells did not grow well the R2 value was 0.7–0.8). The κ constant from the equation y = y0e(κ.t) was the maximal specific growth rate.
Zymolyase sensitivity assays
Zymolyase assays were performed as previous described (Ovalle et al. 1998). Cells were grown to exponential phase in YPD medium, and then washed three times in 1× TE, and resuspended to an OD600 of 0.9 in 1× TE. Triplicate cultures were pipetted in to 96-well plates along with either 10% volume 1× TE (no-enzyme control), or with 10% volume 1× TE containing 100T zymolyase (Zymo Research, Irvine, CA) to a final concentration of 50 μg/ml. The OD600 was measured every 4 min for a total 160 min at room temperature using a GloMax instrument (Promega). Triplicate sample readings were averaged. Each experiment was completed at least three times.
Gene ontology term analysis
Gene ontology (GO) term enrichment analysis was done with the BiNGO plug-in and the GOSlim terms for Cytoscape (Shannon et al. 2003; Maere et al. 2005). The C. glabrata gene identified was assumed to be the ortholog of the most closely related S. cerevisiae gene as identified by BLASTP (Altschul et al. 1990). GO terms for S. cerevisiae genes were downloaded from the GO consortium (http://www.geneontology.org/GO.downloads.annotations.shtml). The hypergeometric test was applied, and derived P-values were adjusted for multiple hypothesis testing by the method of Benjamini and Hochberg (1995). GO terms with adjusted P-values < 0.05 were considered significantly enriched. In addition, the gene list was submitted for analysis by FunSpec with and without Bonferroni correction (Robinson et al. 2002).
Phenotype analysis
Mutants from the Tn7 collection selected for further analysis were stored at –80° in 15% glycerol in 96-well submasters. The mutants, either from the Tn7 collection, or the knockout mutants generated as described below, were diluted via a 96-pin replicator tool to new 96-well microtiter plates containing 200 µl of deionized water per well. Diluted cells were then replica printed onto plates containing solid YPD with and without one of the following: caspofungin, rapamycin (Sigma-Aldrich), Calcofluor white, tunicamycin (Sigma-Aldrich), caffeine (Sigma-Aldrich), Congo red (Thermo Fisher Scientific), or FK-506 monohydrate (Sigma-Aldrich). Specific concentrations used in a given experiment are listed in the corresponding figure legends.
In some experiments, diluted mutant strains were also replica printed onto YPD plates containing different salts including NaCl, KCl, LiCl, or CaCl2, with or without the addition of sorbitol. Effects of lower pH were also tested by preparing YPD plates buffered at pH 3.0 and 4.0 by the addition of phosphate-citrate buffers at pH 2.5 and 3.5, respectively, to YPD prior to autoclaving (Adams et al. 1997). Plates were incubated at 25° for 48 hr, except for plates incubated at 30°, 37°, or 42° to examine temperature sensitivity.
Growth of individual strains was evaluated semiquantitatively by 10-fold serial dilution followed by spotting onto solid medium. No growth even at the highest cell concentration was given a score of 0. Abundant growth even at the highest dilution was given a score of 4. All experiments were performed at least twice, and all data were evaluated by two of the authors.
Based on growth scores, hierarchal clustering analysis was performed using the hclust function of the stats package in the R programming environment, which utilizes the complete-linkage hierarchal clustering method (R Development Core Team 2013). In this agglomerative method, a distance matrix is calculated for the set of elements and each element is then placed in a cluster of its own, and clusters are then sequentially combined into larger clusters, until all elements end up in one cluster. At each step, the two clusters separated by the shortest distance are combined (Hartigan 1975). Heat maps showing expression fold change were generated using the gplots package developed for the R programming environment.
Creation of C. glabrata knockout strains
A PCR-based method was used to create a gene knockout cassette to replace a gene with URA3 in the BG14 background (Kuwayama et al. 2002). In each case, a piece containing the URA3 gene flanked by 20 bp upstream and downstream sequences immediately proximal and distal to the open-reading frame to be replaced was stitched to an ∼500 bp upstream fragment and a ∼500 bp downstream fragment of the target gene to create a linear fragment large enough for homologous recombination to occur at the correct locus. The PCR reactions were performed with PrimeSTAR HS (Takara, Clontech, Mountain View, CA) following the manufacturer’s protocol.
First, amplification of the upstream, downstream, and URA3 fragments was performed using genomic DNA with the appropriate primers designed from sequences in the Candida Genome Database (http://candidagenome.org; Inglis et al. 2012). Each PCR product was then size-fractionated on a 0.8% agarose gel in Tris-borate EDTA buffer (89 mM Tris-borate, pH 8.3, 2 mM EDTA). The corresponding band was excised and purified with GFX MicroSpin columns (GE Healthcare, Fairfield, CT). Second, a fusion PCR reaction was performed with PrimeSTAR HS (Takara) using purified upstream, downstream, and URA3 fragments with primers complementary to the 5′ ends of the upstream and downstream fragments.
After confirming the size of the final fusion fragment, 50–100 ng of each fragment was transformed into C. glabrata strain BG14 (Ito et al. 1983). Transformants were selected on SD medium lacking uracil (Adams et al. 1997). Correct insertion of the fragment was verified by isolation of genomic DNA, followed by diagnostic PCR using an anti-sense oligonucleotide specific to URA3 (5′-ATGTCTGCCCATTCTGCTATT-3′), and a sense oligonucleotide upstream of the outermost 5′ primer was used to construct the upstream fragment.
Cloning of a C. glabrata homolog
In order to clone the MID1 (CAGL0M03597g) gene into a CEN plasmid, the gene sequence with predicted 5′ and 3′ UTR regions was obtained from Candida Genome Database (http://candidagenome.org; Inglis et al. 2012). Primers were designed to amplify the entire gene sequence upstream and downstream of the predicted 5′ and 3′ UTR, and to add a Not1 restriction site to both ends of the resulting PCR product. Genes were amplified using 200 ng of BG14 genomic DNA, and 0.1 μM of each primer with Q5 DNA Polymerase (NEB). The resulting PCR product was purified using the MiniElute Reaction Cleanup Kit (Qiagen), and then digested with Not1 HF restriction enzyme (NEB). pRS410, an ARS CEN plasmid vector (Christianson et al. 1992) (obtained from Addgene, Cambridge, MA), was modified to replace the KanMX resistance cassette with NatMX. This was done by digesting the pCR2.1-NatMX vector with Pme1 and BglII (NEB) to excise the NatMX cassette, which was subsequently ligated to the pRS410 vector digested with BglII and NruI (NEB). After successful ligation creating the pRS410-NatMX plasmid, this vector was then cut with NotI HF, and treated with calf intestinal alkaline phosphatase (NEB). Ligations were performed using T4 DNA ligase, and subsequently electroporated into electrocompetent DH10B cells. Candidate colonies were selected on LB plates containing 100 μg/ml ampicillin (Sigma-Aldrich) and 40 μg/ml X-gal (Sigma-Aldrich).
The plasmid as constructed above containing the MID1 (CAGL0M03597g) gene was transformed into the mid1::URA3 strain (Ito et al. 1983), and positive transformants were selected on YPD with 100 μg/ml nourseothricin/clonNat (Werner BioAgents, Jena, Germany). The empty pRS410-NatMX vector was used as a control.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Identification of genes that, when disrupted, cause altered sensitivity to caspofungin
We screened a collection of 27,000 Tn7 insertion mutants for their responses to caspofungin, a member of the echinocandin family of anti-fungal drugs. We empirically found that two concentrations, 100 ng/ml and 200 ng/ml, in rich medium (YPD) allowed for selection of mutants both more sensitive and more tolerant than wild type (Supplemental Material, Figure S1). The entire collection, contained in 266 96-well plates, was replica-printed onto caspofungin-containing solid medium. The potential positives were screened a second time by streak-out on solid medium containing the same concentrations of caspofungin. From the set of ∼ 250 mutants that passed these initial tests, we prepared genomic DNA and created plasmids including the Tn7 replicon. The modified Tn7 element contains an origin of replication for E. coli as well as selectable markers for E. coli and yeast. Genomic DNA derived from a Tn7 insertion strain can thus be recovered as a plasmid and propagated in E. coli. Sequencing of the plasmid from regions within the Tn7 element into the surrounding genomic DNA identifies the region interrupted by the transposon (Castano et al. 2003). The resulting plasmids were sequenced, and strains where the transposon had inserted between two genes, or where it appeared there was an unusual recombination event, were eliminated, keeping only those strains where the Tn7 replicon had inserted into the middle of an open reading frame.
Many of the strains were found to have insertions in the same gene, often at different sites. Each of these strains had the same phenotype with respect to caspofungin (i.e., either all sensitive or all tolerant). We were therefore confident that these genes represented authentic hits. However, many other genes were represented only by a single example. For these strains, we examined whether there was evidence for nonhomologous recombination by diagnostic PCR. If any strains appeared to have a wild-type copy of the gene of interest, in addition to the disrupted copy, they were eliminated from further analysis. At the end of this screening process, we identified a total of 64 genes that, when disrupted, resulted in a strong phenotype with respect to caspofungin, 48 that caused increased sensitivity, and 16 that caused tolerance (Table 1 and Table 2).
Table 1. Genes whose disruption confers tolerance to 200 ng/ml caspofungin in YPD medium.
| C gla Locus Tag | S. cerevisiae Homolog | Gene Name | Description of Gene Product (Engel et al. 2016) |
|---|---|---|---|
| Cell wall assembly | |||
| CAGL0E02629g | YOR002W | ALG6 | α-1,3 Glucosyltransferase |
| CAGL0L00693g | YIL049w | DFG10 | Polyprenol reductase |
| CAGL0G00286g | YMR307W | GAS1 | GPI-anchored β-1,3 glucanosyltransferase |
| Cytoskeleton and vesicular transport | |||
| CAGL0L11814g | YER166w | DNF1 | Aminophospholipid translocase (flippase) |
| CAGL0M08052g | YEL022w | GEA2 | Arf GEF |
| CAGL0A02629g | YHR108w | GGA2 | Regulates Arf1 and Arf2 to facilitate Golgi trafficking |
| CAGL0B04631g | YOR109W | INP53 | Polyphosphatidylinositol phosphatase |
| Cell signaling/response to stress | |||
| CAGL0K04169g | YGR040w | KSS1 | MAPK involved in filamentous growth and pheromone response |
| CAGL0J01870g | YGL167C | PMR1 | High affinity Ca2+/Mn2+ P-type ATPase/transport into Golgi |
| Transcription regulation/DNA and RNA repair and modification | |||
| CAGL0F05379g | YDR206w | EBS1 | Involved in translation inhibition and nonsense-mediated decay |
| CAGL0K11132g | YDR240c | SNU56 | Component of U1 snRNP required for mRNA splicing via spliceosome |
| Miscellaneous and unknown functions | |||
| CAGL0D05324g | YBR255W | MTC4 | Unknown function; β-1,6 glucan excretion increased in null |
| CAGL0K04147g | YGR038w | ORM1 | Unknown function/response to unfolded protein |
| CAGL0C02717g | YAL009W | SPO7 | Putative regulatory subunit of Nem1p-Spo7p phosphatase holoenzyme |
| CAGL0C04587g | YJR098c | — | Unknown function, found in highly purified mitochondria |
| CAGL0K08008g | YPR089W | — | Unknown function, interacts genetically with ERG11 and physically with Hsp82 |
Table 2. Genes whose disruption confers sensitivity to 100 ng/ml caspofungin in YPD medium.
| C gla Locus Tag | S. cerevisiae Homolog | Gene Name | Description of Gene Product (Engel et al. 2016) |
|---|---|---|---|
| PKC/cell wall integrity pathway | |||
| CAGL0I06512g | YER155c | BEM2 | RhoGAP involved in the control of cytoskeleton organization |
| CAGL0C05599g | YDL240w | LRG1 | Putative GAP appears to specifically regulate β-1,3 glucan synthesis |
| CAGL0J03828g | YPL140C | MKK1 | MAPKK involved in control of cell integrity |
| CAGL0G09559g | YOR188w | MSB1 | Unknown function; may be involved in positive regulation of β-1,3 glucan synthesis and the Pkc1-MAPK pathway |
| CAGL0G01320g | YNL053W | MSG5 | Dual-specificity protein phosphatase/ regulates Slt2 |
| CAGL0E04620g | YDR055w | PST1 | Cell wall protein that contains a putative GPI-attachment site |
| CAGL0J00539g | YHR030c | SLT2 | Serine/threonine MAP kinase; regulates cell wall integrity |
| CAGL0A04565g | YER111c | SWI4 | DNA binding component of the SBF complex |
| Cytoskeleton and vesicular transport | |||
| CAGL0J04312g | YBL017C | PEP1 | Type I transmembrane sorting receptor for multiple vacuolar hydrolases |
| CAGL0E05302g | YOR329c | SCD5 | Protein required for normal actin organization and endocytosis |
| CAGL0G05786g | YDL212w | SHR3 | Endoplasmic reticulum packaging chaperone |
| CAGL0K05291g | YPR032W | SRO7 | Effector of Rab GTP-binding protein Sec4 |
| High affinity Ca2+ influx (HACS) | |||
| CAGL0B02211g | YGR217W | CCH1 | Voltage-gated high-affinity Ca2+ channel |
| CAGL0M00748g | YLR443W | ECM7 | Role in Ca2+ uptake |
| CAGL0M03597g | YNL291C | MID1 | Ca2+-permeable cation channel required for Ca2+ influx |
| Mannan synthesis | |||
| CAGL0K11231g | YDR245w | MNN10 | Subunit of a Golgi mannosyltransferase complex |
| CAGL0M02871g | YJL186w | MNN5 | α-1,2-mannosyltransferase |
| CAGL0J08734g | YAL023c | PMT2 | Protein O-mannosyltransferase |
| CAGL0B02321g | YML115C | VAN1 | Component of the mannan polymerase I |
| Cell signaling/response to stress | |||
| CAGL0L11110g | YLR433c | CNA1 | Calcineurin A; regulates Crz1 |
| CAGL0K01507g | YDL035c | GPR1 | Plasma membrane GPCR; nutritional state sensor |
| Vacuole | |||
| CAGL0F06347g | YMR054W | STV1 | Subunit a of the vacuolar-ATPase V0 domain |
| Transcriptional/translational regulation | |||
| CAGL0M06831g | YNL027w | CRZ1 | TF, activates transcription of stress response genes |
| CAGL0J10120g | YNL068c | FKH2 | Forkhead family transcription factor |
| CAGL0M13431g | YHR187w | IKI1 | Subunit of hexameric RecA-like ATPase Elp456 elongator subcomplex |
| CAGL0C04477g | YDL005c | MED2 | Subunit of the RNA polymerase II mediator complex |
| CAGL0M07029g | YCR077C | PAT1 | Deadenylation-dependent mRNA-decapping factor |
| CAGL0J03322g | YER082C | UTP7 | Nucleolar protein/processing of pre-18S rRNA |
| CAGL0H04367g | YML076C | WAR1 | Homodimeric Zn2Cys6 zinc finger transcription factor |
| DNA replication/repair/cell cycle regulation | |||
| CAGL0D06028g | YJR053w | BFA1 | Component of the GTPase-activating Bfa1-Bub2 complex |
| CAGL0I02222g | YHR164C | DNA2 | Tripartite DNA replication factor |
| CAGL0K09438g | YOR144c | ELG1 | Subunit of a complex important for DNA replication/genome integrity |
| CAGL0A03300g | YGL192w | IME4 | mRNA N6-adenosine methyltransferase required for entry into meiosis |
| CAGL0D05786g | YLL002w | RTT109 | Histone acetyltransferase |
| CAGL0A03432g | YLR032w | RAD5 | DNA helicase |
| CAGL0E02475g | YOL004W | SIN3 | Component of the Sin3-Rpd3 histone deacetylase complex |
| Proteosome | |||
| CAGL0G09493g | YOR191w | ULS1 | Protein involved in proteolytic control of sumoylated substrates |
| Miscellaneous and unknown functions | |||
| CAGL0E03355g | YLL015W | BPT1 | ABC type transmembrane transporter of MRP/CFTR family |
| CAGL0J08910g | YLR213c | CRR1 | Putative glycoside hydrolase of the spore wall envelope |
| CAGL0M05511g | YBR207w | FTH1 | Putative high affinity iron transporter |
| CAGL0E03311g | YGR163w | GTR2 | Putative GTP binding protein that negatively regulates Ran/Tc4 GTPase cycle |
| CAGL0I07447g | YOL103w | ITR2 | Myo-inositol transporter |
| CAGL0J07436g | YNL231c | PDR16 | PITP controlled by the multiple drug resistance regulator Pdr1 |
| CAGL0H04213g | YML081W | TDA1 | Protein kinase with unknown role; localizes to cytosol and nucleus |
| CAGL0J09966g | YNL064c | YDJ1 | Type I HSP40 cochaperone |
| CAGL0M08250g | YKL175w | ZRT3 | Vacuolar membrane zinc transporter |
| CAGL0L05060g | YKL075c | — | Unknown function; localizes to cytosol |
| CAGL0K03377g | YMR102C | — | Unknown function; paralog of Dgr2 |
Few genes in the C. glabrata genome have been annotated beyond pointing to a highly similar gene in the S. cerevisiae genome. The corresponding S. cerevisiae gene names were submitted for GO term analysis using the BinGO package (Shannon et al. 2003; Maere et al. 2005). Table S1A shows the biological processes in which the genes are involved: transport (including vesicle transport), signal transduction, response to chemical stimulus, and importantly, cell wall organization. When the gene names were submitted to the FunSpec ontology tool (Robinson et al. 2002), the resulting output also suggested many of the genes encode modulators of ion homeostasis in cells, especially Ca2+ homeostasis (Table S1B).
Phenotypes of selected Tn7 mutants
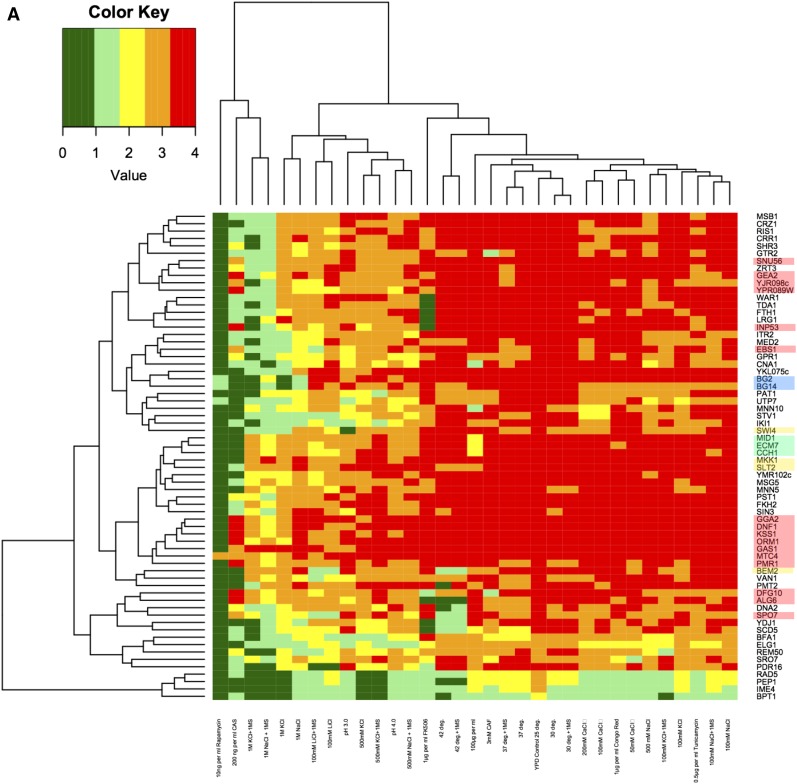
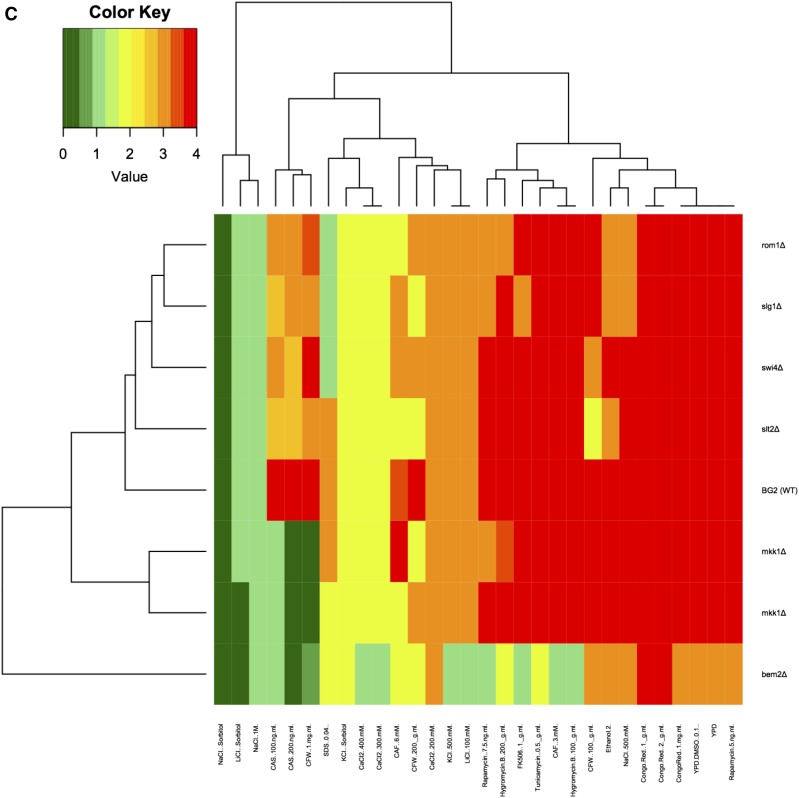
Tn7-disruption mutants in each of the 64 genes identified in the screen were further characterized. The phenotypes tested by replica-printing onto solid medium included sensitivity to a variety of cell wall disrupting agents in addition to caspofungin, including Congo red and Calcofluor white; a number of other drugs including tunicamycin (a glycosylation inhibitor), rapamycin and caffeine (TOR inhibitors), and FK506 (a calcineurin inhibitor); salts including KCl, NaCl, LiCl, and CaCl2; different pH treatments; and different temperatures (summarized in Figure 2A); for complete data set, see Table S3.
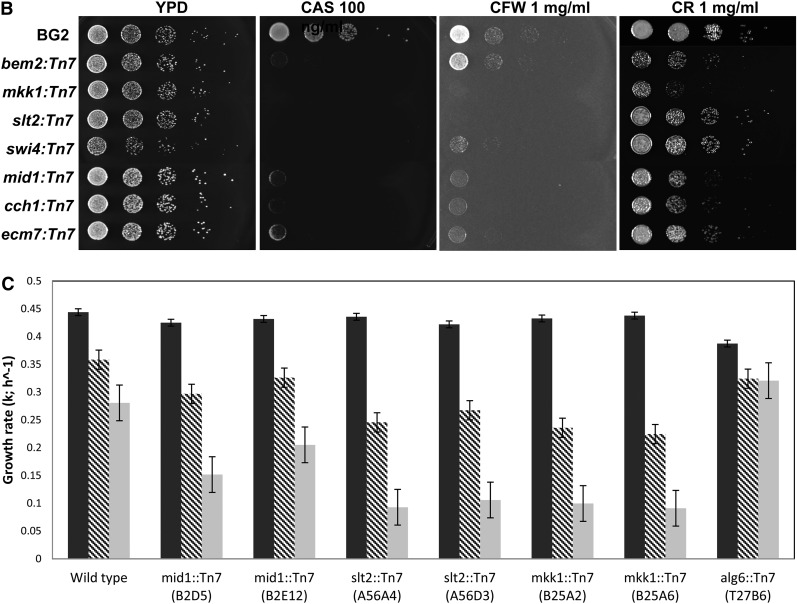
Figure 2.
Phenotypes of the 64 Tn7 disruption mutants. (A) Sixty-four strains, each with one of the genes of interest disrupted by Tn7, were grown, diluted, and then replica-printed onto YPD medium, without or with the addition of a variety of drugs or salts as described in Materials and Methods. All plates were incubated for 2 d, then were photographed and scored. All plates were incubated at 25°, except for plates incubated at 30°, 37°, or 42°, with or without the addition of 1 M sorbitol. Additions included: 100 mM NaCl, with and without 1 M sorbitol, 500 mM NaCl, with and without 1 M sorbitol, 1 M NaCl, with and without 1 M sorbitol, 100 mM LiCl, with and without 1 M sorbitol, 100 mM KCl, with and without 1 M sorbitol, 500 mM KCl, with and without 1 M sorbitol, 1 M KCl, with and without 1 M sorbitol, 50 mM CaCl2, 100 mM CaCl2, 200 mM CaCl2, 1 μg/ml Congo red, 10 ng/ml rapamycin, 0.5 μg/ml tunicamycin, YPD at pH 3.0, YPD at pH 4.0, 100 μg/ml calcofluor white, 3 mM caffeine, 200 ng/ml caspofungin, or 1 μg/ml FK506. Strains were compared to the parental wild-type strains BG2 and BG14 (BG2 ura3). The raw data (in Table S3) were summarized as a heat map. Strains that grew well under a given condition were scored as 4+ and assigned the color red as shown on the schematic; strains that grew poorly were scored as 0 and assigned the color green. The names of disrupted C. glabrata orthologs of S. cerevisiae genes in each strain are shown at the left; the growth conditions that cluster together are shown at the top. The two wild-type strains are highlighted in blue, the genes that, when disrupted, cause caspofungin resistance are highlighted in red (all others are caspofungin sensitive). Members of the HACS complex are highlighted in green, and members of the CWI pathway are highlighted in yellow. Phenotypes of each strain with respect to the different growth conditions were verified at least twice. This figure represents one set of growth experiments. (B) Growth of mutants on solid medium with and without cell wall disrupting agents. Tn7 disruption mutants for putative C. glabrata CWI pathway and HACS complex genes, bem2:Tn7 (A96B09), mkk1:Tn7 (B25A2), slt2:Tn7 (A56A4), swi4Δ:Tn7(A4F9), mid1:Tn7 (B2D5), cch1:Tn7 (T77F2), and ecm7:Tn7 (T66C3) were spotted as 10-fold dilutions on YPD plates containing 100 ng/mL caspofungin (CAS), 1 mg/ml calcofluor white (CFW), 1 mg/ml Congo red (CR). The parental strain BG2 served as a comparison for growth. (C) Effects of caspofungin on growth of selected Tn7 disruption mutants in liquid culture. Log-phase cells were diluted as described in Materials and Methods in YPD alone (black bars) or with caspofungin [4 ng/ml (hatched bars) or 8 ng/ml (gray bars)]. OD600 was measured at 15-min intervals for several hours to measure growth. Growth rate, κ was calculated as described. Cg MID1 encodes a putative member of the HACS complex, while Cg MKK1 and Cg SLT2 encode putative kinases of the CWI pathway. Cg ALG6 encodes a putative α-1,3 glucosyltransferase. Disruptions of Cg MID1, Cg SLT2, and Cg MKK1 all result in sensitivity to caspofungin, while disruption of Cg ALG6 is unaffected by the presence of the drug.
Most of the mutant strains, like the wild-type parental strains, were resistant to both cell wall perturbing dyes, Calcofluor white and Congo red (Figure 2A and Table S3). A few mutant strains exhibited sensitivity to caffeine, while nearly all mutants exhibited sensitivity to rapamycin, except for the strain with an insertion in Cg MTC4 (CAGL0D05324g). Most mutants were sensitive to tunicamycin, and, finally, the majority of mutants were resistant to the concentration of FK506 tested (Table S3).
Since FunSpec analysis revealed that genes involved in ion homeostasis and calcium transport were represented in our gene set, we explored the ion tolerance of mutants by growth on media containing NaCl, KCl, LiCl, and CaCl2. Sorbitol was added as an osmotic stabilizer to examine whether growth of mutants exhibiting sensitive phenotypes could be rescued; however, in most cases, sorbitol exacerbated sensitivity. Most of the mutants exhibited unimpeded growth in the presence of NaCl and KCl at levels of 100 mM and 500 mM. However, in the presence of 1 M NaCl or KCl, a greater diversity in growth effects was observed. A few mutants exhibited moderate to high sensitivity to most ionic conditions while another group of mutants showed a high degree of resistance to most conditions except the presence of caspofungin (Table S3), including the orthologs of the HACS complex genes and the orthologs of some genes encoding members of the CWI pathway.
The results of the 33 different phenotype tests were subjected to clustering analysis (Figure 2A), which revealed several broad groupings. First, the wild type strains (BG2 and its ura3 derivative, BG14 the parent of the Tn7 collection, highlighted in blue on the phylogram) clustered together, demonstrating the robustness of this approach. The smallest group contained mutant strains that were sensitive to most conditions, except for growth at 25° in rich medium (at the bottom of Figure 2A); these four mutants generally grew poorly under most stresses, and were not examined further. The caspofungin-resistant mutants clustered together for the most part (highlighted in red, Figure 2A) while caspofungin-sensitive mutants were dispersed among the clusters.
Disruption of CWI pathway and HACS genes: further characterization of the caspofungin phenotype
Mutant strains with disruptions of genes encoding orthologs of kinases in the cell wall integrity (CWI) pathway (Figure 1), Cg SLT2 (CAGL0J00539g) and Cg MKK1 (CAGL0H05621g), both sensitive to caspofungin, clustered in our analysis (highlighted in yellow, Figure 2A). In addition, we identified two additional genes homologous to S. cerevisiae CWI pathway orthologs, Cg BEM2 (CAGL0I06512g; encoding RhoGAP) and Cg SWI4 (CAGL0A04565g ; a transcription factor) (also highlighted in yellow, Figure 2A). Finally, we observed that strains with disruptions in one of the three genes encoding orthologs of voltage-gated high-affinity Ca2+ channel genes grouped together (Martin et al. 2011) [Cg CCH1 (CAGL0B02211g), Cg ECM7 (CAGL0M00748g), and Cg MID1 (CAGL0M03597g); highlighted in green, Figure 2A].
We next examined the presumptive CWI [Cg bem2:Tn7 (CAGL0I06512g), Cg mkk1:Tn7 (CAGL0J03828g), Cg slt2:Tn7 (CAGL0J00539g), and Cg swi4:Tn7 (CAGL0A04565g)] and HACS [Cg mid1:Tn7 (CAGL0M03597g), Cg cch1:Tn7 (CAGL0B02211g), and Cg ecm7:Tn7 (CAGL0M00748g)] mutant strains in more detail first, by growth on solid medium (Figure 2B) and then by growth in liquid culture (Figure 2C). On solid medium, all of the Tn7 insertion mutants tested grew reasonably well in the absence of drugs [although the Cg swi4:Tn7 (CAGL0A04565g) strain was noticeably slower]. All the mutants were sensitive to caspofungin and Calcofluor white, although on Calcofluor white, it was possible to distinguish a greater variety of phenotypes. Finally, several of these strains were quite sensitive to Congo red, including the Cg bem2:Tn7 (CAGL0I06512g) and Cg mkk1:Tn7 (CAGL0J03828g) strains. In contrast, the Cg slt2:Tn7 (CAGL0J00539g) and Cg swi4:Tn7 (CAGL0A04565g) strains grew as well as the wild-type strain in the presence of Congo red; the remaining strains had intermediate phenotypes (Figure 2B).
We then examined the effect of caspofungin on growth of two independent Cg SLT2 (CAGL0J00539g) and Cg MKK1 (CAGL0J03828g) mutants in liquid medium. As can be seen in Figure 2C, while wild type was little affected by the lower concentration of caspofungin, the growth of all four mutant strains was adversely affected. We also examined two independent strains with insertions into Cg MID1 (CAGL0M03597g), encoding a subunit of the HACS complex, and found these were also more sensitive to caspofungin than wild type. In contrast, a strain with an insertion in Cg ALG6 (CAGL0E02629g), putatively encoding α-1,6 glucosyltransferase, obtained from the screen as a caspofungin-resistant mutant, was unaffected by the presence of caspofungin in this assay.
Phenotypes of C. glabrata deletion mutants
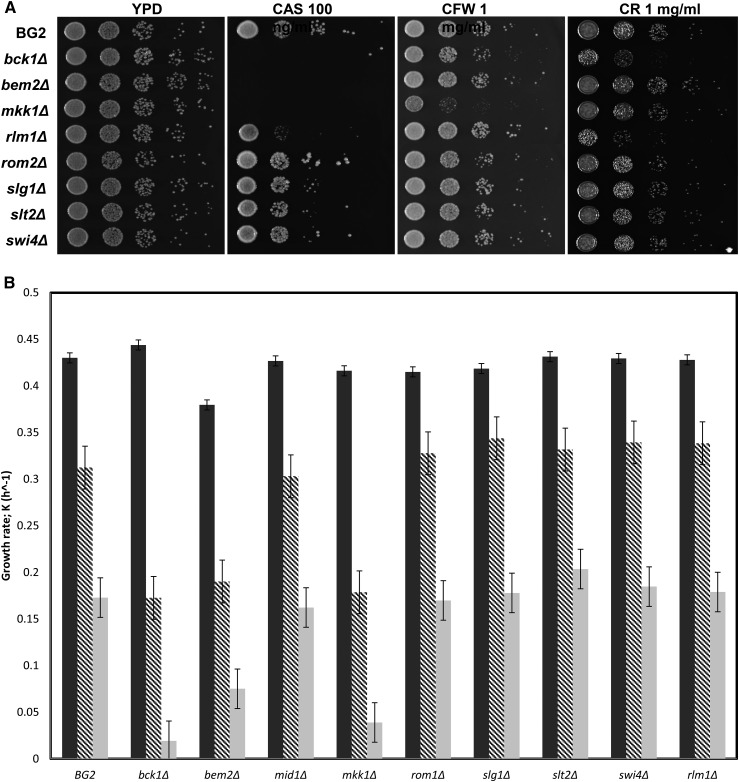
To further explore the CWI pathway in C. glabrata, we constructed deletions of each of the four putative CWI genes obtained in the screen [i.e., Cg BEM2 (CAGL0I06512g), Cg MKK1 (CAGL0J03828g), Cg SLT2 (CAGL0J00539g), and Cg SWI4 (CAGL0A04565g)], and four other presumptive CWI pathway genes, Cg BCK1 (CAGL0L03520g , a third Ser/Thr kinase), Cg RLM1 (CAGL0H05621g , a transcription factor), Cg ROM2 (CAGL0G04873g , Rho GEF), and Cg SLG1 (CAGL0F01507g , a plasma membrane sensor) (Levin 2011). These genes represent all stages of the pathway, from cell-surface sensing to regulation of transcription (Figure 1). We also constructed deletions of Cg MID1 (CAGL0M03597g), Cg CCH1 (CAGL0B02211g), and Cg ECM7 (CAGL0M00748g)—all members of HACS complex (for the complete set of deletion strains constructed see Table S2).
We first tested each of the CWI knockout mutants on solid medium containing caspofungin, as well as in the presence of Calcofluor white and Congo red (Figure 3A). The mutants showed a range of sensitivities but fell more or less into two groups based on the caspofungin phenotype, with the Cg bck1Δ (CAGL0L03520g), Cg bem2Δ (CAGL0I06512g), and Cg mkk1Δ (CAGL0J03828g) mutants showing increased sensitivity relative to the Cg rlm1Δ (CAGL0H05621g), Cg rom2Δ (CAGL0G04873g), Cg slg1Δ (CAGL0F01507g), Cg slt2Δ (CAGL0J00539g), and Cg swi4Δ (CAGL0A04565g) mutant strains. The Cg bck1Δ (CAGL0L03520g), Cg bem2Δ (CAGL0I06512g), and Cg mkk1Δ (CAGL0J03828g) mutants were also slightly more sensitive to Calcofluor white than the others, while the Cg bck1Δ (CAGL0L03520g), and Cg rlm1Δ (CAGL0H05621g), mutant strains were sensitive to Congo red. Under liquid culture conditions (Figure 3B), the Cg bck1Δ (CAGL0L03520g), Cg bem2Δ (CAGL0I06512g), and Cg mkk1Δ (CAGL0J03828g) mutants were quite sensitive to caspofungin, while the other mutants appeared to have growth characteristics similar to wild type. In general however, the knockout mutant strains exhibited milder phenotypes than the Tn7 insertion mutants, suggesting that there may be cis effects stemming from the transposon into neighboring genomic DNA.
Figure 3.
The CWI knockout mutants fall into two distinct groups based on growth in caspofungin. (A) CWI knockouts have altered sensitivity to caspofungin on plates. Complete knockout strains for CWI pathway genes, bck1Δ, bem2Δ, mkk1Δ, rlm1Δ, rom2Δ, slg1Δ, slt2Δ, and swi6Δ were spotted as 10-fold dilutions on YPD plates containing 100 ng/mL caspofungin (CAS), 1 mg/ml calcofluor white (CFW), 1 mg/ml Congo red (CR). The parental strain BG2 served as a comparison for growth. (B) Knockout strains have altered sensitivity to caspofungin in liquid medium. Strains with knockout of genes in the CWI pathway were grown with and without caspofungin as described in the legend to Figure 2C. (C) Phenotypes of the CWI pathway knockout mutants The nine knockout mutants constructed were examined on a variety of different media similar to those used to examine the phenotypes of the 64 Tn7 disruption mutants. These included: YPD, with or without 0.1% DMSO or 2% ethanol, 500 mM KCl, with and without 1 M sorbitol, 500 mM NaCl, with and without 1 M sorbitol, 1 M NaCl, 100 mM LiCl, with and without 1 M sorbitol, CaCl2 (200, 300, and 400 mM), calcofluor white (100 and 200 μg/ml, 1 mg/ml), caffeine (3 and 6 mM), Congo red (1 and 2 μg/ml, 1 mg/ml), rapamycin (5, 7.5, and 10 ng/ml, delivered in 0.1% DMSO), tunicamycin (0.5 μg/ml), FK506 (1 μg/ml), caspofungin (100 and 200 ng/ml), SDS (0.04%), and hygromycin B (100 and 200 μg/ml). The bem2Δ mutant was very sensitive to many of these conditions. Raw data are in Table S4.
We examined the eight CWI knockout mutants under a variety of other conditions, similar to the tests used for the Tn7 disruption mutants isolated from the screen (Figure 3C summarizes these experiments; the complete data set is found in Table S4). As on plates and in liquid culture in the presence of caspofungin, the mutants fell into the same two groups based on these experiments. The Cg rom2Δ (CAGL0G04873g), Cg rlm1Δ (CAGL0H05621g), Cg slg1Δ (CAGL0F01507g), Cg slt2Δ (CAGL0J00539g), and Cg swi4Δ (CAGL0A04565g) mutants were mildly sensitive to many of the treatments, while the Cg bem2Δ (CAGL0I06512g), Cg bck1Δ (CAGL0L03520g), and Cg mkk1Δ (CAGL0J03828g) mutants were more sensitive. The Cg bem2Δ (CAGL0I06512g) mutant in particular was very sensitive to most treatments, including caspofungin, a variety of salts, and caffeine.
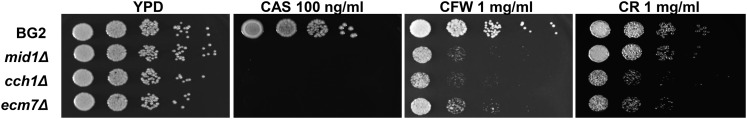
We also characterized strains with knockouts of presumptive HACS complex genes, Cg cch1Δ (CAGL0B02211g), Cg ecm7Δ (CAGL0M00748g), and Cg mid1Δ (CAGL0M03597g) (Figure 4). As with the Tn7 disruption mutants, the knockout strains were sensitive to caspofungin. In addition, the three mutant strains showed differential sensitivities to Calcofluor white [Cg cch1Δ (CAGL0B02211g) ανδ Cg mid1Δ (CAGL0M03597g) were slightly more sensitive than Cg ecm7Δ [CAGL0M00748g)] and Congo red [Cg cch1Δ (CAGL0B02211g) and Cg ecm7Δ (CAGL0M00748g) were more sensitive than Cg mid1Δ (CAGL0M03597g)].
Figure 4.
HACS complex knockouts display sensitivity to caspofungin, calcofluor white, and Congo red. Strains containing complete deletions of the HACS complex genes, mid1Δ, cch1Δ, and ecm7Δ were spotted as 10-fold dilutions on YPD plates containing 100 ng/mL caspofungin (CAS), 1 mg/ml calcofluor white (CFW), 1 mg/ml Congo red (CR). The parental strain BG2 served as a comparison for growth.
To confirm that the gene we deleted was responsible for the phenotype, as proof-of-principle we cloned Cg MID1 (CAGL0M03597g) into a low-copy S. cerevisiae vector, since such vectors function in C. glabrata (Zhou et al. 1994). The plasmid was transformed into the appropriate Cg mid1Δ (CAGL0M03597g) strain and phenotypes were examined. As shown in Figure S2, the cloned MID1 (CAGL0M03597g) gene complemented the phenotype, thus assuring the phenotypes observed were due to the deletion of the gene of interest.
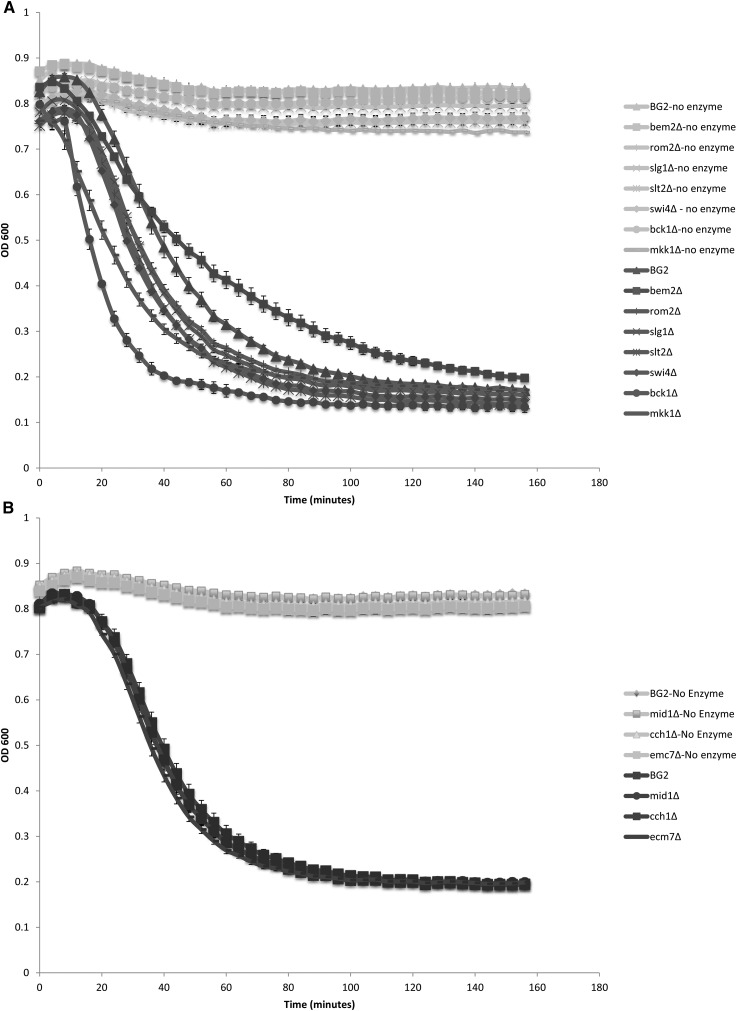
Enzymatic cell wall digestion occurs more rapidly in CWI mutants
The cell wall normally protects the cell from osmotic pressure, but defects in, or damage to, the cell wall can lead to lysis. Cell wall integrity can be challenged by zymolyase, a cocktail of 1,3-β-glucanase and protease activities used to degrade yeast cell walls (Ovalle et al. 1998, 1999). Enzymatic digestion of cell walls in the absence of an osmotic stabilizer such as sorbitol results in lysis due to increased internal turgor pressure (Ovalle et al. 1998, 1999). We compared the sensitivity of the deletion mutant strains to zymolyase (Figure 5). Strains with deletions of CWI orthologs had a greater sensitivity to zymolyase, particularly the bck1Δ (CAGL0L03520g) strain (Figure 5A). Interestingly, the bem2Δ (CAGL0I06512g) strain displayed lower sensitivity (Figure 5A), despite the fact that the bem2Δ (CAGL0I06512g) strain was very sensitive to caspofungin, and other cell-well disrupting agents (Figure 3C). In contrast, strains containing deletions of HACS complex orthologs demonstrated the same level of sensitivity as the wild-type BG2 strain (Figure 5B), suggesting that the changes that result in caspofungin sensitivity in these strains is not the result of changes in glucan content.
Figure 5.
Cell wall mutants display sensitivity to zymolyase. Deletion mutants were tested for their sensitivity to cell wall digestion by treatment with zymolyase as described in Materials and Methods. (A) CWI pathway deletion mutants. (B) HACS deletion mutants
Discussion
The work described here represents a global approach to identifying genes important for cellular responses to caspofungin in C. glabrata. Our results, taken all together, demonstrate that many different genes impact cell wall structure and function in C. glabrata, including members of the CWI pathway and the HACS complex.
Similar screens have been performed with caspofungin in S. cerevisiae (Lesage et al. 2004; Markovich et al. 2004), using the yeast deletion collection (Winzeler et al. 1999), and with micafungin, another member of the echinocandin family in Schizosaccharomyces pombe (Zhou et al. 2013), using the S. pombe deletion collection (Kim et al. 2010). In addition, microarray analysis identified genes in S. cerevisiae that are up or downregulated in response to caspofungin (Reinoso-Martin et al. 2003). Most recently, a large collection of deletion strains (representing 600+ genes) was screened for caspofungin sensitivity in C. glabrata (Schwarzmuller et al. 2014). However, in comparing the lists of genes identified in each instance, numbers of different genes were identified, even between the two screens performed in S. cerevisiae (Lesage et al. 2004; Markovich et al. 2004). Nevertheless, in instances where the same gene was found in different screens, mutant strains were observed to have the same general phenotype. For example, mutation of MNN10, which encodes a subunit of the Golgi apparatus mannosyltransferase complex, was found to result in sensitivity to caspofungin in S. cerevisiae (Lesage et al. 2004; Markovich et al. 2004), as well as in our study, and the deletion collection screen in C. glabrata (Schwarzmuller et al. 2014). The sole exception to this trend was mutation of CRZ1, a transcription factor that is regulated by calcineurin. We found that disruption with Tn7 results in sensitivity to caspofungin in C. glabrata, a result supported by the work of Miyazaki et al. (2010b) in S. cerevisiae, whereas Lesage et al. (2004) found that deletion of CRZ1 in S. cerevisiae results in resistance.
Despite the fact that incompletely overlapping sets of genes were identified in the various screens, in each of these reports, genes controlling the CWI pathway were identified. In our case, these genes included the C. glabrata orthologs of BEM2 (CAGL0I06512g), MKK1 (CAGL0J03828g), SLT2 (CAGL0J03828g), and SWI4 (CAGL0A04565g). Similarly, in the screen of the 600+ members of the C. glabrata deletion collection, the C. glabrata orthologs of MKK1 (CAGL0J03828g), BCK1 (CAGL0L03520g), SLT2 (CAGL0J00539g), and SLG1/WSC1 (CAGL0F01507g) were identified (Schwarzmuller et al. 2014). Further, we here demonstrated that strains with deletions of BCK1 (CAGL0L03520g), RLM1 (CAGL0H05621g), ROM2 (CAGL0G04873g), or SLG1 (CAGL0F01507g) were also sensitive to caspofungin, although to varying degrees.
Other groups previously investigated individual genes involved in the CWI pathway in C. glabrata. First, it was demonstrated that deletion of Cg SLT2 (CAGL0J00539g), encoding a serine/threonine kinase, results in increased sensitivity to cell wall stress, including the presence of caspofungin (Miyazaki et al. 2010a), micafungin (Miyazaki et al. 2010a; Nagayoshi et al. 2014), Calcofluor white (Edlind et al. 2002), and fluconazole (Borah et al. 2011), while overexpression of Cg SLT2 (CAGL0J00539g) resulted in tolerance—findings that correlated with decreased or increased ability to survive in a mouse host, respectively (Edlind et al. 2002; Cota et al. 2008). In addition, the overexpression of Cg SLT2 (CAGL0J00539g) leads to increases in chitin content, correlating with poor killing by caspofungin (Cota et al. 2008). Similarly, deletion of Cg SWI4 (CAGL0A04565g), Cg SWI6 (CAGL0B01144g), and Cg RLM1 (CAGL0H05621g), transcription factors that are activated as a result of the CWI pathway, results in sensitivity to micafungin, another member of the echinocandin family, and deletion of Cg SWI4 (CAGL0A04565g) results in sensitivity to caspofungin (Nagayoshi et al. 2014). Thus it seems clear that mutations in CWI pathway genes enhance the efficacy of caspofungin. Such results suggest that chemical modulators of the CWI pathway may increase the efficacy of caspofungin as well. Indeed, it has been reported that S. cerevisiae strains with deletions of CWI pathway genes Cg BCK1 (CAGL0L03520g) and Cg SLT2 (CAGL0J00539g) are sensitive to the anti-malarial drug chloroquine, and that chloroquine and caspofungin show synergy in S. cerevisiae, C. albicans, and A. fumigatus, as well as C. glabrata (Islahudin et al. 2013).
Regulation of chitin content also appears to be a means of regulating responses to echinocandins. Increased abundance of chitin correlates with echinocandin resistance in C. albicans (Walker et al. 2013). In contrast, deletion of Cg CHS3B (CAGL0I04840g)—a paralog of chitin synthase 3—results in sensitivity to several different echinocandins, including micafungin and nikkomycin Z, as well as caspofungin (Ueno et al. 2011).
Inhibition of Ca2+ influx via the HACS complex may also increase the efficacy of caspofungin. As shown in Figure 2, we obtained Tn7 disruption mutants in each of the three HACS complex members, Cg CCH1 (CAGL0B02211g), Cg ECM7 (CAGL0M00748g), and Cg MID1 (CAGL0M03597g)—results we verified by creating deletion mutants (Figure 4). Others have shown that ECM7 is important in C. albicans (orf19.5643) for responses to oxidative stress and Ca2+ homeostasis (Ding et al. 2013). Previous work in S. cerevisiae demonstrated that FKS2 is regulated not only via the CWI pathway, but also as a result of Ca2+ flux, mediated by calmodulin, calcineurin, and Crz1 (Zhao et al. 1998; Levin 2011). Cross-talk between this pathway and the CWI pathway might occur at some level; for example, it is tempting to speculate that protein kinase C, Pkc1 might also bind and be activated by Ca2+, since it has a C2-like domain (Rizo and Sudhof 1998), but purified S. cerevisiae Pkc1 tested in vitro is unaffected by Ca2+ (Antonsson et al. 1994; Watanabe et al. 1994).
There are two other proteins, Kch1 and Kch2 , which act as low-affinity transporters of K+ (Stefan et al. 2013), that are known to be important for the activity of the HACS complex in S. cerevisiae. The activity of Kch1 and Kch2 may set up a voltage differential across the plasma membrane, thus activating the HACS complex. Kch1 is conserved in C. albicans (orf19.6563) (Stefan and Cunningham 2013), and in C. glabrata (CAGL0D06050g) (Inglis et al. 2012). However, we did not obtain the C. glabrata ortholog in our screen. Similarly, strains with deletions in glycerol channels, Cg FPS1 (CAGL0C03267g) or Cg FPS2 (CAGL0E03894g) (Beese-Sims et al. 2012) are sensitive to caspofungin, yet we did not isolate these in our screen. Thus, although the Tn7 insertion collection is extensive, not every nonessential gene in the genome may be represented. This likely explains why we did not isolate all of the nonessential CWI pathway genes in the screen or any of the chitin synthase genes, for example. Nevertheless, we have identified a number of different pathways and complexes that, when disrupted, lead to altered sensitivity to caspofungin. We plan to further investigate the hits obtained from this study, particularly the genes involved in mannan synthesis, including ALG6 (CAGL0E02629g), DFG10 (CAGL0L00693g), MNN5 (CAGL0M02871g), MNN10 (CAGL0K11231g), PMT2 (CAGL0J08734g), and VAN1 (CAGL0B02321g) (Table 1 and Table 2).
We also identified a number of genes that, when disrupted, result in increased tolerance to caspofungin compared to wild type. None of these genes are currently associated with clinical resistance; rather resistance in the clinic is associated primarily with mutations in FKS1 and FKS2 (Arendrup et al. 2012; Duran-Valle et al. 2012; Singh-Babak et al. 2012; Alexander et al. 2013; Borghi et al. 2014; Pham et al. 2014). Nevertheless, it will be important to know what other mutations can cause resistance to echinocandins, given that these drugs are among the few available to treat many C. glabrata infections.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Brendan Cormack and members of his laboratory for the Tn7 collection and advice. Emily Fink, Natalie Mevedev, and Brad Hilson helped with screening the C. glabrata Tn7 collection. Taylor Craig made several of the cell wall integrity pathway knockout mutants. Matthew Linz helped with the Zymolyase assays. Many of the results shown here were verified by members of the Georgetown University Biology 151 (Genetics) classes of Fall 2013 and Fall 2014. Finally, we thank the members of Rolfes and Rosenwald laboratories, as well as Dr. Richard Calderone and Dr. William Fonzi (Georgetown University Medical Center) for their review of this work. We also thank the Georgetown University Graduate School of Arts and Sciences for a pilot grant that supported this work.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.032490/-/DC1
Communicating editor: J. H. McCusker
Literature Cited
- Adams A., Gottschling D. E., Kaiser C. A., Stearns T., 1997. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Adolph K. W. (Editor), 1996. Microbial Genome Methods, CRC Press, Boca Raton, FL. [Google Scholar]
- Alexander B. D., Johnson M. D., Pfeiffer C. D., Jimenez-Ortigosa C., Catania J., et al. , 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Nephrol. Dial. Transplant. 56: 1724–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Antonsson B., Montessuit S., Friedli L., Payton M. A., Paravicini G., 1994. Protein kinase C in yeast. Characteristics of the Saccharomyces cerevisiae PKC1 gene product. J. Biol. Chem. 269: 16821–16828. [PubMed] [Google Scholar]
- Arendrup M. C., Perlin D. S., Jensen R. H., Howard S. J., Goodwin J., et al. , 2012. Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FKS resistance mutations. Antimicrob. Agents Chemother. 56: 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese-Sims S. E., Pan S. J., Lee J., Hwang-Wong E., Cormack B. P., et al. , 2012. Mutants in the Candida glabrata glycerol channels are sensitized to cell wall stress. Eukaryot. Cell 11: 1512–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bertani G., 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S., Shivarathri R., Kaur R., 2011. The Rho1 GTPase-activating protein CgBem2 is required for survival of azole stress in Candida glabrata. J. Biol. Chem. 286: 34311–34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi E., Andreoni S., Cirasola D., Ricucci V., Sciota R., et al. , 2014. Antifungal resistance does not necessarily affect Candida glabrata fitness. J. Chemother. 26: 32–36. [DOI] [PubMed] [Google Scholar]
- Castano I., Kaur R., Pan S., Cregg R., De Las Penas A., et al. , 2003. Tn7-based genome-wide random insertional mutagenesis of Candida glabrata. Genome Res. 13: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Falkow S., 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota J. M., Grabinski J. L., Talbert R. L., Burgess D. S., Rogers P. D., et al. , 2008. Increases in SLT2 expression and chitin content are associated with incomplete killing of Candida glabrata by caspofungin. Antimicrob. Agents Chemother. 52: 1144–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot P. W., Kraneveld E. A., Yin Q. Y., Dekker H. L., Gross U., et al. , 2008. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 7: 1951–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente N., Portillo F., 2000. The cell wall integrity/remodeling MAPK cascade is involved in glucose activation of the yeast plasma membrane H(+)-ATPase. Biochim. Biophys. Acta 1509: 189–194. [DOI] [PubMed] [Google Scholar]
- Dichtl K., Samantaray S., Wagener J., 2016. Cell wall integrity signaling in human pathogenic fungi. Cell. Microbiol. DOI:10.1111/cmi.12612. [DOI] [PubMed] [Google Scholar]
- Ding X., Yu Q., Xu N., Wang Y., Cheng X., et al. , 2013. Ecm7, a regulator of HACS, functions in calcium homeostasis maintenance, oxidative stress response and hyphal development in Candida albicans. Fungal Genet. Biol. 57: 23–32. [DOI] [PubMed] [Google Scholar]
- Duran-Valle M. T., Gago S., Gomez-Lopez A., Cuenca-Estrella M., Jimenez Diez-Canseco L., et al. , 2012. Recurrent episodes of candidemia due to Candida glabrata with a mutation in hot spot 1 of the FKS2 gene developed after prolonged therapy with caspofungin. Antimicrob. Agents Chemother. 56: 3417–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlind T., Smith L., Henry K., Katiyar S., Nickels J., 2002. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol. Micro. 46: 257–268. [DOI] [PubMed] [Google Scholar]
- Engel S. R., Weng S., Binkley G., Paskov K., Song G., et al. , 2016. From one to many: expanding the Saccharomyces cerevisiae reference genome panel. Database (2016) 2016: article ID baw020; DOI:10.1093/database/baw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B., Bouchier C., Fairhead C., Craig N. L., Cormack B. P., 2012. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557–580. [DOI] [PubMed] [Google Scholar]
- Hartigan, J. A., 1975 Clustering algorithms, Wiley, New York. [Google Scholar]
- Inglis D. O., Arnaud M. B., Binkley J., Shah P., Skrzypek M. S., et al. , 2012. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 40: D667–D674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islahudin F., Khozoie C., Bates S., Ting K. N., Pleass R. J., et al. , 2013. Cell wall perturbation sensitizes fungi to the antimalarial drug chloroquine. Antimicrob. Agents Chemother. 57: 3889–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A., 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung U. S., Sobering A. K., Romeo M. J., Levin D. E., 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46: 781–789. [DOI] [PubMed] [Google Scholar]
- Kale P., Johnson L. B., 2005. Second-generation azole antifungal agents. Drugs Today (Barc) 41: 91–105. [DOI] [PubMed] [Google Scholar]
- Katiyar S. K., Alastruey-Izquierdo A., Healey K. R., Johnson M. E., Perlin D. S., et al. , 2012. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob. Agents Chemother. 56: 6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Domergue R., Zupancic J. L., Cormack B. P., 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8: 378–384. [DOI] [PubMed] [Google Scholar]
- Kim D. U., Hayles J., Kim D., Wood V., Park H. O., et al. , 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F. M., de Koster C. G., Brul S., 2014. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 13: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock C., Dufrene Y. F., Heinisch J. J., 2015. Up against the wall: is yeast cell wall integrity ensured by mechanosensing in plasma membrane microdomains? Appl. Environ. Microbiol. 81: 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krcmery V., Barnes A. J., 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50: 243–260. [DOI] [PubMed] [Google Scholar]
- Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., et al. , 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Bussey H., 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Sdicu A. M., Menard P., Shapiro J., Hussein S., et al. , 2004. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167: 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K., Snyder M., 1998. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52: 687–744. [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M., 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Manning B. D., Padmanabha R., Snyder M., 1997. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol. Biol. Cell 8: 1829–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovich S., Yekutiel A., Shalit I., Shadkchan Y., Osherov N., 2004. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 48: 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. C., Kim H., Mackin N. A., Maldonado-Baez L., Evangelista C. C., Jr, et al. , 2011. New regulators of a high affinity Ca2+ influx system revealed through a genome-wide screen in yeast. J. Biol. Chem. 286: 10744–10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W. W., Jiang W., Daniels L. L., Kim S. K., Haldimann A., et al. , 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35: 1–13. [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Inamine T., Yamauchi S., Nagayoshi Y., Saijo T., et al. , 2010a Role of the Slt2 mitogen-activated protein kinase pathway in cell wall integrity and virulence in Candida glabrata. FEMS Yeast Res. 10: 343–352. [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Yamauchi S., Inamine T., Nagayoshi Y., Saijo T., et al. , 2010b Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 54: 1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. M., Locke E. G., Cunningham K. W., 2001. Differential regulation of two Ca(2+) influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 159: 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayoshi, Y., T. Miyazaki, A. Minematsu, S. Yamauchi, T. Takazono et al., 2014 Contribution of the Slt2-regulated transcription factors to echinocandin tolerance in Candida glabrata. FEMS Yeast Res. 14(7): 1128–1131. [DOI] [PubMed]
- Nasmyth K., Dirick L., 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66: 995–1013. [DOI] [PubMed] [Google Scholar]
- Orlean P., 2012. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192: 775–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovalle R., Lim S. T., Holder B., Jue C. K., Moore C. W., et al. , 1998. A spheroplast rate assay for determination of cell wall integrity in yeast. Yeast 14: 1159–1166. [DOI] [PubMed] [Google Scholar]
- Ovalle R., Spencer M., Thiwanont M., Lipke P. N., 1999. The spheroplast lysis assay for yeast in microtiter plate format. Appl. Environ. Microbiol. 65: 3325–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K., Tanaka K., Imamura H., Hihara T., Kameyama T., et al. , 1996. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15: 2196–2207. [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J., 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10: 11–23. [DOI] [PubMed] [Google Scholar]
- Pham C. D., Iqbal N., Bolden C. B., Kuykendall R. J., Harrison L. H., et al. , 2014. The role of FKS mutations in C. glabrata: MIC values, echinocandin resistance and multidrug resistance. Antimicrob. Agents Chemother. 58: 4690–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2013 R: A language and environment for statistical computing pp, R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Reinoso-Martin C., Schuller C., Schuetzer-Muehlbauer M., Kuchler K., 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2: 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M., Lass-Florl C., 2008. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 14(Suppl 4): 5–24. [DOI] [PubMed] [Google Scholar]
- Rizo J., Sudhof T. C., 1998. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273: 15879–15882. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Grigull J., Mohammad N., Hughes T. R., 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A., Gabaldon T., Schuller C., 2011. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzmuller T., Ma B., Hiller E., Istel F., Tscherner M., et al. , 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog. 10: e1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Perlin D. S., 2015. Coping with stress and the emergence of multidrug resistance in fungi. PLoS Pathog. 11: e1004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund R. F., Nasmyth K. A., 1996. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol. Cell. Biol. 16: 2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Babak S. D., Babak T., Diezmann S., Hill J. A., Xie J. L., et al. , 2012. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 8: e1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C. P., Cunningham K. W., 2013. Kch1 family proteins mediate essential responses to endoplasmic reticulum stresses in the yeasts Saccharomyces cerevisiae and Candida albicans. J. Biol. Chem. 288: 34861–34870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C. P., Zhang N., Sokabe T., Rivetta A., Slayman C. L., et al. , 2013. Activation of an essential calcium signaling pathway in Saccharomyces cerevisiae by Kch1 and Kch2, putative low-affinity potassium transporters. Eukaryot. Cell 12: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher A. J., Chahine E. B., Balcer H. E., 2009. Echinocandins: the newest class of antifungals. Ann. Pharmacother. 43: 1647–1657. [DOI] [PubMed] [Google Scholar]
- Ueno K., Namiki Y., Mitani H., Yamaguchi M., Chibana H., 2011. Differential cell wall remodeling of two chitin synthase deletants Deltachs3A and Deltachs3B in the pathogenic yeast Candida glabrata. FEMS Yeast Res. 11: 398–407. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Schekman R., 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100: 10287–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. A., Munro C. A., de Bruijn I., Lenardon M. D., McKinnon A., et al. , 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4: e1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. A., Gow N. A., Munro C. A., 2013. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 57: 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Chen C. Y., Levin D. E., 1994. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J. Biol. Chem. 269: 16829–16836. [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Zhao C., Jung U. S., Garrett-Engele P., Roe T., Cyert M. S., et al. , 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Szczypka M. S., Young R., Thiele D. J., 1994. A system for gene cloning and manipulation in the yeast Candida glabrata. Gene 142: 135–140. [DOI] [PubMed] [Google Scholar]
- Zhou X., Ma Y., Fang Y., W. gerile, W. Jaiseng et al, 2013. A genome-wide screening of potential target genes to enhance the antifungal activity of micafungin in Schizosaccharomyces pombe. PLoS One 8: e65904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.